Abstract
Small recombinant antibody molecules such as bispecific single-chain diabodies (scDb) possessing a molecular mass of ∼55 kDa are rapidly cleared from circulation. We have recently extended the plasma half-life of scDb applying various strategies including PEGylation, N-glycosylation and fusion to an albumin-binding domain (ABD) from streptococcal protein G. Here, we further analyzed the influence of these modifications on the biodistribution of a scDb directed against carcinoembryonic antigen (CEA) and CD3 capable of retargeting T cells to CEA-expressing tumor cells. We show that a prolonged circulation time results in an increased accumulation in CEA+ tumors, which was most pronounced for scDb-ABD and PEGylated scDb. Interestingly, tumor accumulation of the scDb-ABD fusion protein was ∼2-fold higher compared with PEGylated scDb, although both molecules exhibit similar plasma half-lives and similar affinities for CEA. Comparing half-lives in neonatal Fc receptor (FcRn) wild-type and FcRn heavy chain knock-out mice the contribution of the FcRn to the long plasma half-life of scDb-ABD was confirmed. The half-life of scDb-ABD was ∼2-fold lower in the knock-out mice, while no differences were observed for PEGylated scDb. Binding of the scDb derivatives to target and effector cells was not or only marginally affected by the modifications, although, compared with scDb, a reduced cytotoxic activity was observed for scDb-ABD, which was further reduced in the presence of albumin. In summary, these findings demonstrate that the extended half-life of a bispecific scDb translates into improved accumulation in antigen-positive tumors but that modifications might also affect scDb-mediated cytotoxicity.
Bispecific single-chain diabodies (scDb)2 are recombinant molecules composed of the variable heavy and light chain domains of two antibodies connected by three linkers in the order VHA-VLB-VHB-VLA (1). These domains assemble into molecules with a compact structure and molecular masses of ∼55 kDa. Bispecific single-chain diabodies have been developed for various applications including the retargeting of cytotoxic T lymphocytes to tumor cells for cellular cancer therapy (2).
Although scDb are capable of efficiently retargeting effector cells to tumor cells the small size leads to their rapid elimination after i.v. injection. The terminal half-life of these molecules in mice is only in the range of 5–6 h, compared with several days for whole IgG molecules (3, 4). The fast clearance of such small molecules from circulation hampers therapeutic applications, e.g. requiring infusions or repeated injections to maintain a therapeutically effective dose over a prolonged period of time (5). For example, a bispecific tandem scFv directed against CD19 and CD3 (blinatumomab) having a similar size as an scDb molecule had to be given as an 8-week infusion (maximum dose 60 μg/m2 per day) in a clinical phase I trial for the treatment of B cell lymphoma patients (6).
To extend plasma half-lives of therapeutic proteins and thus to improve pharmacokinetics and pharmacodynamics, several strategies can be applied (7). Strategies such as conjugation of polyethylene glycol chains (PEGylation) or production of hyperglycosylated variants through introduction of additional N-glycosylation sites primarily aim at increasing the hydrodynamic volume of the molecule, thus reducing renal filtration and degradation. Some of these strategies further implement FcRn-mediated recycling processes, e.g. fusion to the IgG Fc region and fusion or binding to serum albumin.
We recently applied several of these strategies to improve the plasma half-life of a scDb molecule. These strategies included site-directed conjugation of a 40-kDa PEG chain (PEGylated scDb, scDb-A′-PEG40k), production of N-glycosylated scDb variants possessing 3, 6, or 9 N-glycosylation sites (scDb-ABC1–7), a scDb-human serum albumin fusion protein (scDb-HSA), and a scDb fused to an albumin-binding domain from streptococcal protein G (scDb-ABD) (3, 4, 8). In these studies we showed that N-glycosylation only moderately increased half-life, while a strong improvement was observed for the PEGylated scDb, scDb-HSA, and scDb-ABD.
In the present study we further analyzed the biodistribution of unmodified scDb as well as three of the scDb derivatives (PEGylated scDb, N-glycosylated scDb, scDb-ABD) in tumor-bearing mice. We show that the modified scDb molecules exhibit a reduced renal clearance and that an extended half-life leads to an increased accumulation in antigen-positive tumors. The strongest improvement was observed for scDb-ABD. Using FcRn knock-out mice we confirmed that FcRn-mediated recycling contributes to the long half-life of scDb-ABD. Affinities of the scDb derivatives for target and effector cells were not or only marginally affected by the modifications, although, compared with scDb, a reduced cytotoxic activity was observed for scDb-ABD, which was further reduced in the presence of albumin. These findings demonstrate that half-life extension of scDb results in increased tumor accumulation but that modifications might also affect scDb-mediated cytotoxicity.
EXPERIMENTAL PROCEDURES
Materials
Horseradish peroxidase (HRP)-conjugated anti-His-tag antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), unconjugated mouse anti-His-tag antibody from Dianova (Hamburg, Germany) and anti-rabbit IgG-FITC or anti-mouse IgG-PE-conjugated antibody from Sigma (Taufkirchen, Germany). Carcinoembryonic antigen was obtained from Europa Bioproducts (Cambridge, UK). HSA was purchased from Sigma. The human colon adenocarcinoma cell line LS174T was purchased from ECACC (Wiltshire, UK) and cultured in RPMI, 5% fetal bovine serum, 2 mm glutamine (Invitrogen, Karlsruhe, Germany). The stably transfected human fibroblast activation protein (FAP)-expressing fibrosarcoma cell line HT1080 FAPhu (kindly provided by W. Rettig, Boehringer Ingelheim Pharma, Vienna, Austria) were grown in RPMI, 5% fetal bovine serum, 2 mm glutamine. Buffy coat from healthy human donors were obtained from the blood bank (Ulm, Germany). IL-2 was purchased from Immunotools (Friesoythe, Germany) and phytohemagglutinin-L (PHA-L) from Boehringer-Mannheim (Mannheim, Germany). FcRn heavy chain knock-out mice (strain B6.129X1-Fcgrttm1Dcr/Dcr) were purchased from Jackson Laboratories (Bar Harbor, ME). Athymic female nude mice were purchased from Harlan Laboratories (Indianapolis, IN). For iodination, 125I and 131I from Perkin Elmer (Boston, MA) were used.
Biodistribution
ScDb and the various scDb derivatives were prepared as described previously (3, 4, 8). Antibody constructs were iodinated by the Iodogen method (Pierce Chemical Co.) yielding a specific activity of 1.4–2.2 μCi/μg. 6-week-old athymic nude mice were xenografted s.c. with 8 × 105 MC-38 murine colon carcinoma cells on the right flank and 106 LS-174T human colon carcinoma cells on the left flank. When the tumors reached a volume of ∼500 mm3, lugol iodine solution was added to the drinking water 1 day before the injection of the radiolabeled biomolecules. 4 groups of 3 mice received intravenous injections of a mix of 5 μg of 125I-scDb and 5 μg of 131I-scDb-ABD or 125I-scDb-A′-PEG40k and 131I-scDb-ABC7. Groups of mice were sacrificed at 2, 24, 48, or 96 h after i.v. injection. Blood, tumors, and tissues were weighed, and the radioactivity counted in a dual channel scintillation counter (CobraTM II, Packard).
ELISA
Carcinoembryonic antigen (CEA) (300 ng/well) was coated overnight at 4 °C, and remaining binding sites were blocked with 2% (w/v) skimmed milk powder/PBS. Purified recombinant antibodies and serum samples were titrated in duplicates and incubated for 1 h at room temperature. Detection was performed either with mouse HRP-conjugated anti-His tag antibody or rabbit antiserum against PEGylated scDb and horseradish peroxidase-conjugated goat anti-rabbit antibody (8) using TMB substrate (1 mg/ml TMB, sodium acetate buffer pH 6.0, 0.006% H2O2). The reaction was stopped with 50 μl of 1 m H2SO4. Absorbance was measured at 450 nm in an ELISA reader.
Affinity Measurements
Affinities of scDb-ABD for HSA at different pH were determined by quartz crystal microbalance measurements (A-100, Attana). HSA was chemically immobilized on a carboxyl sensor chip according to the manufacturer's protocol at a density resulting in a signal increase of 120 Hz. Binding experiments were performed in phosphate-buffered saline, pH 7.4, or 50 mm sodium phosphate buffer, 150 mm NaCl, pH 6.0 at a flow rate of 25 μl/min. The chip was regenerated with 6.3 μl of 10 mm HCl. Before every measurement a baseline was measured which was subtracted from the binding curve. A mass transport model (9) was fitted to the data.
Flow Cytometry
Binding to CEA- and CD3-expressing cells were determined by flow cytometry (10). LS174T or Jurkat-CD3 cells (2 × 105) were incubated with dilution series of antibodies for 3 h at 4 °C. Cells were then washed with phosphate-buffered saline and bound antibodies were detected with mouse anti-His tag antibody and PE-labeled goat anti-mouse antibody or, in case of the PEGylated construct, with rabbit antiserum (8) and FITC-labeled goat anti-rabbit IgG antibody. Data were fitted with GraphPrism software (La Jolla, CA) from three independent binding curves. From these three individual EC50, the mean and standard error was calculated.
Pharmacokinetics
Animal care and all experiments performed were in accordance with federal guidelines and have been approved by university and state authorities. C57BL/6 and BL/6 FcRn heavy chain knock-out mice (20–33 weeks, weight between 21–39 g) received an i.v. injection of 25 μg of scDb-ABD or PEGylated scDb in a total volume of 200 μl. In time intervals of 3 min, 3-h, 1-, 2-, 3-, and 4-day blood samples (100 μl) were taken from the tail and incubated on ice. Clotted blood was centrifuged at 10,000 × g for 10 min, 4 °C, and serum samples stored at −20 °C. Serum concentrations of CEA-binding recombinant antibodies were determined by ELISA (as described above), interpolating the corresponding calibration curves. For comparison, the first value (3 min) was set to 100%. Terminal half-lives (t½β) and AUC were calculated with Excel. For statistics, Student's t test was applied.
In Vitro Cytotoxicity
Cytotoxicity assays were performed according to Asano et al. (11). 15,000 LS174T or 5,000 HT1080 FAPhu cells per well were seeded into 96-well plates grown overnight. Dilution series of antibodies were then added to the target cells. Peripheral blood mononuclear cells (PBMCs) from a healthy donor were isolated from buffy coat as described before (3). PBMCs were thawed the day before and seeded on a cell culture dish to remove monocytes by the attachment to the plastic surface. Cells that remained in suspension were preactivated with 1 μg/ml PHA-L and 100 units/ml IL-2 (3) for at least 3 days. These preactivated PBMCs were added to the target cells in an E:T ratio of 3:1 and incubated for 24 h. After the wells were washed three times with PBS, 100 μl of medium with 50 μg/ml MTT (methylthiazolyldiphenyl-tetrazolium bromide) (Sigma) was added, and cells were incubated for 2 h. Then 100 μl of lysis buffer (10% SDS, 50% N,N-dimethylformamide, pH 4.7) was added, and wells were incubated overnight. A595 nm − A655 nm was measured in an ELISA reader, and data were normalized to the values of the untreated cell.
RESULTS
Biodistribution
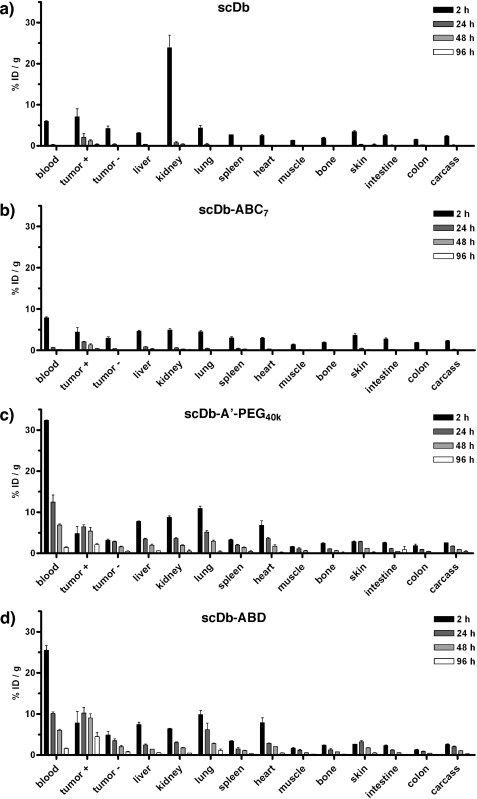
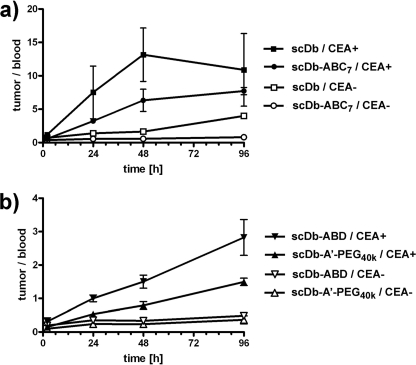
Single-chain diabody scDb CEACD3 and three derivatives thereof (PEGylated scDb, scDb-A′-PEG40k; N-glycosylated scDb possessing 9 NXT sequons, scDb-ABC7; scDb fused to an albumin-binding domain, scDb-ABD) were labeled with 125I or 131I and injected into nude mice bearing CEA+ LS174T and CEA− MC38 tumors. Tissue distribution was monitored over a period of 4 days (Fig. 1 and Table 1). Unmodified scDb was rapidly cleared from the blood and showed strong kidney accumulation after 2 h (Fig. 1a). A selective accumulation within the CEA+ tumor was observed with the highest value (∼7% ID/g) after 2 h. Similar values were found for the N-glycosylated scDb (scDb-ABC7), which, however, showed after 2 h as compared with scDb a strongly reduced accumulation in the kidney (Fig. 1b). PEGylated scDb and scDb-ABD had a strongly increased residence in the blood, which resulted also in increased values in all the other organs (Fig. 1, c and d). Both modified scDb variants exhibited an increased and selective accumulation in the CEA+ tumors reaching maximum values after 24 to 48 h, while concentrations in the CEA− tumors gradually dropped similar to the other organs and tissues analyzed. Enrichment in CEA+ tumors was also evident from increased tumor-to-blood ratios in comparison to CEA− tumors (Fig. 2). Over the period of 4 days, scDb reached a maximal tumor-to-blood ratio in CEA+ tumors of ∼13 after 2 days, while that of scDb-ABC7 reached only a value of ∼8 after 4 days (Fig. 2a). Tumor-to-blood ratios of scDb-A′-PEG40k and scDb-ABD in CEA+ tumors were weaker and gradually increased to a value of 1.5 and 3, respectively, at day 4 (Fig. 2b). The tumor-to-blood ratio in CEA− tumors did not change over the period of 4 days, except for scDb, which was slightly higher than that of scDb-ABC7 reaching a value of ∼4 after 4 days.
FIGURE 1.
Organ distribution of 131I-labeled scDb (a), 131I-labeled glycosylated scDb-ABC7 (b), 125I-labeled scDb-A′-PEG40k (c), and 125I-labeled scDb-ABD (d) in nude mice bearing subcutaneous CEA+ (LS174T) and CEA− (MC38) tumors.
TABLE 1.
AUC of the various scDb constructs in different tissues (% ID/g·h)
| Tissue | scDb | scDb-ABC7 | scDb-A′-PEG40k | scDb-ABD |
|---|---|---|---|---|
| Blood | 77.7 ± 3.1 | 110.0 ± 2.9 | 924.9 ± 34.3 | 767.8 ± 18.3 |
| CEA+ tumor | 167.7 ± 19.8 | 149.9 ± 6.4 | 450.0 ± 13.8 | 752.5 ± 46.6 |
| CEA− tumor | 66.5 ± 14.3 | 45.7 ± 3.8 | 170.9 ± 8.7 | 225.8 ± 23.3 |
| Liver | 50.0 ± 2.9 | 85.3 ± 2.1 | 251.9 ± 11.3 | 202.6 ± 12.8 |
| Kidney | 334.0 ± 28.6 | 81.2 ± 5.4 | 262.7 ± 8.3 | 217.0 ± 7.5 |
| Lung | 64.9 ± 14.3 | 66.7 ± 2.2 | 354.8 ± 15.6 | 377.1 ± 4.3 |
| Spleen | 39.6 ± 0.6 | 56.0 ± 3.4 | 145.7 ± 4.6 | 119.4 ± 6.2 |
| Heart | 33.2 ± 2.4 | 44.9 ± 1.5 | 227.6 ± 15.6 | 239.8 ± 13.2 |
| Muscle | 16.7 ± 0.5 | 24.5 ± 1.2 | 67.7 ± 4.2 | 70.6 ± 9.7 |
| Bone | 25.5 ± 3.7 | 32.1 ± 1.8 | 81.3 ± 1.4 | 89.2 ± 5.1 |
| Skin | 58.7 ± 1.1 | 58.1 ± 4.4 | 147.6 ± 4.2 | 172.0 ± 9.7 |
| Intestine | 32.9 ± 6.5 | 38.3 ± 3.5 | 95.0 ± 20.8 | 78.7 ± 2.2 |
| Colon | 23.6 ± 2.9 | 27.5 ± 1.3 | 63.1 ± 5.9 | 53.8 ± 3.1 |
FIGURE 2.
Tumor-to-blood ratios over the period of 4 days of (a) 131I-labeled scDb and 131I-labeled glycosylated scDb-ABC7, and (b) 125I-labeled scDb-A′-PEG40k and 125I-labeled scDb-ABD in CEA+ (LS174T) and CEA− (MC38) tumors.
Over the period of 4 days, PEGylated scDb showed as compared with scDb a 12-fold increase in AUC in the blood, while that of scDb-ABD was increased 10-fold. The blood AUC of scDb-ABC7 was only increased 1.4-fold. The strongest accumulation in the CEA+ tumor was found for scDb-ABD with a 4.5-fold increase in AUC as compared with scDb. The AUC of the PEGylated scDb in the CEA+ tumor was increased 2.7-fold. Comparing the AUC between CEA+ and CEA− tumors we observed for all constructs 2.5–3.3-fold higher values in the CEA+ tumors. Tumor-to-blood ratios for CEA+ tumors were 2.2, 1.4, 0.5, and 1.0 for scDb, scDb-ABC7, PEGylated scDb, and scDb-ABD, respectively, as determined from the AUC. Tumor-to-blood ratios for CEA− tumors were 0.9, 0.4, 0.2, and 0.3 for scDb, scDb-ABC7, PEGylated scDb, and scDb-ABD, respectively, as determined from the AUC.
Binding to CEA- and CD3-expressing Cells
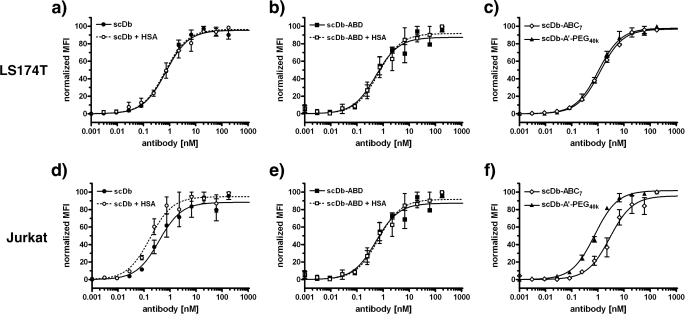
Binding of scDb and the half-life-extended derivatives to CEA-expressing LS174 cells and CD3-expressing Jurkat cells was determined by flow cytometry measurements (Fig. 3 and Table 2). All molecules exhibited a similar EC50 for CEA+ LS174T (in the range of 0.7–1.2 nm). Also, EC50 values of scDb, scDb-ABD, and PEGylated scDb for CD3+ Jurkat cells were in a similar range (between 0.4 and 0.8 nm). ScDb-ABC7 showed a somewhat reduced binding for CD3+ cells (EC50 = 3.4 nm), however, this difference was statistically not significant (p = 0.13). EC50 values of scDb and scDb-ABD for both antigens were not significantly affected by the presence of 1 mg/ml HSA (scDb ± HSA: p = 0.73 for binding to LS174T and p = 0.16 for binding to Jurkat; scDb-ABD ± HSA: p = 0.99 for binding to LS174T and p = 0.5 for binding to Jurkat).
FIGURE 3.
Flow cytometry analysis of binding of scDb and its half-life extended derivatives to CEA-expressing LS174T (a–c) and CD3-positive Jurkat cells (d–f). For scDb and scDb-ABD, the EC50 was determined in the absence or presence of HSA (1 mg/ml) (n = 3).
TABLE 2.
Binding of scDb and its derivatives for cell surface-expressed CEA and CD3
| Construct | HSA | EC50 for LS174T | EC50 for Jurkat |
|---|---|---|---|
| nm | nm | ||
| scDb | – | 0.7 ± 0.1 | 0.4 ± 0.1 |
| scDb | + | 0.8 ± 0.2 | 0.2 ± 0.03 |
| scDb-ABD | – | 0.9 ± 0.4 | 0.6 ± 0.2 |
| scDb-ABD | + | 0.9 ± 0.4 | 1.3 ± 0.8 |
| scDb-A′-PEG40k | – | 0.7 ± 0.1 | 0.8 ± 0.04 |
| scDb-ABC7 | – | 1.2 ± 0.01 | 3.4 ± 1.6 |
Affinity of scDb-ABD for HSA
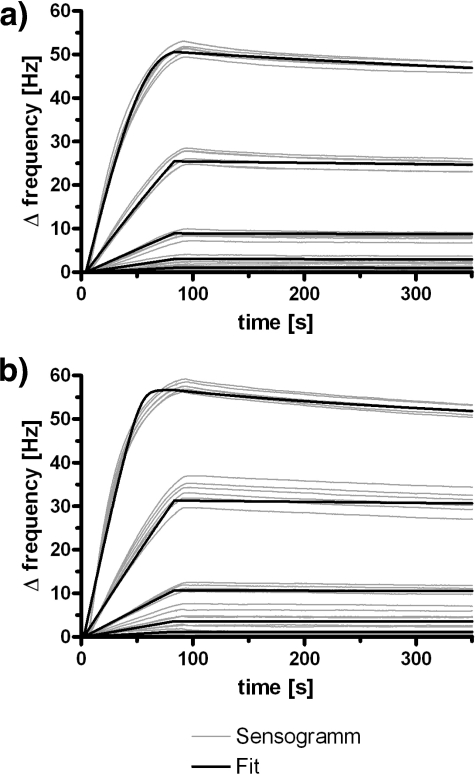
The affinity of scDb-ABD for HSA was determined by quartz crystal microbalance measurements (Fig. 4 and Table 3). At neutral pH (pH 7.4) scDb-ABD exhibited an affinity of 1.7 nm for HSA. At acidic pH (pH 6.0) affinity was 0.9 nm for HSA. These findings confirm that scDb-ABD is capable of binding to serum albumin at both neutral and acidic pH (4).
FIGURE 4.
QCM affinity measurements of scDb-ABD binding to immobilized HSA at pH 7. 4 (a) and pH 6.0 (b) (n = 6). Data were fitted (bold lines) assuming mass-limited transport.
TABLE 3.
Binding of scDb-ABD to HSA at pH 7.4 and 6.0 analyzed by QCM
| pH | kon | koff | KD |
|---|---|---|---|
| m−1s−1 | s−1 | nm | |
| 7.4 | 2.0 × 105 ± 3.0 × 103 | 3.5 × 10−4 ± 4.5 × 10−6 | 1.7 ± 0.04 |
| 6.0 | 4.3 × 105 ± 1.1 × 104 | 4.1 × 10−4 ± 8.7 × 10−6 | 0.9 ± 0.05 |
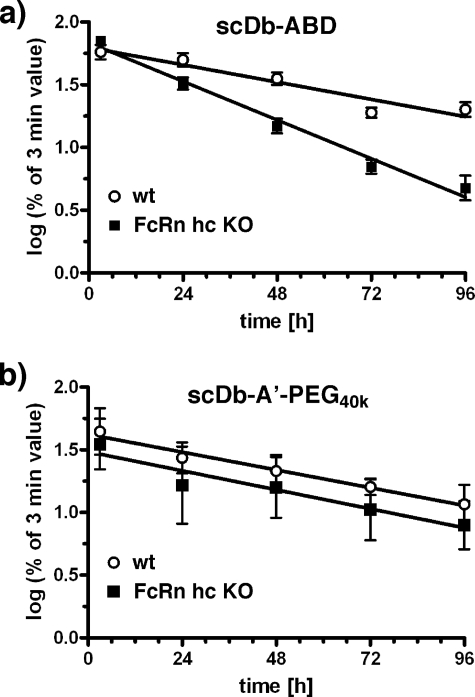
Pharmacokinetics of scDb-ABD in FcRn Heavy Chain Knock-out Mice
Next, we analyzed clearance of scDb-ABD in C57BL/6 wild-type and FcRn heavy chain knock-out mice. After a single dose i.v. injection into the tail vein blood samples were taken and analyzed by ELISA for the presence of active molecules (Fig. 5). A terminal half-life of 53.0 ± 10.0 h was determined for scDb-ABD in wt mice (n = 10), while half-life was reduced to 24.8 ± 2.2 h in the FcRn knock-out mice (n = 10). In contrast, no differences of the terminal half-lives were observed for scDb-A′-PEG40k, included as control, with 47.9 ± 2.7 h in wt mice (n = 2) and 51.4 ± 4.3 h in FcRn heavy chain knock-out mice (n = 3).
FIGURE 5.
Plasma clearance of scDb-ABD (a) or scDb-A′-PEG40k (b) in C57BL/6 wild-type (wt) and FcRn heavy chain knock-out (FcRn hc KO) mice after a single i.v. dose (25 μg) of antibody molecules.
In Vitro Cytotoxicity of scDb and scDb-ABD
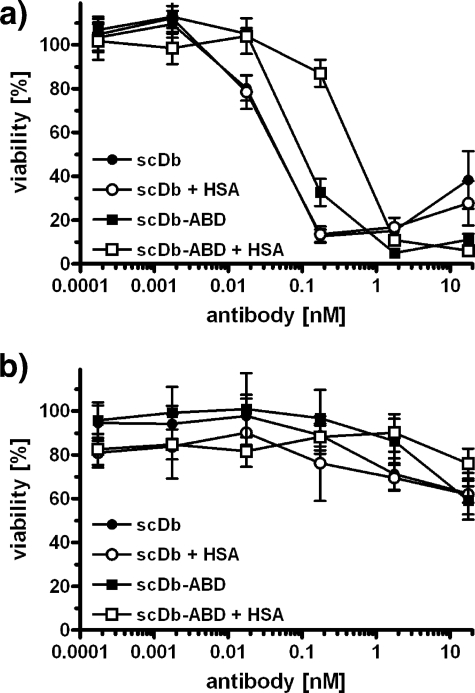
Using pre-activated human PBMCs, we observed a concentration-dependent killing of CEA+ LS174T tumor cells by scDbCEACD3 and scDbCEACD3-ABD (Fig. 6a). At an effector to target (E:T) ratio of 3, half-maximal killing (EC50) was reached at 50 pm scDb and 100 pm scDb-ABD, respectively. The addition of 1 mg/ml HSA had no effect on the EC50 of scDb. In contrast, the EC50 of scDb-ABD was reduced 5-fold to 500 pm. No cytotoxicity of scDb and scDb-ABD was observed toward CEA− HT1080 cells (Fig. 6b).
FIGURE 6.
ScDb-mediated cytotoxicity. CEA+ LS174T cells (a) or CEA− HT1080 cells (b) were incubated with preactivated PBMCs at a ratio of 1:3 and varying concentrations of scDbCEACD3 and scDbCEACD3-ABD in the presence or absence of HSA (1 mg/ml). Remaining viable target cells were determined by MTT assay after 24 h (n = 3).
DISCUSSION
In the present study we determined the biodistribution of an anti-CEA × anti-CD3 bispecific single-chain diabody and showed that a prolonged half-life translates into an increased accumulation in CEA-positive tumors. All modified scDb molecules exhibited similar EC50 values for binding to CEA- and CD3-expressing cell lines indicating that modifications do not interfere with binding to the cell surface-exposed antigens. Thus, the reduced accumulation of scDb and scDb-ABC7 as compared with PEGylated scDb and scDb-ABD is not caused by a reduced affinity for the CEA-expressing tumor cells. Furthermore, binding of scDb and scDb-ABD to these cell lines was not affected in the presence of serum albumin. This finding is in accordance with results obtained for a half-life-extended anti-HER2 Fab 4D5, which was either fused to the same ABD or an albumin-binding peptide (AB.Fab4D5) (12, 13). In contrast, different results were described for PEGylated antibody fragments. Thus, C-terminal conjugation of the scFv 4D5 with a 20-kDa PEG resulted in a 5-fold loss of affinity (14), while, for example, a PEGylated tandem scFv directed against MUC-1 showed a similar binding to MUC-1 as the wild-type tandem scFv (15). A recent study of polysialylated anti-CEA scFv MFE-23 molecules revealed that the conjugation chemistry has a strong effect on immunoreactivity (16). Random polysialylation of in average 1.4 11-kDa PSA chains per scFv resulted as compared with the unmodified scFv in a 20-fold reduction of binding in ELISA. In contrast, no reduction was observed after site-directed conjugation of 1 PSA polymer to a C-terminal cysteine residue, which is similar to our approach of generating PEGylated scDb.
Although binding of scDb to CEA+ tumor cells and CD3+ effector cells was not affected by the various modifications, previous studies showed that they reduce the bispecific antibody-mediated stimulatory activity on PBMCs in a target cell-dependent assay (4, 8). In these studies, a 3–12-fold reduction was observed for scDb-ABC7, PEGylated scDb, and scDb-ABD. The T cell-stimulating activity of scDb-ABD was 3-fold reduced as compared with the unmodified scDb and was further reduced 4-fold in the presence of HSA, while HSA had no effect on the stimulating activity of unmodified scDb. As shown in the present study, this resulted also in a comparable reduction of the cytotoxic effects using preactivated human PBMCs as effector cells. Presumably, these modifications lead to a sterical hindrance of the formation of a close contact between target and effector cells and/or the ability to efficiently activate the T cell receptor through binding to CD3. This reduction might also be influenced by the choice of target antigen and the location of the epitope. Further studies are, therefore, required to investigate the effects of half-life extending modifications on the bioactivity of other bispecific antibodies.
The modifications leading to the longest half-life extension, i.e. PEGylation and fusion to an ABD, resulted in increased accumulation in CEA+ tumors. The highest tumor accumulation was found for the scDb-ABD fusion protein, which was ∼5-fold increased as compared with the unmodified scDb. Interestingly, PEGylated scDb showed a ∼2-fold lower accumulation in CEA+ tumors as compared with scDb-ABD, although possessing a slightly higher plasma half-life and a similar cell binding activity. In a previous study, we showed that the hydrodynamic radius of scDb-ABD bound to HSA is 4.8 nm compared with 7.9 nm of the PEGylated scDb, which might explain the better tumor penetration of scDb-ABD (8). Increased tumor accumulation was also observed for various other half-life-extended recombinant antibody molecules, such as PEGylated anti-HER2 scFv 4D5 and anti-HER2 AB.Fab4D5 (13, 14). The AB.Fab showed a 5–6-fold higher tumor accumulation than the Fab without the binding peptide, and the accumulation was much faster in case of the AB.Fab than in case of the parental IgG (13). The latter finding also indicates that the size of the molecule (the MW of AB.Fab bound to albumin is ∼130 kDa, that of IgG 150 kDa) influences tumor accumulation. Furthermore, the noncovalent binding of scDb-ABD (and AB.Fab) to albumin, allowing the dissociation of the antibody moiety from albumin, might facilitate tumor penetration. The site-directed conjugation of an scFv directed against HER2 with PEG 20 kDa caused a 8.5-fold better tumor accumulation (14). Recently an anti-EGFR tandem nanobody was fused to an anti-albumin nanobody. The nanobody construct revealed a similar circulation half-life (∼48 h) in mice as our scDb-ABD construct. Compared with the anti-EGFR IgG cetuximab, the nanobody construct showed a similar tumor accumulation, but distribution within the tumor tissue was more homogeneous (17). This might be due to the smaller size of the nanobody construct (∼50 kDa), which is similar to the size of the scDb-ABD, as compared with 150 kDa for the IgG molecule. Site-directed polysialylation of anti-CEA scFv MFE-23 (MFE-23-Cys-PSA) with an apparent molecular mass of >300 kDa as determined by size exclusion chromatography showed a 10-fold increased tumor accumulation as compared with unmodified MFE-23, leading to a maximum accumulation of ∼10% ID/g tumor (16). These data compare very well with our own data obtained for PEGylated scDb and scDb scDb-ABD. Thus, these experiments further confirm that an extended half-life translates into increased tumor accumulation.
The long circulation time of scDb-ABD is caused by an increased hydrodynamic radius of the scDb-ABD albumin complex, which is ∼2-fold increased as compared with scDb (4.8 nm versus 2.7 nm) (8), as well as recycling by the FcRn. In the present study we confirmed that recycling by FcRn contributes to the long half-life of scDb-ABD. In FcRn heavy chain knock-out mice half-life of scDb-ABD was reduced ∼2-fold. Similar values were reported for the half-life of radioiodinated mouse albumin, which was reduced from 39 h in wild-type mice to 24 h in heavy chain knock-out mice (18). HSA binds pH-dependent to the FcRn with an affinity of ∼5 μm at pH 6.0 (19, 20). A prerequisite of the recycling of scDb-ABD via the FcRn is that the complex between scDb-ABD, albumin and FcRn remains stable in the acidic environment of the endosome. In QCM studies we found that the affinity of scDb-ABD is not decreased at pH 6.0 and that the measured affinities are similar to those determined by others for the binding of the single ABD domain to HSA at neutral pH (21–23).
Our results and data from others clearly demonstrate that PEGylation, N-glycosylation, and fusion to an ABD can improve pharmacokinetic properties of recombinant antibodies to various extent. However, these improvements might not only be dependent on the applied half-life extension strategy but also from the antibody format itself, which directly affects protein size and stability (24, 25). Furthermore, it is likely that antibody affinity and valency but also other factors such as tumor vascularization, vessel permeability, and the kind of the target antigen as well as its density on target cell and within the tumor plays a determinant role in biodistribution (26–30). Thus, different results might be obtained for scDb recognizing other target cell antigens. In this context, it is noteworthy that no experimental data are currently available to which extend binding to CD3-positive T cells affects plasma half-life and biodistribution. A physiological-based pharmacokinetic model suggests that rather than guiding the T cells to the target tissue, the bispecific antibody is being dragged around the system by the T cell (31). Experimental data of a bispecific tandem scFv directed against CD19 and CD3 analyzed in chimpanzees revealed a half-life of 2 h, indicating that the binding to T-cells has no beneficial impact on the circulation time (32). The scDb CEACD3 used in our study is human-specific and, therefore, does not bind to mouse T cells complicating the analysis of this aspect, for instance in mice. This question can be answered using a bispecific scDb directed against mouse CD3. A respective scFv (2C11) is available (33) and has been already used by others to study antitumor effects of bispecific tandem scFv molecules possessing a similar size as scDb molecules (34). In an immunocompetent mouse model the antitumor effect of these small bispecific molecules on solid tumors could be demonstrated (35). Further studies are therefore planned to convert our scDb and its derivatives into bispecific anti-CEA × anti-mouse CD3 molecules.
In summary, we showed for our scDb that an extended half-life translates into increased tumor accumulation and that fusion of an albumin-binding domain to a scDb is superior compared with a PEGylated scDb prepared by site-directed conjugation of a 40-kDa branched PEG chain. Further studies in immunocompetent mice are now planned to investigate the antitumor activity of these bispecific T cell-recruiting antibody derivatives to elucidate if the prolonged circulation time compensates for the reduced bioactivity observed for the target cell-dependent activation of cellular cytotoxicity in vitro.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Ko1461/2).
- scDb
- single-chain diabody
- ABD
- albumin-binding domain
- AUC
- area under the curve
- CEA
- carcinoembryonic antigen
- FcRn
- neonatal Fc receptor
- HSA
- human serum albumin
- PBMC
- peripheral blood mononuclear cells
- PEG
- polyethylene glycol
- QCM
- quartz crystal microbalance
- scFv
- single-chain fragment variable
- MTT
- methylthiazolyldiphenyl-tetrazolium bromide
- wt
- wild type.
REFERENCES
- 1.Kontermann R. E. (2005) Acta Pharmacol. Sin. 26, 1–9 [DOI] [PubMed] [Google Scholar]
- 2.Müller D., Kontermann R. E. (2007) Curr. Opin. Mol. Ther. 9, 319–326 [PubMed] [Google Scholar]
- 3.Müller D., Karle A., Meissburger B., Höfig I., Stork R., Kontermann R. E. (2007) J. Biol. Chem. 282, 12650–12660 [DOI] [PubMed] [Google Scholar]
- 4.Stork R., Müller D., Kontermann R. E. (2007) Protein Eng. Des. Sel. 20, 569–576 [DOI] [PubMed] [Google Scholar]
- 5.Batra S. K., Jain M., Wittel U. A., Chauhan S. C., Colcher D. (2002) Curr. Opin. Biotechnol. 13, 603–608 [DOI] [PubMed] [Google Scholar]
- 6.Bargou R., Leo E., Zugmaier G., Klinger M., Goebeler M., Knop S., Noppeney R., Viardot A., Hess G., Schuler M., Einsele H., Brandl C., Wolf A., Kirchinger P., Klappers P., Schmidt M., Riethmüller G., Reinhardt C., Baeuerle P. A., Kufer P. (2008) Science 321, 974–977 [DOI] [PubMed] [Google Scholar]
- 7.Kontermann R. E. (2009) BioDrugs 23, 93–109 [DOI] [PubMed] [Google Scholar]
- 8.Stork R., Zettlitz K. A., Müller D., Hanisch F.-G., Kontermann R. E. (2008) J. Biol. Chem. 283, 7804–7812 [DOI] [PubMed] [Google Scholar]
- 9.Myszka D. G. (1997) Curr. Opin. Biotechnol. 8, 50–57 [DOI] [PubMed] [Google Scholar]
- 10.Benedict C. A., MacKrell A. J., Anderson W. F. (1997) J. Immunol. Methods 201, 223–231 [DOI] [PubMed] [Google Scholar]
- 11.Asano R., Sone Y., Makabe K., Tsumoto K., Hayashi H., Katayose Y., Unno M., Kudo T., Kumagai I. (2006) Clin. Cancer Res. 12, 4036–4042 [DOI] [PubMed] [Google Scholar]
- 12.Schlapschy M., Theobald I., Mack H., Schottelius M., Wester H. J., Skerra A. (2007) Protein Eng. Des. Sel. 20, 273–284 [DOI] [PubMed] [Google Scholar]
- 13.Dennis M. S., Jin H., Dugger D., Yang R., McFarland L., Ogasawara A., Williams S., Cole M. J., Ross S., Schwall R. (2007) Cancer Res. 67, 254–261 [DOI] [PubMed] [Google Scholar]
- 14.Kubetzko S., Balic E., Waibel R., Zangemeister-Wittke U., Plückthun A. (2006) J. Biol. Chem. 281, 35186–35201 [DOI] [PubMed] [Google Scholar]
- 15.Xiong C. Y., Natarajan A., Shi X. B., Denardo G. L., Denardo S. J. (2006) Protein Eng. Des. Sel. 19, 359–367 [DOI] [PubMed] [Google Scholar]
- 16.Constantinou A., Epenetos A. A., Hreczuk-Hirst D., Jain S., Wright M., Chester K. A., Deonarain M. P. (2009) Bioconjugate Chem. 20, 924–931 [DOI] [PubMed] [Google Scholar]
- 17.Tijink B. M., Laeremans T., Budde M., Stigter-van Walsum M., Dreier T., de Haard H. J., Leemans C. R., van Dongen G. A. (2008) Mol. Cancer Ther. 7, 2288–2297 [DOI] [PubMed] [Google Scholar]
- 18.Chaudhury C., Mehnaz S., Robinson J. M., Hayton W. L., Pearl D. K., Roopenian D. C., Anderson C. L. (2003) J. Exp. Med. 197, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen J. T., Dee Qian J., Sandlie I. (2006) Eur. J. Immunol. 36, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 20.Chaudhury C., Brooks C. L., Carter D. C., Robinson J. M., Anderson C. L. (2006) Biochemistry 45, 4983–4990 [DOI] [PubMed] [Google Scholar]
- 21.Jonsson A., Dogan J., Herne N., Abrahmsén L., Nygren P. A. (2008) Protein Eng. Des. Sel. 21, 515–527 [DOI] [PubMed] [Google Scholar]
- 22.Linhult M., Binz H. K., Uhlén M., Hober S. (2002) Protein Sci. 11, 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson M. U., Frick I. M., Nilsson H., Kraulis P. J., Hober S., Jonasson P., Linhult M., Nygren P. A., Uhlén M., Björck L., Drakenberg T., Forsén S., Wikström M. (2002) J. Biol. Chem. 277, 8114–8120 [DOI] [PubMed] [Google Scholar]
- 24.Colcher D., Pavlinkova G., Beresford G., Booth B. J., Choudhury A., Bastra S. K. (1998) Q. J. Nucl. Med. 42, 225–241 [PubMed] [Google Scholar]
- 25.Borsi L., Balza E., Bestagno M., Castellani P., Carnemolla B., Biro A., Leprini A., Sepulveda J., Burrone O., Neri D., Zardi L. (2002) Int. J. Cancer 102, 75–85 [DOI] [PubMed] [Google Scholar]
- 26.Adams G. P., Schier R., Marshall K., Wolf E. J., McCall A. M., Marks J. D., Weiner L. M. (1998) Cancer Res. 58, 485–490 [PubMed] [Google Scholar]
- 27.Adams G. P., Tai M. S., McCartney J. E., Marks J. D., Stafford W. F., 3rd, Houston L. L., Huston J. S., Weiner L. M. (2006) Clin. Cancer Res. 12, 1599–1605 [DOI] [PubMed] [Google Scholar]
- 28.Wu A. M., Chen W., Raubitschek A., Williams L. E., Neumaier M., Fischer R., Hu S. Z., Odom-Maryon T., Wong J. Y., Shively J. E. (1996) Immunotechnology 2, 21–36 [DOI] [PubMed] [Google Scholar]
- 29.Juweid M., Neumann R., Paik C., Perez-Bacete M. J., Sato J., van Osdol W., Weinstein J. N. (1992) Cancer Res. 52, 5144–5153 [PubMed] [Google Scholar]
- 30.Shockley T. R., Lin K., Nagy J. A., Tompkins R. G., Yarmush M. L., Dvorak H. F. (1992) Cancer Res. 52, 367–376 [PubMed] [Google Scholar]
- 31.Friedrich S. W., Lin S. C., Stoll B. R., Baxter L. T., Munn L. L., Jain R. K. (2002) Neoplasia 4, 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlereth B., Quadt C., Dreier T., Kufer P., Lorenczewski G., Prang N., Brandl C., Lippold S., Cobb K., Brasky K., Leo E., Bargou R., Murthy K., Baeuerle P. A. (2006) Cancer Immunol. Immunother. 55, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao K. W., Lo Y. C., Roffler S. R. (2000) Gene Ther. 7, 339–347 [DOI] [PubMed] [Google Scholar]
- 34.Amann M., Friedrich M., Lutterbuese P., Vieser E., Lorencewski G., Petersen L., Brischwein K., Kufer P., Kischel R., Baeuerle P. A., Schlereth B. (2009) Cancer Immunol. Immunother. 58, 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlereth B., Kleindienst P., Fichtner I., Lorenczewski G., Brischwein K., Lippold S., da Silva A., Locher M., Kischel R., Lutterbüse R., Kufer P., Baeuerle P. A. (2006) Cancer Immunol Immunother. 55, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]