Abstract
The adapter protein MyD88 adapter-like (Mal), encoded by TIR-domain containing adapter protein (Tirap) (MIM 606252), is the most polymorphic of the five adapter proteins involved in Toll-like receptor signaling, harboring eight non-synonymous single nucleotide polymorphisms in its coding region. We screened reported mutations of Mal for activity in reporter assays to test the hypothesis that variants of Mal existed with altered signaling potential. A TIR domain variant, Mal D96N (rs8177400), was found to be inactive. In reconstituted cell lines, Mal D96N acted as a hypomorphic mutation, with impaired cytokine production and NF-κB activation upon lipopolysaccharide or PAM2CSK4 stimulation. Moreover, co-immunoprecipitation studies revealed that Mal D96N is unable to interact with MyD88, a prerequisite for downstream signaling to occur. Computer modeling data suggested that residue 96 resides in the MyD88 binding site, further supporting these findings. Genotyping of Mal D96N in three different cohorts suggested that it is a rare mutation. We, thus, describe a rare variant in Mal that exerts its effect via its inability to bind MyD88.
TLRs5 are the first line of defense against a diverse range of pathogens, recognizing pathogen-associated molecular patterns of microbes and initiating an innate immune response (1). TLR2 recognizes lipopeptide from the cell walls of bacteria, whereas TLR4 (together with MD-2) is the receptor for LPS (2). The adapter protein Mal/TIRAP (hereafter referred to as Mal) is involved in the MyD88-dependent pathway downstream of TLR2 and TLR4 (3, 4). It acts as a bridging adapter between the receptor and the sorting adapter MyD88 (5). Upon activation of TLR2 or TLR4, a signaling cascade is initiated which leads to the activation of transcription factors such as nuclear factor-κ B (NF-κB), interferon regulatory factor 5 (IRF5), and activator protein (AP-1), ultimately culminating in the production of pro-inflammatory cytokines (6).
The importance of single nucleotide polymorphisms (SNPs) in TLR-related proteins and their association with various infectious and inflammatory diseases have recently emerged (7, 8). Notable among them is a study by Khor et al. (9), who reported that individuals heterozygous for the polymorphism S180L in Mal are protected against pneumococcal disease, bacteremia, malaria, and tuberculosis (9). Within the coding region of a gene, SNPs can alter the characteristics and affect the ability of a protein to function (10). Interestingly, Mal is the most polymorphic of all adapter proteins, harboring at least eight non-synonymous variants in its coding region. It, thus, appears to be an important candidate for studying genetic variation in relation to TLR2 and TLR4 signaling.
In the present study we characterized the known polymorphisms in the coding region of Mal for effects on the function of Mal. We report that one of the mutations in the TIR domain of Mal, D96N, is broadly defective in TLR2 and the MyD88 branch of the TLR4 signaling pathway. We show by biochemical and computer modeling data that the basis of this abnormality is the inability of Mal D96N to bind MyD88. Although the incidence of this lesion in humans is not known, we screened three small populations for the defect and found a single heterozygous individual, suggesting that D96N is a rare mutation. We, thus, identify a mutation in Mal that causes a structural change, affecting the ability of the protein to bind the downstream effector MyD88.
EXPERIMENTAL PROCEDURES
Plasmids and Site-directed Mutagenesis
Most of the constructs described in this work have been described elsewhere. These include pEF-BOS-Mal-FLAG (11), pCDNA3-MyD88CFP (12), pCMV-IRF5-FLAG (13), NF-κB-luciferase, interferon-stimulated response element (ISRE)-luciferase, and Renilla-luciferase (14), and the retroviral vector pMSCV2.2-IRES-GFP (15). pEF-BOS-Mal-FLAG and pMSCV2.2-IRES-GFP-human-Mal with the different mutations in the Tirap gene were generated by using a site-directed mutagenesis QuikChange kit (Stratagene) per the manufacturer's instructions.
Reconstitution of Immortalized Mal Knock-out Macrophages
The immortalized Mal-deficient macrophage-like cell line was transduced with the retrovirus MSCV2.2. Equal expression of Mal or the Mal variants in selected clones was confirmed by Western blotting with anti-FLAG antibody. A Mal antibody (a gift from S. Akira (16)) was used to compare the expression of endogenous protein in wild type cells with that of the transduced cell lines.
Luciferase Reporter Assay
HEK293T cells were seeded into 96-well plates at a density of 20,000 cells per well and transfected 16 h later with 40 ng of the indicated luciferase reporter genes and the indicated amounts of Mal (WT or polymorphic) using GeneJuice (EMD Biosciences, San Diego, CA). In the case of Fig. 1B, IRF5 construct (10 ng) was co-transfected. The thymidine kinase Renilla-luciferase reporter was co transfected (40 ng) to normalize the data for transfection efficiency. Two days later reporter gene activity was measured using the Dual Luciferase assay system (Promega).
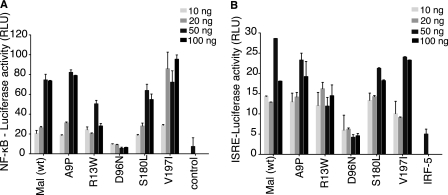
FIGURE 1.
Human Mal carrying the D96N mutation is unable to activate either NF-κB or IRF5. HEK293T cells were transfected with different variants of Mal and either NF-κB luciferase (A) or IRF5 and ISRE-luciferase reporters (B). 48 h post-transfection, lysates were analyzed for luciferase activity. Renilla-luciferase activity was used to normalize for transfection efficiency. Results are reported as the mean of triplicate determinations ±S.D. Each graph is representative of three (A) or two (B) independent experiments. RLU, relative luciferase units.
Enzyme-linked Immunosorbent Assay
Cells were seeded into 96-well plates at a density of 20,000 cells per well and stimulated overnight with the indicated concentrations of the TLR ligands. Cell culture supernatants were assayed for TNFα with enzyme-linked immunosorbent assay kits from R&D systems, according to the manufacturer's instructions.
Quantitative Real-time PCR
Wild type, Mal-deficient, or D96N-expressing macrophages were plated at a density of 2 × 106 cells per well in a 6-well plate. They were stimulated for 100 ng/ml LPS for 2 h. RNA extraction and IFN-β mRNA induction analysis by quantitative Real-time PCR was carried out as described elsewhere (17).
IkB Degradation Assay
2 × 106 cells were seeded per well into a 6-well plate. After 24 h the cells were treated with 10 nm PAM2CSK4 or 100 ng/ml LPS for the indicated time intervals. Cell lysates were made and run on an 8–16% SDS gel (NuSep), transferred to nitrocellulose membrane, and probed with anti-IkB. (Cell Signaling Technology).
Immunoprecipitations and Immunoblots
HEK293T cells were transfected with TLR2-YFP, TLR4-CFP, or MyD88-CFP and FLAG-tagged Mal (WT or D96N). Two days post-transfection, cells were harvested, and the lysates were incubated overnight with polyclonal anti-GFP antibody (Invitrogen) and poly-A-Sepharose beads. Rabbit IgG was used as a control. The processed samples were run on an SDS gel, transferred to nitrocellulose membrane, and probed with a horseradish peroxidase-conjugated anti-FLAG antibody (SIGMA, MO, USA). The immunoprecipitation of TLR2-YFP, TLR4-CFP and MyD88-CFP was checked by immunoblotting the same membrane with monoclonal anti-GFP antibody (Invitrogen, CA, USA). Expression level of the proteins in whole cell lysates was checked by immunoblotting with horseradish peroxidase-conjugated anti-FLAG antibody and monoclonal anti-GFP antibody.
Molecular Modeling of Mal D96N
The amino acid sequences and three-dimensional structures of the TIR domains of TLR1 and TLR2, homologous proteins used as templates for comparative modeling, were obtained from the Protein Data Bank. Initial alignments between Mal and its templates were obtained using the program FUGUE (18). Models were produced using the program MODELLER (19) as previously described (20). Site-directed Mutator (21) was used to calculate a stability score for a specific point mutation (96N) in the Mal TIR model. Three-dimensional structure visualization and image generation was carried out using Pymol (38).
Genotyping of TIRAP D96N in Human Subjects
Genetic studies were approved by the Ethical Committee of the Radboud University Nijmegen, The Netherlands. Genomic DNA from 91 African Tanzanians, 97 Chinese Han, and 188 Caucasian Dutch individuals (22) were screened for the presence of TIRAP D96N (which encodes for Mal D96N). Screening was performed by PCR amplification and sequencing. For amplification, the forward primer 5′-AGTGAGAGGGCACCTGGTAA-3′ and the reverse primer 5′-CACAGCTCGGACACTATAGCGCC-3′ were used. Sequencing was performed with the same primers on a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) by the DNA Sequencing Facility, Radboud University.
RESULTS
Transient Expression of D96N Does Not Activate NF-κB and ISRE Reporters
The non-synonymous SNPs occurring in the coding region of human Mal were selected from the NCBI SNP data base. These were examined for their ability to drive two signaling pathways known to function downstream of Mal. Transient expression of WT Mal in HEK293T cells triggers activation of NF-κB- and IRF5-dependent ISRE luciferase reporters (23, 24). Notably, IRF5 is activated downstream of Mal and MyD88 but not TRIF or TRAM (24). As expected, WT Mal could drive both reporters. Moreover, four of the five polymorphisms screened were comparable to wild type Mal in their ability to drive both reporters (Fig. 1, A and B). However, Mal D96N was severely compromised in driving either the NF-κB or the IRF5-ISRE reporter. IRF5 by itself can weakly activate the ISRE reporter, but the co-expression of wild type Mal leads to a much greater activation above this background value (Fig. 1B). In the case of Mal D96N, this increase in activity over the background level was not seen. These results suggest that Mal D96N does not drive signal transduction upon overexpression.
The Expression of WT Mal Functionally Complements Mal-deficient Macrophages
To study these polymorphisms in phagocytes with a clean genetic background, we immortalized bone marrow-derived macrophages from wild type and Mal-deficient mice using a retrovirus encoding several oncogenes (15, 25). These cell lines behave similar to their respective primary bone marrow-derived macrophages (supplemental Fig. S1). We generated retroviruses expressing FLAG-tagged wild type Mal in tandem with an internal ribosomal entry site (IRES)-encoded green fluorescent protein (GFP). This construct expresses a bicistronic mRNA that allows the translation of both Mal and GFP in the Mal-deficient immortalized cell line. The transduced cells were sorted for GFP and then cloned by limiting dilution. In Mal-deficient macrophages, TNFα production in response to LPS was significantly reduced, whereas in response to PAM2CSK4, it was completely abrogated. We examined TNFα production in the clone obtained from transducing Mal-deficient cell line with retrovirus expressing WT Mal. The single cell clone could completely restore responsiveness to LPS and PAM2CSK4 stimulation (Fig. 2A). Similarly, cells lines expressing each of the variant forms of Mal were generated. Clones with similar levels of expression of Mal, as assessed by Western blot, were selected for further study (Fig. 2B).
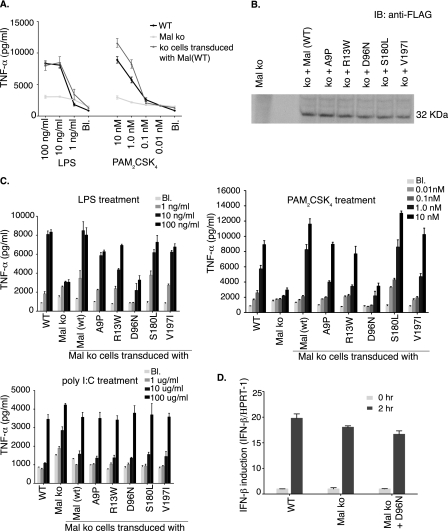
FIGURE 2.
D96N has impaired cytokine production in response to TLR2 or TLR4 ligands. A, functional restoration of Mal-deficient immortalized macrophage cell line. Immortalized Mal knock-out (ko) cells were transduced with a retrovirus carrying FLAG-tagged Mal. Single cell clones were selected and tested for their responsiveness to LPS and PAM2CSK4. B, the same strategy was used to generate cell lines expressing the different variants of Mal. For each cell line clones were chosen for further study based on similar levels of expression of Mal or the Mal variant. IB, immunoblot. C, immortalized Mal-deficient macrophage cell lines expressing either WT Mal or one of the variants were stimulated with LPS, PAM2CSK4, and poly I:C overnight. The supernatants were then analyzed for TNFα levels. Poly I:C, a TLR3/TRIF ligand, was used as a control for Mal-independent signaling. Results are reported as the mean of triplicate determinations ±S.D. Each graph is representative of three independent experiments. D, immortalized WT and Mal-deficient (Mal ko)- and Mal-deficient macrophages (Mal ko + D96N)-expressing D96N were stimulated for 2 h with LPS (100 ng/ml). Total RNA was extracted. Levels of mRNA for IFN-β were determined by quantitative real-time-PCR and normalized to hypoxanthine phosphoribosyltransferase 1 (HPRT1) level of expression. Results are reported as the mean of duplicate determinations ±S.D. This graph is representative of one of two independent experiments. Bl, blank.
Mal D96N Has Impaired Cytokine Production
Reconstituted cell lines were stimulated with LPS and PAM2CSK4, and the pro-inflammatory cytokine TNFα was measured in cell supernatants by enzyme-linked immunosorbent assay. The variants A9P, R13W, S180L, and V197I were fully functional and produced comparable levels of TNFα as WT cells. However, in agreement with our earlier experiments, D96N was significantly compromised in its ability to produce pro-inflammatory cytokines upon stimulation (Fig. 2C). The production of another proinflammatory cytokine, interleukin-6, was similar to TNFα (data not shown). As a control, TNFα production by the double-stranded RNA mimetic poly I:C stimulation was also measured. Poly(I·C) is a TLR3 ligand whose signaling is independent of Mal (26). In this case, TNFα production in cells expressing the D96N mutation was comparable with WT macrophages as well as to cells expressing the different variants of Mal. To test whether the MyD88-independent TLR4 signaling is intact in D96N-expressing cells, we measured induction of IFN-β mRNA upon LPS stimulation using real-time PCR (Fig. 2D). D96N expressing macrophages induced comparable amounts of IFN-β mRNA as WT and Mal-deficient cells. Thus, macrophages expressing the D96N variant of Mal are defective in signaling through TLR2 and the MyD88 branch of TLR4, whereas signaling initiated through other TLRs occurs normally.
IκB Degradation Assays
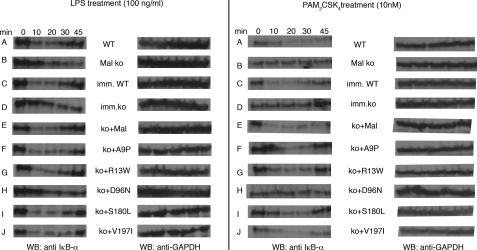
NF-κB is the predominant transcription factor responsible for pro-inflammatory cytokine production downstream of TLR2- and MyD88-dependent TLR4-signaling pathways. We, therefore, examined the activation of NF-κB (via IκB degradation) after LPS and PAM2CSK4 treatment in the reconstituted cell lines mentioned above. In Mal-deficient macrophages, IκB degradation is abrogated upon PAM2CSK4 treatment, whereas for LPS stimulation Mal-deficient macrophages undergo delayed degradation (27). In WT bone marrow-derived macrophages, IκB was completely degraded at 10 min after LPS or PAM2CSK4 treatment (Fig. 3, row A). As expected, Mal-deficient bone marrow-derived macrophages had a delayed degradation profile in the case of LPS (Fig. 3, left panel, row B) and exhibited no degradation in the case of PAM2CSK4 treatment (Fig. 3, right panel, row B). Immortalized wild type and Mal-deficient cell lines mimicked their primary cell types (Fig. 3, rows C and D). Mal-deficient cell lines expressing wild type Mal, A9P, R13W, S180L, or V197I degraded IκB at 10 min, like the wild type immortalized cells (Fig. 3, rows E--G, I, and J). In contrast, however, and in agreement with the cytokine data, cells expressing the D96N mutant behaved like the Mal-deficient cells (Fig. 3, row H), with no degradation in the case of PAM2CSK4 and a delayed degradation profile after LPS treatment. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal loading control. Thus, Mal D96N is a hypomorphic, if not a null variant of Mal, fully defective in NF-κB activation.
FIGURE 3.
Mal D96N fails to degrade IκB-α effectively. NF-κB activation in the reconstituted cell lines was examined via IκB-α degradation assay. The cells were stimulated with LPS (left panel) or PAM2CSK4 (right panel) for the indicated time points, and the whole cell lysates loaded on a denaturing gel, transferred to nitrocellulose membrane, and blotted with IκB-α antibody. Levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) act as an internal loading control. WT, wild type; ko, knock out; WB, Western blot.
Interaction Studies with TLR4, TLR2, and MyD88
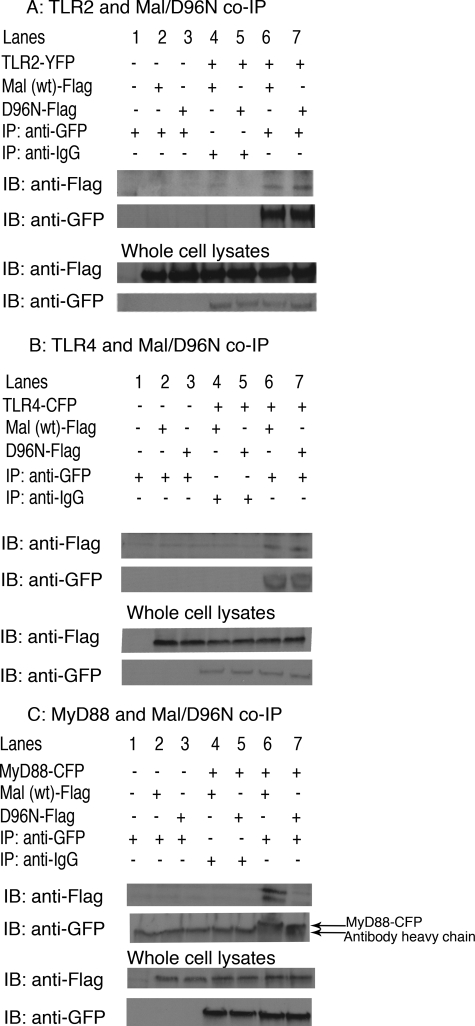
Having established the failure of Mal D96N to signal, we next sought an explanation for the inability of D96N to transduce the signal downstream of TLR2 and TLR4. It is thought that Mal acts as a bridging adapter between the receptor (TLR2 or TLR4) and MyD88 and, thus, physically interacts with both (28). We, therefore, examined whether the D96N mutation affected the ability of Mal to interact with TLR2, TLR4, or MyD88. Transient transfections in HEK293T cells followed by immunoprecipitation assays were carried out to address this question (Fig. 4). As expected, wild type Mal interacted with the receptors TLR2 (Fig. 4A) and TLR4 (Fig. 4B) and with MyD88 (Fig. 4C, lane 6). Like wild type Mal, D96N could be co-immunoprecipitated with both TLR2 and TLR4, indicating that this mutation does not affect the ability of Mal to interact with either of these receptors. However, D96N was severely compromised in its ability to interact with MyD88. Although wild type Mal could co-immunoprecipitate with MyD88, D96N could not (Fig. 4C, lane 7). Thus, the hypomorphic phenotype of D96N appeared to result from its inability to interact with MyD88.
FIGURE 4.
D96N fails to interact with MyD88. HEK293T cells were plated to half confluence in six-well plates. After attachment, cells were transiently transfected with either TLR2-YFP (A), TLR4-CFP (B), or MyD88-CFP (C) and FLAG-tagged Mal WT or Mal D96N. Two days after transfection, supernatants were removed, and cells were lysed. Anti-GFP polyclonal antibody was used for immunoprecipitation (IP). Immunoprecipitates were loaded on a denaturing gel, transferred to nitrocellulose membrane, and visualized with a monoclonal anti-FLAG-horseradish peroxidase antibody. The immunoprecipitation of TLR2-YFP, TLR4-CFP, and MyD88-CFP was confirmed by immunoblotting the membrane with anti-GFP antibody. The same antibodies were used to check the level of expression of the proteins in whole cell lysates. IB, immunoblot.
Modeling Studies for Mal Suggest That a Change in Surface Charge Accounts for the D96N Phenotype
We generated high quality models of the TIR domains of both WT and D96N Mal using the resolved crystal structures of TLR1 and -2 as templates (29). The Mal TIR domain was predicted to have good geometry, with none of its ϕ-ψ dihedral angles within the disallowed regions of a Ramachandran plot and 90.6% in geometrically favored regions. JOY and Verify3D outputs (20) revealed that the environment of residues in the model of Mal are similar to those of the TLR1 and TLR2 TIR domains used as templates and were not energetically unfavorable. Residue Asp-96 was predicted to lie on the surface of the TIR domain in a highly negatively charged region. Mal D96N was modeled using the same approach as the wild type protein and exhibited no significant changes in structure or geometry compared with wild type Mal, as might have been expected from a change in a surface residue. This prediction was further supported using software capable of estimating the level of structural destabilization encoded by point mutations (site-directed mutator or SDM) (21). This program uses a statistical potential energy function that predicts the effect that a specific amino acid substitution may have on the stability of that protein and produces the pseudo-free energy difference between a wild type and mutant protein (21, 30). The D96N substitution was shown to induce structural alterations with corresponding pseudo ΔΔG values of 0.729 kcal/mol, implying that there is no significant change to the overall stability of the Mal TIR domain caused by this mutation.
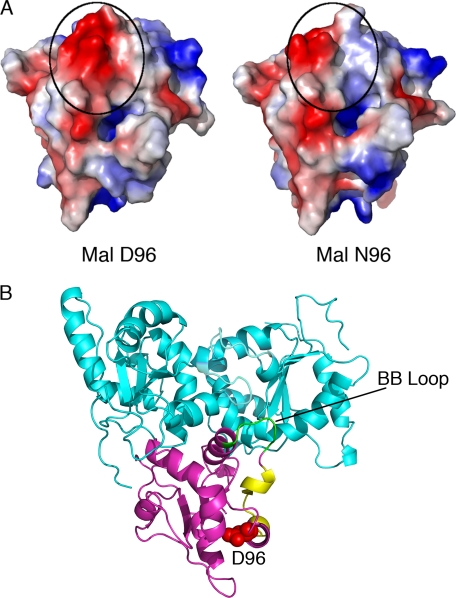
The loss of a surface-exposed negatively charged side chain resulting from an aspartic acid to asparagine substitution (Asp → Asn) (Fig. 5A) would be predicted to be responsible for the experimentally observed disruption of the Mal/MyD88 protein interaction. Fig. 5B shows a model of the TLR4-TIR homodimer (cyan)-Mal TIR domain (magenta) complex. The change in electrostatic charge resides near a surface region (pattern 108–122; shown in yellow) that is predicted to interact with MyD88 (Fig. 5B and Ref. 20). Moreover, residue 96 lies distal to the BB loop region of Mal (green) that has previously been proposed to interact with TLR4 (Fig. 5B and Ref. 20), consistent with the immunoprecipitation studies in Fig. 4 indicating that Mal D96N retains its ability to bind to TLRs 2 and 4 even though it is no longer capable of binding MyD88.
FIGURE 5.
The D96N mutation results in a loss of negative charge on the surface of the Mal-TIR domain in a region predicted to be involved in MyD88, but not TLR interactions. A, comparison of the electrostatic surfaces of wild-type (left) and Asn-96 (right) Mal-TIR models, indicating a loss of negative charge (red) in the region of residue 96 (circled). B, model of the TLR4-TIR homodimer (cyan)/Mal TIR domain (magenta) complex (20), indicating the location of Mal-D96 (red, space fill), its proximity to the predicted MyD88 interaction surface (pattern 108–122, yellow), and its distance from the BB-loop region (green) that is vital for the interaction of Mal with TLR4 (cyan).
Genotyping of TIRAP in Human Populations
Mal D96N is an inactivating mutation that is unable to transduce signaling downstream of TLR2 and TLR4. We next addressed the question of what the frequency of this polymorphism was in the general population. We sequenced Tirap in African Tanzanian (n = 91), Chinese Han (n = 97), and Caucasian Dutch (n = 188) individuals. The African Tanzanian and the Chinese individuals all apparently expressed wild type Mal. However, one individual in the Caucasian Dutch cohort was heterozygous for the D96N mutation. Although this experiment is not an exhaustive assessment of the frequency of MalD96N in all populations, we can conclude that this variation of Mal has a very low prevalence in the populations studied. This is understandable given the strong phenotype in innate signaling that we observed in our experiments. Indeed, individuals homozygous for MalD96N would be expected to have a phenotype similar to what has been reported for MyD88 deficiency, i.e. hypersusceptible to common life-threatening pathogens such as Streptococcus pneumoniae (31).
DISCUSSION
TLRs are the key sensors of microbial infection in mammals. Upon encountering a pathogen, they elicit an inflammatory response involving an intricate network of signaling molecules. Changes in the structure of the TLR signaling components can have dramatic effects on the outcome of various infectious diseases. Some of these changes may be caused by the presence of a SNP in the coding region of the protein. The effects of SNPs on the structure of proteins and the consequent effect on TLR signaling have been described in many studies (32–35).
TIR domain interactions between the TLR components are a necessary event for downstream signaling to occur. SNPs in adapter proteins could, thus, be influential in determining the progression of infection. Incidentally, Mal is the most polymorphic of all the adapter proteins. Of the eight known coding polymorphisms in Mal, only one, S180L, has been previously described (9). Khor et al. (9) found that the heterozygotes carrying this polymorphism were protected from malaria, tuberculosis, and bacteremia. In view of these observations we hypothesized that SNPs in Mal could affect TLR2 and TLR4 signaling.
The data presented herein make a strong case that a variant of Mal, D96N, is essentially nonfunctional. Our model would indicate that Mal D96N does not bind MyD88 because of a critically located change in charge on its surface and, hence, fails to function as a bridging adapter between the receptors TLR2 and TLR4 and the sorting adapter MyD88. Its inability to engage MyD88 prevents activation of NF-κB, IRF5, and most likely activator protein (AP-1), all of which are required to trigger pro-inflammatory cytokine production.
A noteworthy point is that Mal-deficient macrophages from mice were all reconstituted with human Mal. The TIR domains of murine and human Mal are 71% identical, and indeed, human Mal was capable of fully restoring function in these cells. As expected, D96N was defective in pro-inflammatory cytokine production upon stimulation. A surprising finding was the absence of a phenotype for Mal S180L in our studies, an outcome that conceivably could be because of differences in species of the reconstituted protein and cell types, although this explanation would not hold for the ability of transfected Mal S180L to drive reporter constructs in HEK293T cells. In our experiments Mal S180L was fully active, and its effect upon stimulation was comparable with that of wild type Mal. It is true that we have no truly satisfactory explanation for the discrepancy between our findings and that of Khor et al. (9). One explanation for our findings, at least in Mal knock-out macrophages complemented with the various mutant Mal constructs, is that Mal S180L may be a hypomorphic lesion rather than a null mutation. Hence, sufficient overexpression may overcome its relative inability to signal. Indeed, Western blots of the transduced Mal as well as the variants using an established anti-Mal antibody suggested that the recombinant proteins were at least 10–100-fold more abundant in the immortalized cell lines than endogenous wild type Mal (data not shown). Clearly, this discrepancy from previously published results needs to be better analyzed. To this effect, a knock-in mouse expressing the S180L lesion has been engineered (data not shown) and will need to be characterized to determine the nature of this mutation in vivo.
Residue 96 and the surrounding 10–12 amino acid region is fairly well conserved in the known orthologs of TIRAP. The presence of an aspartic acid (Asp) at position 96 creates a negatively charged surface that is conducive to protein-protein interactions. Conversion to uncharged asparagine would, thus, be expected to change the binding potential of the protein. Modeling of the TIR domains of Mal and Mal D96N suggested that the residue 96 lies away from the TLR4 or TLR2 binding site but is in very close proximity to the MyD88 binding site (Fig. 5). These predictions were confirmed in co-immunoprecipitation assays and show that Mal D96N retains the ability to bind TLR2 and TL4 but is unable to bind MyD88. Hence, this characterization of a natural Mal mutant identifies with apparent pinpoint precision a single residue in the TIR domain of Mal that is critical for its interaction with MyD88.
The frequency of D96N in the populations we analyzed was very low. In the initial screen we genotyped individuals from three different populations. However, we found just one individual (of 376) who was heterozygous for D96N. Considering the deleterious phenotype we observe, the low frequency of this polymorphism is not unexpected. As the mutation exists, it is nearly certain that there are sporadic individuals born who are homozygous for D96N. Because the data suggest the failure to bind MyD88 is the reason for the phenotype of D96N, we believe that individuals carrying this mutation in a homozygous state would be extremely susceptible to both Gram-negative and Gram-positive infections. In view of the reported serious clinical significance of MyD88 and IRAK4 deficiencies, two gene products that are downstream of Mal and which should be similar to Mal D96N expression, we believe that Mal D96N represents a rare, potentially life-threatening mutation (36, 37). Indeed, individuals occasionally present for medical case with sepsis in the absence of any known risk factors; we would suggest that MalD96N should be considered under such circumstances as a potential cause of such a clinical presentation.
In contrast to what one might expect for the rare individual with two mutant alleles, the fact that this amino acid variant exists in the population, albeit at a very low frequency, suggests that heterozygous individuals have a survival advantage under some, but still undefined, circumstances. The key to determining the nature of this hypothetical heterozygous advantage is to analyze enough individuals from enough different populations to define clinical correlates.
Supplementary Material
Acknowledgment
We thank Dr. Shizuo Akira for the gift of anti-Mal/TIRAP antiserum and sharing of the Tirap knockout mice.
This work was supported, in whole or in part, by National Institutes of Health Grants AI52455 and GM54060 (to D. T. G. and K. N.) and AI067497 (to K. A. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TLR
- Toll-like receptor
- Mal
- MyD88 adapter-like
- TIRAP
- TIR domain-containing adapter protein
- SNP
- single nucleotide polymorphism
- GFP
- green fluorescent protein
- LPS
- lipopolysaccharide
- IRES
- internal ribosomal entry site
- GFP
- encoded green fluorescent protein
- IRF
- interferon regulatory factor
- WT
- wild type
- TNF
- transforming growth factor
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- ISRE
- interferon-stimulated response element.
REFERENCES
- 1.O'Neill L. A. (2008) Immunity 29, 12–20 [DOI] [PubMed] [Google Scholar]
- 2.Hong-Geller E., Chaudhary A., Lauer S. (2008) Curr. Drug Discov. Technol. 5, 29–38 [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 4.Horng T., Barton G. M., Medzhitov R. (2001) Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 5.Kagan J. C., Medzhitov R. (2006) Cell 125, 943–955 [DOI] [PubMed] [Google Scholar]
- 6.Vogel S. N., Fitzgerald K. A., Fenton M. J. (2003) Mol. Interv. 3, 466–477 [DOI] [PubMed] [Google Scholar]
- 7.Hawn T. R., Dunstan S. J., Thwaites G. E., Simmons C. P., Thuong N. T., Lan N. T., Quy H. T., Chau T. T., Hieu N. T., Rodrigues S., Janer M., Zhao L. P., Hien T. T., Farrar J. J., Aderem A. (2006) J. Infect. Dis. 194, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurfel M. M., Gordon A. C., Holden T. D., Radella F., Strout J., Kajikawa O., Ruzinski J. T., Rona G., Black R. A., Stratton S., Jarvik G. P., Hajjar A. M., Nickerson D. A., Rieder M., Sevransky J., Maloney J. P., Moss M., Martin G., Shanholtz C., Garcia J. G., Gao L., Brower R., Barnes K. C., Walley K. R., Russell J. A., Martin T. R. (2008) Am. J. Respir. Crit. Care Med. 178, 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khor C. C., Chapman S. J., Vannberg F. O., Dunne A., Murphy C., Ling E. Y., Frodsham A. J., Walley A. J., Kyrieleis O., Khan A., Aucan C., Segal S., Moore C. E., Knox K., Campbell S. J., Lienhardt C., Scott A., Aaby P., Sow O. Y., Grignani R. T., Sillah J., Sirugo G., Peshu N., Williams T. N., Maitland K., Davies R. J., Kwiatkowski D. P., Day N. P., Yala D., Crook D. W., Marsh K., Berkley J. A., O'Neill L. A., Hill A. V. (2007) Nat. Genet. 39, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasl J., Prohinar P., Gioannini T. L., Weiss J. P., Jerala R. (2008) J. Immunol. 180, 6107–6115 [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 12.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. (2004) Nat. Immunol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
- 13.Schoenemeyer A., Barnes B. J., Mancl M. E., Latz E., Goutagny N., Pitha P. M., Fitzgerald K. A., Golenbock D. T. (2005) J. Biol. Chem. 280, 17005–17012 [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawley R. G., Lieu F. H., Fong A. Z., Hawley T. S. (1994) Gene Ther. 1, 136–138 [PubMed] [Google Scholar]
- 16.Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., Takeda K., Akira S. (2002) Nature 420, 324–329 [DOI] [PubMed] [Google Scholar]
- 17.Charrel-Dennis M., Latz E., Halmen K. A., Trieu-Cuot P., Fitzgerald K. A., Kasper D. L., Golenbock D. T. (2008) Cell Host Microbe 4, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J., Blundell T. L., Mizuguchi K. (2001) J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 19.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 20.Núñez Miguel R., Wong J., Westoll J. F., Brooks H. J., O'Neill L. A., Gay N. J., Bryant C. E., Monie T. P. (2007) PLoS ONE 2, e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topham C. M., Srinivasan N., Blundell T. L. (1997) Protein Eng. 10, 7–21 [DOI] [PubMed] [Google Scholar]
- 22.Ferwerda B., McCall M. B., Alonso S., Giamarellos-Bourboulis E. J., Mouktaroudi M., Izagirre N., Syafruddin D., Kibiki G., Cristea T., Hijmans A., Hamann L., Israel S., ElGhazali G., Troye-Blomberg M., Kumpf O., Maiga B., Dolo A., Doumbo O., Hermsen C. C., Stalenhoef A. F., van Crevel R., Brunner H. G., Oh D. Y., Schumann R. R., de la Rúa C., Sauerwein R., Kullberg B. J., van der Ven A. J., van der Meer J. W., Netea M. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16645–16650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miggin S. M., Pålsson-McDermott E., Dunne A., Jefferies C., Pinteaux E., Banahan K., Murphy C., Moynagh P., Yamamoto M., Akira S., Rothwell N., Golenbock D., Fitzgerald K. A., O'Neill L. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3372–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paun A., Reinert J. T., Jiang Z., Medin C., Balkhi M. Y., Fitzgerald K. A., Pitha P. M. (2008) J. Biol. Chem. 283, 14295–14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., Tschopp J. (2004) Nat. Immunol. 5, 503–507 [DOI] [PubMed] [Google Scholar]
- 27.McGettrick A. F., O'Neill L. A. (2004) Mol. Immunol. 41, 577–582 [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald K. A., Chen Z. J. (2006) Cell 125, 834–836 [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. (2000) Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 30.Worth C. L., Bickerton G. R., Schreyer A., Forman J. R., Cheng T. M., Lee S., Gong S., Burke D. F., Blundell T. L. (2007) J. Bioinform. Comput. Biol. 5, 1297–1318 [DOI] [PubMed] [Google Scholar]
- 31.von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C. L., Chrabieh M., Mustapha I. B., Ghandil P., Camcioglu Y., Vasconcelos J., Sirvent N., Guedes M., Vitor A. B., Herrero-Mata M. J., Aróstegui J. I., Rodrigo C., Alsina L., Ruiz-Ortiz E., Juan M., Fortuny C., Yagüe J., Antón J., Pascal M., Chang H. H., Janniere L., Rose Y., Garty B. Z., Chapel H., Issekutz A., Maródi L., Rodriguez-Gallego C., Banchereau J., Abel L., Li X., Chaussabel D., Puel A., Casanova J. L. (2008) Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davila S., Hibberd M. L., Hari Dass R., Wong H. E., Sahiratmadja E., Bonnard C., Alisjahbana B., Szeszko J. S., Balabanova Y., Drobniewski F., van Crevel R., van de Vosse E., Nejentsev S., Ottenhoff T. H., Seielstad M. (2008) PLoS Genet. 4, e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson C. M., Lyle E. A., Omueti K. O., Stepensky V. A., Yegin O., Alpsoy E., Hamann L., Schumann R. R., Tapping R. I. (2007) J. Immunol. 178, 7520–7524 [DOI] [PubMed] [Google Scholar]
- 34.Schumann R. R., Tapping R. I. (2007) Eur. J. Immunol. 37, 2059–2062 [DOI] [PubMed] [Google Scholar]
- 35.Seabury C. M., Cargill E. J., Womack J. E. (2007) Genomics 90, 502–515 [DOI] [PubMed] [Google Scholar]
- 36.Medvedev A. E., Lentschat A., Kuhns D. B., Blanco J. C., Salkowski C., Zhang S., Arditi M., Gallin J. I., Vogel S. N. (2003) J. Exp. Med. 198, 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picard C., Puel A., Bonnet M., Ku C. L., Bustamante J., Yang K., Soudais C., Dupuis S., Feinberg J., Fieschi C., Elbim C., Hitchcock R., Lammas D., Davies G., Al-Ghonaium A., Al-Rayes H., Al-Jumaah S., Al-Hajjar S., Al-Mohsen I. Z., Frayha H. H., Rucker R., Hawn T. R., Aderem A., Tufenkeji H., Haraguchi S., Day N. K., Good R. A., Gougerot-Pocidalo M. A., Ozinsky A., Casanova J. L. (2003) Science 299, 2076–2079 [DOI] [PubMed] [Google Scholar]
- 38.DeLano W. (2002) Pymol, DeLano Scientific, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.