Abstract
Retinoic acid (RA) causes HL-60 human myeloblastic leukemia cell myeloid differentiation that is dependent on MAPK signaling. The process is propelled by c-Cbl, which binds the CD38 receptor as part of a signaling complex generating MAPK signaling. Here we report that the capability of c-Cbl to do this is lost in the G306E tyrosine kinase-binding domain mutant. Unlike wild-type (WT) c-Cbl, the G306E mutant c-Cbl fails to propel RA-induced differentiation, and disrupts the normal association with CD38. The G306E mutant does, like WT c-Cbl, co-immunoprecipitate with Vav, Slp-76, and p38. But unlike WT c-Cbl, does not cause MAPK signaling. In contrast, the C381A Ring finger domain mutant functions like WT c-Cbl. It binds CD38 and is part of the same apparent c-Cbl/Slp-76/Vav/p38 signaling complex. The C381A mutant causes MAPK signaling and propels RA-induced differentiation. In addition to HL-60 cells and their WT or mutant c-Cbl stable transfectants, the c-Cbl/Vav/Slp-76 complex is also found in NB4 cells where c-Cbl was previously also found to bind CD38. The data are consistent with a model in which the G306E mutant c-Cbl forms a signaling complex that includes Slp-76, Vav, and p38; but does not drive MAPK signaling because it fails to bind the CD38 receptor. Without the G306E mutation the c-Cbl unites CD38 with the signaling complex and delivers a MAPK signal that drives RA-induced differentiation. The results demonstrate the importance of the Gly306 residue in the ability of c-Cbl to propel RA-induced differentiation.
Retinoic acid (RA),2 a form of vitamin A, can cause activation of MAPK signaling leading to induced cell differentiation and G0 cell cycle arrest (1–2). Because of this, RA has been used therapeutically for the chemoprevention and treatment of cancer (3), notably acute promyelocytic leukemia, making its mechanism of action of significant interest. The HL-60 human myeloblastic leukemia cell line serves as a model for studying differentiation induction therapy and the mechanism of RA action (4–6). The HL-60 cell line was established from peripheral blood leukocytes of a patient originally diagnosed with acute promyelocytic leukemia (4), which was retrospectively re-evaluated to be acute myeloblastic leukemia (7). HL-60 cells undergo G0 cell cycle arrest and myeloid differentiation in response to RA or monocytic differentiation in response to 1,25-dihydroxyvitamin D3. Induced differentiation depends on hyperactive MAPK signaling (1, 8). c-Cbl contributes propulsion to this process (9, 10). It is part of a signaling complex connected to the CD38 receptor that activates the RAF/MEK/ERK MAPK signaling axis to drive differentiation and G0 cell cycle arrest (9).
The 120-kDa product of the c-Cbl protooncogene is a prominent component of tyrosine kinase signaling cascades downstream of activated cell surface receptors, including the epidermal growth factor receptor, the B cell receptor, Fc receptor, and cytokine receptors (11, 12). Cbl proteins have a highly conserved N-terminal domain termed the tyrosine kinase-binding (TKB) domain, which binds to phosphotyrosines on activated receptor-tyrosine kinases and other signaling proteins (13, 14), a short linker region, and a Ring finger domain that binds to ubiquitin-conjugating enzymes (15, 16). The Ring finger domain of c-Cbl presents the most conserved region among Cbl family proteins, and it is implicated as an important element in the function of Cbl proteins (17–19). The TKB domain contains a four-helix bundle, EF-hand calcium-binding domain, and a variant SH2 domain that binds to phosphotyrosine residues (13, 20). Several features of the Cbl/Zap-70 complex (13) suggest that the four-helix bundle, EF-hand, and SH2 domains together form an interactive structure that is crucial for phosphoprotein recognition. Studies by Thien et al. (21) indicated that the TKB mutation, G306E, fails to bind phosphotyrosine residues and is a loss of function mutation in the c-Cbl TKB domain.
Cbl interacts with a number of intracellular signaling molecules including various receptors, adaptors, cytoskeletal proteins, ubiquitin, and structural proteins via its various domains (22, 23). One of the receptors is CD38. The human cell surface antigen CD38 can be found in lipid rafts and causes RAF and ERK activation (24, 25). Kontani et al. (26) suggested that CD38 causes tyrosine phosphorylation of target molecules, including the Cbl adaptor. Vav is another signal regulator that plays a role in several cellular functions, including cell proliferation and maturation, cytoskeletal reorganization, regulation of gene expression, and apoptosis (27, 28). Bertagnolo et al. (29) reported that Vav is a potential target of Syk and that the SH3-SH2-SH3 fragment of Vav can associate with Cbl and Slp76. Slp76 was first identified as a substrate of the protein-tyrosine kinases that are engaged by activated T-cell receptors. It lacks intrinsic enzymatic activity but contains multiple protein-binding domains. Slp76 can interact with Cbl (30, 31). p38 is a member of the MAPK signaling family and is associated with cellular stress responses and apoptosis (32, 33). Studies by Frey and colleagues (34) have shown that epidermal growth factor receptor ubiquitinylation and Cbl activation are dependent on p38 activity. It is not known if an interaction between c-Cbl and p38 is induced by RA to support RA-induced differentiation and G0 arrest.
Although the biological importance of the adaptor function of the Cbl protein and E3 ligase activity is now apparent, the precise role of specific residues of Cbl in cell differentiation and cell growth, as well as in the interaction of c-Cbl with other cellular proteins still remains to be well elucidated. We have targeted two residues of c-Cbl located in the Ring finger domain and the TKB domain (see Fig. 1A). This study evaluates if these mutations affect the ability of c-Cbl to drive RA-induced differentiation by altering its interactions with partners. Binding to the CD38 receptor was determined. The interactions of c-Cbl and Vav, c-Cbl and Slp76, as well as c-Cbl and p38 were also investigated. The interaction studies were carried out in HL-60 cells as well as NB4 cells, which was derived from an APL patient and bears the t(15,17) promyelocytic leukemia-retinoic acid receptor α translocation (35, 36). The data are consistent with a model in which Cbl is in a complex with Vav, Slp76, and p38 and this subassembly MAPK signaling complex is recruited to CD38 by an interaction dependent on Cbl Gly306 to propel RA-induced differentiation through activation of MAPK signaling.
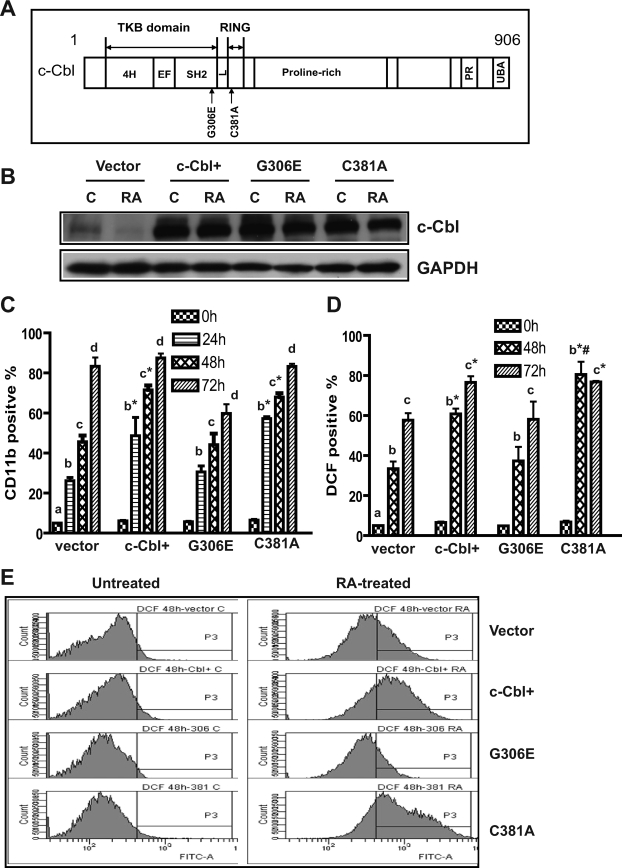
FIGURE 1.
c-Cbl TKB mutant failed to enhance CD11b expression and RA-induced cell differentiation compared with WT c-Cbl stably transfected cells (c-Cbl+). A, diagrammatic representation of the major domains of c-Cbl and mutations within the TKB and Ring finger domains used in this study. The c-Cbl protein consists of the TKB, the Ring finger domain (RING), proline-rich region, a PX(P/A)XXR motif (PR), and a ubiquitin-associated domain (UBA) at its C terminus. The TKB domain containing a four-helix bundle (4H), EF-hand calcium binding, and a variant SH2 domain is separated from the RING domain by a short linker (L) region. B, in stably transfected cells WT c-Cbl (c-Cbl+) or two c-Cbl mutants (G306E and C381A) were strongly overexpressed compared with c-Cbl in vector control cells. G306E and C381A were transfected into HL-60 cells. Western blots of c-Cbl (upper) and GAPDH (lower) expression in vector control, c-Cbl+, and G306E and C381C stable transfectant cells were untreated control (C) or RA treated (RA, 48 h). C, compared with vector controls, c-Cbl+ and C381A transfectants showed significantly enhanced expression of CD11b after RA treatment; however, the G306E mutant did not. Vector controls, WT c-Cbl, G306E, and C381A stable transfectants were treated with RA for the indicated times, and stained with allophycocyanin-conjugated anti-CD11b antibody. Bars are means ± S.E. of 3 repeats. D, RA-induced expression of the functional differentiation marker inducible oxidative metabolism was accelerated by WT c-Cbl and the C381A mutant, but not the G306E mutant. Vector control, c-Cbl+, G306E, and C381A stable transfectants were treated with RA for the indicated times, and the percentage of cells capable of inducible oxidation metabolism detected by 2′,7′-dichlorohydrofluorescein diacetate (DCF) fluorescence was analyzed by flow cytometry. E, representative DCF fluorescence histograms of untreated and (48 h) RA-treated vector control, WT c-Cbl, G306E, and C381A stable transfectants. The different letters indicate different time points. Asterisk indicates that CD11b or DCF expression levels from WT c-Cbl and C381A transfectants were significantly (p ≤ 0.05) different from vector controls and G306E transfectants. # denotes that C381A transfectants were significantly (p ≤ 0.05) different from c-Cbl+ transfectants.
EXPERIMENTAL PROCEDURES
Cell Culture
Human myeloblastic leukemia cells (HL-60) and their derivatives (WT c-Cbl, c-Cbl mutants G306E and C381A stable transfectants) were grown in a humidified atmosphere of 5% CO2 at 37 °C and maintained in RPMI 1640 supplemented with 5% fetal bovine serum (Invitrogen). The cells were cultured in constant exponential growth as previously described (37). The experimental cultures were initiated at a cell density of 0.2 × 106 cells/ml. To maintain c-Cbl high expression in stable transfectants, the cells were periodically resorted for high expression of fluorescent EGFP using fluorescence-activated cell sorting (FACSAria flow cytometer, BD Biosciences).
Chemicals
RA (Sigma) was dissolved in 100% ethanol with a stock concentration of 5 mm, and used at a final concentration of 1 μm as previously described (37).
Construction of c-Cbl Mutants
WT c-Cbl plasmid construction was described previously (9). For c-Cbl mutants, G306E and C381A, constructs were generated by site-directed mutagenesis using the QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Glycine 306 and cysteine 381 were mutated to glutamic acid 306 and alanine 381 using the primers, 5′-GGT CAG TGG GCT ATT GaG TAT GTT ACT GCT GAT G-3′ for G306E and 5′-G ATG GGC TCC ACA TTC CAA CTA gcT AAA ATA TGT GCT G-3′ for C381A, and c-Cbl knock-in plasmid (9) as template. All inserts were confirmed by sequencing.
CD11b and CD38 Expression Studies by Flow Cytometry
HL-60 cells (0.5 × 106) were harvested at 120 × g for 5 min. Cells were resuspended in 100 μl of PBS containing 5 μl of allophycocyanin-conjugated anti-CD11b antibody and phycoerythrin-conjugated anti-CD38 antibody (BD Biosciences). Following incubation for 1 h at 37 °C, cells were analyzed by flow cytometry (LSRII flow cytometer, BD Biosciences) using 633-nm red laser and 488-nm blue laser excitations. The threshold to determine the percent increase of expression was set at the highest 5% of control cells.
ERK Phosphorylation
Cells (0.5 × 106) were fixed by resuspension in 100 μl of PBS with 2% paraformaldehyde (Alfa Aesar, Ward Hill, MA) for a 10-min incubation at room temperature and then permeabilized by addition of 900 μl of methanol for 20 min at −20 °C. Following incubation and two washes, cells were stained with Alexa 647-conjugated anti-phospho-p44/42 MAPK antibody (Cell Signaling, Beverly, MA) for 1 h and analyzed by flow cytometry using 633-nm red laser excitation with emitted fluorescence reflected from a dichroic 735 long-pass through a 660/20 band-pass. The gate to determine the percent of increased expression was set to exclude 5% of HL-60 control cells, which represents basal levels of pERK.
Measurement of Inducible Oxidative Metabolism
0.5 × 106 cells were harvested by centrifugation and resuspended in 200 μl of PBS at 37 °C containing 10 μm 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (H2-DCF, Molecular Probes, Eugene, OR) and 0.4 μg/ml 12-O-tetradecanoylphorbol-13-acetate (TPA, Sigma) with incubation for 20 min in a humidified atmosphere of 5% CO2 at 37 °C. Flow cytometric analysis was done (BD LSRII flow cytometer) using 488-nm laser excitation and emission collected through 505 long-pass dichroic and 530/30 band-pass filters. The shift in fluorescence intensity in response to TPA was used to determine the percent of cells with the capability of generating inducible superoxide. Gates to determine the percent of positive cells were set to exclude 95% of control cells. Control cells with or without TPA and RA-treated cells without TPA typically showed indistinguishable DCF fluorescence histograms.
Cytology and Morphological Evaluation
RA-treated cells (vector control, c-Cbl+, G306E, and C381A transfectants) were harvested after 4-day RA treatments, and untreated cells were used as controls. 5 × 104 cells were cytospun on slides and stained with Wrights stain (Seimens, Tarrytown, NY). More than 50 cells were inspected and observed microscopically (Olympus BX41 microscope). Images were taken using a ×100 objective.
Cell Cycle Analysis
0.5 × 106 cells were collected by centrifugation and resuspended in 200 μl of cold hypotonic staining solution containing 50 μg/ml propidium iodine, 1 μl/ml Triton X-100, and 1 mg/ml sodium citrate. Cells were incubated at room temperature for 1 h and analyzed by flow cytometry (BD LSRII) using 488-nm excitation and collected through 550 long-pass dichroic and a 576/26 band-pass filters.
WT c-Cbl, Mutants G306E and C381A Stable Transfection
Fifty micrograms of plasmid DNA was transfected into HL-60 cells as previously described (38), and selected with G418 (1 mg/ml) for 2–3 weeks. Stable transfectants underwent 3 cycles of cell sorting and amplification to select cells expressing high fluorescent EGFP using fluorescence-activated cell sorting (FACSAria, BD Biosciences).
Fluorescence Resonance Energy Transfer (FRET)
0.5 × 106 cells were fixed in paraformaldehyde and permeabilized by methanol as described above. After two washes, cells were resuspended in 200 μl of PBS containing 5 μl of primary antibodies and then stained with Alexa 350- and 430-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies (Invitrogen). The immunocomplexes were analyzed using a BD LSRII flow cytometer. The FRET signal was measured using 325-nm excitation (from a UV laser) of Alexa 350 to excite Alexa 430 measuring the emitted fluorescence through 505 long-pass dichroic and 530/30 band-pass filters. Controls with secondary antibody(s) only or secondary(s) plus donor or acceptor primary antibody were included. Cells stained with just one primary antibody and Alexa 350 or 430, respectively, were used for compensation controls for spillover into all fluorescence collection channels. FRET signals were corrected by subtraction of background fluorescence of negative controls with just secondary antibodies and compensation controls. Rabbit anti-c-Cbl (Santa Cruz, CA) and mouse anti-CD38 (BD Biosciences) antibodies were used in Cbl/CD38 FRET measurements. The antibodies of mouse anti-c-Cbl (BD Biosciences) and rabbit anti-Vav (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-SLP76 or anti-p38 (Cell Signaling, Beverly, MA) were used in c-Cbl/Vav or c-Cbl/Slp76 or c-Cbl/p38 FRET measurements.
Immunoprecipitation (IP) and Western Blot
2.5 × 107 cells were lysed using 200 μl of lysis buffer (Pierce) and lysates were cleared by centrifugation at 16,950 × g in a microcentrifuge for 20 min at 4 °C. 10 μl of primary antibody was added and maintained with gentle shaking at 4 °C overnight. Anti-c-Cbl (Santa Cruz Biotechnology, Inc.), anti-Vav, anti-Slp76, or anti-p38 (Cell Signaling, Beverly, MA) was used in IPs. A negative control using an equivalent amount of normal rabbit IgG (Santa Cruz Biotechnology, Inc.) was included. 50 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology, Inc.) were then added and the final mixture was incubated at 4 °C under rotary agitation overnight. The protein/antibody/protein A/G slurry was centrifuged at 9,500 × g for 2 min and washed 3 times with co-IP buffer (Pierce). The immunoprecipitate was resolved by SDS-PAGE and Western blotted using anti-c-Cbl, anti-Vav, anti-Slp76, or anti-p38 primary antibodies, and horseradish peroxidase-linked anti-rabbit or anti-mouse IgG secondary antibodies (BD Biosciences). For Western blot analyses, equal amounts of protein lysates (8 μg) were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and probed with the same antibodies as in the IP. GAPDH antibody (Cell signaling, Beverly, MA) was used to check uniform loading.
Statistics
Three or more independent repeats were conducted for all experiments. Error bars represent mean ± S.E. of the repeats. StatView statistical package (SAS Institute Inc., version 5.0.1) was used to analyze the data via analysis of variance, Fisher's protected least significant difference.
RESULTS
c-Cbl TKB Mutant G306E Failed to Enhance CD11b Expression and Cell Differentiation Compared with WT c-Cbl Stable Transfectants
Our previous study (9) demonstrated that c-Cbl bound CD38, and that stable transfectants ectopically expressing c-Cbl underwent myeloid differentiation faster than wild-type cells when treated with RA. To determine whether a G306E TKB domain or a C381A Ring finger domain-mutated Cbl retained this ability to propel cell differentiation, we created two stable transfectant cell lines expressing these c-Cbl mutations (G306E or C381A). Western blots verified that c-Cbl protein expression was appropriately enhanced in WT c-Cbl (c-Cbl+), G306E and C381A stable transfectants (Fig. 1B). The ability of these different stable transfectant cell lines (vector control, c-Cbl+, G306E, and C381A) to differentiate in response to RA was measured using a cell surface marker and a functional differentiation marker. First, a cell surface differentiation marker, CD11b, was measured by immunofluorescence using allophycocyanin-conjugated anti-CD11b antibody and flow cytometry. We compared CD11b expression in vector control, c-Cbl+, G306E, and C381A stable transfectants treated with RA for 24, 48, and 72 h. Compared with vector controls, c-Cbl+ and C381A transfectants showed significantly enhanced expression of CD11b after RA treatment; however, the G306E mutant did not (Fig. 1C), indicating that substitution of Gly306 with glutamic acid is a loss of function mutation.
To confirm that the G306E mutation crippled the ability of c-Cbl to promote RA-induced cell differentiation, a functional differentiation marker, the inducible oxidative metabolism was also measured. Inducible oxidative metabolism is a hallmark of terminally differentiated myeloid cells. Consistent with CD11b expression, in RA-treated cells c-Cbl and its C381A mutant promoted functional differentiation, enhancing the percentage of cells capable of inducible oxidative metabolism, but the G306E mutant did not (Fig. 1, D and E). The ability of c-Cbl to promote RA-induced differentiation was thus lost in the G306E mutant, but not the C381A mutant. An intact Gly306 in the TKB domain of c-Cbl thus appears to be needed for c-Cbl to promote RA-induced differentiation, whereas the Cys381 in the Ring finger domain can be mutated without affecting this capability.
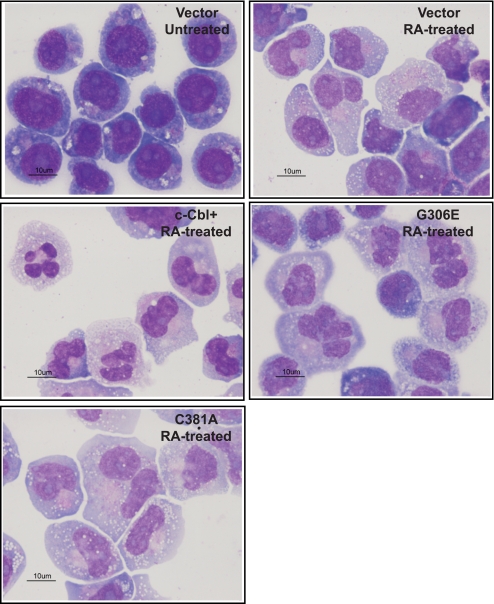
To visualize the differentiation process, morphological evaluation of vector control, c-Cbl+, G306E, and C381A cells treated with RA for 4 days was done using Wrights stain (Fig. 2). The morphology of all untreated cells was typical of immature blasts, displayed round nuclei, prominent nucleoli, and very small amounts of deep blue cytoplasm. The RA-treated cells showed more lobulated nuclei with less frequent nucleoli and a more abundant lighter blue cytoplasm, compatible with maturation toward a myeloid lineage. The morphological shift was more prominent in c-Cbl+ and C381A RA-treated cells. Morphology was consistent with the differentiation assays, indicating that c-Cbl and C381A promoted RA-induced differentiation, but the G306E mutant did not.
FIGURE 2.
Cytological observation of Wrights-stained untreated cells, and RA-treated vector control, c-Cbl+, G306E, and C381A stable transfectants. Cells were treated with 1.0 μm RA for 4 days and stained with Wrights stain. More than 50 cells were inspected, and images were taken using the ×100 objective. Cytological data are consistent with differentiation assays.
The G306E c-Cbl Mutant Failed to Enhance ERK Activation and RA-induced Cell Cycle Arrest
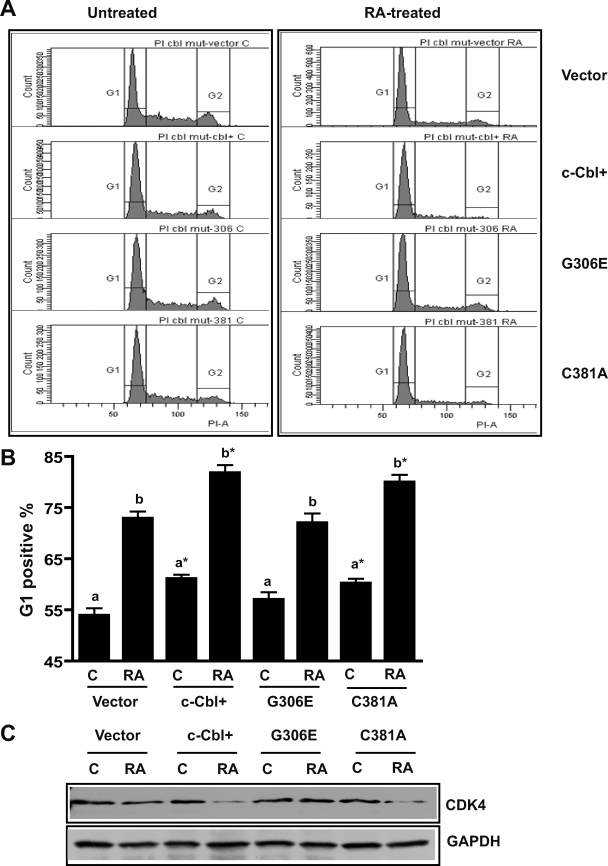
c-Cbl stable transfectants enhanced RA-induced G1/0 cell cycle arrest (9). To determine whether the c-Cbl mutants (G306E and C381A) could also promote RA-induced G1/0 arrest, the percentage of cells (vector, c-Cbl+, G306E, and C381A stable transfectants) in G1/0 was measured using flow cytometry (Fig. 3, A and B). Induced G1/0 arrest would be revealed by an enrichment of cells with G1 DNA. c-Cbl WT and the C381A mutant enhanced G1/0 arrest, whereas the G306E mutant transfectants failed to enhance RA-induced G1/0 arrest. This was consistent with differentiation assays, indicating that the G306E mutant lost the ability to promote RA-induced differentiation and cell cycle arrest. We also compared the expression of cyclin-dependent kinase 4 (CDK4) by RA-treated and untreated vector control, c-Cbl+, G306E, and C381A stable transfectants (Fig. 3C). Consistent with enhanced RA-induced growth arrest the c-Cbl WT and the C381A transfectants had less CDK4 expression, potentially retarding the cell cycle, than the G306E mutant, which did not exhibit enhanced cell cycle arrest.
FIGURE 3.
c-Cbl G306E failed to promote RA-induced cell cycle arrest. A, representative DNA histograms of untreated and (72 h) RA-treated vector control, c-Cbl+, G306E, and C381A stable transfectants. B, percentage of vector control, c-Cbl+, G306E, and C381A transfectant cells with G1/G0 DNA after a 72-h RA treatment. Cells stained with hypotonic propidium iodine (PI) staining solution were analyzed by flow cytometry. Bars represent means ± S.E. of 8 repeats. The different letters indicate significant differences at the p ≤ 0.05 levels between untreated (C) and RA treated (RA) among the same cell lines. Asterisk indicates a significant (p ≤ 0.05) difference compared with vector control cells. C, Western blots of CDK4 for vector control, c-Cbl+, G306E, and C381A stable transfectants. The WT c-Cbl and the C381A transfectants had less CDK4 expression than vector control or G306E transfectants after RA treatment (72 h). GAPDH was used to check protein loading. The data from the CDK4 Western blots were consistent with G1/0 arrest assays, indicating that the G306E mutant failed to promote cell cycle arrest.
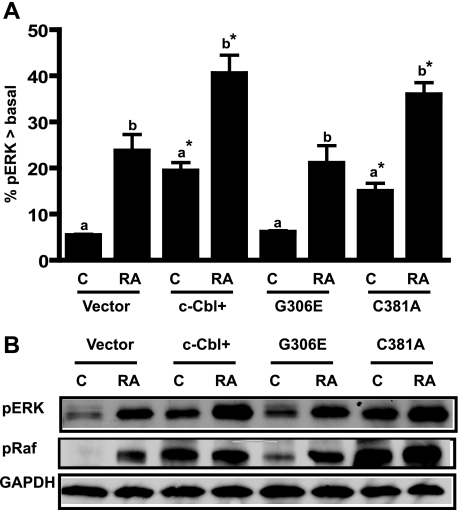
RA-induced differentiation and G1/0 arrest are propelled by activation of ERK (1). c-Cbl+ transfectants showed increased ERK activation, suggesting that overexpression of c-Cbl enhanced ERK MAPK signaling and accelerated cell differentiation and cell cycle arrest (9). To address whether failure of the G306E mutant to enhance RA-induced differentiation was attributable to a failure to enhance ERK activation, we compared ERK (dual phosphorylated ERK (203TEY205) activation among vector control, c-Cbl+, G306E, and C381A stable transfectants. Fig. 4A shows that the amount of activated ERK per cell was significantly increased by ectopic expression of c-Cbl and mutant C381A, but not by the G306E mutant, suggesting that overexpression of the G306E c-Cbl mutant failed to enhance RA-induced ERK activation, which resulted in a loss of function in cell differentiation and cell cycle arrest. Western blotting was performed for the same cases to confirm the flow cytometric data. The results from the Western blots of pERK were consistent with the flow cytometric measurements. We previously reported that RA caused a Raf activation necessary to induce differentiation and G0 arrest (39). To investigate how the G306E mutant affected RAF, phosphorylation of RAF (Ser621) was compared among different cell lines. Consistent with ERK activation data, the TKB mutant failed to enhance RA-induced Raf kinase activation (Fig. 4B), suggesting that the TKB mutation affects RA-induced Raf/ERK signaling.
FIGURE 4.
c-Cbl TKB mutant G306E failed to increase RAF/ERK activation. A, percentage of cells with pERK exceeding basal levels in vector control, c-Cbl+, and G306E or C381A mutant stable transfectants that were untreated (C) or RA treated. Ectopic high expression of c-Cbl or mutant C381A enhanced activated ERK expression; however, the G306E mutant did not. Induced ERK activation upon RA treatment for 24 h was indicated by a shift in the percentage of cells with activated ERK above the basal levels of control cells. Bars represent means ± S.E. of 3 repeats. The different letters indicate a significant difference (p ≤ 0.05) between untreated and RA treated among the same cell lines. Asterisks indicates a significant (p ≤ 0.05) difference compared with untreated (a) or RA-treated (b) vector control cells. B, Western blots of activated phospho-ERK and phospho-RAF for vector control, c-Cbl+, G306E, and C381A transfectants. GAPDH was used to check protein loading. RA treatment was for 24 h. The data from Western blots were consistent with flow cytometry results, indicating that c-Cbl+ and C381A enhanced activated ERK expression, but G306E mutant did not.
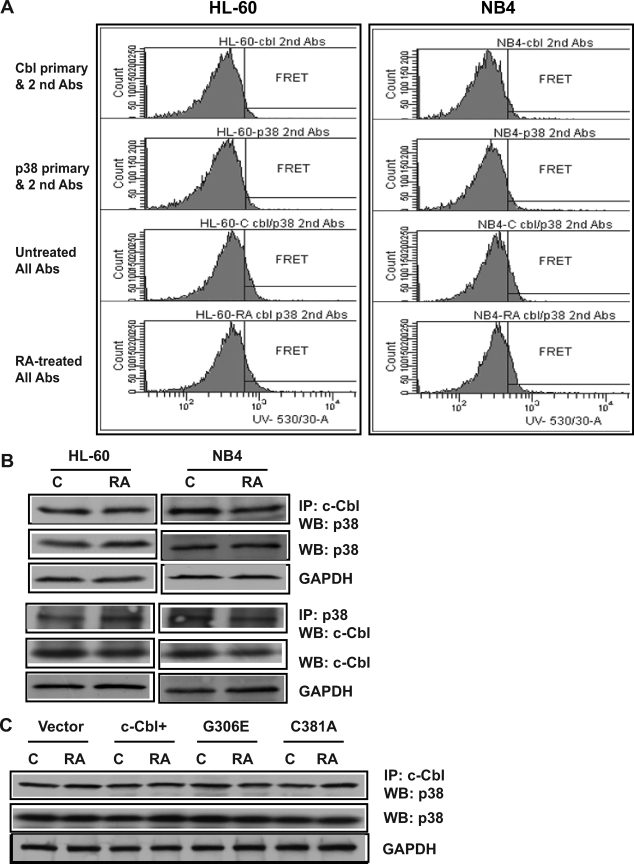
The G306E c-Cbl Mutation Did Not Affect p38 Binding or Expression
p38 is a MAP kinase that can potentially affect Cbl activation. We used FRET and IPs to determine whether c-Cbl and p38 interact in RA-treated (24 h) or untreated HL-60 and NB4 cells. FRET is a process by which an excited donor chromophore nonradiatively transfers energy to an acceptor chromophore in close proximity (typically <10 nm) resulting in a detectable emission. It is a sensitive method to identify protein-protein interactions in intact cells (40). FRET signals between c-Cbl and p38 were detected in untreated and RA-treated HL-60 and NB4 cells, indicating an interaction between Cbl and p38. The FRET signal between c-Cbl and p38 was not altered by RA treatment (Fig. 5A). The c-Cbl and p38 association was confirmed by co-immunoprecipitation. c-Cbl and p38 co-immunoprecipitated in untreated or RA-treated HL-60 and NB4 cells (Fig. 5B). RA did not affect the interaction. Reciprocal IPs confirmed the results. Negative controls using nonspecific IgG as an isotype-specific control did not immunoprecipitate c-Cbl or p38 (data not shown). To determine whether c-Cbl expression or RA affected p38 expression, the expression levels of p38 among vector control, RA-treated or untreated c-Cbl+ and Cbl mutant stable transfectants were measured (Fig. 5C). The expression levels of p38 were similar, suggesting that neither overexpression of c-Cbl nor c-Cbl mutants enhances p38 expression. Nor did RA affect expression. To determine whether the G306E or C381A c-Cbl mutations disrupted c-Cbl/p38 binding, we immunoprecipitated c-Cbl from vector control, c-Cbl+, G306E, and C381A transfectants and probed for p38 in Western blots (Fig. 5C). Neither the G306E nor C381A mutation changed the binding of c-Cbl with p38. c-Cbl and p38 co-immunoprecipitation thus occurs independent of G306E, C381A, or RA, suggesting a constitutive association.
FIGURE 5.
The G306E c-Cbl mutation did not affect p38 binding or expression. A, representative histograms of FRET signals of untreated and (24 h) RA-treated HL-60 and NB4 cells. c-Cbl and p38 interaction was observed in both HL-60 and NB4 cells. Cells fixed in paraformaldehyde and permeabilized were incubated in PBS containing mouse anti-c-Cbl and rabbit anti-p38 primary antibodies and then stained with Alexa 350- and 430-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies. B, p38 or c-Cbl were co-immunoprecipitated in both HL-60 and NB4 cells. Co-IP using anti-c-Cbl or anti-p38 antibody pulled down c-Cbl or p38 from protein lysates of WT-HL60 and NB4 (untreated and RA-treated for 24 h) and Western blotted (WB) using anti-p38 or anti-c-Cbl. Nonspecific immunoglobulin was used as a negative control and no signal was detected. Western blots of total protein lysates show the expression levels of p38 or c-Cbl in untreated or RA-treated HL-60 or NB4 cells. Anti-GAPDH was used to show consistently the input of total cellular proteins in the co-IP reactions. C, c-Cbl was immunoprecipitated from protein lysates of vector control, c-Cbl+, G306E, and C381A transfectants and probed for p38 in Western blots. Total cell lysates from different transfectants were also Western blotted with p38 to check p38 expression.
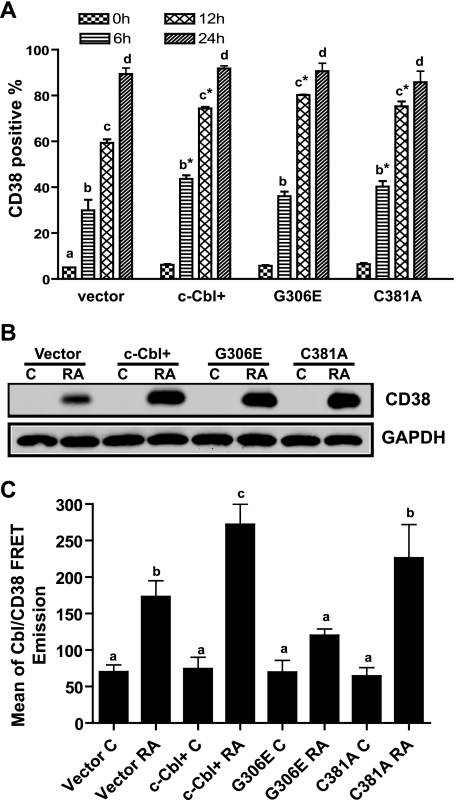
c-Cbl G306E TKB Mutant Disrupted the Binding between c-Cbl and CD38
Because the G306E mutant failed to enhance expression of later differentiation markers of CD11b and the inducible oxidative metabolism, its effect on an earlier marker became of interest. CD38 is among the earliest membrane receptors up-regulated by RA. It causes RAF/ERK activation and propels RA-induced differentiation and cell cycle arrest, making it of functional significance to induced differentiation. To evaluate the ability of mutant c-Cbl to affect the expression of CD38, we compared CD38 expression levels in WT c-Cbl and mutant G306E and C381A stable transfectants treated with RA. RA-induced CD38 expression was similarly enhanced for WT c-Cbl, the G306E and C381A transfectants at different time points (Fig. 6A), indicating that the c-Cbl mutants (G306E and C381A) behaved like WT c-Cbl and enhanced RA-induced CD38 expression compared with vector controls. To confirm flow cytometric measurements, Western blotting of CD38 from cell lysates with untreated and RA-treated (12 h) cells was performed. The Western blots (Fig. 6B) showed that RA-treated WT c-Cbl, G306E, and C381A transfectants had similar enhanced CD38 expression levels compared with vector controls.
FIGURE 6.
c-Cbl G306E TBK mutant disrupted the binding between c-Cbl and CD38. A, percentage of vector control, WT c-Cbl, G306E, and C381A transfectants expressing CD38 after RA treatment for the indicated times. RA-treated c-Cbl+, G306E, and C381A transfectants had increased expression of CD38 compared with vector controls. The different letters indicate significant (p ≤ 0.05) differences at different treated times. Asterisks indicate that CD38 expression levels from c-Cbl+, G306E, and C381A transfectants were significantly (p ≤ 0.05) different from vector controls. B, Western blot of CD38 for vector control, c-Cbl+, G306E, and C381A stable transfectants (upper panel). GAPDH (lower panel) was used to check protein loading. The RA treatment is for 12 h. The data from Western blots were consistent with flow cytometry results. C, the FRET signals between c-Cbl and CD38 for control and RA-treated vector control cells, c-Cbl+, and mutant G306E and C381A transfectants. The FRET signal from RA-treated G306E transfectants was significantly lower than that from vector control, WT c-Cbl, or C381A transfectants. The different letters indicate significant differences between untreated and other groups of cells. Bars represent means ± S.E. of 3 repeats.
c-Cbl is known to bind CD38 (9). To determine whether the G306E or C381A mutations disrupt binding between Cbl and CD38, FRET was used to assay CD38 interaction with WT and mutant c-Cbl in the c-Cbl, G306E, and C381A transfectants. c-Cbl or its mutants were used as the donor and CD38 was used as acceptor molecules in RA-treated and untreated cells. The FRET signal from RA-treated G306E transfectants was significantly lower than that from vector control, WT c-Cbl, or C381A transfectants (Fig. 6C). The G306E mutation thus perturbed the binding of the TKB domain and disrupted the interaction of c-Cbl with CD38. In contrast, the C381A Ring finger mutation did not prevent Cbl from binding to CD38. We do not know the explicit character of the disruption. The simplest would be a failure to bind; but an unknown function of the mutant cannot be excluded.
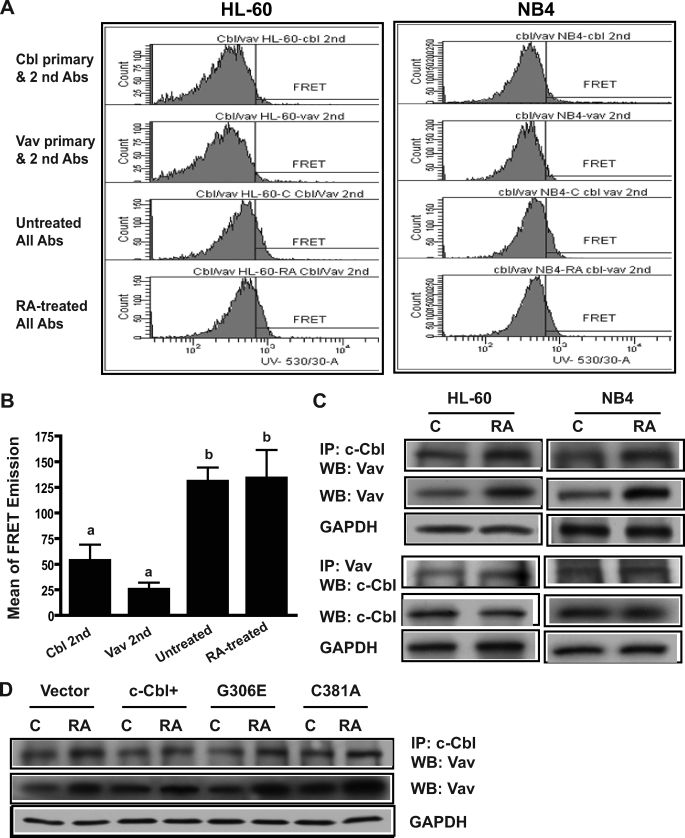
The Interaction of c-Cbl G306E and C381A Mutants with Vav Proteins
Cbl-b has been reported to associate with Vav upon T-cell receptor stimulation of primary murine lymphocytes and Jurkat T cells (41). FRET and immunoprecipitation were used to determine whether c-Cbl and Vav interact in both HL-60 and NB4 cells that were RA-treated (24 h) or untreated. FRET was measured using c-Cbl and Vav as donor and acceptor, respectively. Fig. 7, A and B, shows that FRET between c-Cbl and Vav was observed in untreated and RA-treated HL-60 and also NB4 cells. The FRET signals were comparable for control and RA-treated cells, suggesting that there was an RA-independent interaction between Cbl and Vav. Immunoprecipitation was used to confirm the binding between c-Cbl and Vav. Vav and c-Cbl co-immunoprecipitated in untreated and RA-treated HL-60 and NB4 cells (Fig. 7C). As expected, the reciprocal co-IPs for Cbl or Vav and probed for Vav or c-Cbl, respectively, using untreated or RA-treated HL-60 or NB4 cells, corroborated the c-Cbl/Vav interaction. Negative controls using nonspecific IgG as an isotype-specific control did not immunoprecipitate c-Cbl or Vav (data not shown). Western blotting of total cell lysates (Fig. 7C) showed that RA up-regulated Vav expression and down-regulated Cbl expression as previously reported (9). To determine whether the G306E or C381A c-Cbl mutations disrupted c-Cbl/Vav binding, we also immunoprecipitated c-Cbl from protein lysates of vector control, Cbl+, G306E, and C381A transfectants and probed for Vav in Western blots (Fig. 7D). Neither G306E nor C381A mutations abrogated binding to Vav. Western blots of total cell lysate using anti-Vav paralleled the co-IP Vav.
FIGURE 7.
The interaction of c-Cbl G306E and C381A with Vav proteins. A, histograms of FRET signals showing an interaction of c-Cbl and Vav in HL-60 and NB4 cells. B, the FRET signals between c-Cbl and Vav for control and RA-treated (24 h) HL-60 cells. Bars represent means ± S.E. of 3 repeats. The different letters indicate significant differences from cells stained with donor or acceptor and both secondary antibodies. C, immunoprecipitation confirms the binding between Cbl and Vav in both HL-60 and NB4 cells that were untreated (C) or RA treated (24 h). Co-IP using anti-c-Cbl or anti-Vav antibody pulled down c-Cbl or Vav from protein lysates of WT-HL60 and NB4 that were Western blotted (WB) using anti-Vav or anti-c-Cbl. No signal was detected in a negative control. Western blots of total protein lysates show the expression levels of Vav or c-Cbl in untreated or RA-treated HL-60 or NB4 cells. Anti-GAPDH was used to show consistently the input of total cellular proteins in the co-IP reactions. D, c-Cbl was immunoprecipitated from protein lysates of vector control, WT c-Cbl, and mutant G306E and C381A transfectants and co-immunoprecipitated Vav detected in Western blots. Total cell lysates from these cells were also Western blotted for Vav to check Vav expression.
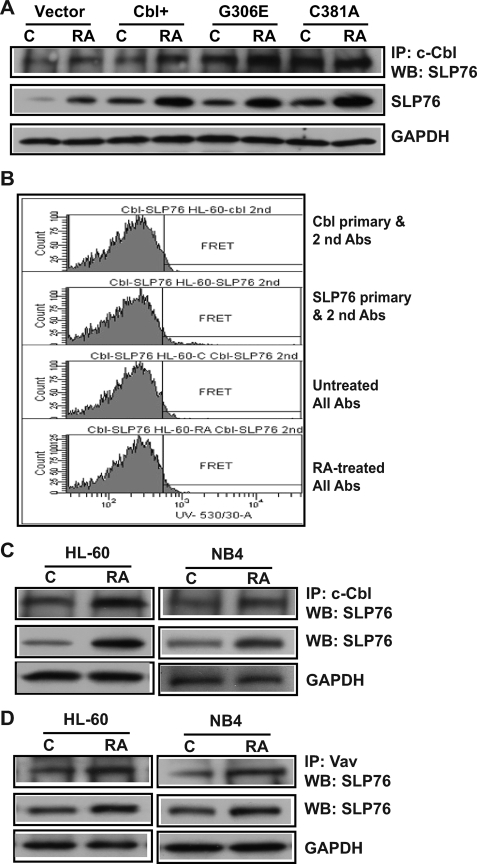
The Interaction of c-Cbl G306E and C381A Mutants with Slp76
The potential interaction between c-Cbl and Slp76 was assessed. Slp76 co-immunoprecipitating with c-Cbl in vector control, c-Cbl+, G306E, and C381A transfectants was measured. In untreated and RA-treated cells stably transfected with WT c-Cbl or G306E or C381A mutants, Slp76 was associated with c-Cbl and the mutants (Fig. 8A), suggesting that the mutations did not eliminate interaction between c-Cbl and Slp76. Ectopic expression of c-Cbl or the mutants also similarly enhanced the expression of Slp76 compared with vector control for untreated or RA-treated cells. FRET between c-Cbl and Slp76 was measured in RA-treated (24 h) HL-60 cells. Fig. 8B showed that no FRET signal between Cbl and Slp76 was detectable. Likewise, corroborating this, no FRET emissions were detected for NB4 cells too (data not shown). The FRET measurement suggested either that there was no direct interaction between Cbl and Slp76 or that there was an indirect interaction where the distance between two molecules was more than 10 nm or that something intervenes to block the energy transfer between Cbl and Slp76. To confirm the interaction of c-Cbl and Slp76, immunoprecipitation was performed using HL-60 and NB4 cells. Slp76 protein was co-immunoprecipitated with c-Cbl protein in both HL-60 and NB4 cells, which were untreated and RA treated (Fig. 8C). RA increased the amount of co-immunoprecipitated Slp76. Negative controls using IgG as an isotype-specific control were included and did not co-immunoprecipitate Slp76 (data not shown). Taken together, IP and FRET data suggest that c-Cbl and Slp76 are connected through an intermediate, possibly Vav, which binds c-Cbl. The binding between Vav and Slp76 was also investigated. Slp76 was co-immunoprecipitated with Vav in both HL-60 and NB4 cells. RA treatment increased the amount of co-immunoprecipitated Slp76. Slp76 levels in total cell lysates paralleled this (Fig. 8D). c-Cbl and Slp76 thus appear to reside in a Vav-containing complex where something intervenes to block FRET between c-Cbl and Slp76.
FIGURE 8.
The interaction of c-Cbl G306E and C381A mutants with Slp76. A, immunoprecipitation of c-Cbl and Western blots of Slp76 in vector control, WT c-Cbl, G306E, and C381A stable transfectants. No signal was detected in a negative control using nonspecific immunoglobulin. B, histograms of FRET signals showing no c-Cbl and Slp76 FRET signal in HL-60 cells. C, Slp76 protein was co-immunoprecipitated with c-Cbl in untreated or (24 h) RA-treated HL-60 and NB4 cells. Proteins immunoprecipitated with c-Cbl from protein lysates of HL60 and NB4 were Western blotted using anti-Slp76 (top) antibody. Nonspecific immunoglobulin was used as a negative control, and no signal was detected. Western blots (WB) of total protein lysates using anti-Slp76 (middle panel) and anti-GAPDH (lower panel) were used to show Slp76 expression levels and consistency of input of total cellular proteins in the co-IP reactions. D, Slp76 protein was co-immunoprecipitated with Vav in untreated or (24 h) RA-treated HL-60 and NB4 cells (top). Slp76 expression and consistency of input of total proteins in the co-IP reactions are shown in the middle and lower panels. To better show Slp76 expression levels, the exposure is darker than that in A.
DISCUSSION
Our previous studies (9) showed that c-Cbl small interfering RNA knockdowns diminished RA-induced cell differentiation and G0 arrest, and ectopically overexpressing c-Cbl caused enhanced RA-induced differentiation and cell growth, indicating that c-Cbl is required for RA-induced myeloid differentiation and G0 arrest. RA treatment gradually down-regulated c-Cbl expression and loss of c-Cbl resulted in the failure of c-Cbl/CD38 binding, suggesting that the role of c-Cbl in driving differentiation is relatively early in the RA-induced differentiation process. In this study, we have directly addressed the possibility that Gly306 in the TKB domain could contribute to cell differentiation and cell cycle arrest. The G306E mutation failed to promote ERK activation, cell differentiation, and cell cycle arrest, whereas the Ring finger domain mutant C381A did not affect the ability of c-Cbl to enhance ERK activation and cell differentiation. This provides direct evidence that enhancement of cell differentiation and growth arrest caused by c-Cbl depends on an intact Gly306 in the TKB domain. This study is the first to indicate the importance of the TKB domain of c-Cbl in RA-induced HL-60 cell differentiation and cell cycle arrest.
We have previously shown that Raf-driven MAPK signaling propels RA-induced differentiation (39). In contrast to WT c-Cbl or its C381A Ring mutant, the G306E mutant failed to drive RAF/ERK MAPK activation, which is needed to promote cell differentiation. Another MAP kinase, p38, regulates epidermal growth factor receptor activity and is required for EGF-stimulated Cbl activation (34). Our studies showed that WT c-Cbl and its mutants (G306E or C381A) similarly co-immunoprecipitated p38. The c-Cbl G306E mutation causing failure to propel RA-induced differentiation thus did not affect p38/c-Cbl binding. Nor did expression of c-Cbl or its mutant affect p38 expression. Furthermore, RA did not affect p38 expression levels or binding to c-Cbl. Regulation of RA-induced differentiation through c-Cbl thus does not seem to be via regulating p38 although it is part of the signaling complex. This is consistent with our previous report showing independence of RA-induced differentiation on p38 activity (2).
RA-induced differentiation can be propelled by the binding of c-Cbl to CD38. CD38 mediates the production of proinflammatory cytokines, proliferation, and protection from apoptosis in lymphocytes (42–44). Lamkin et al. (45) showed that RA induces the early expression of CD38, which signals through MAPK to promote RA-induced cell differentiation. The WT c-Cbl and mutant G306E and C381A stable transfectants all had comparable, enhanced RA-induced CD38 expression compared with vector controls. However, the FRET between CD38 and G306E was significantly lower than that between CD38 and WT c-Cbl or C381A in RA-treated cells. This suggested that the TKB mutation disrupted the binding of c-Cbl to CD38. The TKB mutant G306E failed to promote cell differentiation, suggesting that failure of the G306E mutant to propel cell differentiation reflects loss of binding to CD38 with consequential failure to activate MAPK signaling. This supports our previous study (9) suggesting that direct binding of Cbl and CD38 is important in propulsion of RA-induced differentiation.
The present results suggest that Gly306 in the domain c-Cbl TKB identifies a high affinity binding site for CD38. It has been reported that the c-Cbl TKB domain has a critical role in targeting proteins for ubiquitination (46), as well as being unique to Cbl proteins and is often referred to as a specialized or “embedded” SH2 domain (47). G306E may thus affect important c-Cbl functions in phosphoprotein recognition and interaction with other proteins. Studies by Thien et al. (21) showed that the c-Cbl TKB domain is essential for c-Cbl to negatively regulate ZAP-70, and implied a direct interaction between the TKB domain and phosphorylated tyrosine in ZAP-70. The G306E in the TKB domain abolishes c-Cbl interaction with negative regulatory tyrosine 292 in ZAP70 (48). Structural studies by Meng et al. (13) have shown that Gly306 lies near the universally conserved arginine present in all SH2 domains that hydrogen bond with the phosphate group of the incoming tyrosine. The substitution of Gly306 with glutamic acid could form a buried salt bridge with Arg294 and prevent c-Cbl interaction with phosphotyrosine.
c-Cbl interacts with multiple proteins to regulate signaling events via their protein-protein interaction domains (17, 22, 23). The discovery of novel partners of c-Cbl is important to an understanding of c-Cbl protein function. In this study, we found interactions of c-Cbl with Vav, Slp76, and p38. Both FRET and immunoprecipitation revealed the interaction of c-Cbl with Vav or c-Cbl with p38. Consistent with our finding that c-Cbl interacts with Vav, it has been previously reported that Cbl forms complexes with Vav as well as other molecules containing a guanine nucleotide exchange factor domain (49, 50). Vav1 has been reported to play a crucial role in overcoming the maturation blockage of APL promyelocytes, and overexpression of Vav1 promotes cell differentiation (51). It is possible that c-Cbl links Vav and CD38 to result in enhancement of cell differentiation and cell cycle arrest. p38 may inhibit phosphatases acting on Cbl activity and epidermal growth factor receptor ubiquitinylation/degradation (34). Our results showed that c-Cbl bound to p38. We also demonstrated an association between c-Cbl and Slp76. Stable transfectants ectopically expressing c-Cbl caused enhanced Slp76 expression, and c-Cbl mutations G306E or C381A did not abolish this. Immunoprecipitation showed an association between c-Cbl and Slp76; however, no FRET signal was observed between c-Cbl and Slp76. This suggests an association such that either the distance between c-Cbl and Slp76 is too great for FRET or something between them might block the energy transfer. Slp76 is a substrate for ZAP70 and Syk tyrosine kinase and is reported to associate with the SH2 domain of Vav in antigen-stimulated T cells (52). We observed that Vav associated with Cbl and Slp76. Thus, one possibility is that c-Cbl does not directly bind Slp76, but they are connected through Vav. The G306E mutant did not bind to CD38, but still bound to Slp76, Vav, and p38, suggesting that the c-Cbl G306E mutant does not globally disrupt the c-Cbl structure while abrogating CD38 binding. It is possible that the G306E mutation disrupts the binding of Cbl with some other cellular protein(s), and that there are also other Cbl partners involved in c-Cbl-mediated signal transduction for RA-induced differentiation.
Together these data indicated that c-Cbl enhanced cell differentiation and bound CD38, Slp76, Vav, and p38. It is becoming evident that the binding of c-Cbl and CD38 drives MAPK signaling and propels RA-induced differentiation; however, the full role of Vav and Slp76 in the c-Cbl/CD38/Vav/Slp76 containing complex still remains to be fully elucidated. The complex of c-Cbl with CD38, Vav, Slp76, and p38 could play an important role in the function of c-Cbl in promoting cell differentiation and growth; however, we do not know explicitly how c-Cbl interacts and functions with these molecules to form an active signaling complex. Our current study used immunoprecipitation and FRET to show that CD38 interacts with Vav and Slp76 and that ectopically overexpressing CD38 by stable transfection (data not shown) enhances the amount of CD38 interacting Vav and Slp76. c-Cbl interacts with Vav and Slp76 as well as p38. Hence it appears that there is a signaling complex containing c-Cbl, Vav, Slp76, and p38 that binds to CD38 to give rise to a MAPK signal dependent on Gly306 whose mutation prevents CD38 binding and MAPK signaling. However, the other interactions detected are not lost and hence c-Cbl may be a nucleating factor for forming the signaling complex to bring to CD38. Although we have detected c-Cbl, Vav, Slp76, and p38 in the putative signaling complex binding CD38 in a Gly306 dependent way, there may be other molecules in the complex that affect CD38 binding and signaling. Our general thesis is that Cbl helps sustain a long MAPK activation that causes cell differentiation and cell cycle arrest. This conforms to the emerging paradigm that a hyperactivated MAPK signal drives arrest and differentiation in contrast to the canonical mitogenic signal. This paradigm was originally suggested by Cohen and co-workers (53) in studies of the contrasting effects of epidermal growth factor and nerve growth factor. It has been validated now in other contexts as well (54). In our model, we find that both Vav and Slp-76, which are molecules known to facilitate MAPK signaling, bound c-Cbl. The active CD38-c-Cbl complex in our studies thus appears to also utilize these to drive the hyperactive MAPK that has been shown to cause differentiation and cell cycle arrest (1, 2, 53, 54).
Acknowledgments
We thank Dr. James Smith for expert assistance in flow cytometry, especially in the design and execution of the FRET experiments. We are especially grateful to Deanna M. W. Schaefer from the clinical pathology laboratory at the Cornell University College of Veterinary Medicine for excellent assistance in obtaining the cytology images.
This work was supported, in whole or in part, by grants from the National Institutes of Health of the USPHS and New York State Stem Cell Science.
- RA
- retinoic acid
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- TKB domain
- tyrosine kinase-binding domain
- IP
- immunoprecipitation
- FRET
- fluorescence resonance energy transfer
- CDK4
- cyclin-dependent kinase 4
- DCF
- 2′,7′-dichlorohydrofluorescein diacetate
- SH2
- Src homology domain 2
- WT
- wild-type
- PBS
- phosphate-buffered saline
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Yen A., Roberson M. S., Varvayanis S., Lee A. T. (1998) Cancer Res. 58, 3163–3172 [PubMed] [Google Scholar]
- 2.Yen A., Roberson M. S., Varvayanis S. (1999) In Vitro Cell Dev. Biol. 35, 527–532 [DOI] [PubMed] [Google Scholar]
- 3.Mann G., Reinhardt D., Ritter J., Hermann J., Schmitt K., Gadner H., Creutzig U. (2001) Ann. Hematol. 80, 417–422 [DOI] [PubMed] [Google Scholar]
- 4.Collins S. J., Gallo R. C., Gallagher R. E. (1977) Nature 270, 347–349 [DOI] [PubMed] [Google Scholar]
- 5.Breitman T. R., Selonick S. E., Collins S. J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen A. (1990) Hematol. Rev. 4, 5–46 [Google Scholar]
- 7.Dalton W. T., Jr., Ahearn M. J., McCredie K. B., Freireich E. J., Stass S. A., Trujillo J. M. (1988) Blood 71, 242–247 [PubMed] [Google Scholar]
- 8.Battle T. E., Roberson M. S., Zhang T., Varvayanis S., Yen A. (2001) Eur. J. Cell Biol. 80, 59–67 [DOI] [PubMed] [Google Scholar]
- 9.Shen M., Yen A. (2008) Cancer Res. 68, 8761–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelstein L. D., Shimizu Y. (2000) Biochem. J. 345, 385–392 [PMC free article] [PubMed] [Google Scholar]
- 11.Lupher M. L., Jr., Andoniou C. E., Bonita D., Miyake S., Band H. (1998) Int. J. Biochem. Cell Biol. 30, 439–444 [DOI] [PubMed] [Google Scholar]
- 12.Liu Y. C., Altman A. (1998) Cell Signal. 10, 377–385 [DOI] [PubMed] [Google Scholar]
- 13.Meng W., Sawasdikosol S., Burakoff S. J., Eck M. J. (1999) Nature 398, 84–90 [DOI] [PubMed] [Google Scholar]
- 14.Sanjay A., Houghton A., Neff L., DiDomenico E., Bardelay C., Antoine E., Levy J., Gailit J., Bowtell D., Horne W. C., Baron R. (2001) J. Cell Biol. 152, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. (1999) Science 286, 309–312 [DOI] [PubMed] [Google Scholar]
- 16.Yokouchi M., Kondo T., Houghton A., Bartkiewicz M., Horne W. C., Zhang H., Yoshimura A., Baron R. (1999) J. Biol. Chem. 274, 31707–31712 [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan G., Tsygankov A. Y. (2006) J. Cell. Physiol. 209, 21–43 [DOI] [PubMed] [Google Scholar]
- 18.Ota S., Hazeki K., Rao N., Lupher M. L., Jr., Andoniou C. E., Druker B., Band H. (2000) J. Biol. Chem. 275, 414–422 [DOI] [PubMed] [Google Scholar]
- 19.Visser G. D., Lill N. L. (2005) Exp. Cell Res. 311, 281–293 [DOI] [PubMed] [Google Scholar]
- 20.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 21.Thien C. B., Scaife R. M., Papadimitriou J. M., Murphy M. A., Bowtell D. D., Langdon W. Y. (2003) J. Exp. Med. 197, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M. H., Dikic I. (2005) Nat. Rev. Mol. Cell Biol. 6, 907–918 [DOI] [PubMed] [Google Scholar]
- 23.Dikic I., Szymkiewicz I., Soubeyran P. (2003) Cell Mol. Life Sci. 60, 1805–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deaglio S., Mehta K., Malavasi F. (2001) Leuk. Res. 25, 1–12 [DOI] [PubMed] [Google Scholar]
- 25.Zubiaur M., Fernández O., Ferrero E., Salmerón J., Malissen B., Malavasi F., Sancho J. (2002) J. Biol. Chem. 277, 13–22 [DOI] [PubMed] [Google Scholar]
- 26.Kontani K., Kukimoto I., Nishina H., Hoshino S., Hazeki O., Kanaho Y., Katada T. (1996) J. Biol. Chem. 271, 1534–1537 [DOI] [PubMed] [Google Scholar]
- 27.Clevenger C. V., Ngo W., Sokol D. L., Luger S. M., Gewirtz A. M. (1995) J. Biol. Chem. 270, 13246–13253 [DOI] [PubMed] [Google Scholar]
- 28.Raab M., da Silva A. J., Findell P. R., Rudd C. E. (1997) Immunity 6, 155–164 [DOI] [PubMed] [Google Scholar]
- 29.Bertagnolo V., Marchisio M., Brugnoli F., Bavelloni A., Boccafogli L., Colamussi M. L., Capitani S. (2001) Cell Growth Differ. 12, 193–200 [PubMed] [Google Scholar]
- 30.Chu J., Liu Y., Koretzky G. A., Durden D. L. (1998) Blood 92, 1697–1706 [PubMed] [Google Scholar]
- 31.Chiang Y. J., Sommers C. L., Jordan M. S., Gu H., Samelson L. E., Koretzky G. A., Hodes R. J. (2004) J. Exp. Med. 200, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. (1995) Science 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 33.Daly J. M., Olayioye M. A., Wong A. M., Neve R., Lane H. A., Maurer F. G., Hynes N. E. (1999) Oncogene 18, 3440–3451 [DOI] [PubMed] [Google Scholar]
- 34.Frey M. R., Dise R. S., Edelblum K. L., Polk D. B. (2006) EMBO J. 25, 5683–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biondi A., Rambaldi A., Alcalay M., Pandolfi P. P., Lo Coco F., Diverio D., Rossi V., Mencarelli A., Longo L., Zangrilli D. (1991) Blood 77, 1418–1422 [PubMed] [Google Scholar]
- 36.Fagioli M., Alcalay M., Pandolfi P. P., Venturini L., Mencarelli A., Simeone A., Acampora D., Grignani F., Pelicci P. G. (1992) Oncogene 7, 1083–1091 [PubMed] [Google Scholar]
- 37.Brooks S. C., 3rd, Kazmer S., Levin A. A., Yen A. (1996) Blood 87, 227–237 [PubMed] [Google Scholar]
- 38.Wightman J., Roberson M. S., Lamkin T. J., Varvayanis S., Yen A. (2002) Mol. Cancer Ther. 1, 493–506 [PubMed] [Google Scholar]
- 39.Wang J., Yen A. (2008) J. Biol. Chem. 283, 4375–4386 [DOI] [PubMed] [Google Scholar]
- 40.Bhaumik S. R. (2006) Methods 40, 353–359 [DOI] [PubMed] [Google Scholar]
- 41.Bustelo X. R., Crespo P., López-Barahona M., Gutkind J. S., Barbacid M. (1997) Oncogene 15, 2511–2520 [DOI] [PubMed] [Google Scholar]
- 42.Albeniz I., Demir O., Türker-Sener L., Yalçintepe L., Nurten R., Bermek E. (2007) Hematology 12, 409–414 [DOI] [PubMed] [Google Scholar]
- 43.Damle R. N., Temburni S., Calissano C., Yancopoulos S., Banapour T., Sison C., Allen S. L., Rai K. R., Chiorazzi N. (2007) Blood 110, 3352–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.March S., Graupera M., Rosa Sarrias M., Lozano F., Pizcueta P., Bosch J., Engel P. (2007) Am. J. Pathol. 170, 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamkin T. J., Chin V., Varvayanis S., Smith J. L., Sramkoski R. M., Jacobberger J. W., Yen A. (2006) J. Cell. Biochem. 97, 1328–1338 [DOI] [PubMed] [Google Scholar]
- 46.Ng C., Jackson R. A., Buschdorf J. P., Sun Q., Guy G. R., Sivaraman J. (2008) EMBO J. 27, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B. A., Jablonowski K., Raina M., Arcé M., Pawson T., Nash P. D. (2006) Mol. Cell 22, 851–868 [DOI] [PubMed] [Google Scholar]
- 48.Rao N., Lupher M. L., Jr., Ota S., Reedquist K. A., Druker B. J., Band H. (2000) J. Immunol. 164, 4616–4626 [DOI] [PubMed] [Google Scholar]
- 49.Marengère L. E., Mirtsos C., Kozieradzki I., Veillette A., Mak T. W., Penninger J. M. (1997) J. Immunol. 159, 70–76 [PubMed] [Google Scholar]
- 50.Reedquist K. A., Fukazawa T., Panchamoorthy G., Langdon W. Y., Shoelson S. E., Druker B. J., Band H. (1996) J. Biol. Chem. 271, 8435–8442 [DOI] [PubMed] [Google Scholar]
- 51.Bertagnolo V., Marchisio M., Volinia S., Caramelli E., Capitani S. (1998) FEBS Lett. 441, 480–484 [DOI] [PubMed] [Google Scholar]
- 52.Tuosto L., Michel F., Acuto O. (1996) J. Exp. Med. 184, 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traverse S., Gomez N., Paterson H., Marshall C., Cohen P. (1992) Biochem. J. 288, 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos S. D., Verveer P. J., Bastiaens P. I. (2007) Nat. Cell Biol. 9, 324–330 [DOI] [PubMed] [Google Scholar]