Abstract
The cGMP-stimulated PDE2A hydrolyzes both cyclic nucleotides, cGMP and cAMP. Three splice variants have been cloned from several species. Whereas PDE2A1 is soluble, PDE2A2 and PDE2A3 are membrane-bound enzymes of rat and bovine origin, respectively. To date it is unclear whether one species expresses all three variants. The splice variants only differ in their N termini, which likely determine the subcellular localization. However, the mechanism for membrane attachment remains unknown. Here, we show that myristoylation underlies membrane targeting of PDE2A3. The myristoylated enzyme was bound to plasma membranes, whereas mutation of the myristoyl recipient Gly2 prevented incorporation of [3H]myristate and turned PDE2A3 completely soluble. Additionally, Cys5 and to a minor extent Cys11 are required for targeting of PDE2A3. Substitution of the putatively palmitoylated cysteines partially solubilized the enzyme and led to an accumulation in the endoplasmic reticulum/Golgi compartment, as shown by fluorescence microscopy in HEK 293 and PC12 cells. In vivo, PDE2A is expressed in many tissues. By using newly generated antibodies selectively detecting the splice variants PDE2A3 or PDE2A1, respectively, we demonstrate on the protein level PDE2A3 expression in mouse brain where it is entirely membrane-associated and a widespread expression of soluble PDE2A1 in mouse tissues. We show that PDE2A localizes to synaptosomal membranes and in primary cultures of hippocampal neurons partially overlaps with the presynaptic marker synaptophysin as demonstrated by immunofluorescence. In sum, these results demonstrate dual acylation as mechanism targeting neuronal PDE2A3 to synapses thereby ensuring local control of cyclic nucleotides.
The widespread occurrence of cyclic nucleotide second messengers in mammalian signal transduction pathways necessitates a tight control of their intracellular concentration. Regulation of cyclic nucleotide levels not only occurs at the level of synthesis but also at the level of degradation. The enzymes responsible for the elimination of cAMP and cGMP are the cyclic nucleotide phosphodiesterases (PDE).2 So far, 11 members of the PDE family (PDE1 to PDE11) each with distinct properties have been identified (1). PDE2A is a homodimer with an approximate molecular mass of 210 kDa. Each monomer consists of an N-terminal domain followed by a tandem of so-called GAF domains (GAF A and GAF B) and a C-terminal catalytic domain. The enzyme belongs to the dual substrate PDEs and hydrolyzes both cyclic nucleotides with positively cooperative kinetics (2). A characteristic feature of PDE2A is the severalfold stimulation of cAMP hydrolysis by micromolar concentrations of cGMP (2–4). The enzyme thereby allows a cross-talk between cGMP- and cAMP-mediated signaling pathways. One of the physiological consequences is the ANP-dependent inhibition of aldosterone secretion in adrenal glomerulosa cells (5).
The cGMP-stimulated PDE activity, first observed in rat liver (6), has since been purified from a number of mammalian tissues, i.e. bovine heart, adrenal gland, liver, and brain (2, 7, 8). PDE2A was found in the cytosol but also in particulate fractions (8–10), where it exhibited a slightly higher molecular weight. Proteolytic digestion resulted in two peptides that differed between the cytosolic and particulate enzyme suggesting that at least two variants exist (9). Three PDE2 variants with completely different N termini (supplemental Fig. S1) have been cloned from bovine adrenal (PDE2A1), rat brain (PDE2A2), and human brain (PDE2A3) tissues (11–13). The three different N termini result from alternative splicing of a single gene (PDE2A) and determine whether the isoforms are soluble (PDE2A1) or associated to membranes (PDE2A2 and -A3). However, it remains unclear how the latter isoforms are targeted to membranes and whether they coexist in one species.
In contrast to the other PDE2A isoforms, the PDE2A3 N terminus features a potential motif for N-myristoylation and in the vicinity possesses two cysteine residues (Cys5/Cys11) as possible targets for palmitoylation. In the current report, we demonstrate dual acylation as the mechanism underlying membrane targeting of PDE2A3. Using radiolabeling PDE2A3 is shown to be N-terminally myristoylated at Gly2. Biochemical analysis as well as fluorescence microscopy revealed that mutation of the acylation sites shifts PDE2A3 from membranes to the cell cytosol in HEK 293 and PC12 cells. By using antibodies selectively detecting the PDE2A3 or PDE2A1 isoform, respectively, predominant expression of membrane-bound PDE2A3 in mouse brain is demonstrated on the protein level for the first time, and expression of soluble PDE2A1 in a wide variety of mouse tissues is confirmed. In accordance with a special function in synaptic transmission, high amounts of PDE2A are detected in isolated synaptosomal membranes, and PDE2A co-localizes with the synaptic marker synaptophysin in primary hippocampal neurons.
MATERIALS AND METHODS
Cloning and Site-directed Mutagenesis
The PDE2A1 and PDE2A3 sequences were amplified by PCR using mouse brain cDNA as template. Sense oligonucleotides were chosen to match sequences in the 5′-untranslated regions of the two splice variants (5′-GGTTCCTATGCCTGCTGGGAGTGCC-3′ and 5′-CGGGGCCAGCAGGTCTTCC-3′, respectively), and the antisense oligonucleotide corresponded to the common coding region (5′-CCTGCTGCAGATATTGAAGGACTTTG-3′). The initial PCR products were then used as template for a second round of PCR using sense primers that introduced NheI restriction sites (5′-GCTAGCACCATGGGGCAGGC-3′ and 5′-GCTAGCACCATGCGCCGAC-3′, respectively) and an antisense primer matching the coding region (5′-GATCTTTCGGTCATGGTCGGTGTA-3′). PCR products were subcloned into the NheI site of the pcDNA3.1zeo (Invitrogen) multicloning site and an internal OliI site of PDE2A sequences that were cloned to this vector previously. The QuikChange site-directed mutagenesis kit (Stratagene) was used to make point mutations in the PDE2A3 construct. Mutagenic oligonucleotides were ordered from Sigma-Aldrich. To generate double and triple mutants (e.g. PDE2A3 G2A/C5S/C11S) the corresponding single and double mutants were again used as template for site-directed mutagenesis. Sequences of all oligonucleotides are available on request.
For CFP constructs the pcDNA3.1zeo-vector was equipped with PCR-amplified sequences coding for ECFP (Clontech). For cloning, the NheI/NotI sites were used and an additional restriction site (BsmBI) was introduced in front of the CFP starter methionine. This site was used for in-frame cloning of PCR-amplified sequences coding for the divergent N-terminal amino acids of PDE2A1 and -2A3 and the respective mutants. To ensure in-frame cloning and the presence of desired mutations, all manipulated DNAs were sequenced (SeqLab, Göttingen, Germany).
For GST-tagged clones used in metabolic labeling, sequences coding for amino acids 1–46 of PDE2A3 or PDE2A3-G2A were PCR-amplified and cloned via NheI/BamHI into the pEYFP-N1 vector (Clontech). Subsequently YFP-encoding sequences were exchanged for GST-encoding ones (derived from pGEX 4T3, Amersham Biosciences) via restriction enzymes AgeI/NotI.
Cell Culture
HEK 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% heat-inactivated fetal calf serum. PC12 cells were sustained in Ham's F-12 medium with 2 mm l-glutamine and 1.5 g/liter sodium bicarbonate supplemented with 15% horse serum and 2.5% fetal calf serum in poly-d-lysine-coated culture dishes. All cells were grown in the presence of 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 atmosphere.
Expression and Analysis of Subcellular Distribution of PDE2A
Transfection of PDE2A cDNA into HEK 293 cells was performed using FuGENE6 transfection reagent (Roche Diagnostics) according to the manufacturer's protocol. Cells were grown in 6-well plates and harvested 48–72 h post transfection. All following steps were carried out at 4 °C or on ice. Cells were washed twice with 2 ml of phosphate-buffered saline (PBS), resuspended in 0.5 ml of lysis buffer (150 mm NaCl (unless stated otherwise), 1 mm EDTA, 2 mm dl-dithiothreitol, 50 mm triethanolamine/hydrochloride, pH 7.4, containing the protease inhibitors phenylmethylsulfonyl fluoride (0.4 mm), benzamidine (0.2 mm), and Pepstatin A (1 μm)) and lysed by sonication (two 5-s pulses). To separate cytosolic and membrane fractions, 300 μl of the homogenate was centrifuged (125,000 × g, 40 min, 4 °C). The membranes were washed once before resuspension in 300 μl of lysis buffer supplemented with 1% Triton X-100.
Western Analysis
HEK 293 homogenate, cytosolic proteins, and membrane fractions (4 μl/lane) were separated by SDS-PAGE and transferred to nitrocellulose membranes (Protran BA-85, Schleicher & Schuell/Whatman) using standard procedures. After blocking (Roti-Block, Carl Roth, Germany), PDE2A was detected with polyclonal goat anti-PDE2A antibody (1:1000, sc-17228, Santa Cruz Biotechnology, Santa Cruz, CA) unless stated otherwise. This antibody is directed against a region common to all PDE2A splice variants. Secondary peroxidase-labeled anti-goat IgG was obtained from Sigma-Aldrich. For detection, SuperSignal West Dura chemiluminescent substrate (Pierce) and a charge-coupled device camera (GDS 8000, UVP) were used. Quantitative analysis of Western signals was carried out with LabWorks 4.0 software (UVP) setting cytosol plus membrane to 100%. In Western analysis of tissue preparations, glyceraldehyde-3-phosphate dehydrogenase was detected with Abcam antibody ab8245 (diluted 1:1000) and secondary peroxidase-labeled anti-mouse IgG (Sigma-Aldrich). To discriminate between PDE2A1 and PDE2A3, antibodies raised against unique N-terminal peptides (amino acids 9–23 of PDE2A1 and amino acids 11–25 of PDE2A3) were used at a 1:1000 dilution, respectively. Specificity of the antisera is shown using HEK 293 cells overexpressing PDE2A1, -A2, or -A3 (supplemental Fig. S2A). In mouse brain, the antibodies recognized a band at the expected molecular weight of PDE2A, which was suppressed by the respective blocking peptide (supplemental Fig. S2B).
Inhibition of Myristoylation
The myristoylation inhibitor 2-hydroxymyristic acid (2-HMA, Alexis Biochemicals) was dissolved in DMSO to a final concentration of 100 mm and further diluted to 1 mm in Dulbecco's modified Eagle's medium, containing 1% defatted bovine serum albumin (Sigma-Aldrich). Before addition to cells, the 1 mm 2-HMA solution was sonicated briefly and filtered to remove any undissolved 2-HMA. After incubation for 2 h at 37 °C (1 ml/well of a 6-well plate), 1 ml/well Dulbecco's modified Eagle's medium, containing 5% fetal calf serum, was added, followed by overnight incubation.
Metabolic Labeling and GST Precipitation
HEK 293 cells grown in 75-cm2 flasks were transfected with PDE2A3 wt- or G2A-GST constructs as described above. After 1 day, cells were rinsed twice with Dulbecco's modified Eagle's medium, containing 5 mm pyruvate, and subsequently cultured for an additional 4 h in the presence of 300 μCi/ml [3H]myristic acid (PerkinElmer Life Sciences, supplied at 1 mCi/ml ethanol and concentrated in a nitrogen stream to 10 mCi/ml ethanol). Cells were then harvested using trypsin and washed twice with PBS. For lysis, cells were resuspended in 500 μl of lysis buffer containing 50 mm NaCl and subjected to three freeze/thaw cycles (−20 °C). The lysate was cleared of debris (800 g, 10 min, 4 °C) and incubated with 100 μl of glutathione Sepharose 4B (Amersham Biosciences) for 1 h at 20 °C on an overhead rotator. Sepharose was then collected (800 g, 10 min, 4 °C), washed five times with 1 ml of lysis buffer, resuspended in 100 μl of Laemmli sample buffer, and centrifuged (15,000 × g, 5 min, 20 °C). Supernatants were resolved on 9% SDS-polyacrylamide gels and blotted. For detection, an imaging plate (BAS-TR, Fujifilm) was exposed to the blotting membrane for 3 days and read in a phosphorimaging device (FLA-3000, Fujifilm). Raw logarithmic imaging data were linearized using the formula of the manufacturer and ImageJ.3
Assay of PDE Activity
PDE activity was measured by the conversion of [32P]cGMP into [32P]GMP as described previously (14). Reaction mixtures contained 0.1–0.25 μl of the cell fraction. All measurements were carried out in the presence of 1 μm [32P]cGMP (∼2 kBq) and 3 mm MgCl2. Data presented represent means ± S.D. of at least three independent experiments performed in duplicates.
Fluorescence Imaging
For fluorescence microscopy, HEK 293 cells were seeded on glass coverslips 24 h prior to transfection. PC12 cells were seeded on poly-d-lysine-coated coverslips, respectively, and transfected with Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. 48 h post transfection, cells were washed twice with PBS buffer and either fixed in 4% paraformaldehyde for 20 min and stored in PBS until signal detection or imaged directly as live cells. Fluorescence signals were visualized using an inverted microscope (Zeiss Axiovert 200) equipped with the following components: a Polychrome V xenon-lamp based monochromator (TILL Photonics, Grafelfing, Germany), an LD Achroplan 40×/0.6 Korr Ph2 objective, a MultiSpec Micro-Imager (DUAL-view, Optical Insights) containing a dichroic splitter (505dcxr) and emission filters (D465/30 and HQ535/30), and a cooled digital charge-coupled device camera (PCO Sensicam). Offline adjustments were made using the TILLvisION 4.0 software and ImageJ. The plasmids pECFP-Golgi and pECFP-ER (Clontech) were transfected for comparison of cellular localization of PDE2A.
Localization Studies in Hippocampal Neurons
Rat hippocampal neurons were isolated from newborn rats (P0) as described previously (15). Thirty thousand cells were seeded on 12-mm poly-l-lysine-coated coverslips and processed for immunodetection 7 days after plating (P7). Cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, briefly permeabilized with 1% Triton X-100, and incubated with blocking buffer containing 3% fetal calf serum, 1% bovine serum albumin, 0.5% Tween 20 in PBS for 1 h. Polyclonal goat anti-PDE2A (sc-17228; Santa Cruz Biotechnology) and monoclonal mouse anti-synaptophysin (Clone SVP-38; Sigma) antibodies were diluted 1:200 in the same buffer and incubated for 16 h at 4 °C followed by extensive washing with PBS, including 0.5% Tween 20. Immunodetection was performed with donkey anti-goat IgG Alexa Fluor 488 nm and goat anti-mouse IgG Alexa Fluor 568 nm antibodies (Invitrogen) diluted 1:3000 in blocking buffer. After 1 h cells were washed, and nuclei were stained with 4′,6-diamidino-2-phenylindole and mounted using Prolong Antifade (Invitrogen). Staining controls were performed by omitting the primary PDE2A antibody or after blocking with peptide sc-17228 as recommended by the manufacturer (Santa Cruz Biotechnology). An LSM510 Meta system (Zeiss, Jena, Germany) equipped with UV/argon/helium lasers and a Plan Apochromat 63×/1.4 oil objective was used to visualize each specific fluorescence in multitrack mode. The 405/488/543 nm laser lines (15–20% power), main beam splitters set at 405/488/548 nm, and band pass filters at 411–464 nm, 505–530 nm, and 585–615 nm were used. All adjustments with respect to gain, pinhole size, or background were optimized using the LSM510 Meta software. Pictures were exported as 8-bit color images and processed with Adobe Photoshop CS3.
Tissue Preparation
All experiments were performed using adult mice of the C57BL/6 strain. After sacrificing the animals, the indicated organs were immediately homogenized in 10 volumes (w/v) of ice-cold lysis buffer (with 50 mm NaCl) by 15 strokes in a glass-Teflon Potter-Elvehjem homogenizer. The lysate was cleared by centrifugation at 1000 × g (10 min, 4 °C) before sonication. The homogenate was then centrifuged at 125,000 × g for 40 min at 4 °C, and the resulting supernatant was saved as the cytosolic fraction. The particulate fraction (membranes) was washed once and finally resuspended in the original volume of lysis buffer. Equal volumes of all fractions (corresponding to 25 μg of protein in the homogenate) were applied to SDS-PAGE.
Preparation of Synaptosomal Cytosol and Membranes
Synaptosomal cytosol and membranes were isolated as described (16).
RESULTS
The PDE2A3 Isoform Is Firmly Attached to Membranes in HEK 293 Cells
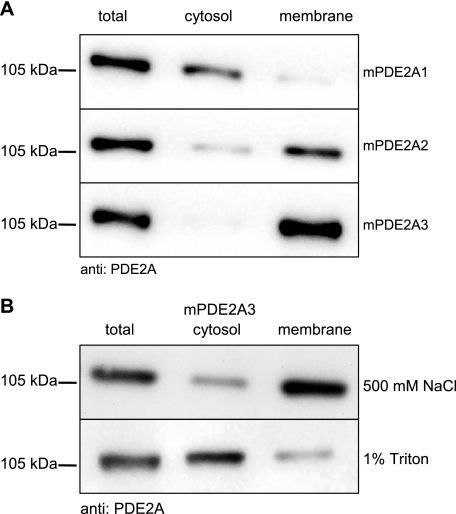
Three splice variants of the PDE2A family have been described. To analyze their subcellular localization, all three variants were cloned from mouse cDNA and expressed in HEK 293 cells. The compartmental localization of the isoforms was then assessed by subcellular fractionation. As shown by Western immunodetection, PDE2A1 was found in the cytosol, whereas PDE2A2 and PDE2A3 were detected in the membrane fractions (Fig. 1A). These results are in accordance with several previous reports (11–13, 17). For PDE2A2, membrane association via its hydrophobic N-terminal end has been suggested (12). However, the mechanism of PDE2A3 targeting must be somewhat different, because its N terminus contains comparably few hydrophobic amino acids. In a first set of experiments, high salt conditions (500 mm NaCl) did not extract PDE2A3 from the membrane, but 1% Triton X-100 solubilized most of the enzyme (Fig. 1B). Because PDE2A3 contains no predicted membrane-spanning region these results argue for membrane anchoring by lipid modification. In fact, sequence analysis of the PDE2A3 N terminus revealed a possible myristoylation motif (Table 1).
FIGURE 1.
Subcellular localization of PDE2A splice variants. A, HEK 293 cells were transiently transfected with the PDE2A splice variants. Cells were homogenized and separated into cytosolic and membrane fractions as described under “Materials and Methods.” Western blot analysis was performed using anti-PDE2A antibody. B, PDE2A3-expressing HEK 293 cells were homogenized in the presence of 500 mm NaCl (top) or 1% Triton X-100 (bottom). Cytosolic and membrane fractions were analyzed in Western blots. Shown are representative results of three independent experiments.
TABLE 1.
Myristoylation/palmitoylation motifs
Listed are the amino acid sequences of a few examples of N-terminally myristoylated/palmitoylated signaling proteins. Myristoylated glycine residues are double underlined, and palmitoylated cysteines are single underlined. For PDE2A3 the residues in question for modifications are italicized.
| Protein | Sequence | Role of modification | Ref. |
|---|---|---|---|
| p59fyn | MGCVQCKDKEATKLTE | Targeting to plasma membrane and lipid rafts; regulation of cell signaling | 38, 39 |
| Gαi | MGCTLSAEDKAAVERS | Modulation of G-protein assembly and activity | 40 |
| eNOS | MGNLKSVAQEPGPPCGLGLGLGLGLCG | Targeting to cell membranes and caveolae | 25, 26 |
| cGK II | MGNGSVKPKHSKHPDG | Membrane binding of cGK II | 41 |
| AKAP18 | MGQLCCFPFSRDEGKISE | Membrane anchoring of PKA and assembly of signaling proteins | 42 |
| PDE2A3 | MGQACGHSILCRSQQYPA | Targeting to cell membranes | This study |
The N-terminal Region of PDE2A3 Is Myristoylated in HEK 293 Cells
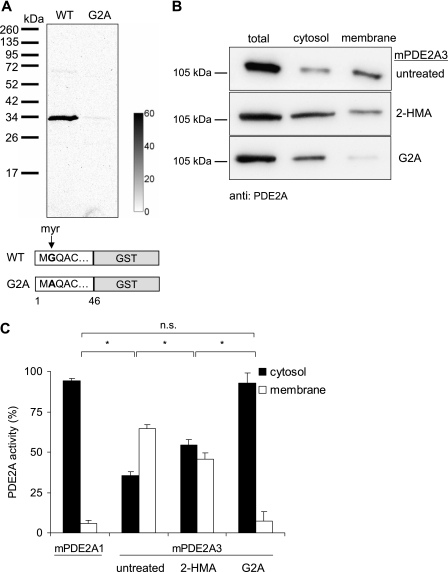
Myristoylation of PDE2A3 was examined by metabolic labeling of HEK 293 cells expressing the unique PDE2A3 N terminus (amino acids 1–46) with a C-terminal GST tag. As a control, a point-mutated PDE2A3 containing a Gly2 to Ala exchange (G2A) was used. Transfected cells were incubated with 300 μCi/ml [3H]myristic acid for 4 h. PDE2A3 was then precipitated using the GST tag, subjected to SDS-PAGE, and transferred to nitrocellulose membranes. Incorporation of the radiolabel was examined by autoradiography (Fig. 2A). A 3H signal was detected on PDE2A3 but not on the PDE2A3 G2A mutant, indicating that PDE2A3 is indeed myristoylated at the second glycine. Because the G2A control did not show any signal, metabolic interconversion between [3H]fatty acids, e.g. myristate to palmitate can be ruled out.
FIGURE 2.
N-Myristoylation of PDE2A3. A, GST fusion proteins of the N-terminal 46 amino acids of PDE2A3 or its G2A mutant, respectively, were labeled with [3H]myristic acid in HEK 293 cells. After precipitation, electrophoresis, and blotting, labeled proteins were detected using a phosphorimaging device. The calibration bar depicts mPSL units (photostimulated luminescence units, Fujifilm). B, Western blot of cytosolic and membrane fractions of PDE2A3-expressing HEK 293 cells. Top, untreated cells. Middle, cells treated with the myristoylation inhibitor 2-HMA (0.5 mm). Bottom, untreated cells expressing the myristoylation deficient G2A mutant of PDE2A3. C, PDE activities in subcellular fractions of HEK 293 cells expressing PDE2A1, PDE2A3, or the G2A mutant of PDE2A3. PDE2A3-expressing cells were treated with 0.5 mm 2-HMA as indicated. Shown are means ± S.D. of at least three independent experiments performed in duplicate; n.s., not significant (p > 0.05); *, significant (p < 0.003), two-tailed t test assuming unequal variances.
Non-myristoylated PDE2A3 Is Soluble
Next, the importance of myristoylation for subcellular localization was investigated. For that purpose, non-myristoylated PDE2A3 was generated by expression of full-length PDE2A3 in the presence of the N-myristoyl transferase inhibitor 2-HMA (0.5 mm) in HEK 293 cells. Cells where then fractionated, and PDE2A was detected by Western analysis. In addition, the cellular distribution of the myristoylation-deficient PDE2A3 G2A mutant was examined. As shown in Fig. 2B, inhibition of myristoylation resulted in a (partial) shift of the otherwise membrane-bound enzyme to the cytosol. This effect was enhanced by the G2A mutation, which rendered the enzyme completely soluble (Fig. 2B). To quantify the subcellular distribution, enzyme activity in the respective cell fractions was determined and expressed as percentage of total activity measured in the cytosol plus membranes (Fig. 2C). The total activities in the different samples were similar. Because neither in the presence of the PDE2-specific inhibitor BAY 60-7550 (50 nm) nor in mock transfected cells any cGMP-degrading activity was detected (data not shown), all activity was ascribed to PDE2A. For comparison, the distribution of the soluble PDE2A1 was analyzed. The data obtained support the Western analysis of subcellular localization, i.e. the myristoylation inhibitor or the G2A mutation shifted PDE2A3 (at least partly) to the cytosol. Thus, the results demonstrate the essential role of Gly2 myristoylation for membrane targeting.
Identification of Cysteines 5 and 11 as Likely Palmitoylation Sites
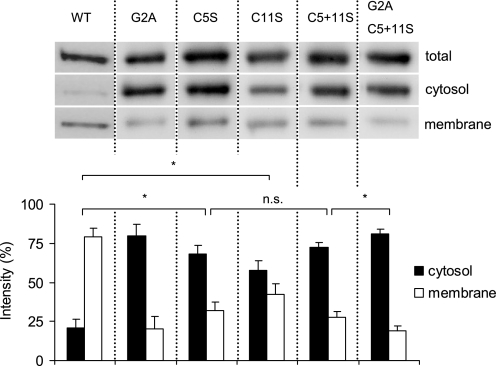
Normally, myristoylation of a protein is not sufficient to allow stable membrane association and therefore is often accompanied by palmitoylation of nearby cysteine residues as a secondary reversible modification (18). This mechanism of membrane targeting has been shown for several other proteins involved in cyclic nucleotide signaling (for references see Table 1) and appears plausible for PDE2A3 membrane anchoring, because the enzyme contains cysteines at positions 5 and 11. To examine the impact of palmitoylation, single and double mutants were created in which the cysteines in question were changed to serine residues. As judged by quantitative analysis of Western signals in cytosol and membrane fractions, elimination of Cys5 increased PDE2A3 solubility 3-fold (cytosolic content of WT PDE2A3 27% versus C5S 74%), and elimination of Cys11 increased solubility 2.5-fold (to 65%). The combined substitution of both cysteines had the same effect as exchange of Cys5 alone, whereas the additional elimination of the myristoylation site (G2A/C5S/C11S) further increased PDE2A3 solubility (to 85%) (Fig. 3).
FIGURE 3.
Cysteines-5 and -11 contribute to membrane attachment of PDE2A3. PDE2A3 carrying point mutations at positions 2 (G2A), 5 (C5S), and 11 (C11S) or a combination thereof, were transiently transfected into HEK 293 cells. Equal volumes of separated cell fractions were then blotted and probed with the PDE2A-specific antibody. Shown is a representative blot of three independent experiments. For comparison, the PDE2A3 WT is shown on the left. The lower panel shows the quantitative analysis of PDE2A3 signal intensities in cytosol and membrane fractions, which together were set to 100%. Bars are means ± S.D. of three independent experiments; n.s., not significant (p > 0.05); *, significant (p < 0.01), two-tailed paired t test.
The N Termini of the PDE2A Isoforms Suffice for Targeting to Different Cellular Compartments in HEK 293 and PC12 Cells
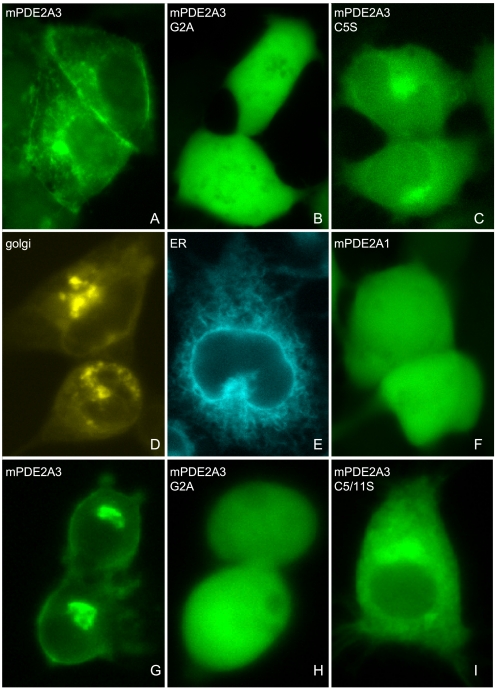
We next sought independent confirmation for our findings by fluorescence microscopy (Fig. 4). To this end, the divergent N-terminal sequences of PDE2A1 and -A3 and the mutants thereof were subcloned as fusion proteins with CFP and expressed in HEK 293 cells. As shown in Fig. 4A, the WT PDE2A3 N terminus was excluded from the nucleus and targeted the CFP fluorescence to the plasma membrane. Also, a considerable staining of the perinuclear region, most likely the Golgi complex and ER, was observed (compare Fig. 4, A, D, and E). Mutation of the myristoylation site (G2A) completely abolished this clearly localized pattern leading to evenly distributed fluorescence throughout the nucleus and cell cytosol (Fig. 4B). Mutation of the palmitoylation site (C5S) led to a partial redistribution of the enzyme from the plasma membrane to the cytosol while retaining the pronounced incidence in the Golgi complex and the exclusion from the nucleus (Fig. 4C). CFP fluorescence of the PDE2A1 fusion protein that was expressed as an example for a soluble protein was spread throughout the nucleus and cell cytosol (Fig. 4F), thus resembling the distribution of the myristoylation-deficient G2A mutant. Identical observations were made when expressing the CFP fusion proteins in a PC12 cell line used as a model for neuronal cells (Fig. 4, G–I). In sum, the above results suggest that myristoylation of glycine-2 is required for membrane targeting of PDE2A3 and that additional palmitoylation of cysteines-5/-11 enhances membrane association.
FIGURE 4.
Membrane targeting of a PDE2A3/CFP fusion protein is disrupted by G2A and C5S mutations in HEK 293 and PC12 cells. Cells were grown on coverslips and transfected with the PDE2A3 N terminus fused to CFP. A–F, expression in HEK 293 cells. G–I, distribution of the denoted CFP fusion proteins in PC12 cells. A, wild-type PDE2A3 N terminus was fused to CFP. B, Gly2 of the PDE2A3 N terminus was changed to Ala. C, Cys5 of the PDE2A3 N terminus was changed to Ser. D, Golgi marker vector pECFP-Golgi (Clontech). E, ER marker pECFP-ER (Clontech). F, as an example of a soluble enzyme a PDE2A1/CFP fusion protein was expressed.
Predominant Expression of PDE2A3 in Brain
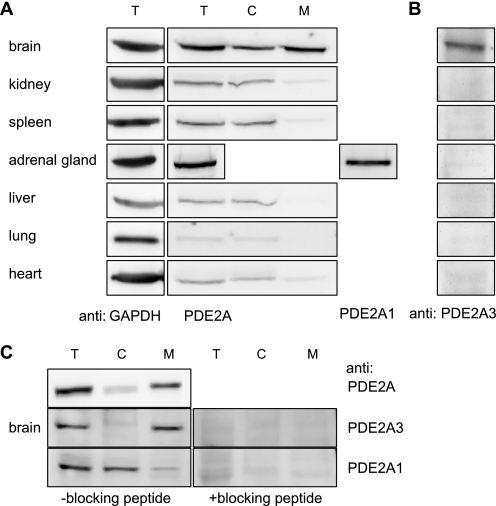
To identify tissues in which membrane targeting of PDE2A3 occurs, distribution of PDE2A was assessed in cytosolic and membrane fractions of mouse tissues. Using a pan-specific PDE2A antibody recognizing all splice variants equally well, highest expression of PDE2A was detected in adrenal gland and brain (Fig. 5A). Furthermore, only in brain tissue substantial amounts of PDE2A were found in the membrane fraction, whereas in peripheral tissues only soluble PDE2A was detectable. Because a newly generated antibody specific for the soluble isoform, PDE2A1, recognized a similar distribution in peripheral tissues (not shown) the signals detected in cytosolic fractions can be ascribed to PDE2A1.
FIGURE 5.
Tissue distribution of PDE2A splice variants in mouse. A, mouse organs were processed by means of a glass-Teflon homogenizer and fractionated by centrifugation. PDE2A localization was then analyzed by SDS-PAGE and Western blotting using the PDE2A-specific antibody. Due to the very limited amount of material, adrenal gland homogenate was not fractionated but probed with isoform-specific antibodies detecting PDE2A1 or PDE2A3 (see B). For an internal standard glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected in the homogenates. B, homogenates of mouse organs were probed using an antibody specifically detecting PDE2A3. C, mouse brain was processed as described above using buffer containing 150 mm NaCl. 25 μg of total protein was blotted, and PDE2A was detected using antibodies as indicated. On the right, isoform-specific detection in the presence of the respective blocking peptide is shown. T, total; C, cytosol; and M, membrane.
The identity of the membrane-bound PDE2A in brain was clarified with an antibody raised against a unique N-terminal sequence of PDE2A3. Using this antibody, expression of PDE2A3 exclusively in brain was demonstrated (Fig. 5B). To analyze the content of PDE2A1 versus PDE2A3 isoforms in brain, the isoform-specific antibodies were used (Fig. 5C). Cytosolic and membrane fractions were probed with antibodies detecting either PDE2A3 (middle) or PDE2A1 (bottom) as well as a pan-specific antibody detecting all splice variants (top). Obviously, both isoforms are expressed in brain. The signal intensities demonstrate that PDE2A3 is the predominant isoform in brain.
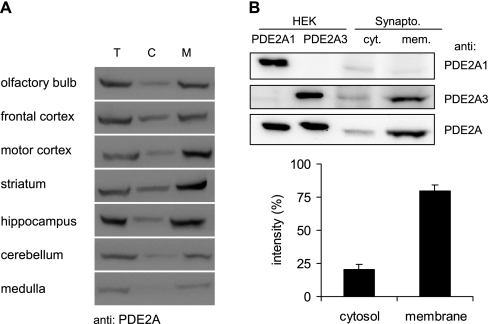
Closer analysis of PDE2A distribution in different areas of the brain revealed comparable enzyme levels for olfactory bulb, frontal cortex, motor cortex, striatum, and hippocampus, whereas cerebellum and medulla displayed little expression of PDE2A (Fig. 6A). Because the membrane-bound PDE2A3 is the primary form in all areas of the brain, the myristoylation-dependent membrane attachment might play an important role in synaptic transmission. Therefore synaptic localization of PDE2A3 was studied. We prepared crude synaptosomal fractions from whole mouse brains and separated them into cytosol and membranes. Subsequent Western analysis revealed high amounts of PDE2A in synaptosomes, with 80% of the synaptosomal PDE2A being particulate (Fig. 6B). Using isoform-specific antibodies, this membrane-bound PDE2 was identified as PDE2A3.
FIGURE 6.
Neuronal expression of PDE2A. A, the denoted areas of mouse brains were separated into soluble and particulate fractions. 20 μg of total protein per lane were separated by SDS-PAGE. For Western detection, the PDE2A-specific antibody was used. T, total; C, cytosol; M, membrane. B, crude synaptosomal vesicles obtained from mouse brains were separated into cytosol and membranes and subjected to Western blots using isoform-specific antibodies against PDE2A1 and PDE2A3 and a pan-specific antibody (PDE2A) recognizing both isoforms. For comparison, homogenates of HEK cells expressing either PDE2A1 or PDE2A3 are shown. Quantitative analysis of pan-specific PDE2A signals from three independent experiments is displayed graphically with the signal intensity of the cytosol plus membrane set to 100%. Bars are means ± S.D. of three independent experiments; p < 0.01 (two-tailed paired t test).
In Hippocampal Neurons PDE2A Is Located at Presynaptic Terminals
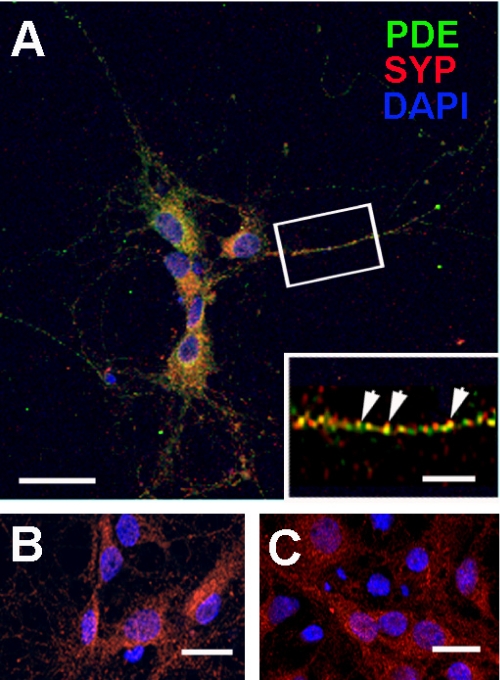
Finally, using immunofluorescence analysis in primary cultures of hippocampal neurons we found a synaptic localization of PDE2A as shown by the partially overlapping localization with the presynaptic marker synaptophysin (Fig. 7). Both, synaptophysin and PDE2A were expressed in a punctate pattern throughout the neurons (see Fig. 7A, inset), arguing for localization in transport vesicles and synaptic terminals of processes. Thus, it stands to reason that dual acylation of PDE2A3 serves to transport the enzyme from the area of synthesis to the synaptic ends, where it likely provides a compartmentalized interplay between cGMP- and cAMP-dependent signaling pathways.
FIGURE 7.
PDE2A and synaptophysin co-localization in primary hippocampal neurons. A, rat hippocampal neurons isolated as described under “Materials and Methods” and fixed for immunohistochemistry were double labeled with PDE2A (PDE, green), synaptophysin (SYP, red), and Alexa Fluor-conjugated secondary antibodies. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Neurons were examined by confocal microscopy. The inset demonstrates the distribution of PDE2A and SYP within a single neuronal process and indicates co-localizing puncta (arrowheads). B, the PDE2A-specific signal was blocked by incubation with blocking peptide. C, control with secondary antibody alone. Scale bars represent 30 μm (A), 6 μm (inset), and 20 μm (B and C).
DISCUSSION
The present study demonstrates the existence of PDE2A3 protein in mouse brain and addresses the basis of its membrane attachment. Since Beavo et al. (3) in 1971 first described that cGMP-stimulated PDE activity could be found both in soluble and particulate fractions of rat liver, increasing evidence for the existence of more than one PDE2A enzyme has accumulated. To date, the PDE2A splice variants 1, 2, and 3 have been cloned from different species. The three variants are identical except for their most N-terminal sequences, which appear to determine the subcellular localization of the enzyme. PDE2A1 was isolated as soluble protein from bovine heart. It remains the only splice variant whose amino acid sequence was determined by sequencing the purified protein (17). The splice variant 2 was cloned from a rat brain cDNA library and characterized as membrane-bound enzyme (12). The third splice variant, PDE2A3, has been isolated from bovine and human cDNA libraries (8, 13). However, as yet it remained unclear which of the membrane-bound isoforms is expressed on the protein level in a given tissue or species because isoform-specific antibodies that recognize the divergent N termini have not been used.
In the past, the properties of the N termini of the splice variants have been analyzed by several approaches. Sequence analysis and mass spectrometry of digests of the soluble PDE2A1 protein from bovine heart have revealed that the N terminus is blocked by Nα-acetylation, but aside from that no additional post-translational modifications were detected (17). In the rat PDE2A2, a very hydrophobic N-terminal segment was identified (12), and depalmitoylation experiments led to partial solubilization of the enzyme (19). These results point at a mechanism for PDE2A2 membrane association involving hydrophobic interactions as well as palmitoylation of the protein. For PDE2A3, it was shown that in bovine brain up to 85% of cGMP-stimulated PDE activity is associated with particulate fractions (8). Of this activity very little could be released with buffers containing 0–500 mm NaCl or 5 mm EDTA. The cytosolic distribution of PDE2A1 and the membrane association of PDE2A2 and -3 was confirmed in this study in HEK 293 cells. Because PDE2A3 was solubilized by Triton X-100 but not by NaCl, ionic interactions of the protein with the membrane appeared to be unlikely.
Inspection of the amino acid sequence of the PDE2A3 N terminus with regards to possible mechanisms for membrane association does not reveal a particularly hydrophobic domain, as described for the rat PDE2A2. However, PDE2A3 contains a glycine at position 2, making the protein a candidate for myristoylation by N-myristoyl transferase. Myristoylation is known to occur exclusively to N-terminal glycine residues after cleavage of the starter methionine, and the myristoyl moiety provides an affinity (relatively weak) for membranes (18). PDE2A3 indeed incorporated [3H]myristate, a feature that was lost upon mutation of Gly2. Because the G2A mutant did not incorporate the radioactive fatty acid, metabolic interconversion of myristate to [3H]palmitate can be ruled out. Furthermore, inhibition of myristoylation or mutation of Gly2 increased the soluble portion of the enzyme in Western blots and activity measurements. Total PDE2A3 activity was not affected by the loss of the myristoyl group. In these experiments, inhibitor treatment did not completely relocate PDE2A3 to the cytosol, which is explicable by insufficient entry of the inhibitor into the cells or slow turnover of myristoylated protein synthesized before 2-HMA application.
Because the N-terminal 46 amino acids fused to CFP targeted fluorescence to membranes, it is concluded that the N terminus of PDE2A3 containing the myristoylated Gly2 is sufficient to determine localization of the enzyme. Mutation of Gly2 rendered the fusion protein completely soluble. Together, above results indicate that N-terminal myristoylation is a prerequisite for membrane binding of PDE2A3.
It is well established that myristoylation alone provides only weak affinity for membranes, whereas tandem lipid modifications are sufficient for tight membrane interactions (20, 21). So far, several members of signaling protein families have been shown to be sequentially modified. Often, N-myristoylated proteins are subsequently palmitoylated on one or more nearby cysteine residues (22, 23). To the examples for proteins carrying such modifications belong Src tyrosine kinase family members as well as members of the Gαi subfamily (see Table 1). Palmitoylation, as a secondary modification, occurs at cysteine residues up to 20 amino acids away from the N terminus (24) and in the case of endothelial nitric-oxide synthase even further away (Cys15 and Cys26) (25). To date, no well defined consensus sequence for palmitoylation other than a requirement for cysteine has been identified. PDE2A3 exhibits two N-terminal cysteines (see Table 1). Mutation of the respective cysteines shifted the protein to the cytosol and caused a diffuse distribution of the otherwise clearly localized enzyme in fluorescence microscopy (see Fig. 4, compare A to C (HEK 293) and G to I (PC12)). Whereas the N terminus of WT PDE2A3, including the N-myristoylation and palmitoylation sites, targeted CFP fluorescence to the ER, Golgi, and plasma membrane, the non-palmitoylatable C5S mutation led to an accumulation of fluorescence in the Golgi region and cytosol. These results are in support of a mechanism similar to that suggested for membrane targeting of endothelial nitric-oxide synthase. There, N-myristoylation is required for endothelial nitric-oxide synthase to get into the Golgi compartment, where the protein is then palmitoylated and targeted via the trans-Golgi network to caveolae (26).
In vivo, PDE2A has been shown to play important roles in many signal transduction pathways as a regulator of both cGMP and cAMP levels. For example, several groups observed PDE2A-dependent modulation of L-type Ca2+ channel function in cardiac myocytes (27–30). In adrenal glomerulosa cells, PDE2A plays a role in ANP-dependent inhibition of aldosterone release (5, 31). Our analyses of PDE2A distribution in different tissues demonstrate a widespread expression of the soluble PDE2A1 with highest levels found in adrenal gland and brain. These results are in good agreement with most previously published studies. However, in heart we could not find membrane-associated PDE2A, which stands in contrast to a study by Mongillo et al. (30) describing PDE2A as entirely confined to a membrane compartment in cardiac myocytes. We have no explanation for these discrepancies, perhaps the differences result from looking at whole heart versus isolated cardiac myocytes. Also species differences might be considered.
In brain, both cyclic nucleotides have been implicated in various forms of learning and memory formation. Yet, the precise mechanisms underlying these processes are not fully understood. At excitatory synapses in the hippocampus CA1-region, nitric oxide is discussed as a retrograde messenger increasing presynaptic (and post synaptic) cGMP synthesis by guanylyl cyclase activation (32). PDE2 may well be involved in regulation of synaptic cGMP levels, because the PDE2-specific inhibitor BAY 60-7550 significantly increased long-term potentiation and improved the object memory performance (33). Also, pre and post synaptic roles for cAMP/protein kinase A signaling pathways are currently being discussed (34, 35). The involvement of cyclic nucleotides in synaptic function necessitates a tight control of cyclic nucleotide levels by cyclic nucleotide-degrading PDEs in synapses.
In line with a role for PDE2A in synaptic transmission, we show that PDE2A is highly expressed in mouse brain. Being the first to use isoform-specific antibodies, we unambiguously identify soluble PDE2A1 and membrane-bound PDE2A3 proteins, with PDE2A3 being the predominant isoform in brain. Its localization to synaptosomal membranes, as shown here, places it in an appropriate position to rapidly hydrolyze the cyclic nucleotides at synapses. Furthermore, PDE2A is present in synapses of primary hippocampal neurons where it co-localizes with synaptophysin, a presynaptic marker. Yet, an additional post synaptic function, as proposed in a recent publication that suggested a PDE2-mediated cross-talk of cAMP and cGMP signals (36) is also compatible with the immunofluorescence images.
In sum, PDE2A isoforms are distributed in a specific manner. The soluble PDE2A1 is expressed in a variety of peripheral tissues, whereas the membrane-associated PDE2A3 is found exclusively in the brain. Here, its expression is similarly high for most analyzed regions, only cerebellum and medulla display lower expression levels, matching results obtained by previous analysis of mRNA expression (37). In line with a role for PDE2A3 in synaptic plasticity, the enzyme is detected primarily in synaptosomal membrane fractions and co-localizes with the synaptic marker synaptophysin in primary hippocampal neurons. Our results suggest that PDE2A3 requires N-myristoylation together with palmitoylation to be targeted to synapses allowing control and cross-talk of cyclic nucleotides.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of the RUBION team (Zentrale Einrichtung für Ionenstrahlen und Radionuklide der Ruhr-Universität Bochum) and thank Arkadius Pacha, Ulla Krabbe, Fred Eichhorst, and Erika Mannheim for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (Grant Ko1157) and the Kommission für Finanzautonomie und Ergänzungsmittel of the Medical Faculty.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
W. S. Rasband (1997–2008) ImageJ, National Institutes of Health, Bethesda, MD, rsb.info.nih.gov/ij.
- PDE
- phosphodiesterase
- CFP
- cyan fluorescent protein
- 2-HMA
- 2-hydroxymyristic acid
- ER
- endoplasmic reticulum
- ECFP
- enhanced CFP
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Bender A. T., Beavo J. A. (2006) Pharmacol. Rev. 58, 488–520 [DOI] [PubMed] [Google Scholar]
- 2.Martins T. J., Mumby M. C., Beavo J. A. (1982) J. Biol. Chem. 257, 1973–1979 [PubMed] [Google Scholar]
- 3.Beavo J. A., Hardman J. G., Sutherland E. W. (1971) J. Biol. Chem. 246, 3841–3846 [PubMed] [Google Scholar]
- 4.Erneux C., Couchie D., Dumont J. E., Baraniak J., Stec W. J., Abbad E. G., Petridis G., Jastorff B. (1981) Eur. J. Biochem. 115, 503–510 [DOI] [PubMed] [Google Scholar]
- 5.MacFarland R. T., Zelus B. D., Beavo J. A. (1991) J. Biol. Chem. 266, 136–142 [PubMed] [Google Scholar]
- 6.Beavo J. A., Hardman J. G., Sutherland E. W. (1970) J. Biol. Chem. 245, 5649–5655 [PubMed] [Google Scholar]
- 7.Yamamoto T., Manganiello V. C., Vaughan M. (1983) J. Biol. Chem. 258, 12526–12533 [PubMed] [Google Scholar]
- 8.Murashima S., Tanaka T., Hockman S., Manganiello V. (1990) Biochemistry 29, 5285–5292 [DOI] [PubMed] [Google Scholar]
- 9.Pyne N. J., Cooper M. E., Houslay M. D. (1986) Biochem. J. 234, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whalin M. E., Strada S. J., Thompson W. J. (1988) Biochim. Biophys. Acta 972, 79–94 [DOI] [PubMed] [Google Scholar]
- 11.Sonnenburg W. K., Mullaney P. J., Beavo J. A. (1991) J. Biol. Chem. 266, 17655–17661 [PubMed] [Google Scholar]
- 12.Yang Q., Paskind M., Bolger G., Thompson W. J., Repaske D. R., Cutler L. S., Epstein P. M. (1994) Biochem. Biophys. Res. Commun. 205, 1850–1858 [DOI] [PubMed] [Google Scholar]
- 13.Rosman G. J., Martins T. J., Sonnenburg W. K., Beavo J. A., Ferguson K., Loughney K. (1997) Gene 191, 89–95 [DOI] [PubMed] [Google Scholar]
- 14.Mullershausen F., Russwurm M., Thompson W. J., Liu L., Koesling D., Friebe A. (2001) J. Cell Biol. 155, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoidl G., Meier C., Petrasch-Parwez E., Zoidl C., Habbes H. W., Kremer M., Srinivas M., Spray D. C., Dermietzel R. J. (2002) J. Neurosci. Res. 69, 448–465 [DOI] [PubMed] [Google Scholar]
- 16.Russwurm M., Wittau N., Koesling D. (2001) J. Biol. Chem. 276, 44647–44652 [DOI] [PubMed] [Google Scholar]
- 17.Trong H. L., Beier N., Sonnenburg W. K., Stroop S. D., Walsh K. A., Beavo J. A., Charbonneau H. (1990) Biochemistry 29, 10280–10288 [DOI] [PubMed] [Google Scholar]
- 18.Towler D. A., Gordon J. I., Adams S. P., Glaser L. (1988) Annu. Rev. Biochem. 57, 69–99 [DOI] [PubMed] [Google Scholar]
- 19.Noyama K., Maekawa S. (2003) Neurosci. Res. 45, 141–148 [DOI] [PubMed] [Google Scholar]
- 20.Peitzsch R. M., McLaughlin S. (1993) Biochemistry 32, 10436–10443 [DOI] [PubMed] [Google Scholar]
- 21.Shahinian S., Silvius J. R. (1995) Biochemistry 34, 3813–3822 [DOI] [PubMed] [Google Scholar]
- 22.Dunphy J. T., Linder M. E. (1998) Biochim. Biophys. Acta 1436, 245–261 [DOI] [PubMed] [Google Scholar]
- 23.Resh M. D. (1999) Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 24.Navarro-Lérida I., Alvarez-Barrientos A., Gavilanes F., Rodriguez-Crespo I. (2002) J. Cell Sci. 115, 3119–3130 [DOI] [PubMed] [Google Scholar]
- 25.Robinson L. J., Michel T. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11776–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-Cardeña G., Oh P., Liu J., Schnitzer J. E., Sessa W. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivet-Bastide M., Vandecasteele G., Hatem S., Verde I., Bénardeau A., Mercadier J. J., Fischmeister R. (1997) J. Clin. Invest. 99, 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verde I., Vandecasteele G., Lezoualc'h F., Fischmeister R. (1999) Br. J. Pharmacol. 127, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandecasteele G., Verde I., Rücker-Martin C., Donzeau-Gouge P., Fischmeister R. (2001) J. Physiol. 533, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mongillo M., Tocchetti C. G., Terrin A., Lissandron V., Cheung Y. F., Dostmann W. R., Pozzan T., Kass D. A., Paolocci N., Houslay M. D., Zaccolo M. (2006) Circ. Res. 98, 226–234 [DOI] [PubMed] [Google Scholar]
- 31.Nikolaev V. O., Gambaryan S., Engelhardt S., Walter U., Lohse M. J. (2005) J. Biol. Chem. 280, 1716–1719 [DOI] [PubMed] [Google Scholar]
- 32.Bon C. L., Garthwaite J. (2003) J. Neurosci. 23, 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boess F. G., Hendrix M., van der Staay F. J., Erb C., Schreiber R., van Staveren W., de Vente J., Prickaerts J., Blokland A., Koenig G. (2004) Neuropharmacology 47, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 34.Frey U., Huang Y. Y., Kandel E. R. (1993) Science 260, 1661–1664 [DOI] [PubMed] [Google Scholar]
- 35.Blitzer R. D., Wong T., Nouranifar R., Iyengar R., Landau E. M. (1995) Neuron 15, 1403–1414 [DOI] [PubMed] [Google Scholar]
- 36.Makhinson M., Opazo P., Carlisle H. J., Godsil B., Grant S. G., O'Dell T. J. (2006) Neuroscience 140, 415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Repaske D. R., Corbin J. G., Conti M., Goy M. F. (1993) Neuroscience 56, 673–686 [DOI] [PubMed] [Google Scholar]
- 38.Alland L., Peseckis S. M., Atherton R. E., Berthiaume L., Resh M. D. (1994) J. Biol. Chem. 269, 16701–16705 [PubMed] [Google Scholar]
- 39.Koegl M., Zlatkine P., Ley S. C., Courtneidge S. A., Magee A. I. (1994) Biochem. J. 303, 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degtyarev M. Y., Spiegel A. M., Jones T. L. (1994) J. Biol. Chem. 269, 30898–30903 [PubMed] [Google Scholar]
- 41.Vaandrager A. B., Ehlert E. M., Jarchau T., Lohmann S. M., de Jonge H. R. (1996) J. Biol. Chem. 271, 7025–7029 [DOI] [PubMed] [Google Scholar]
- 42.Fraser I. D., Tavalin S. J., Lester L. B., Langeberg L. K., Westphal A. M., Dean R. A., Marrion N. V., Scott J. D. (1998) EMBO J. 17, 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.