Abstract
The lipopolysaccharide of Vibrio cholerae has been reported to contain a single 3-deoxy-d-manno-octulosonic acid (Kdo) residue that is phosphorylated. The phosphorylated Kdo sugar further links the hexa-acylated V. cholerae lipid A domain to the core oliogosaccharide and O-antigen. In this report, we confirm that V. cholerae possesses the enzymatic machinery to synthesize a phosphorylated Kdo residue. Further, we have determined that the presence of the phosphate group on the Kdo residue is necessary for secondary acylation in V. cholerae. The requirement for a secondary substituent on the Kdo residue (either an additional Kdo sugar or a phosphate group) was also found to be critical for secondary acylation catalyzed by LpxL proteins from Bordetella pertussis, Escherichia coli, and Haemophilus influenzae. Although three putative late acyltransferase orthologs have been identified in the V. cholerae genome (Vc0212, Vc0213, and Vc1577), only Vc0213 appears to be functional. Vc0213 functions as a myristoyl transferase acylating lipid A at the 2′-position of the glucosamine disaccharide. Generally acyl-ACPs serve as fatty acyl donors for the acyltransferases required for lipopolysaccharide biosynthesis; however, in vitro assays indicate that Vc0213 preferentially utilizes myristoyl-CoA as an acyl donor. This is the first report to biochemically characterize enzymes involved in the biosynthesis of the V. cholerae Kdo-lipid A domain.
Lipopolysaccharide (LPS),2 the major surface molecule in the outer membrane of Gram-negative bacteria, is composed of three domains: lipid A, core oligosaccharide, and O-antigen (1). The core oligosaccharide is further divided into two distinct regions: inner and outer core. The inner core consists of the Kdo sugars, which are responsible for linking the core region to the lipid A moiety of LPS. Lipid A is the hydrophobic anchor of LPS and is the only portion of LPS required for activating the host innate immune response by interacting with Toll-like receptor 4 and the accessory molecule, MD2.
Kdo-lipid A biosynthesis is a well conserved and ordered process among Gram-negative bacteria; however, not all Gram-negative bacteria produce similar lipid A structures (2). In Escherichia coli, the biosynthesis of the Kdo-lipid A domain occurs via a nine-step process, resulting in the production of a hexa-acylated lipid A structure known as Kdo2-lipid A. Kdo2-lipid A has long been thought to be essential for the viability of E. coli; however, viable suppressor strains have been isolated that lack the Kdo sugar (3). The late steps of the biosynthetic pathway involve the addition of the Kdo sugars and the secondary or “late” acyl chains. The enzyme responsible for the addition of the Kdo sugars is the Kdo transferase (WaaA). In E. coli, this enzyme is bifunctional, transferring two Kdo sugars to the lipid A precursor, lipid IVA (4); however, other Gram-negative bacteria have been shown to possess a monofunctional or trifunctional WaaA, as is the case in Haemophilus influenzae (5) or Chlamydia trachomatis (6), respectively.
Previous reports have shown that in E. coli, the addition of the Kdo sugars is critical for the functionality of the secondary acyltransferases (LpxL, LpxM, and LpxP). The E. coli late acyltransferase LpxL catalyzes the transfer of laurate (C12:0) to the acyl chain linked at the 2′-position of Kdo2-lipid IVA (7). LpxM then catalyzes the addition of a myristate (C14:0) to the 3′-linked acyl chain of the penta-acylated lipid A precursor (8). When E. coli experience cold shock conditions (temperatures below 20 °C), the late acyltransferase LpxP transfers a palmitoleate (C16:1) to the 2′-position of Kdo2-lipid IVA, replacing the C12:0 acyl chain transferred by LpxL (9). Lipid A secondary acyltransferases have been shown to primarily utilize acyl-acyl carrier proteins (acyl-ACPs) as their acyl chain donor; however, a recent report by Six et al. (10) has shown that purified E. coli LpxL is capable of utilizing acyl-coenzyme A (acyl-CoA) as an alternative acyl donor at a lesser rate.
The Gram-negative bacteria Vibrio cholerae is the causative agent of the severe diarrheal disease cholera. Cholera is transmitted via the fecal-oral route by ingestion of contaminated drinking water or food. The World Health Organization reported ∼130,000 cases of cholera in 2005 with the majority occurring in Africa. There are two serogroups of V. cholerae capable of epidemic and pandemic disease: O1 and O139 (11). Previous structural analyses have revealed that these serogroups possess very different lipid A structures. The V. cholerae O1 lipid A structure was reported as hexa-acylated, bearing secondary acyl chains at the 2- and 2′-positions of phosphorylated Kdo-lipid A (11–13); however, V. cholerae O139 was reported as having an octa-acylated lipid A (see Fig. 1) (11, 14).
FIGURE 1.
Comparison of E. coli K12 lipid A species to V. cholerae O1 and V. cholerae O139 lipid A species. The covalent modifications of lipid A are indicated with dashed bonds, and the lengths of the acyl chains are indicated below each structure. The lipid A of E. coli K12 is a hexa-acylated structure, bearing two secondary acyl chains at the 2′- and 3′-positions. The E. coli lipid A structure is glycosylated at the 6′-position with two Kdo moieties and is phosphorylated at the 1- and 4′-positions of the disaccharide backbone. Similar to E. coli, the lipid A species of V. cholerae serogroup O1 is hexa-acylated, but with a symmetrical acyl chain distribution. The proposed lipid A structure of V. cholerae O139 is the octa-acylated structure. Both V. cholerae serogroups O1 and O139 reported lipid A species have a single Kdo sugar that is phosphorylated (red) and a phosphoethanolamine (magenta) attached to the 1-phosphate.
Our report focuses on V. cholerae O1 El Tor, which is the predominant disease-causing strain worldwide. Because little attention has been given to the Kdo-lipid A domain of V. cholerae, we investigated the assembly of the inner core structure of V. cholerae O1 LPS and the late acylation steps. This report demonstrates the importance of a secondary negative charge on the primary Kdo sugar of lipid A for late acyltransferase activity in V. cholerae and in other Gram-negative bacteria. Also, we have identified the putative V. cholerae late acyltransferase, Vc0213 as the LpxL homolog, transferring a myristate (C14:0) to the 2′-position of V. cholerae lipid A. These initial findings provide us with the groundwork needed to investigate the modifications of the V. cholerae Kdo-lipid A structure, which may serve as attractive vaccine targets in future research.
EXPERIMENTAL PROCEDURES
Chemicals and Other Materials
[γ-32P]ATP and 32Pi were purchased from PerkinElmer Life Sciences. Triton X-100 and bicinchoninic acid were obtained from Pierce. Silica gel 60 (0.25 mm) thin layer plates were purchased from EM Separation Technology (Merck). Luria-Bertani (LB) agar and LB broth were from EMD Chemicals. M9 minimal salts were from Difco. All other chemicals were reagent grade and were purchased from either Sigma or Fisher.
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in the study are summarized in supplemental Table S1. E. coli were typically grown at 37 °C in LB broth. The E. coli late acyltransferase mutant, MKV13, was grown as described previously (15) in M9 minimal medium at 30 °C. When required for selection of plasmids, the cells were grown in the presence of ampicillin (100 μg/ml).
Recombinant DNA Techniques
Plasmids were isolated using the QIAprep spin miniprep kit (Qiagen). Custom primers were obtained from Invitrogen and Integrated DNA Technologies. PCR reagents were purchased from New England BioLabs and Roche Applied Science. PCR clean-up was performed using the QIAquick PCR purification kit (Qiagen). DNA fragments were isolated from agarose gels using the QIAquick gel extraction kit (Qiagen). Restriction endonucleases, T4 DNA ligase, and Antarctic phosphatase were purchased from New England BioLabs. All of the modifying enzymes were used according to the manufacturers' instructions.
Cloning and Overexpression of the Late Acyltransferases (Vc0212, Vc0213, Vc1577, Bp3073, and Hi1527) behind a T7lac Promoter
The putative late acyltransferase genes of V. cholerae O1 N16961 (vc0212, vc0213, and vc1577) and the lpxL homologs of Bordetella pertussis and H. influenzae (bp3073 and hi1527, respectively) were separately subcloned into either pET21a or pET28a (Novagen) behind the T7lac promoter. The genes were PCR-amplified using the appropriate genomic DNA as template. Sequences of primers are shown in supplemental Table S2. vc0212, vc0213, vc1577, bp3073, and hi1527 PCR products and pET21 were digested with the indicated restriction enzymes (supplemental Table S2). The vc1577 PCR product and pET28 were digested with NheI and BamHI. PCR products were ligated into their respective vectors to give pVc0212, pVc0213, pET28Vc1577, pBp3073, and pHi1527. pET28Vc1577 was digested with BamHI and XbaI, and the gene fragment was ligated into pET21a to give pVc1577. Each pET21a construct was transformed into XL-1 Blue (Stratagene) for propagation of the plasmids. pVc0212, pVc0213, pVc1577, pBp3073, and pHi1527 were then transformed into MKV15 (DE3) for overexpression of the protein.
Cloning of Vc0213 into pBluescript SK II(+)
The vc0213 coding region, along with the pET21 ribosomal binding site, were excised from the plasmid pVc0213 using XbaI and XhoI. The gene fragment was inserted into pBluescript SK II(+) to give pBSVc0213 and was transformed into XL-1 Blue for propagation. pBSVc0213 was then transformed into MKV13 to be used to isolate lipid A species for mass spectrometry.
Cloning and Overexpression of the V. cholerae O1 N16961 Kdo Transferase (Vc0233) and Kdo Kinase (Vc0227) behind a T7lac Promoter
vc0233 and vc0227 were PCR-amplified using genomic DNA as template. Primer sequences are shown in supplemental Table S2. The PCR products and pET21a were digested with the indicated restriction enzymes (supplemental Table S2). vc0227 and vc0233 were ligated separately into pET21a using T4 DNA Ligase, to give pVc0227 and pWaaA-Vc, respectively. pWaaA-Vc and pVc0227 were transformed into XL-1 Blue for propagation of the plasmids. pWaaA-Vc and pVc0227 were transformed into NovaBlue (DE3) and HMS174 (DE3), respectively, for overexpression of the proteins.
Preparation of Cell-free Extracts, Membrane-free Cytosol, and Washed Membrane
Typically, 200-ml cultures of E. coli were grown to mid-log phase (A600 of ∼0.6–0.7) at 37 °C upon which the cells were induced with 1 mm isopropoyl 1-thio-β-d-galactopyranoside. After induction, the cells were allowed to grow for an additional 4 h or until cell growth declined. The cells were harvested by centrifugation at 6000 × g for 30 min. All of the samples were prepared at 4 °C. Cell-free extract, membrane-free cytosol, and washed membranes were prepared as described previously (16) and were stored in aliquots at −20 °C. Protein concentration was determined by the bicinchoninic acid method (17), using bovine serum albumin as the standard.
Preparation of Radiolabeled Substrates
The substrate [4′-32P]lipid IVA was generated from 125 μCi of [γ-32P]ATP and the tetraacyl-disaccharide 1-phosphate lipid acceptor, using the overexpressed 4′-kinase present in membranes of E. coli BLR (DE3)/pLysS/pJK2 as described previously (16). Kdo2-[4′-32P]lipid IVA was prepared by adding purified E. coli Kdo transferase (WaaA) immediately after the 4′-kinase, as described previously (18, 21). Hexa-acylated Kdo2-[4′-32P]lipid A was prepared by adding membranes of E. coli BLR (DE3)/LpxL and BLR (DE3)/LpxM and C12:0-ACP immediately after the Kdo transferase reaction as described previously (16, 21).
Kdo-[4′-32P]lipid IVA was prepared similarly to Kdo2-[4′-32P]lipid IVA except that NovaBlue (DE3) membranes expressing the V. cholerae WaaA (0.001 mg/ml) were added to the 4′-kinase reaction. The Kdo transferase reaction proceeded for 5 min at room temperature, followed by inactivation of the enzyme at 65 °C for 20 min. Phosphorylated-Kdo-[4′-32P]lipid IVA was prepared by adding 50 mm Hepes (pH 7.5), 0.1% Triton X-100, 5 mm ATP, and HMS174 (DE3) membranes (0.05 mg/ml) expressing the V. cholerae KdkA immediately after the completion of the Kdo transferase reaction. The Kdo kinase reaction proceeded for 30 min at room temperature. Phosphorylated Kdo-myristoyl-[4′-32P]lipid IVA was prepared by adding E. coli MKV15 (DE3) Vc0213 membranes (0.01 mg/ml) and C14:0-CoA (5 μm) to phosphorylated Kdo-[4′-32P]lipid IVA (100,000 cpm, 2.5 μm). The reaction proceeded for 1 h at 30 °C. Vc0213 activity was inactivated for 20 min at 65 °C.
TLC and Phosphorimaging Analysis
When [4′-32P]lipid IVA was employed as the substrate, the reaction products were separated using the solvent chloroform, pyridine, 88% formic acid, water (50:50:16:5, v/v). The reaction products generated from substrates having the Kdo moiety were separated using the solvent chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v). TLC plates were exposed overnight to a PhosphorImager screen, and product formation was detected and analyzed using a Bio-Rad personal molecular imager equipped with Quantity One software. The enzyme activity was calculated by determining the percentage of the substrate converted to product.
Assay of Kdo Transferase (WaaA) Activity
Kdo transferase (WaaA) activity was assayed under optimized conditions as described previously by Brozek et al. (19, 20). Reaction mixtures (10 μl) contained 50 mm Hepes (pH 7.5), 4 mm Kdo, 10 mm CTP, 10 mm MgCl2, 1:4 dilution of purified CMP-Kdo synthase (0.05 total units), 0.1% Triton X-100, and 10 μm [4′-32P]lipid IVA (∼5,000 cpm/nmol). Purified CMP-Kdo synthase was prepared as described previously (21). During assay, washed membranes at 0.001 mg/ml were employed as the enzyme source, as indicated. Enzymatic reactions were incubated at 30 °C for 1 h and were terminated by spotting 4.5 μl of the mixtures onto silica gel 60 TLC plates.
Assay of V. cholerae Kdo Kinase (Vc0227) Activity
V. cholerae Kdo kinase activity was assayed under optimized conditions based upon the method of White et al. (22). Reaction mixtures (10 μl) contained 50 mm Hepes (pH 7.5), 0.1% Triton X-100, 10 mm MgCl2, 5 mm ATP, and 2.5 μm Kdo-[4′-32P]lipid IVA (∼5,000 cpm/nmol). Membranes expressing Vc0227 at 0.1 mg/ml were employed as the enzyme source, as indicated. Enzymatic reactions were incubated at 30 °C for 1 h and terminated by spotting 4.5-μl portions of the mixture onto silica gel 60 TLC plates.
Assay of Late Acyltransferase Activity
Late acyltransferase activity was assayed under optimized conditions (7) in a 10-μl reaction mixture containing 50 mm Hepes (pH 7.5), 0.1% Triton X-100, 50 mm NaCl, 5 mm MgCl2, 0.1 mg/ml bovine serum albumin, and 2.5 μm of the indicated lipid A substrate (∼5,000 cpm/nmol) and 5 μm of the indicated acyl donor. Washed membranes were employed as the enzyme source at concentrations indicated in figure legends. Enzymatic reactions were incubated at 30 °C for the times indicated in figure legends. The reactions were terminated by spotting 4.5 μl of the mixtures onto silica gel 60 TLC plates.
Mass Spectrometry of Lipid A Species
Typically, 200-ml cultures of each strain were grown at 30 °C until cultures reached an A600 of ∼1.0. Lipid A was released from cells and purified as described previously (23). The lipid A species were analyzed by the UT-Austin Analytical Instrumentation Facility Core using a MALDI-TOF/TOF (ABI 4700 Proteomics Analyzer) mass spectrometer equipped with a Nd:YAG laser (355 nm) using a 200-Hz firing rate. The spectra were acquired in negative ion linear mode, and each spectrum represented the average of a minimum of 4000 shots. The matrix used was a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, v/v). The samples were dissolved in chloroform-methanol (4:1, v/v) and deposited on the sample plate, followed by an equal portion of matrix solution (0.3 μl).
RESULTS
Putative V. cholerae Late Acyltransferases Display No Activity When Assayed with Kdo-[4′-32P]Lipid IVA
The proposed lipid A structure of V. cholerae serogroup O1 is hexa-acylated, having secondary acyl chains at the 2- and 2′-positions of the glucosamine disaccharide (Fig. 1) (11–13). Three putative late acyltransferases (vc0212, vc0213, and vc1577) were identified in the V. cholerae O1 genome by the Clusters of Orthologous Groups Data Base (24). The putative late acyltransferases of V. cholerae were cloned into the pET21 expression vector behind the T7lac promoter and expressed in E. coli MKV15 (DE3) (supplemental Fig. S1 and Table S1). MKV15 (DE3) lacks functional copies of lpxL, lpxM, and lpxP eliminating endogenous E. coli late acyltransferase activity (25). MKV15 (DE3) membranes containing overexpressed Vc0212, Vc0213, or Vc1577 (supplemental Fig. S1) were isolated and assayed for late acyltransferase activity (supplemental Fig. S2). The proposed Kdo-lipid A domain structure of V. cholerae LPS contains a single Kdo sugar (Fig. 1); thus Kdo-[4′-32P]lipid IVA was the chosen lipid acceptor for our initial study. Acyl donors were provided in the form of an acyl-ACP mix that contained C12:0-ACP, C14:0-ACP, C16:0-ACP, and C18:0-ACP. In the chloroform:pyridine:formicacid:water (30:70:16:10, v/v/v/v) TLC system employed, the addition of an acyl chain to the substrate would create a more hydrophobic lipid, which would migrate faster than Kdo-[4′-32P]lipid IVA. However, as shown in supplemental Fig. S2, the putative V. cholerae late acyltransferases were nonfunctional when assayed with Kdo-[4′-32P]lipid IVA.
Biochemical Assays Confirm V. cholerae WaaA and KdkA Activity
Previous structural analyses have demonstrated that V. cholerae possess a phosphorylated Kdo sugar at the 6′-position of lipid A (11–13, 26). Therefore, we hypothesized that the putative V. cholerae late acyltransferases may require the addition of a phosphate group to the Kdo sugar for activity. To examine this further, we determined whether V. cholerae possessed the necessary enzymatic machinery to synthesize a phosphorylated Kdo-lipid A domain.
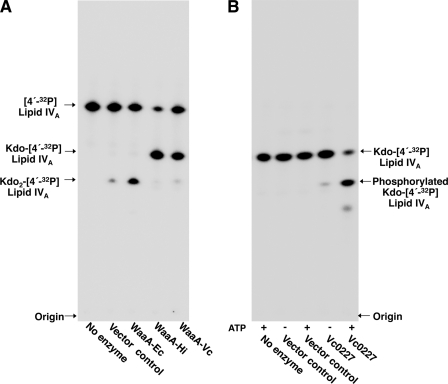
Membranes isolated from E. coli overexpressing the V. cholerae homolog of E. coli WaaA, Vc0233 (E value < 10−84), were assayed for Kdo transferase activity. Upon assay (Fig. 2A), the V. cholerae WaaA functioned similarly to the WaaA of H. influenzae (5), transferring a single Kdo sugar from CMP-Kdo to the tetra-acylated lipid acceptor, lipid IVA. As previously reported, the E. coli WaaA was bifunctional, transferring two Kdo sugars to [4′-32P]lipid IVA substrate (Fig. 2A) (18). The single Kdo sugar of the H. influenzae lipopolysaccharide is phosphorylated by a dedicated kinase, KdkA (22). Vc0227, a putative homolog of KdkA (E value < 10−58), was heterologously expressed in E. coli and assayed for its ability to modify Kdo-[4′-32P]lipid IVA. In the presence of ATP, Vc0227-containing membranes catalyzed the formation of a slower migrating lipid species (Fig. 2B) that was attributed to the addition of a phosphate group to the Kdo sugar. Phosphorylated Kdo-[4′-32P]lipid IVA is less hydrophobic than Kdo-[4′-32P]lipid IVA and would migrate more slowly in the employed TLC system (50:50:16:5, v/v), indicating that Vc0227 functions as a Kdo kinase (Fig. 2B).
FIGURE 2.
Enzymatic assay of the V. cholerae Kdo transferase and Kdo kinase. A, E. coli NovaBlue (DE3) membranes containing either pET21 or WaaA plasmids from E. coli, H. influenzae, or V. cholerae were assayed for Kdo transferase activity at 0.001 mg/ml for 1 h at 30 °C with [4′-32P]lipid IVA substrate. The small amount Kdo2-l4′-32P]lipid IVA in the vector control lane results from endogenous levels of E. coli WaaA. B, E. coli HMS174 (DE3) membranes expressing Vc0227 (0.1 mg/ml) were assayed for Kdo kinase activity using Kdo-[4′-32P]lipid IVA substrate in the presence or absence of ATP.
Vc0213 Is a V. cholerae Lipid A Late Acyltransferase
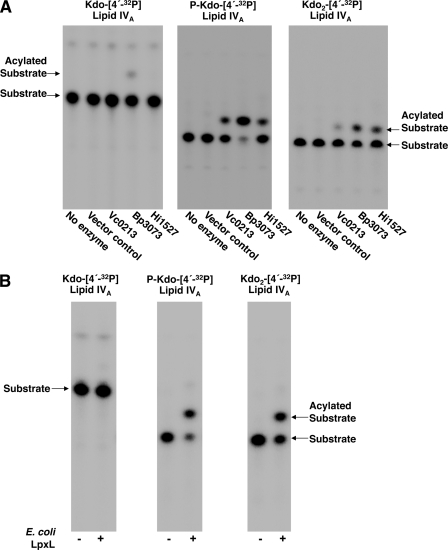
Membranes expressing the WaaA and KdkA of V. cholerae were utilized to synthesize a phosphorylated Kdo-[4′-32P]lipid IVA substrate. In vitro assays using phosphorylated Kdo-[4′-32P]lipid IVA and a mixture of acyl-ACP donors with MKV15 (DE3) membranes overexpressing Vc0212, Vc0213, or Vc1577 demonstrated that Vc0213 is a functional lipid A late acyltransferase (Fig. 3A). Additionally, Vc0213 displayed acyltransferase activity when assayed using Kdo2-[4′-32P]lipid IVA as the lipid acceptor (Fig. 3B). Similarly to phosphorylated Kdo-[4′-32P]lipid IVA, Kdo2-[4′-32P]lipid IVA has an additional negative charge on the primary Kdo sugar, indicating that Vc0213 is active when two negative charges are present on the inner Kdo residue.
FIGURE 3.
Assay of the V. cholerae late acyltransferases in the presence of phosphorylated Kdo-lipid IVA (P-Kdo-lipid IVA), or Kdo2-lipid IVA. E. coli MKV15 (DE3) membranes expressing the V. cholerae late acyltransferases (Vc0212, Vc0213, or Vc1577) were assayed for activity at 0.01 mg/ml for 1 h at 30 °C with a mixture of acyl-ACPs (C12:0, C14:0, C16:0, and C18:0) and either P-Kdo-[4′-32P]lipid IVA (A) or Kdo2-[4′-32P]lipid IVA (B).
Vc0212 and Vc1577 were nonfunctional when assayed using the late acyltransferase assay conditions and either phosphorylated Kdo-[4′-32P]lipid IVA or Kdo2-[4′-32P]lipid IVA (Fig. 3). Previous reports have shown that the LpxM of E. coli preferentially transfers an acyl chain to the 3′-position of a penta-acylated lipid A precursor. In an attempt to demonstrate that Vc0212 and Vc1577 were LpxM homologs requiring prior acylation at the 2′-position, membranes containing Vc0212 or Vc1577 were assayed with a penta-acylated lipid A substrate (phosphorylated Kdo-myristoyl-[4′-32P]lipid IVA) and a mixture of acyl-ACPs as donors. However, both Vc0212 and Vc1577 remained nonfunctional when assayed with the penta-acylated substrate.3 LpxP of E. coli functions during cold shock. Interestingly, Vorachek-Warren et al. (15) demonstrated that the acyltransferase activity of E. coli LpxP overexpressed at 37 °C was greatly reduced as compared with LpxP assayed from cells grown at 12 °C, even though the level of LpxP expression was similar (15). However, expression of Vc0212 or Vc1577 at 12 °C did not result in acyltransferase activity even in the presence of unsaturated acyl-ACPs.3
LpxL Homologs in Other Gram-negative Bacteria Also Require an Additional Negative Charge for Optimal Activity
Similarly to V. cholerae, both B. pertussis (27) and H. influenzae (28, 29) have been reported to synthesize a phosphorylated Kdo-lipid A structure. The LpxL homologs of B. pertussis (Bp3073, E value < 10−31) and H. influenzae (Hi1527, E value < 10−96) were separately cloned into pET21a and the proteins expressed in E. coli MKV15 (DE3). Membranes overexpressing Bp3073 and Hi1527 (supplemental Fig. S3) were assayed for late acyltransferase activity with either Kdo-[4′-32P]lipid IVA, phosphorylated Kdo-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid IVA. As demonstrated in Fig. 4A, these LpxL homologs catalyzed an acylation event when either phosphorylated Kdo-[4′-32P]lipid IVA or Kdo2-[4′-32P]lipid IVA were utilized as the lipid acceptor. However, when Kdo-[4′-32P]lipid IVA was employed as the lipid acceptor, Bp3073 and Hi1527 possess little to no late acyltransferase activity (< 10%). Similar results were also obtained when E. coli LpxL membranes were assayed with C12:0-ACP and either Kdo-[4′-32P]lipid IVA, phosphorylated Kdo-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid IVA (Fig. 4B). Taken together, these results indicate that the presence of the additional negative charge on the primary Kdo sugar is critical for late acyltransferase activity in a number of Gram-negative bacteria.
FIGURE 4.
H. influenzae and B. pertussis LpxL proteins also require phosphorylated-Kdo lipid IVA for optimal activity. A, E. coli MKV15 (DE3) membranes expressing Vc0213, Bp3073, or Hi1527 were assayed for activity at 0.01 mg/ml for 1 h at 30 °C using a mixture of acyl-ACPs (C12:0, C14:0, C16:0, and C18:0) and either Kdo-[4′-32P]lipid IVA, phosphorylated Kdo-[4′-32P]lipid IVA (P-Kdo-[4′-32P]lipid IVA), or Kdo2-[4′-32P]lipid IVA as the lipid acceptor. B, E. coli LpxL membranes (0.001 mg/ml) were assayed at 30 °C for 1 h using C12:0-ACP and either Kdo-[4′-32P]lipid IVA, P-Kdo-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid IVA. The origin and solvent front are not shown.
Vc0213 Transfers a Myristate (C14:0) to Phosphorylated Kdo-lipid IVA
We have demonstrated that Vc0213 functions as a lipid A late acyltransferase when assayed with phosphorylated Kdo-[4′-32P]lipid IVA or Kdo2-[4′-32P]lipid IVA and a mixture of acyl-ACP donors (Figs. 3 and 4A). To differentiate the preferred Vc0213 acyl donor, membranes containing Vc0213 were assayed with individual acyl-ACPs. Vc0213 was shown to favorably transfer a myristate (C14:0) to the lipid acceptor (Fig. 5A). This result was expected because the proposed V. cholerae O1 lipid A contains secondary acyl chains that are 14 carbons in length (Fig. 1) (11, 13, 14). Detailed analysis with its preferred acyl donor (C14:0-ACP) and either Kdo-[4′-32P]lipid IVA, phosphorylated Kdo-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid IVA (supplemental Fig. S4) clearly shows that a lipid A acceptor with a phosphorylated Kdo is the preferred substrate. Under the specified assay conditions, the specific activity of the enzyme was 2.6 μmol/min/mg when phosphorylated Kdo-[4′-32P]lipid IVA was used as the substrate. A specific activity of 1.6 μmol/min/mg was obtained using substrate with two Kdo sugars. However, the specific activity decreased to only 0.1 μmol/min/mg with Kdo-[4′-32P]lipid IVA (Fig. 4 and supplemental Fig. S4).
FIGURE 5.
Vc0213 is a myristoyl (C14:0) acyltransferase and utilizes C14:0-CoA as its preferred acyl donor. A, MKV15 (DE3) membranes (0.01 mg/ml) expressing either Bp3073, Hi1527, or Vc0213 were assayed with Kdo2-[4′-32P]lipid IVA and individual acyl-ACPs (C12:0, C14:0, C16:0, or C18:0) for 30 min. The percentage of conversion was calculated based on the amount of Kdo2-[4′-32P]lipid IVA converted to the acylated form. B, membranes expressing Vc0213, pBp3073, or pHi1527 (0.01 mg/ml) were assayed for 15 min. with either C14:0-ACP or C14:0-CoA using phosphorylated Kdo-[4′-32P]lipid IVA as the lipid acceptor. For assays containing E. coli LpxL, the concentration of membranes was 0.001 mg/ml and either C12:0-ACP or C12:0-CoA and Kdo2-[4′-32P]lipid IVA as the lipid acceptor.
Previous studies have shown that lipid A late acyltransferases preferentially employ acyl-ACPs as acyl donors; however, a recent report by Six et al. (30) showed that purified E. coli LpxL utilizes C12:0-CoA as an alternate acyl donor at ∼5% of the specific activity of C12:0-ACP. We obtained a similar result upon assay of membranes overexpressing E. coli LpxL (Fig. 5B and supplemental Fig. S5). Similarly, the acyltransferases of B. pertussis and H. influenzae that preferentially utilize C14:0-ACP during in vitro assay were unable to efficiently utilize acyl-CoA (Fig. 5B). Surprisingly, the V. cholerae acyltransferase showed a strong preference for C14:0-CoA compared with C14:0-ACP (Fig. 5B). There was an approximate 7-fold decrease in the specific activity of the acyltransferase when acyl-ACP served as the fatty acyl donor (supplemental Fig. S5). This is the first report of a lipid A late acyltransferase that preferentially utilizes acyl-CoA rather than acyl-ACP as an acyl donor in vitro.
MALDI-TOF Analysis Confirms Vc0213 Adds to the 2′-Position of Lipid A
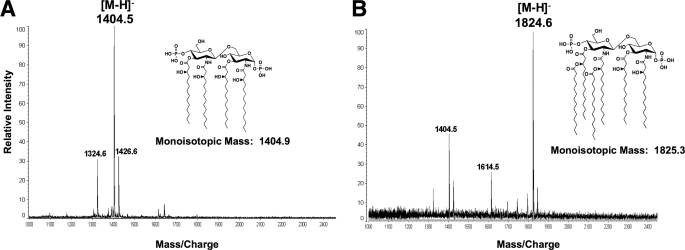
To confirm the location of the Vc0213 acyl chain addition, the lipid A of E. coli strain MKV13 containing either pBluescript or pBSVc0213 (supplemental Table S1) was isolated and analyzed by MALDI-TOF mass spectrometry in the negative ion mode. MKV13 is a temperature-sensitive E. coli strain that lacks functional copies of LpxL and LpxP. The predominant lipid species synthesized by MKV13 is tetra-acylated lipid A because LpxM catalyzed acylation is dependent upon secondary acylation at the 2′-position of lipid A (25). The mass spectrometry results of the lipid A isolated from MKV13 containing pBluescript gave an expected peak at m/z 1404.5 atomic mass units (amu). This result is indicative of the major tetra-acylated species synthesized by MKV13 E. coli (Fig. 6A) (25). However, the mass spectrometry results of MKV13 expressing Vc0213 revealed a major peak at m/z 1824.6 atomic mass units (Fig. 6B). This result is consistent with the molecular weight of a hexa-acylated lipid A containing a myristate (C14:0) at both the 2′- and 3′-positions of the glucosamine disaccharide. This result indicates that Vc0213 is the LpxL homolog, transferring a myristate (C14:0) to the 2′-position of Kdo2-lipid IVA. Endogenous LpxM activity is responsible for the addition of the second C14:0 to the 3′-position of the penta-acylated lipid A precursor.
FIGURE 6.
MALDI-TOF mass spectrometry confirms Vc0213 is the E. coli LpxL homolog. Lipid A of E. coli MKV13 (lpxL and lpxP) expressing pBluescript (A) or pBSVc0213 (B) were analyzed by MALDI-TOF mass spectrometry in the negative ion mode. The major ion peak in A is m/z 1404.5 amu, which corresponds to the expected mass of lipid IVA. The major peak in B is m/z 1824.6 amu, indicating that Vc0213 is adding a myristate (C14:0) to the 2′-position of the glucosamine disaccharide. Endogenous MKV13 LpxM adds a C14:0 to the 3′-position, thus producing the hexa-acylated lipid A species with a predicted mass of 1825.3 (inset structure). Minor peaks are explained below: at 1614.5 amu, the addition of a single C14:0; at 1324.6 amu, the loss of phosphate group at 1-position of the glucosamine disaccharide from parent ion; and at 1426.6 amu, the addition of a sodium adduct.
DISCUSSION
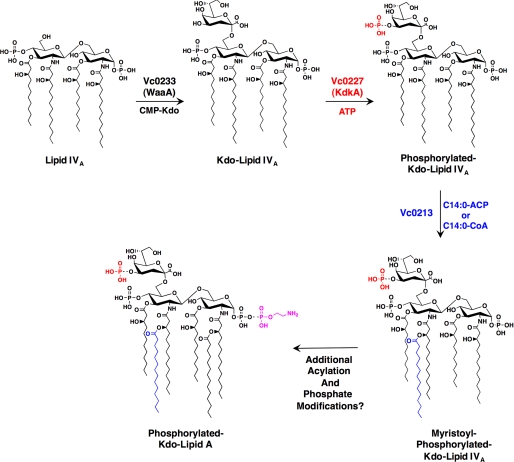
The Kdo-lipid A domain of LPS has been shown to vary widely among Gram-negative bacteria. Previous structural analyses have demonstrated the lipid A of V. cholerae serogroup O1 differs greatly from the lipid A of V. cholerae serogroup O139 (Fig. 1) (11–14). This report is the first to biochemically characterize the late enzymatic steps involved in the biosynthesis of the V. cholerae Kdo-lipid A domain (Fig. 7). Understanding the assembly of V. cholerae lipid A will allow for future engineering of strains with decreased endotoxic properties providing a safer vaccine alternative for those living in cholera endemic areas.
FIGURE 7.
Proposed pathway of V. cholerae inner core biosynthesis and secondary acylation of lipid A. The V. cholerae WaaA transfers a single Kdo moiety to the 6′-position of V. cholerae lipid IVA (Fig. 2A). Vc0227, the V. cholerae KdkA, catalyzes the addition of a phosphate group to the Kdo sugar (Fig. 2B). Vc0213 then adds an acyl chain to the 2′-position, using either C14:0-ACP or C14:0-CoA as an acyl donor (Figs. 5 and 6). Additional lipid A modifying enzymes and late acyltransferases are necessary to generate the hexa-acylated lipid A structure of V. cholerae O1 previously reviewed by Chatterjee and Chaudhuri (11).
In the present study, we identify only one of three V. cholerae putative late acyltransferases to be functional using in vitro assays. Vc0213 is homologous to the E. coli LpxL and transfers a myristate to the 2′-position of either phosphorylated Kdo-[4′-32P]lipid IVA or Kdo2-[4′-32P]lipid IVA (Fig. 4). One unusual property of the V. cholerae late acyltransferase is that it favorably employs C14:0-CoA as an acyl donor in vitro (Fig. 5B and supplemental Fig. S5). Because the content of CoAs in bacteria such as E. coli have been found to be approximately eight times larger than the ACP pool (31), it may be physiologically significant for some Gram-negative bacteria to possess lipid A late acyltransferases that utilize acyl-CoA as an acyl donor rather than an acyl-ACP donor.
Furthermore, the current work has shed light on the importance of phosphorylation of the Kdo group of bacterial LPS. Until now, it was unknown whether the phosphate group and the secondary Kdo sugar linked to the primary Kdo functioned similarly in Kdo-lipid A biosynthesis. Both substituents provide an additional negative charge, which we have shown is necessary for secondary acylation in vitro by the late acyltransferases of V. cholerae (Vc0213), B. pertussis (Bp3073), H. influenzae (Hi1527), and E. coli (LpxL) (Fig. 4). Although the additional negative charge may be critical for the secondary acylation of lipid A in some Gram-negative bacteria, it should be noted that a secondary acyltransferase of Pseudomonas aeruginosa (32) and Helicobacter pylori (33) function independently of the Kdo sugars and transfer an acyl chain to lipid IVA.
Previous work by Hood et al. (34) reported that a mutation in the open reading frame orfZ of H. influenzae strain Rd caused attenuation of the strain when introduced in the infant rat model. orfZ was later identified as the kdkA of H. influenzae, and it was hypothesized that the lack of the phosphate group on the Kdo was responsible for the reduced virulence of the H. influenzae strain (22). Together, our results suggest that the decrease in virulence of the H. influenzae kdkA mutant may arise from lack of secondary acylation of the H. influenzae lipid A. Mutations in either lpxL or lpxM have been shown to cause attenuation in Salmonella typhimurium (35), H. influenzae (36), E. coli (37, 38), and Neisseria meningitidis (39).
Previous reports supporting our findings that phosphorylation of the Kdo residue is important for complete acylation of lipid A can be found within the published literature. Although waaA is essential for E. coli growth, temperature-sensitive knock-out mutations have been successfully isolated (40). Complementation of the E. coli waaATS mutant with the mono-functional B. pertussis waaA does not alleviate the temperature-sensitive phenotype, whereas introduction of both the Kdo transferase and Kdo kinase of H. influenzae restores growth at higher temperatures (40–42). Because the B. pertussis WaaA is monofunctional, our findings suggest that it would be unable to transfer secondary acyl chains to Kdo-lipid IVA (Fig. 4A), leading to a reduction in growth rate caused by incomplete Kdo-lipid A biosynthesis.
Two of the proposed lipid A late acyltransferase homologs, Vc0212 and Vc1577, were nonfunctional when assayed using optimized late acyltransferase conditions. In vitro assays were also done to determine whether Vc0212 or Vc1577 were functional as either the LpxM or LpxP homologs; however, both putative V. cholerae late acyltransferase remained nonfunctional.3 As noted under “Experimental Procedures,” membranes expressing the V. cholerae late acyltransferases were isolated from an E. coli strain. The nonfunctional V. cholerae late acyltransferases may utilize an acyl donor that is solely present in the V. cholerae membrane. Alternatively, unidentified lipid A acyltransferases are present in V. cholerae, which account for the proposed hexa-acylated and octa-acylated structures of serogroups O1 and O139 (Fig. 1).
In this report, we have partially described the late stages of lipid A biosynthesis in V. cholerae O1 (Fig. 7) and have determined the importance of an additional negatively charged substituent, whether it be a second Kdo sugar or a phosphate group, on the primary Kdo residue in E. coli, B. pertussis, H. influenzae, and V. cholerae. Further lipid A modifications and acylations are necessary to achieve the proposed V. cholerae O1 lipid A structure. Current studies are ongoing in our laboratory to fully understand the enzymes involved in the biosynthesis of the phosphorylated Kdo-lipid A domain of V. cholerae LPS.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI076322 and AI064184 (to M. S. T.).

The on-line version of this article (available at http://www.jbc.org) contains Tables S1 and S2 and Figs. S1–S5.
J. V. Hankins and M. S. Trent, unpublished results.
- LPS
- lipopolysaccharide
- ACP
- acyl carrier protein
- amu
- atomic mass units
- CoA
- coenzyme A
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight.
REFERENCES
- 1.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trent M. S., Stead C. M., Tran A. X., Hankins J. V. (2006) J. Endotoxin Res. 12, 205–223 [DOI] [PubMed] [Google Scholar]
- 3.Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. (2006) ACS Chem. Biol. 1, 33–42 [DOI] [PubMed] [Google Scholar]
- 4.Clementz T., Raetz C. R. (1991) J. Biol. Chem. 266, 9687–9696 [PubMed] [Google Scholar]
- 5.White K. A., Kaltashov I. A., Cotter R. J., Raetz C. R. (1997) J. Biol. Chem. 272, 16555–16563 [DOI] [PubMed] [Google Scholar]
- 6.Belunis C. J., Mdluli K. E., Raetz C. R., Nano F. E. (1992) J. Biol. Chem. 267, 18702–18707 [PubMed] [Google Scholar]
- 7.Clementz T., Bednarski J. J., Raetz C. R. (1996) J. Biol. Chem. 271, 12095–12102 [DOI] [PubMed] [Google Scholar]
- 8.Clementz T., Zhou Z., Raetz C. R. (1997) J. Biol. Chem. 272, 10353–10360 [DOI] [PubMed] [Google Scholar]
- 9.Carty S. M., Sreekumar K. R., Raetz C. R. (1999) J. Biol. Chem. 274, 9677–9685 [DOI] [PubMed] [Google Scholar]
- 10.Six D. A., Carty S. M., Guan Z., Raetz C. R. (2008) Biochemistry 47, 8623–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee S. N., Chaudhuri K. (2003) Biochim. Biophys. Acta 1639, 65–79 [DOI] [PubMed] [Google Scholar]
- 12.Broady K. W., Rietschel E. T., Lüderitz O. (1981) Eur. J. Biochem. 115, 463–468 [DOI] [PubMed] [Google Scholar]
- 13.Villeneuve S., Souchon H., Riottot M. M., Mazie J. C., Lei P., Glaudemans C. P., Kovác P., Fournier J. M., Alzari P. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8433–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutonnier A., Villeneuve S., Nato F., Dassy B., Fournier J. M. (2001) Infect. Immun. 69, 3488–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorachek-Warren M. K., Carty S. M., Lin S., Cotter R. J., Raetz C. R. (2002) J. Biol. Chem. 277, 14186–14193 [DOI] [PubMed] [Google Scholar]
- 16.Trent M. S., Pabich W., Raetz C. R., Miller S. I. (2001) J. Biol. Chem. 276, 9083–9092 [DOI] [PubMed] [Google Scholar]
- 17.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 18.Belunis C. J., Raetz C. R. (1992) J. Biol. Chem. 267, 9988–9997 [PubMed] [Google Scholar]
- 19.Brozek K. A., Hosaka K., Robertson A. D., Raetz C. R. (1989) J. Biol. Chem. 264, 6956–6966 [PubMed] [Google Scholar]
- 20.Brozek K. A., Raetz C. R. (1992) Methods Enzymol. 209, 476–485 [DOI] [PubMed] [Google Scholar]
- 21.Tran A. X., Karbarz M. J., Wang X., Raetz C. R., McGrath S. C., Cotter R. J., Trent M. S. (2004) J. Biol. Chem. 279, 55780–55791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White K. A., Lin S., Cotter R. J., Raetz C. R. (1999) J. Biol. Chem. 274, 31391–31400 [DOI] [PubMed] [Google Scholar]
- 23.Tran A. X., Whittimore J. D., Wyrick P. B., McGrath S. C., Cotter R. J., Trent M. S. (2006) J. Bacteriol. 188, 4531–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000) Nucleic Acids Res. 28, 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R. (2002) J. Biol. Chem. 277, 14194–14205 [DOI] [PubMed] [Google Scholar]
- 26.Brade H. (1985) J. Bacteriol. 161, 795–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caroff M., Aussel L., Zarrouk H., Martin A., Richards J. C., Thérisod H., Perry M. B., Karibian D. (2001) J. Endotoxin Res. 7, 63–68 [PubMed] [Google Scholar]
- 28.Helander I. M., Lindner B., Brade H., Altmann K., Lindberg A. A., Rietschel E. T., Zähringer U. (1988) Eur. J. Biochem. 177, 483–492 [DOI] [PubMed] [Google Scholar]
- 29.Phillips N. J., Apicella M. A., Griffiss J. M., Gibson B. W. (1992) Biochemistry 31, 4515–4526 [DOI] [PubMed] [Google Scholar]
- 30.Six D. A., Carty S. M., Ziqiang G., Raetz C. R. H. (2008) Biochemistry 65, 4778–4783 [Google Scholar]
- 31.Magnuson K., Jackowski S., Rock C. O., Cronan J. E., Jr. (1993) Microbiol. Rev. 57, 522–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan S., Raetz C. R. (1994) J. Bacteriol. 176, 6944–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stead C. M., Beasley A., Cotter R. J., Trent M. S. (2008) J. Bacteriol. 190, 7012–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood D. W., Cox A. D., Gilbert M., Makepeace K., Walsh S., Deadman M. E., Cody A., Martin A., Månsson M., Schweda E. K., Brisson J. R., Richards J. C., Moxon E. R., Wakarchuk W. W. (2001) Mol. Microbiol. 39, 341–350 [DOI] [PubMed] [Google Scholar]
- 35.Jones B. D., Nichols W. A., Gibson B. W., Sunshine M. G., Apicella M. A. (1997) Infect. Immun. 65, 4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols W. A., Raetz C. R. H., Clementz T., Smith A. L., Hanson J. A., Ketterer M. R., Sunshine M. G., Apicella M. A. (1997) J. Endotoxin Res. 4, 163–172 [Google Scholar]
- 37.Somerville J. E., Jr., Cassiano L., Bainbridge B., Cunningham M. D., Darveau R. P. (1996) J. Clin. Invest. 97, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somerville J. E., Jr., Cassiano L., Darveau R. P. (1999) Infect. Immun. 67, 6583–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Ley P., Steeghs L., Hamstra H. J., ten Hove J., Zomer B., van Alphen L. (2001) Infect. Immun. 69, 5981–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belunis C. J., Clementz T., Carty S. M., Raetz C. R. (1995) J. Biol. Chem. 270, 27646–27652 [DOI] [PubMed] [Google Scholar]
- 41.Isobe T., White K. A., Allen A. G., Peacock M., Raetz C. R., Maskell D. J. (1999) J. Bacteriol. 181, 2648–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brabetz W., Müller-Loennies S., Brade H. (2000) J. Biol. Chem. 275, 34954–34962 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.