Abstract
Glucocorticoids are important regulators of lipid homeostasis, and chronically elevated glucocorticoid levels induce hypertriglyceridemia, hepatic steatosis, and visceral obesity. The occupied glucocorticoid receptor (GR) is a transcription factor. However, those genes regulating lipid metabolism under GR control are not fully known. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor), a protein inhibitor of lipoprotein lipase, is synthesized and secreted during fasting, when circulating glucocorticoid levels are physiologically increased. We therefore tested whether the ANGPTL4 gene (Angptl4) is transcriptionally controlled by GR. We show that treatment with the synthetic glucocorticoid dexamethasone increased Angptl4 mRNA levels in primary hepatocytes and adipocytes (2–3-fold) and in the livers and white adipose tissue of mice (∼4-fold). We tested the mechanism of this increase in H4IIE hepatoma cells and found that dexamethasone treatment increased the transcriptional rate of Angptl4. Using bioinformatics and chromatin immunoprecipitation, we identified a GR binding site within the rat Angptl4 sequence. A reporter plasmid containing this site was markedly activated by dexamethasone, indicative of a functional glucocorticoid response element. Dexamethasone treatment also increased histone H4 acetylation and DNase I accessibility in genomic regions near this site, further supporting that it is a glucocorticoid response element. Glucocorticoids promote the flux of triglycerides from white adipose tissue to liver. We found that mice lacking ANGPTL4 (Angptl4−/−) had reductions in dexamethasone-induced hypertriglyceridemia and hepatic steatosis, suggesting that ANGPTL4 is required for this flux. Overall, we establish that ANGPTL4 is a direct GR target that participates in glucocorticoid-regulated triglyceride metabolism.
Glucocorticoids are steroid hormones that act as key transcriptional regulators of human metabolism during the fasted state, when their levels are physiologically increased. In particular, glucocorticoids facilitate the mobilization of triglycerides (TG)2 from the white adipose tissue (WAT) for use by the liver in processes such as gluconeogenesis, TG synthesis, and very low density lipoporotein synthesis and secretion (1, 2). However, the full set of genes that mediate this effect via transcriptional control by glucocorticoids is not known.
Fasting also increases circulating levels of angiopoietin-like 4 (ANGPTL4, a fasting-induced adipose factor). ANGPTL4 is a protein secreted by the liver and WAT that can inhibit lipoprotein lipase (LPL) activity and stimulate WAT lipolysis (3–5). LPL hydrolyzes lipoprotein TG, promoting fatty acid storage in the WAT. This activity is counterbalanced by that of lipases, which hydrolyze stored TG, promoting fatty acid release by adipocytes. Therefore, one could predict that reducing ANGPTL4 activity would promote WAT TG storage, whereas increasing it would favor lipolysis. This prediction is consistent with the results of genetic and physiological studies. First, mice lacking ANGPTL4 (Angptl4−/−) have decreased plasma TG levels and an increased capacity for weight gain (6). By contrast, mice overexpressing Angptl4 in the WAT have a dramatically limited capacity for TG storage and increased levels of plasma TG, fatty acids, and glycerol (7). Plasma TG and fatty acid levels are similarly increased in mice by adenoviral overexpression of Angptl4 in the liver (8) and by systemic injection of recombinant ANGPTL4 (9, 10). Finally, a recent large population-based study uncovered sequence variations in Angptl4 that are associated with loss of function and reduced plasma TG levels in humans (11). In summary, these data suggest that transcriptional modulation of Angptl4 expression could serve as an important regulatory mechanism in TG homeostasis.
We previously showed that Angptl4 mRNA levels are increased by glucocorticoids in A549 lung epithelial cells, suggesting that glucocorticoids may exert transcriptional control over Angptl4 expression (12). Several lines of evidence support this hypothesis in metabolic tissues. First, excess glucocorticoids promote hypertriglyceridemia, as also seen in models where ANGPTL4 levels are increased (1, 13, 14). Second, reducing the ratio of active to inactive glucocorticoids by pharmacologically inhibiting 11β-hydroxysteroid dehydrogenase type I increased plasma TG clearance and decreased liver TG synthesis, two components of the phenotype seen in Angptl4−/− mice (6, 15). Therefore, it is possible that both physiologic and pathophysiologic responses to glucocorticoids may involve the regulation of Angptl4 expression in metabolic tissues. However, whether Angptl4 expression is indeed directly regulated by glucocorticoids in the liver and WAT remains unexplored.
Given the prominent role of ANGPTL4 in systemic TG metabolism, we explored the regulation of Angptl4 by glucocorticoids. We first examined whether glucocorticoids regulate Angptl4 expression in primary hepatocytes and adipocytes, mouse livers and WAT, and established cell lines. We then dissected the mechanism of glucocorticoid-regulated Angptl4 expression using a rat hepatoma cell line, H4IIE. Finally, we used Angptl4−/− mice to investigate the potential role of ANGPTL4 in glucocorticoid-regulated TG homeostasis.
EXPERIMENTAL PROCEDURES
Cell Culture
H4IIE rat hepatoma cells (a gift from Dr. Daryl Granner, Vanderbilt University) were cultured in Dulbecco's modified Eagle's medium with 5% fetal bovine serum (Invitrogen). When cells were treated with dexamethasone (DEX), Dulbecco's modified Eagle's medium with 5% charcoal stripped fetal bovine serum (J R Scientific) was used. Rat primary hepatocytes were purchased from Cambrix. Human primary adipocytes were purchased from ZenBio.
ChIP
H4IIE cells (1 × 108 to 2 × 108 cells) were cross-linked by using formaldehyde at a final concentration of 1% at room temperature for 5 or 10 min. The chromatin immunoprecipitation (ChIP) protocols were otherwise as previously described (16, 17).
Nuclear Run-on
H4IIE cells were grown to confluence and treated with DMSO or DEX for different times. Cells were then harvested and incubated in lysis buffer (10 mm Tris-HCl, pH 7.4, 3 mm MgCl2, 10 mm NaCl, 150 mm sucrose, and 0.5% Nonidet P-40) at 4 °C for 5 min. Nuclei were then isolated by microcentrifuge at 170 × g at 4 °C for 5 min. The total nuclei from each of the DMSO- or DEX-treated samples were counted, and equal numbers of nuclei were used for in vitro transcription. We split the samples into two aliquots. One was incubated in 100 μl of 2× in vitro transcription buffer (200 mm KCl, 20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 4 mm dithiothreitol, 4 mm each of ATP, GTP, and CTP, 200 mm sucrose, and 20% glycerol) plus 8 μl of biotin-UTP (Roche), and the other in 100 μl of 2× in vitro transcription buffer plus 8 μl of UTP (negative control) for 30 min at 29 °C. 6 μl of 250 mm CaCl2 and 6 μl of RNase-free DNase (Roche) (10 units/μl) were then added to stop the reactions. Total RNA was then isolated using Nucleospin RNA II (E&K).
Dyna beads M-280 (Invitrogen) were washed twice in solution A (0.1 mm NaOH, 0.5 m NaCl) for 5 min, once in solution B (0.1 m NaCl) for 5 min, and then resuspended in binding/wash buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 2 m NaCl) plus 1 μl (40 units) of RNasin (Promega) per 100 μl of beads. 50 μl of beads (in binding/wash buffer) were then added to RNA, incubated at 42 °C for 20 min, and then shaken for 2 h at room temperature. Afterward, the beads were centrifuged and the supernatant discarded. The beads were then washed once (5 min) with 500 μl of 15% formamide plus 2× saline/sodium citrate buffer and twice with 1 ml of 2× saline/sodium citrate buffer. The beads were then resuspended in 30 μl of RNase- and DNase-free water. 10 μl of beads were used for each reverse transcription reaction prior to quantitative PCR (qPCR).
qPCR
Total RNA was isolated from cells by using QIAshredder and RNeasy kits (Qiagen). Total RNA from liver and epididymal WAT was isolated by using Tri-reagent (Molecular Research Center Inc.). To synthesize random-primed cDNA, 0.5 μg of total RNA (10 μl), 4 μl of 2.5 mm dNTP, and 2 μl of random primers (New England Biolabs) were mixed and incubated at 70 °C for 10 min. A 4-μl mixture containing 25 units of Moloney murine leukemia virus reverse transcriptase (New England Biolabs), 10 units of RNasin, and 2 μl of 10× reaction buffer (New England Biolabs) was then added and incubated at 42 °C for 1 h. The reaction was then incubated at 95 °C for 5 min.
The resultant cDNA was diluted to 200 μl, and 2.5 μl was used to perform qPCR in a 25-μl reaction containing Taq DNA polymerase (1.25 units; Promega), 1× reaction buffer, 1.5 mm MgCl2, 0.5 mm dNTP (Invitrogen), 0.2× SYBR Green I dye (Molecular Probes), and 250 nm of each primer. Alternatively, EVA QPCR SuperMix Kit (Biochain) was used per the manufacturer's protocol. qPCR was performed in either an Opticon-2 DNA Engine (MJ Research) or a 7900HT PCR System (Applied Biosystems) and analyzed by using the δ-δ Ct method as supplied by the manufacturer. Rpl19 expression was used for internal normalization. All primer sequences are presented in supplemental Table S1.
Western Blot
Livers and Epididymal WAT were collected, frozen in liquid nitrogen, and stored at −80 °C. Samples were thawed, suspended, and homogenized in RIPA buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 150 mm NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100) supplemented with protease inhibitors. Tissue lysates were cleared by centrifugation (17,000 × g for 15 min at 4 °C). Lysates were then resolved by SDS-PAGE, and proteins were transferred to nitrocellulose membranes (Amersham Biosciences) using semidry transfer (Bio-Rad). Membranes were blocked for 8 h at 22 °C with 5% (w/v) nonfat milk in TBS (50 mm Tris base, 200 mm NaCl, pH 7.5). Membranes were then incubated overnight at 4 °C in TBS with 5% milk containing primary antibody, followed by washes with TBS plus 0.5% Tween 20 at pH 7.5 (TBST). Membranes were then incubated for 2 h at 22 °C in TBS with 5% milk containing secondary antibody followed by washes with TBST. Proteins were detected by chemiluminescence (Western Lighting Plus-ECL, PerkinElmer Life Sciences). Membranes were stripped for 0.5 h at 22 °C in PBS with 7 μl/ml of β-mercaptoethanol, washed with PBS for 0.5 h, and blocked for 4 h in TBS with 10% milk before re-probing with other primary antibodies. The following antibodies were used: ANGPTL4 (Rb polyclonal antibodies to ARP4, ab2920; Abcam Inc.), β-actin (C4) mouse monoclonal IgG1 (sc-47778; Santa Cruz Biotechnology), anti-rabbit IgG1-horseradish peroxidase (Cell Signaling), and anti-mouse IgG1-horseradish peroxidase (sc-2060; Santa Cruz Biotechnology).
Blots were scanned as TIFF files to Adobe Photoshop CS4 (version 11.0) and quantified using Image J software. The optical density of the ANGPTL4 band divided by that of the respective β-actin band are presented.
Cloning and Site-directed Mutagenesis
The rat Angptl4 genomic region containing the predicted GR binding site was amplified by PCR using specific primers. The PCR fragment was restriction-digested and subcloned into the pGL4-TATA reporter. A mutagenesis kit (QuikChange) was used to make site-directed mutations per the manufacturer's instructions (Stratagene).
DNase I Accessibility Assay
Nuclei from H4IIE cells treated with vehicle or DEX for 1 h were digested with 6.25–200 units/ml of DNase I (Qiagen) for 5 min at 22 °C. The reaction was stopped by treatment with Proteinase K for 1 h at 65 °C. DNA samples were purified using PCR purification columns (Qiagen). The samples underwent qPCR analysis to determine the relative amounts of cleaved products (see supplemental Table 1 for primer sequences), which were used to express the percent of DNA cleavage by DNase I.
Transfection
Transfection in H4IIE cells was done with Lipofectamine 2000 (Invitrogen) according to the technical manual. 24 h post-transfection, cells were treated with either DMSO or 0.5 mm DEX overnight. Cells were then harvested, and luciferase activity was measured by a dual-luciferase assay kit (Promega) according to the technical manual.
Electrophoretic Mobility Shift Assay (EMSA)
Serial dilutions of purified GR DNA binding domain (DBD) protein (provided by Dr. Miles Pufall, University of California, San Francisco) were mixed with complementary oligonucleotides (2 × 10−8 m) containing either wild-type or mutant Angptl4 GREs end-labeled with Cy5 in a solution comprised of 20 mm Tris, pH 7.5, 2 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 0.3 mg/ml bovine serum albumin, 4 mm dithiothreitol, and 0.1 μg/μl dI-dC. The mixtures reacted for 30 min to reach equilibrium and were then run out on 8% native gels and scanned by a Typhoon phosphorimager (Amersham Biosciences).
Animals
C57BL/6J (B6) wild-type mice were purchased from The Jackson Laboratory. Angptl4−/− mice were provided by the laboratories of Andras Nagy (Samuel Lunenfeld Research Institute, Mount Sinai Hospital) and Jeff Gordon (Washington University). Heterozygous mice (Angptl4+/−) on a mixed B6:129/Sv background were generated as described (6), and Angptl4+/+, Angptl4+/−, and Angptl4−/− littermates, obtained by crossing Angptl4+/− mice were compared. The PCR protocols for genotyping animals were previously described (6).
Mice (3–4 months old) were injected intraperitoneally daily with 40 mg/kg body weight of water-soluble DEX (Sigma) in PBS for 4 consecutive days. 20 h after the final injection, mice were fasted for 4 h (17) and then used for blood collection and tissue harvest.
Liver and Serum TG Analyses
Liver TG Analysis
Liver samples were pulverized in liquid nitrogen and homogenized in a buffer consisting of 50 mm Tris-HCl, pH 7.4, 250 mm sucrose, and protease inhibitors. Lipids were extracted in chloroform/methanol (2:1) and separated by TLC on Silica Gel G-60 plates with the solvent hexane/ethyl ether/acetic acid (v/v/v, 80:20:1). The TG bands were visualized by exposure to iodine, and then scraped and analyzed as described (18), with triolein (Sigma) as a standard, and expressed per tissue weight.
Serum Analysis
Serum was isolated from whole blood immediately after collection, and a colorimetric kit (Roche Diagnostics) was used to measure serum TG levels.
Statistics
Data are expressed as mean ± S.E. for each group and comparisons were analyzed by Student's t test.
RESULTS
Glucocorticoids Increase Angptl4 Expression in Vitro and in Vivo
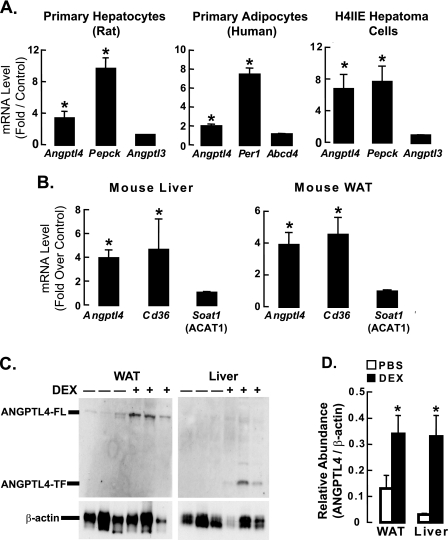
To determine whether Angptl4 expression is regulated by glucocorticoids in primary rat hepatocytes and human adipocytes, we treated these cells with the synthetic glucocorticoid DEX for 5 h. After harvesting the cells to prepare total RNA, cDNA was synthesized and qPCR was performed using specific primers. We first confirmed that DEX treatment increased the expression of Pepck and Per1 in hepatocytes and adipocytes, respectively (positive controls), and did not affect mRNA levels of Angptl3 and Abcd4, two genes not known to be regulated by glucocorticoids (Fig. 1A). Interestingly, DEX treatment significantly increased Angptl4 mRNA levels in both hepatocytes and adipocytes, indicating that Angptl4 expression is regulated by glucocorticoids in vitro. (Fig. 1A).
FIGURE 1.
ANGPTL4 gene and protein expression are regulated by glucocorticoids in vivo and in vitro. A, rat primary hepatocytes, human primary adipocytes, and rat H4IIE hepatoma cells (n = 3–5) were treated with either DMSO or DEX (0.5 μm) for 5 h, after which mRNA levels of Angptl4 and positive and negative control genes were measured by qPCR. Data show fold-induction of gene expression (DEX/DMSO) from at least three separate experiments (*, p < 0.05). B, mice (n = 6) were treated with either PBS or DEX (40 mg/kg) for 4 days then fasted for 4 h, after which their livers and epididymal fat (WAT) were harvested to perform qPCR as in A (*, p < 0.05). C, liver and WAT samples from control and DEX-treated mice in B were also collected for analysis of protein expression by Western blot (6.25 μg of tissue per sample run for WAT and 100 μg for liver). For each blot, the first 3 lanes are samples from individual mice treated with PBS (−), and the last 3 lanes are from DEX-treated mice (+). Full-length ANGPTL4 (ANGPTL4-FL) was the main band in WAT, whereas the truncated form (ANGPTL4-TF) predominated in the liver. β-Actin served as the internal loading control. D, quantification of the intensity of ANGPTL4 bands (see “Experimental Procedures”) from Western blots as in C (n = 3). Data represent relative optical density (ANGPTL4/β-actin; *, p < 0.05). The error bars represent the S. E. for the fold induction and the relative abundance.
We then tested whether glucocorticoids regulate Angptl4 expression in the model rat hepatoma cell line, H4IIE, by treating these cells with DEX for 5 h. As for primary hepatocytes, DEX treatment increased Angptl4 mRNA levels (Fig. 1A). We chose to conduct subsequent in vitro studies using H4IIE cells because the effect of DEX on Pepck, Angptl3, and Angptl4 mRNA expression in these cells replicated what was seen in primary hepatocytes.
To investigate whether glucocorticoids increase Angptl4 expression in vivo, we dissected the livers and epididymal WAT from C57BL6/J mice injected daily for 4 days with either DEX or PBS. We then used these tissues to prepare total RNA, synthesize cDNA, and perform qPCR to measure mRNA levels. DEX treatment increased the levels of CD36 mRNA (positive control) but had no effect on Soat1 (ACAT1) mRNA levels (negative control) in both liver and WAT (Fig. 1B). However, as it did in primary hepatocytes and adipocytes, DEX treatment of mice increased Angptl4 mRNA levels in both the liver and WAT (Fig. 1B), indicating that Angptl4 is regulated by glucocorticoids in vivo.
We next sought to determine whether the increase in Angptl4 mRNA expression induced by DEX treatment is associated with an increase in ANGPTL4 protein levels. We performed Western blots on tissue lysates from control and DEX-treated mice, and found that DEX treatment for 4 days markedly elevated the levels of ANGPTL4 protein in both the WAT and liver (Fig. 1, C and D). Previous studies (19, 20) showed that the WAT mostly produces an uncleaved full-length version of ANGPTL4 (ANGPTL4-FL), whereas the liver produces more of a truncated version of ANGPTL4. Our results from the WAT and livers of DEX-treated mice agree with these observations (Fig. 1C).
Glucocorticoids Increase the Rate of Rat Angptl4 Transcription
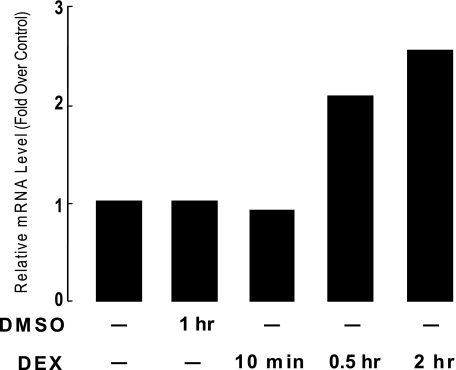
We next investigated whether the elevation of Angptl4 mRNA by glucocorticoid treatment is due to an increase in the rate of Angptl4 transcription in H4IIE cells by using nuclear run-on assays. Cells were treated with DEX in DMSO for 10, 30, or 120 min. Control cells were either untreated or treated with DMSO alone for 120 min. Afterward, nuclear extracts were prepared from these cells, and in vitro transcription was performed by adding biotin-UTP. Newly synthesized RNA was then isolated on streptavidin beads. After random-primed cDNA was synthesized, qPCR was performed to detect changes in transcription rates using Angptl4 specific primers. No effect was seen after 10 min of DEX treatment (Fig. 2). However, treatments of 30 and 120 min both markedly increased the rate of Angptl4 transcription (Fig. 2). These results demonstrate that glucocorticoids can regulate Angptl4 at the transcriptional level.
FIGURE 2.
DEX treatment increases the rate of Angptl4 gene transcription in H4IIE cells. H4IIE cells were untreated, treated with DMSO for 120 min, or with DEX (0.5 μm) for 10, 30, or 120 min as shown. Nuclei from these cells were used for in vitro transcription with biotin UTP. Newly synthesized RNA was isolated on streptavidin beads. cDNA was then synthesized and qPCR performed to monitor for changes in transcription rates using primers specific to Angptl4 and β-actin (control). Shown is the fold-induction in transcription of DEX-treated samples (DEX/untreated) from one of two independent experiments showing similar results.
Identification of a Rat Angptl4 GRE
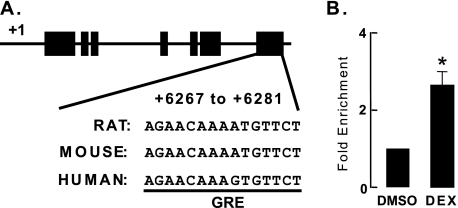
To investigate whether transcription of Angptl4 is directly regulated by the GR, we sought to identify GR binding sites within the Angptl4 genomic sequence. We applied a bioinformatic approach (BioProspector motif), with which we identified specific sequences within 64 kb of rat Angptl4 (which extended 32 kb upstream and downstream of the transcriptional start site) that were highly similar (width of the first motif block was set at 14) to 79 previously identified GR binding sites (21). Because many important transcriptional regulatory regions within genes are conserved between multiple species, we next employed a BLAST search of the University of California Santa Cruz genome browser to filter and isolate sequences conserved across the rat, mouse, and human genomes. This comparison yielded a single species-conserved palindromic sequence (AGAACATTTTGTTCT) located at the 3′-untranslated region of Angptl4 between 6267 and 6281 bp downstream of the transcriptional start site (Fig. 3A). We used a ChIP assay to investigate GR recruitment to this putative GR binding site in H4IIE cells. Cells were treated for 1 h with DEX and then harvested for ChIP (“Experimental Procedures”). Using a control IgG antibody for ChIP, the number of genomic DNA fragments that were amplified in the predicted GR binding site was similar between control (DMSO) and DEX-treated samples (data not shown). By contrast, using a GR-specific antibody for ChIP, we found that DEX treatment resulted in a ∼2.3-fold enrichment of genomic DNA fragments that amplified the predicted GR binding site (Fig. 3B). This finding indicates that GR occupancy of this palindromic sequence occurs in a DEX-dependent manner, suggesting that this sequence may be a functional GRE.
FIGURE 3.
Identification of an Angptl4 GRE. A, schematic diagram of rat Angptl4, including the location of the predicted GR binding site. Black boxes represent exons, and lines connecting them represent introns. The predicted GR binding site sequences (AGAACATTTTGTTCT) and their conserved counterparts in human and mouse genome are shown. B, ChIP experiments confirming the recruitment of GR to its predicted binding site. H4IIE cells were treated with either DMSO or DEX (0.5 μm) for 30 min, after which they were harvested for ChIP as described (“Experimental Procedures”). Shown is the fold-enrichment of GR binding by DEX treatment (DEX/DMSO) from three independent experiments (*, p < 0.05). The error bars represent the S. E. for the fold enrichment.
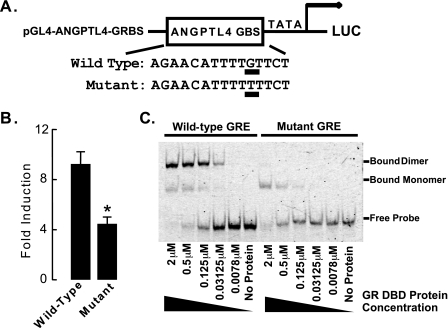
To test whether this identified genomic GR binding site is sufficient to mediate cellular responsiveness to glucocorticoids, we inserted the rat Angptl4 genomic DNA containing this sequence into a reporter plasmid that drove expression of the firefly luciferase gene (pGL4-ANGPTL4-GRBS, Fig. 4A). This plasmid was transfected into H4IIE cells, and 24 h later, cells were treated with either DMSO (control) or DEX overnight. Treatment with DEX strongly induced luciferase activity in transfected H4IIE cells (Fig. 4B), indicating that this predicted GRE confers glucocorticoid responsiveness. To test the specificity of this sequence in mediating the glucocorticoid response, we made a single nucleotide mutation within it (Fig. 4A, the mutated nucleotide is underlined). This nucleotide was chosen for mutation as previous studies demonstrated that it directly contacts GR and is an important mediator of glucocorticoid responses (22, 23). Mutation of this nucleotide reduced the glucocorticoid response by more than 50% (Fig. 4B), consistent with this sequence being a functional GRE.
FIGURE 4.
The GR binding region of Angptl4 confers glucocorticoid responsiveness. A, a sequence containing either the wild-type or a mutated Angptl4 GR binding site was inserted into the pGL4-TATA reporter to create pGL4-ANGPTL4-GRBS (mutation from Gly to Thr underlined). B, glucocorticoid responsiveness of GR-binding sites in rat Angptl4. Reporter plasmids and wild-type or mutant pGL4-ANGPTL4-GRBS (75 ng) were cotransfected with pcDNA3-hGR (150 ng) and pRL (100 ng) into H4IIE cells in a 24-well plate (n = 4 per group). pRL plasmid provided Renilla luciferase expression to document transfection efficiency. Transfected cells were left overnight, then washed with PBS and treated with 0.5 μm DEX for an additional 16–20 h. Cells were then lysed and assayed for firefly and Renilla luciferase activities. Shown is the fold-induction of luciferase activity (DEX-treated/ethanol-treated) in cells from at least three experiments (*, p < 0.05). The error bars represent the S. E. for the fold induction. C, EMSA on mixtures of purified GR DBD and Cy5 end-labeled oligonucleotides containing either wild-type or mutant Angptl4 GRE, confirming direct binding of GR to the GRE. Reactions lasted 30 min, and the mixtures were then run on 8% native gels and scanned by a phosphorimager. The concentration of GR DBD protein mixed with wild-type or mutant GREs ranged from 0 (no protein) to 2 μm, as shown. Data are representative of two independent experiments.
Finally, we performed EMSA to confirm the direct binding of GR to the Angptl4 GRE. We found that 0.03125 μm purified GR DBD was able to bind efficiently to Cy5-labeled Angptl4 GRE and that this binding increased with the concentration of GR DBD protein in the reaction mixture, until, by 2 μm, it bound all of the labeled GRE (Fig. 4C). Notably, most GR DBD bound to the Angptl4 GRE as a dimer (Fig. 4C), a requirement for transactivation. Mutant Angptl4 GRE, which contains a single nucleotide change in one of the two GRE half-sites (Fig. 4A), had a compromised interaction with GR DBD (Fig. 4C). Although GR monomers could bind to mutant GRE, presumably at the wild-type half-site, GR dimers could not, consistent with decreased transactivation of mutant Angptl4 GRE by GR (Fig. 4B). Even 2 μm GR DBD only bound to mutant GRE as a monomer (Fig. 4C). Together these findings confirm that GR binds directly and specifically to the identified Angptl4 GRE.
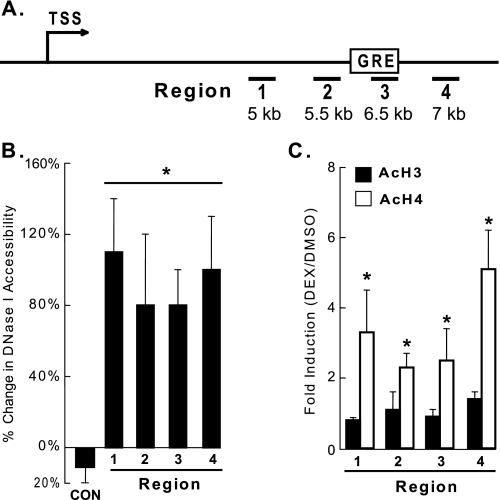
Glucocorticoids Increase DNase I Accessibility and Histone Acetylation in Genomic Regions Near the Angptl4 GRE
Chromatin architecture can control eukaryotic gene expression in vivo by modulating the accessibility of genomic sequences to transcriptional activation machinery (24). Changes in the structure of chromatin are detectable by the way they alter the sensitivity of DNA to cleavage by DNase I (25, 26). Increased accessibility to DNase I cleavage indicates a relatively open chromatin conformation, whereas protection from cleavage by DNase I points to a more compact chromatin structure. The former is associated with transcriptional activation and the latter with repression.
We tested whether glucocorticoid treatment affects DNase I accessibility of genomic regions near the identified Angptl4 GRE. H4IIE cells were treated with either DMSO or DEX for 30 min, the earliest time point at which DEX increased the rate of Angptl4 transcriptional activation in nuclear run-on assays. After DEX treatment, nuclei were isolated and treated with DNase I. Total DNA was then purified, and qPCR was performed to monitor for cleavage of specific genomic sequences by DNase I. DEX treatment markedly increased the accessibility of the Angptl4 GRE (region 3) and surrounding genomic regions to DNase I (Fig. 5B). These results suggest that glucocorticoid treatment opens up the structure of chromatin in the vicinity of the Angptl4 GRE, consistent with the observed increase in transcription activation.
FIGURE 5.
DEX treatment increases DNase I accessibility and histone H4 acetylation in rat Angptl4. A, schematic diagram of rat Angptl4, with TSS indicating the transcriptional start site, amplified genomic regions are underlined and numbered (1–4), and the position of the GRE shown. B, H4IIE cells were treated with DMSO or DEX (0.5 μm) for 30 min, and DNase I accessibility was analyzed as described (“Experimental Procedures”), showing increased cleavage of DNA in DEX-treated cells. C, H4IIE cells (n = 4 per group) were treated with DMSO or DEX (0.5 μm) for 30 min, and the levels of acetylated histones H3 and H4 were measured in regions 1–4 by ChIP. The fold-enrichment (DEX/DMSO) was used to show increased enrichment of AcH4 but not AcH3 in DEX-treated cells (*, p < 0.05). Data in B and C were normalized to a control genomic region in rat Rpl19, and each experiment was done at least 3 times. The error bars represent the S. E. for the percent change and the fold induction.
Nucleosomes form the fundamental repeating units of eukaryotic chromatin and include a core particle of DNA (∼147 bp) wrapped in an octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4. Increased histone acetylation, especially H3 and H4, correlates well with transcriptional activation (27). We therefore investigated whether glucocorticoids affect the acetylation status of rat Angptl4. H4IIE cells were treated with DEX for 30 min, after which ChIP was performed to monitor levels of acetylated histone H3 (AcH3) and histone H4 (AcH4) in genomic regions of Angptl4. Treatment with DEX for 30 min greatly increased levels of AcH4 in all 4 genomic regions tested, including region 3, which contained the GRE (Fig. 5, A and C). Interestingly, in contrast to those of AcH4, the levels of AcH3 were not affected by dexamethasone treatment. Overall, these results suggest that glucocorticoids specifically induce acetylation of histone H4 in activating Angptl4 transcription.
Effects of Glucocorticoids on Lipid Metabolism Are Impaired in Angptl4−/− Mice
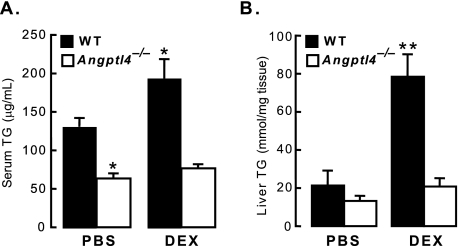
Plasma TG levels in mice are elevated by transgenic overexpression of Angptl4 (7) and by injection of mice with recombinant ANGPTL4 protein (9, 10). In the liver, adenoviral overexpression of Angptl4 results in both hyperlipidemia and hepatic steatosis (8). These features are also seen in states of chronic glucocorticoid excess, such as Cushing syndrome. We therefore investigated whether ANGPTL4 participates in the effects of glucocorticoids on lipid metabolism in mice genetically lacking ANGPTL4 (Angptl4−/−) (6). Angptl4−/− mice and wild-type littermates were treated with either PBS or 40 mg/kg DEX daily for 4 days. On day 5, their blood and livers were collected to measure TG levels. As expected, DEX treatment of wild-type mice increased serum TG levels by ∼50% (Fig. 6A). On the other hand, serum TG levels in Angptl4−/− mice were lower at baseline and were much less responsive to DEX treatment (Fig. 6A), indicating a requirement for ANGPTL4 in this aspect of glucocorticoid-mediated lipid metabolism. DEX treatment also increased liver TG levels ∼4-fold in wild-type mice (Fig. 6B). In Angptl4−/− mice, however, DEX-induced accumulation of hepatic TG was minimal (Fig. 6B). Thus, Angptl4−/− livers were protected from the pro-steatotic effects of glucocorticoids. Overall, these experiments establish ANGPTL4 as an important participant in glucocorticoid-regulated lipid homeostasis in vivo.
FIGURE 6.
The DEX-stimulated increase in serum and liver TG is impaired in Angptl4−/− mice. A, wild-type and Angptl4−/− mice were treated daily with PBS (control; n = 4–5 per group) or DEX (n = 5 per group) for 4 days as described (“Experimental Procedures”), after which serum TG levels were measured. Shown is the fold-increase in serum TG of DEX-treated mice (*, p < 0.05 versus WT PBS) for both wild-type and Angptl4−/− mice. B, liver TG content measured by TLC from the same mice as in A (**, p < 0.05 versus WT PBS). The error bars represent the S. E. for the TG concentration.
DISCUSSION
One important component of the physiologic response to fasting is an increase in the level of circulating glucocorticoids, which maintain levels of energy substrates in part by promoting the flux of TG from the WAT to the liver. Fasting also increases synthesis and secretion of ANGPTL4 by the WAT and liver, prompting the hypothesis that ANGPTL4 may mediate aspects of the regulatory effect of glucocorticoids on metabolism. We therefore investigated whether murine Angptl4 is transcriptionally regulated by glucocorticoids. We establish that Angptl4 mRNA levels are regulated by DEX treatment in cultured adipocytes and hepatocytes, and in the WAT and livers of mice. Using bioinformatics and ChIP, we identified a GR binding site in the genomic region of Angptl4. By finding that DEX treatment stimulated reporter activity in cells expressing this binding site linked to luciferase, we determined that this site is a functional GRE. Furthermore, we used EMSA to show that this GRE interacts directly with GR. Our data suggest that the mechanism by which GR controls Angptl4 transcription involves the modulation of DNase I accessibility and histone acetylation within the gene. We linked these effects to in vivo physiology by studying mice lacking ANGPTL4. These mice had reductions in DEX-induced hypertriglyceridemia and hepatic steatosis, indicating that ANGPTL4 is required for these effects. Overall, we show that Angptl4 is a direct transcriptional target of GR and a contributor to the regulation of TG homeostasis by glucocorticoids in mice.
Other signals have also been shown to modulate Angptl4 expression. For example, Angptl4 transcription is increased by hypoxia in endothelial cells (28, 29). By contrast, microbiota suppress intestinal levels of Angptl4 mRNA in mice (6). In addition to being regulated by glucocorticoids, as shown here, Angptl4 transcription is also controlled by peroxisome proliferator-activated receptors α and γ (PPARα and PPARγ), which function as nutritional sensors in hepatocytes and adipocytes, respectively (19, 20). In particular, PPARα increases the expression of key genes involved in the metabolic response to fasting (30). However, the effect of fasting on Angptl4 mRNA levels is not altered in mice lacking PPARα (19), suggesting that other pathways, including ones under GR regulation as shown here, may mediate this effect. Notably, Pparα mRNA levels are also increased by glucocorticoids (31). Thus, it is conceivable that both Angptl4 and Pparα are part of a GR-regulated gene network that controls the metabolic adaptation to fasting.
Using rat H4IIE hepatoma cells, we analyzed the mechanisms governing the stimulation of Angptl4 mRNA expression by glucocorticoids. We found that glucocorticoids increased the transcriptional rate of Angptl4 and identified a GR binding site between 6267 and 6281 bp downstream of the transcriptional start site within the 3′-untranslated region. We confirmed the recruitment of GR to this site by ChIP and showed that this site can bind GR dimers efficiently. Furthermore, in cells transfected with a reporter plasmid containing this GR binding site, we were able to stimulate reporter activity with DEX treatment, indicating that this site functions as a GRE. Notably, this element is conserved between mouse, rat, and human, highlighting its importance as a mammalian regulatory element. Regarding the sequence location, it is not unusual for a GRE to be far away from the transcriptional start site. Recently, GREs were shown to be distributed both 5′ and 3′ to the transcriptional start site at distances over 5 kb (23). Responsive elements in estrogen receptor- and androgen receptor-regulated genes have been found to be distributed similarly (16, 32, 33). Recently, a study found a GR binding region in human Angptl4 to be located as far as 8 kb upstream of the transcriptional start site using a ChIP on chip approach (23). The exact sequences mediating glucocorticoid responsiveness in this human GR binding region have not been identified, and we were unable to find sequences in the rat or mouse genomes resembling this specific region. Regardless, these human data coupled with our findings suggest the possibility that distinct transcriptional mechanisms may be employed to regulate Angptl4 expression in rodents and humans.
Chromatin remodeling regulates the activity of several GREs (23, 25). To determine how glucocorticoids activate Angptl4 transcription, we therefore tested whether DEX treatment affects the chromatin structure and histone acetylation of Angptl4. We found that DEX treatment increased the DNase I accessibility of genomic regions near the Angptl4 GRE, indicating that activation by GR may open up the structure of chromatin in these regions and facilitate the binding of transcriptional co-regulators. Glucocorticoids also specifically induced the acetylation of histone H4 in genomic regions near the Angptl4 GRE, suggesting that acetylation of key regulatory elements within these regions may serve as another modulator of glucocorticoid-activated Angptl4 transcription. That histone H4, but not H3, was acetylated by DEX treatment suggests the occupied GR recruits only a subset of histone acetyltransferases in activating Angptl4 transcription, although it is possible that histone H3 acetylation is induced at other genomic regions not tested here. Overall, the increased DNase I accessibility and histone H4 acetylation in the identified GRE offer insight into the specific machinery important for regulating Angptl4 transcription.
As seen previously (34), glucocorticoid treatment significantly increased TG levels in the serum (∼1.5-fold) and livers (∼4-fold) of mice (Fig. 6A, B). However, this response was significantly compromised in Angptl4−/− mice (Fig. 6, A and B). Thus, ANGPTL4, potentially by inhibiting LPL, plays a role in the mechanism by which glucocorticoids increase serum and hepatic TG levels. Our DEX treatment protocol matched that of Dolinsky et al. (34), who found that glucocorticoid treatment increased the rate of hepatic TG synthesis, but not very low density lipoporotein secretion. This could explain how the mice we treated developed higher TG levels in the liver than in the plasma. Although we did not observe a significant change in the epididymal fat mass of mice treated for 4 days with DEX (data not shown), it is well known that long-term glucocorticoid treatment promotes a re-partitioning of lipids between different WAT depots (1, 35). Future work should determine whether ANGPTL4 plays a role in this process.
The mechanisms(s) underlying how ANGPTL4 modulates glucocorticoid-stimulated hepatic steatosis are unclear. A likely possibility is that without ANGPTL4, a lipoprotein lipase inhibitor and a stimulator of WAT lipolysis, glucocorticoids cannot efficiently mobilize TG from WAT for use in hepatic TG synthesis. Alternatively, this process may involve an activity of ANGPTL4 distinct from its ability to inhibit LPL. A recent report showed that the fibrinogen-like domain of ANGPTL4, located at its carboxyl terminus, can suppress the basic fibroblast growth factor-induced activation of ERK1/2 MAP kinase in endothelial cells (36). Furthermore, analysis of mRNA expression showed that genes involved in fatty acid oxidation in the muscle were down-regulated in Angptl4−/− mice (37). It is unclear if the ERK1/2 MAP kinase or fatty acid oxidation pathways are similarly modulated in the livers and WAT of Angptl4−/− mice, and investigating this possibility is an important topic for future investigation.
It is worth noting that Angptl4 is not the only GR target that participates in the regulation of liver TG metabolism. Recently, GR was shown to inhibit transcription of the hairy enhancer of split 1 gene (Hes1), which encodes a transcriptional repressor involved in glucocorticoid-induced hepatic steatosis (38). Inhibition of Hes1 coordinately represses the expression of pancreatic lipase (PNL) and pancreatic lipase-related protein (PNLRP) 2, both of which encode TG hydrolases. Down-regulation of these two lipolytic genes in the liver can result in the accumulation of TG (38). These findings along with ours suggest that glucocorticoids potentially regulate the transcription of multiple networks of genes that can exert overlapping effects on hepatic lipid metabolism. Additionally, these GR target genes are likely tissue-specific, as glucocorticoid-dependent effects on lipid metabolism in the liver and WAT are quite different. Thus, gaining a complete picture of how glucocorticoids affect lipid homeostasis will require the systematic identification of all direct GR targets in both the WAT and liver. Our identification of Angptl4 as a direct GR target is an important first step toward the systematic dissection of this metabolic regulatory process.
Our findings mark ANGPTL4 as a potential key mediator of the effects of glucocorticoids on TG homeostasis during physiologic fasting. Fasting TG levels are routinely measured in humans and are often elevated in patients with insulin resistance and type 2 diabetes. Recently, genetic sequence variations interfering with either the production of ANGPTL4 or with its capacity to inhibit LPL were associated with low plasma TG levels in up to 4% of individuals in a large population when combined with mutations in other members of the ANGPTL family (39). These findings, coupled with the work shown here, suggest that ANGPTL4 may mediate the metabolic effects of glucocorticoids on TG metabolism similarly in both in humans and mice.
Supplementary Material
Acknowledgments
We thank Dr. Keith Yamamoto for support on this project; Dr. Andras Nagy for providing Angptl4−/− mice; Drs. Jeff Gordon and Peter Crawford for sharing the experience of breeding Angptl4−/− mice; Dr. Sebastiaan Meijsing for EMSA assays; Dr. Miles Pufall for purified GR DBD; and Dr. Donald Scott for the nuclear run-on assay protocol.
This work was supported by American Heart Association Beginning Grant-in-aid 0565157Y (to J. C. W.) and an A. P. Giannini Foundation Fellowship (to S. K. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- TG
- triglyceride
- WAT
- white adipose tissue
- DEX
- dexamethasone
- LPL
- lipoprotein lipase
- DMSO
- dimethyl sulfoxide
- ChIP
- chromatin immunoprecipitation
- PPAR
- peroxisome proliferator-activated receptor
- ERK
- extracellular signal-regulated kinase
- qPCR
- quantitative PCR
- PBS
- phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay
- DBD
- DNA binding domain
- MAP kinase
- mitogen-activated protein kinase
- Pepck
- phosphoenolpyruvate carboxykinase
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid receptor element.
REFERENCES
- 1.Macfarlane D. P., Forbes S., Walker B. R. (2008) J. Endocrinol. 197, 189–204 [DOI] [PubMed] [Google Scholar]
- 2.Seckl J. R., Morton N. M., Chapman K. E., Walker B. R. (2004) Recent Prog. Horm. Res. 59, 359–393 [DOI] [PubMed] [Google Scholar]
- 3.Hato T., Tabata M., Oike Y. (2008) Trends Cardiovasc. Med. 18, 6–14 [DOI] [PubMed] [Google Scholar]
- 4.Kersten S. (2005) Biochem. Soc. Trans. 33, 1059–1062 [DOI] [PubMed] [Google Scholar]
- 5.Li C. (2006) Curr. Opin. Lipidol. 17, 152–156 [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., Gordon J. I. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandard S., Zandbergen F., van Straten E., Wahli W., Kuipers F., Müller M., Kersten S. (2006) J. Biol. Chem. 281, 934–944 [DOI] [PubMed] [Google Scholar]
- 8.Xu A., Lam M. C., Chan K. W., Wang Y., Zhang J., Hoo R. L., Xu J. Y., Chen B., Chow W. S., Tso A. W., Lam K. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6086–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyama T. E., Lambert G., Nicol C. J., Matsusue K., Peters J. M., Brewer H. B., Jr., Gonzalez F. J. (2004) J. Biol. Chem. 279, 20874–20881 [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K., Shimizugawa T., Ono M., Furukawa H. (2002) J. Lipid Res. 43, 1770–1772 [DOI] [PubMed] [Google Scholar]
- 11.Romeo S., Pennacchio L. A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H. H., Cohen J. C. (2007) Nat. Genet. 39, 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J. C., Derynck M. K., Nonaka D. F., Khodabakhsh D. B., Haqq C., Yamamoto K. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15603–15608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker B. R. (2006) Diabet. Med. 23, 1281–1288 [DOI] [PubMed] [Google Scholar]
- 14.Walker B. R. (2007) Eur. J. Endocrinol. 157, 545–559 [DOI] [PubMed] [Google Scholar]
- 15.Desai U., Lee E. C., Chung K., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K. A., Sands A. T., Schneider M., Van Sligtenhorst I., Suwanichkul A., Vogel P., Wilganowski N., Wingert J., Zambrowicz B. P., Landes G., Powell D. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11766–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolton E. C., So A. Y., Chaivorapol C., Haqq C. M., Li H., Yamamoto K. R. (2007) Genes Dev. 21, 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J. C., Shah N., Pantoja C., Meijsing S. H., Ho J. D., Scanlan T. S., Yamamoto K. R. (2006) Genes Dev. 20, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder F., Stephens N. (1959) Biochim. Biophys. Acta 34, 244–245 [DOI] [PubMed] [Google Scholar]
- 19.Mandard S., Zandbergen F., Tan N. S., Escher P., Patsouris D., Koenig W., Kleemann R., Bakker A., Veenman F., Wahli W., Müller M., Kersten S. (2004) J. Biol. Chem. 279, 34411–34420 [DOI] [PubMed] [Google Scholar]
- 20.Yoon J. C., Chickering T. W., Rosen E. D., Dussault B., Qin Y., Soukas A., Friedman J. M., Holmes W. E., Spiegelman B. M. (2000) Mol. Cell. Biol. 20, 5343–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.So A. Y., Cooper S. B., Feldman B. J., Manuchehri M., Yamamoto K. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. (1991) Nature 352, 497–505 [DOI] [PubMed] [Google Scholar]
- 23.So A. Y., Chaivorapol C., Bolton E. C., Li H., Yamamoto K. R. (2007) PLoS Genet 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 25.Fletcher T. M., Ryu B. W., Baumann C. T., Warren B. S., Fragoso G., John S., Hager G. L. (2000) Mol. Cell. Biol. 20, 6466–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John S., Sabo P. J., Johnson T. A., Sung M. H., Biddie S. C., Lightman S. L., Voss T. C., Davis S. R., Meltzer P. S., Stamatoyannopoulos J. A., Hager G. L. (2008) Mol. Cell 29, 611–624 [DOI] [PubMed] [Google Scholar]
- 27.Shahbazian M. D., Grunstein M. (2007) Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 28.Wang B., Wood I. S., Trayhurn P. (2007) Pflugers Arch. 455, 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Jan S., Amy C., Cazes A., Monnot C., Lamandé N., Favier J., Philippe J., Sibony M., Gasc J. M., Corvol P., Germain S. (2003) Am. J. Pathol. 162, 1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desvergne B., Michalik L., Wahli W. (2006) Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 31.Lemberger T., Staels B., Saladin R., Desvergne B., Auwerx J., Wahli W. (1994) J. Biol. Chem. 269, 24527–24530 [PubMed] [Google Scholar]
- 32.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 33.Deblois G., Giguère V. (2008) Mol. Endocrinol. 22, 1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolinsky V. W., Douglas D. N., Lehner R., Vance D. E. (2004) Biochem. J. 378, 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuzaki H., Paterson J., Shinyama H., Morton N. M., Mullins J. J., Seckl J. R., Flier J. S. (2001) Science 294, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 36.Yang Y. H., Wang Y., Lam K. S., Yau M. H., Cheng K. K., Zhang J., Zhu W., Wu D., Xu A. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 835–840 [DOI] [PubMed] [Google Scholar]
- 37.Bäckhed F., Manchester J. K., Semenkovich C. F., Gordon J. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemke U., Krones-Herzig A., Berriel Diaz M., Narvekar P., Ziegler A., Vegiopoulos A., Cato A. C., Bohl S., Klingmüller U., Screaton R. A., Müller-Decker K., Kersten S., Herzig S. (2008) Cell. Metab. 8, 212–223 [DOI] [PubMed] [Google Scholar]
- 39.Romeo S., Yin W., Kozlitina J., Pennacchio L. A., Boerwinkle E., Hobbs H. H., Cohen J. C. (2009) J. Clin. Invest. 119, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.