Abstract
The glutamine transporter SLC38A3 (SNAT3) plays an important role in the release of glutamine from brain astrocytes and the uptake of glutamine into hepatocytes. It is related to the vesicular GABA (γ-aminobutyric acid) transporter and the SLC36 family of proton-amino acid cotransporters. The transporter carries out electroneutral Na+-glutamine cotransport-H+ antiport. In addition, substrate-induced uncoupled cation currents are observed. Mutation of asparagine 76 to glutamine or histidine in predicted transmembrane helix 1 abolished all substrate-induced currents. Mutation of asparagine 76 to aspartate rendered the transporter Na+-independent and resulted in a gain of a large substrate-induced chloride conductance in the absence of Na+. Thus, a single residue is critical for coupled and uncoupled ion flows in the glutamine transporter SNAT3. Homology modeling of SNAT3 along the structure of the related benzyl-hydantoin permease from Microbacterium liquefaciens reveals that Asn-76 is likely to be located in the center of the membrane close to the translocation pore and forms part of the predicted Na+ -binding site.
The amino acid and auxin permease superfamily comprises a wide variety of transport proteins. In mammals, three distinct solute carrier families (SLC) belong to this superfamily, namely SLC32, SLC36, and SLC38 (1). Despite belonging to the same superfamily, the three solute carrier families have different transport mechanisms. The SLC32 family has only one member, the vesicular inhibitory amino acid transporter, which supposedly carries out a H+-GABA (γ-aminobutyric acid) antiport (2). The SLC36 family comprises four members, two of which have been characterized in more detail. These are the proton amino acid cotransporters 1 and 2 (PAT1 and 2) that carry out glycine and proline uptake in kidney and intestine and are mutated in iminoglycinuria (3, 4). The SLC38 family is comprised of 11 members, 5 of which have been characterized in more detail (5). Two different transport mechanisms are found within this family, namely the Na+-amino acid cotransporters SNAT1, SNAT2, and SNAT4 and the Na+-amino acid cotransporters-H+-antiporters SNAT3 and SNAT5. Transporters of the superfamily play a key role in inhibitory and excitatory neurotransmission, metabolite absorption, and liver metabolism. Despite their important roles in mammalian physiology, relatively little is known about the structure and function of these transporters.
The activity of ion-coupled membrane transporters is frequently associated with currents which de- or hyperpolarize the cell membrane. These currents may be due to electrogenic transport stoichiometry and/or to a non-stoichiometric ion conductance (6). Transport-associated ion conductances have been identified in a number of transporters but have been particularly well studied in several Na+-coupled neurotransmitter transporters (7–11). Transport-associated conductances have also been observed in electroneutral transporters that do not carry out net charge movement (8, 12–15). The glutamine transporter SNAT3, for instance, has a transport mechanism in which glutamine uptake is coupled to the cotransport of 1Na+ and the antiport of 1H+ and, hence, is unaffected by changes of the membrane potential (13, 16). Despite the electroneutral transport mechanism, substrate uptake is accompanied by inward currents, which are carried by cations below pH 7 and by protons at alkaline pH. In addition, a substrate-independent cation conductance and a Na+/H+ exchange activity has been observed (17). Non-stoichiometric currents can be mediated by the same ions that are involved in the coupled transport process, such as in the case of SNAT3, but may also be carried by different ions. Stoichiometric glutamate transport, for instance, involves Na+, H+, and K+ ions, whereas the glutamate transport-associated conductance is carried by chloride (18).
A crucial question concerning transporter-associated ion conductances is whether the conducting pore coincides with the translocation pathway of the substrate and whether both use the same critical residues. In the case of the glutamate transporters, evidence has been presented suggesting that different residues are critical for the anion conductance than for substrate transport (19, 20) but that they all line the same pathway (21). Here we show that asparagine 76 of SNAT3 is critical for substrate-induced ion conductance and affects binding of the cosubstrate Na+. In addition we show that this residue is likely to be localized in the translocation pore in the center of the membrane.
EXPERIMENTAL PROCEDURES
Oocytes and Injections
Xenopus laevis females were purchased from the South African Xenopus facility (Knysna, Republic of South Africa) and from Xenopus Express, Vernassal, France. Oocytes (stages V and VI) were isolated by collagenase treatment as described (22) and allowed to recover overnight. The surgical removal of ovarian tissue was performed under anesthesia (20-min immersion in 1% MS-222) and was approved by the animal ethics committee of the University of Kaiserslautern and the Australian National University. Rat SNAT3 was used as described previously (17). Oocytes were microinjected with 20 nl of rSNAT3 cRNA in water at a concentration of 1 μg/μl by using a microinjection device (WPI, Sarasota, Fl); control oocytes were injected with equivalent amounts of diethyl pyrocarbonate-H2O or remained uninjected.
pH Measurements with Microelectrodes
Double-barrel pH-sensitive microelectrodes, to measure intracellular pH (pHi) and membrane potential in frog oocytes, were prepared as previously described (23). Briefly, the electrodes were pulled in two stages and silanized by filling a drop of 5% tri-N-butylchlorosilane in 99.9% pure carbon tetrachloride into the prospective ion-selective barrel and then baking the pipette on a hot plate at 475 °C for 4.5–5 min.
For pH-selective microelectrodes a small amount of H+-ionophore mixture (Fluka 95291) was backfilled into the tip of the silanized barrel and the remainder filled with 0.1 m sodium citrate, pH 6.0. The reference barrel was filled with 2 m potassium acetate. Electrodes were accepted for experiments if their response exceeded 50 mV per unit change in pH and if they reacted faster in the bath during calibration before and after each experiment than the fastest pHi changes recorded upon substrate addition. On average, electrodes responded with a change of 54 mV to a change in pH by one unit.
The recording arrangement was the same as described previously (24). The central and the reference barrel were connected by chloride silver wires to the headstages of an electrometer amplifier.
As described previously (25), optimal pH changes were detected when the electrode was located near the inner surface of the plasma membrane. This was achieved by carefully rotating the oocyte with the impaled electrode. All experiments were carried out at room temperature (22–25 °C). Only oocytes with a membrane potential more negative than −25 mV were used for experiments.
Electrophysiological Recording
A borosilicate glass capillary 1.5 mm in diameter was pulled to a micropipette and backfilled with 3 m KCl. The resistance of the electrode measured in oocyte saline was around 1 megaohm. For voltage-clamp recordings, both electrodes were connected to the head stages of an Axoclamp-2B amplifier (Axon Instruments, Foster City, CA). The experimental bath was grounded with a chloride-treated silver wire coated by agar dissolved in oocyte saline. Oocytes were clamped at −40 mV unless indicated otherwise. To measure ion conductances associated with SNAT3 expression, short voltage jumps were applied during recordings in increments of 20 mV ranging from −100 to + 20 mV. The slopes of these curves were used to calculate the ion conductance (slope conductance).
Site-directed Mutagenesis
Site-directed mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as recommended by the manufacturer. The following primers were used (only sense primers shown, amino acid changing mutation in uppercase): N76D, ttc aat ctc agc Gac gcc atc atg ggc; N76E, ttc aat ctc agc GaG gcc atc atg ggc; N76H, ttc aat ctc agc Cac gcc atc atg ggc; N76Q, ttc aat ctc agc CaA gcc atc atg ggc; N76S, ttc aat ctc agc TCc gcc atc atg ggc.
Surface Biotinylation
To determine plasma membrane expression, 15 oocytes were washed 3 times with 4 ml of ice-cold phosphate-buffered saline, pH 8.0. Subsequently, oocytes were incubated in 1.5 ml of Sulfo-NHS-lc-Biotin (0.5 mg/ml; Pierce) solution in phosphate-buffered saline for 20 min at room temperature. The reagent was removed by washing oocytes 4 times with 4 ml of ice-cold phosphate-buffered saline, pH 8.0. Subsequently, oocytes were lysed by incubation in 1 ml of lysis buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1% Triton X-100) for 60 min on ice. The lysate was centrifuged at top speed in a tabletop centrifuge for 15 min at 4 °C, and the supernatant was mixed with 50 μl of streptavidin-coated-agarose particles (Pierce). The suspension was incubated at 4 °C overnight with slight agitation. Agarose particles were washed 4 times with 1 ml of lysis buffer, and the pellet was subsequently resuspended in 10 μl of 5 × SDS-PAGE sample buffer. Samples were boiled for 5 min, and an aliquot of 30 μl was loaded on the gel. After gel electrophoresis, proteins were blotted onto nitrocellulose membranes. SNAT3 was detected using an anti-peptide antibody at a 1:2500 dilution that was raised against a rat SNAT3 peptide (Zymed Laboratories Inc., South San Francisco, CA). Antibody binding was detected by enhanced chemiluminescence using the ECL system according to the manufacturer's instructions and using the provided secondary antibody at a dilution of 1:2500 (Amersham Biosciences).
Homology Model
Distantly related sequences with sequence homology below 20% are often not identified by BLAST search algorithms. More recently, sensitivity of homology searches has been improved by using profiles built from alignments of multiple sequences and Hidden Markov Models. In these algorithms only the conserved residues are used for searches and compared with profiles generated from aligned data base sequences. The fidelity of these search algorithms can be improved further by incorporation of secondary structure prediction methods (26). To identify whether SNAT3 is related to known structures, we used the HHPred server (27), which is based on these improved methods to find related structures. The server identified the hydantoin transporter Mhp1 (Protein Data Bank 2jln) as the closest relative with a known structure (E = 1.6 × 10−6). Other related structures, such as LeuT, showed considerably worse scores. To verify the relatedness of the two structures, we analyzed the sequences of rat SNAT3 and Mhp1 from Microbacterium liquefaciens (DD136890) using the Topcons topology prediction algorithm. The resulting predicted position of each residue relative to the center of the membrane (Z-coordinate) (28) and the free energy of insertion into the Sec61 translocon (29) were calculated for both sequences. We then plotted the alignment as generated by the HHPred server by introducing the corresponding gaps into the two topology prediction graphs. A homology model was subsequently built using the “Modeler” (30) as associated with the HHPred server.
RESULTS
We have recently analyzed alignments of functionally characterized members of the SLC32, SLC36, and SLC38 families to identify possible residues involved in ion conductances of the SNAT3 transporter (17). We focused on residues that have been identified in ion binding sites in high resolution structures of ion channels and transporters (31) and that were conserved in the Na+-dependent transporters of the SLC38 family but not in the proton-dependent SLC32 or SLC36 families (see alignment in Ref 17). Using these criteria we identified Asn-76, Gly-210, Ser-215, His-300, Thr-343, and Thr-380 as candidates. In this study asparagine 76 was mutated to different amino acids to study its role in ion translocation.
Substrate-dependent Cation Conductances Are Abolished by Mutation of Asparagine 76 to Glutamine or Histidine
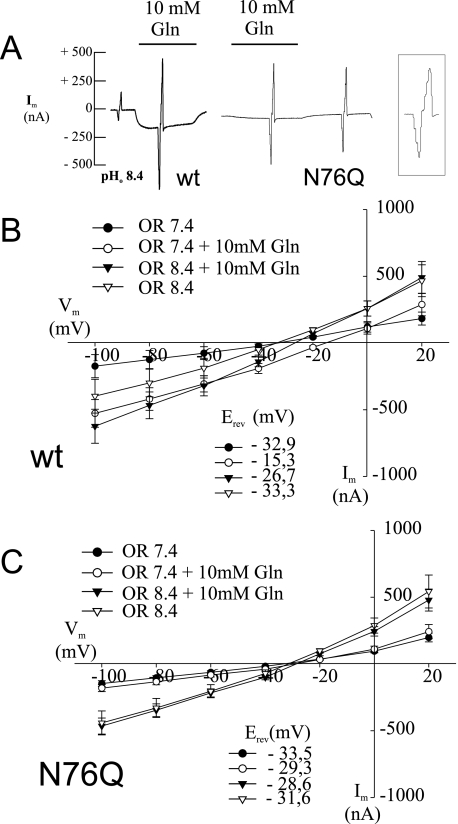
When expressed in X. laevis oocytes, SNAT3 carries out an electroneutral transport of glutamine, asparagine, and histidine coupled to the cotransport of 1Na+ and an antiport of 1H+ (13). At the same time, an inward current (indicated by the downward deflection of the tracing in Fig. 1A and described in detail in Refs. 17 and 12) is observed that at neutral pH is carried largely by cations but at more alkaline pH is carried by protons (17). In addition to these substrate-dependent currents, a substrate-independent alkaline pH-induced conductance is observed. To measure ion conductances associated with SNAT3 expression, short voltage jumps were applied during recordings in increments of 20 mV ranging from −100 to + 20 mV. The size of the resulting current peaks corresponds to the size of the conductance and was measured in the absence and presence of substrate (Fig. 1A). The voltage jumps were used to generate I/V plots (Fig. 1, B and C), and the slopes of the I/V plots were used to derive the conductance for each condition (Table 1). Mutation of asparagine 76 to glutamine (N76Q) completely abolished the glutamine-induced conductance (compare responses to voltage jumps in the presence and absence of glutamine in Fig. 1A). The corresponding I/V plots are shown in Fig. 1, B and C. Fig. 1, B and C, also demonstrate that the mutation abolishes the conductance at both pH 7.4, when it is carried by cations, and 8.4, when it is carried by protons. This suggests that the different cations use the same permeation pathway at both pH values and that this pathway is blocked by the mutation. The substrate-independent conductance at alkaline pH, by contrast, increased in the mutant (compare voltage responses in the absence of substrate in Fig. 1A and see Table 1). No significant difference was observed between asparagine and glutamine-induced currents in N76Q mutants (data not shown). Mutant N76S behaved very similarly to N76Q and also showed no substrate-induced currents at pH 7.4 or 8.4 (data not shown).
FIGURE 1.
Glutamine-induced currents in SNAT3 and its mutant N76Q. Oocytes were injected with 20 ng of cRNA encoding SNAT3 or its mutant N76Q, and glutamine-induced (10 mm) currents were analyzed after 3–6 days of expression at pH 7.4 or pH 8.4 (as indicated). Conductance and current-voltage relationships were measured by applying voltage jumps in increments of 20 mV from −100 mV to +20 mV at different stages of the recording; in the intervening periods oocytes were held at −40 mV. The responses to the voltage jumps generate the spikes in the recordings (shown on an extended time scale in the boxed tracing). The size of the spikes is proportional to the size of the conductance. Typical recordings at pH 8.4 are depicted from oocytes expressing wild type (wt) SNAT3 and its mutant N76Q (A). Current-voltage relationships for SNAT3 and the N76Q mutant were derived from the responses to the voltage jumps and are depicted in (wild type, n = 8) (B) and (N76Q, n = 6) (C), respectively. OR, oocyte ringer.
TABLE 1.
Slope conductance of SNAT3 and its mutants
Ion conductance was calculated from the slope of I/V curves ranging from −40 to 0 mV. Slope conductances are given in microsiemens. ND, not determined.
| cDNA | Na+ |
−Na+ |
||||||
|---|---|---|---|---|---|---|---|---|
| pH 7.4 | pH 8.4 | Gln |
pH 7.4 | pH 8.4 | Gln |
|||
| pH 7.4 | pH 8.4 | pH 7.4 | pH 8.4 | |||||
| Microsiemens | Microsiemens | |||||||
| None | 2.51 | 1.76 | 1.72 | 1.76 | ND | 1.49 | 1.41 | 1.62 |
| SNAT3 | 1.67 | 1.82 | 4.0 | 4.56 | ND | 2.33 | 1.3 | 2.14 |
| N76Q | 3.16 | 6.15 | 6.94 | 10.15 | ND | 5.11 | 1.77 | 4.70 |
| N76H | 3.71 | 13.1 | 3.14 | 6.96 | ND | 9.72 | 2.78 | 7.97 |
| N76D | 2.71 | 5.38 | 1.69 | 3.56 | ND | 12.51 | 1.93 | 56.96 |
| N76E | 3.43 | 16.83 | 5.98 | 12.46 | ND | 22.62 | 6.19 | 78.43 |
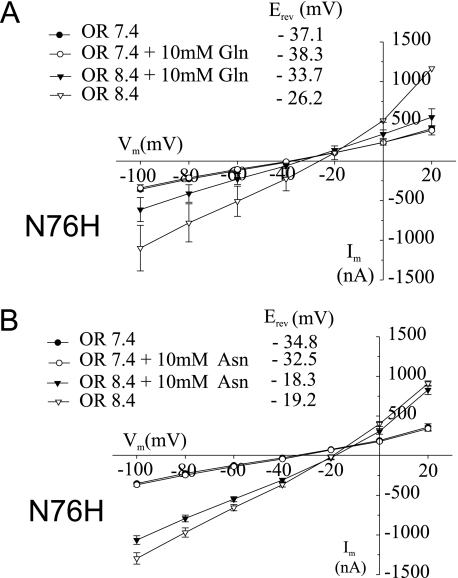
Because mutation of Asn-76 to other neutral residues already changed the conductance properties, we replaced asparagine with histidine, which introduces an ionizable and possibly positively charged side chain. Similar to N76Q and N76S, no substrate-induced currents were observed at pH 7.4 for both glutamine (Fig. 2A) and asparagine (Fig. 2B). The N76H mutation also showed a prominent alkali-induced substrate independent conductance (Fig. 2 and Table 1). This conductance was significantly suppressed by the addition of glutamine (Fig. 2A and Table 1) but almost unaffected by asparagine (Fig. 2B). This difference suggested that the nature of the substrate side chain also influenced the characteristics of the conductance.
FIGURE 2.
Glutamine-induced currents in SNAT3 mutant N76H. Oocytes were injected with 20 ng of cRNA encoding SNAT3 mutant N76H, and glutamine-induced (10 mm) or asparagine-induced (10 mm) currents were analyzed after 4–5 days of expression at pH 7.4 and 8.4. Current-voltage relationships are depicted for glutamine-induced currents (A, n = 8) and asparagine-induced currents (B, n = 3). OR, oocyte ringer.
Mutants N76D and N76E Gain a Large Chloride Conductance
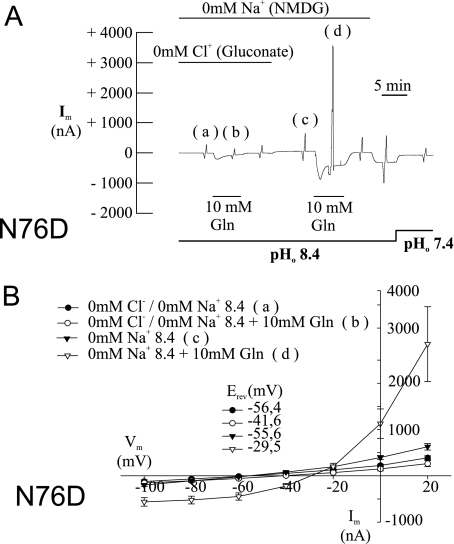
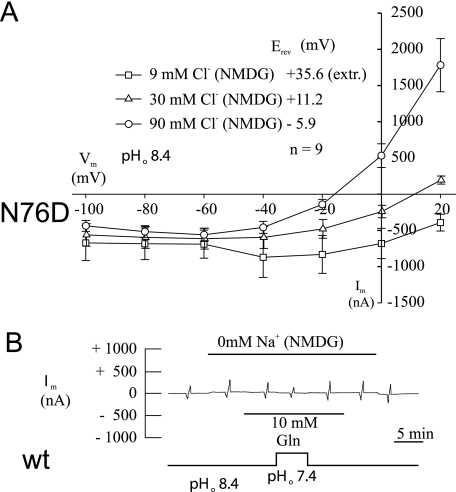
Mutation of Asn-76 to histidine introduced a possible positive charge. To analyze the effect of introducing a negative charge at this position, we exchanged Asn-76 to aspartate or glutamate. Similar to the other mutants, N76D and N76E showed a significantly higher substrate-independent conductance at pH 8.4 (Table 1). Because the transporter is Na+-dependent, we measured conductances in all mutants in the presence and absence of sodium ions (Table 1). As reported previously (12, 17) and shown here (Table 1), substrate-induced currents in the wild type are Na+-dependent. Mutants N76D and N76E by contrast showed very large, reversible substrate-induced currents of up to 8 μA in the absence of Na+ (Fig. 3A, compare responses at positions c and d). This conductance was not observed at pH 7.4 and also not in the presence of Na+ (not shown). At −40 mV and pH 8.4, the currents were inwardly directed. Under these conditions, the driving forces of both K+ and H+ were outwardly directed, excluding a participation of these two ions (Fig. 3B). This suggested the presence of a chloride conductance in the absence of Na+. Replacement of chloride by gluconate (in the absence of Na+) indeed completely abolished the conductance (Fig. 3A compare responses at positions a and b). When three different chloride concentrations were used, the shift of the reversal potential of this current was >50 mV for a 10-fold change in the external chloride concentration and, hence, was close to prediction of the Nernst equation for a pure chloride conductance (Fig. 4A). In addition, the conductance increased with the chloride concentration from 4.7 ± 1.8 microsiemens (μS) (at 9 mm Cl−) to 8.9 ± 1.8 μS (at 30 mm Cl−) and 24.7 ± 5.1 μS (at 90 mm Cl−). Thus, introduction of a nominal negative charge generates a chloride conductance, whereas in the wild type all currents appear to be carried by cations. The current showed a strong outward rectification (Figs. 3, A and B, and 4A) and a significant run-down upon repeated substrate additions (not shown). Similar to the other mutants, N76D and N76E also did not show substrate-dependent conductances in the presence of Na+ (not shown). The wild type did not show any substrate-induced currents in the absence of Na+, consistent with the Na+ dependence of the transporter (Fig. 4B).
FIGURE 3.
Glutamine-induced currents in SNAT3 mutant N76D. Oocytes were injected with 20 ng of cRNA encoding SNAT3 mutant N76D, and glutamine-induced (10 mm) currents were analyzed after 3–6 days of expression at pH 7.4 and pH 8.4 and under different ionic conditions as indicated. Sodium ions were replaced by NMDG+, and chloride ions were replaced by gluconate−. Typical recordings are depicted from oocytes expressing SNAT3 mutant N76D (A); the corresponding current-voltage relationship for SNAT3 mutant N76D is depicted in B (n = 8). The responses to voltage jumps (A) at the indicated positions (a, b, c, and d) were converted into I/V plots (B) and labeled accordingly. NMDG, N-methyl-d-glucamine.
FIGURE 4.
Reversal potential of glutamine-induced currents in SNAT3 mutant N76D. Oocytes were injected with 20 ng of cRNA encoding SNAT3 or its mutant N76D, and glutamine-induced (10 mm) currents were analyzed after 3–6 days of expression at pH 7.4 and 8.4. The reversal potential was recorded at three different chloride concentrations (A). The reversal potential at 9 mm chloride was extrapolated (extr.); chloride ions were replaced by gluconate ions. Wild type (wt) SNAT3 expressing oocytes did not show glutamine-induced currents in the absence of Na+ as shown in a typical recording (B). NMDG, N-methyl-d-glucamine.
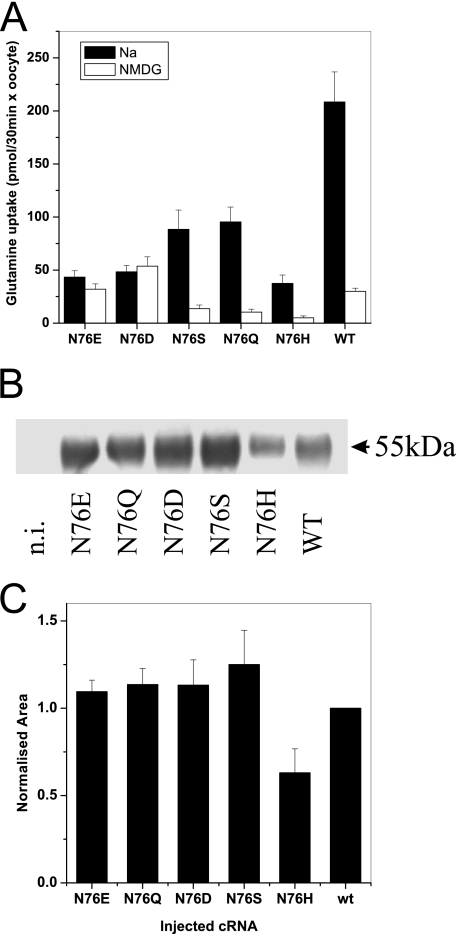
The crucial importance of Asn-76 for the ion conductance in the SNAT3 protein suggested that it might also be involved in the coupled substrate transport process. To assay glutamine transport in the mutants, we used flux assays. Uptake of [14C]glutamine ranged from 20 to 50% of the wild type for the different Asn-76 mutants (Fig. 5A). In agreement with an important role of Asn-76 in Na+-coupled substrate transport, we found that N76D and N76E carried out a reduced uptake of glutamine, which was, however, Na+-independent, whereas substrate transport of the wild type and the other mutants was largely Na+-dependent (Fig. 5A). The transport activity of the mutants at pH 7.4 was too low to be evaluated (data not shown). Thus, Asn-76 appears to be crucial for ion binding of both coupled and uncoupled transport. To further analyze coupled transport, we also analyzed substrate-dependent proton antiport. In agreement with the flux experiments, all mutants showed residual proton translocation (Table 2), with mutant N76H showing the slowest proton antiport activity. Apart from the N76H mutant, all mutants were expressed at the cell surface in similar amounts as the wild type, suggesting that the mutations changed the functional properties of the transporter rather than the folding and trafficking (Fig. 5, B and C).
FIGURE 5.
Transport activity and surface expression of oocytes expressing SNAT3 and its mutants. Oocytes were injected with 20 ng of cRNA encoding SNAT3 or its mutants. After 5 days of expression, uptake of [14C]glutamine was measured over an incubation period of 30 min, pH 8.4. The mean ± S.D. activity of 10 oocytes was determined in the presence (black bars) and absence (open bars) of Na+ (A). The experiment was repeated three times with similar results. Surface proteins in oocytes of the same batch were biotinylated, and membrane proteins were isolated by binding to streptavidin-coated agarose particles. Surface expression of SNAT3 and its mutants was subsequently determined by Western blotting (B). The complete series of Western blots was quantitatively evaluated (C; n = 3). n.i., non-injected oocytes. WT, wild type; NMDG, N-methyl-d-glucamine.
TABLE 2.
Changes of intracellular pH induced by substrates in SNAT3 and its mutants
The change of [H+]/min was recorded immediately after the addition of 10 mm glutamine for SNAT3 wild type and its mutants at pH 8.4, and the S.E. was calculated (n = 12 experiments). ND, not detectable; NI, non-injected oocytes.
| SNAT3 | N76D | N76E | N76Q | N76H | NI | |
|---|---|---|---|---|---|---|
| nm H+/min | nm H+/min | nm H+/min | nm H+/min | nm H+/min | nm H+/min | |
| 100 mm Na+ | −33.3 ± 11 | −8.5 ± 2.7 | −12.4 ± 3.6 | −9.6 ± 2.9 | −3.1 ± 0.4 | 0.015 ± 0.015 |
| 0 Na+ | −9.1 ± 5 | −4.7 ± 1.5 | −2.2 ± 0.6 | −0.9 ± 0.4 | −0.63 ± 0.3 | ND |
Residue Asn-76 Is Likely to Be Located in the Substrate Translocation Pore
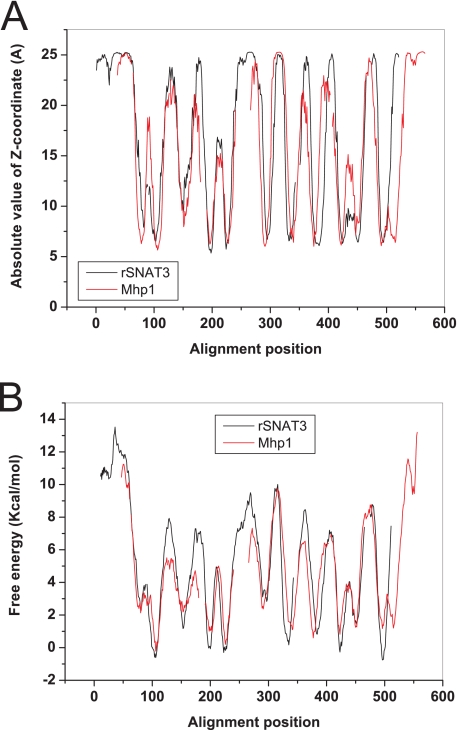
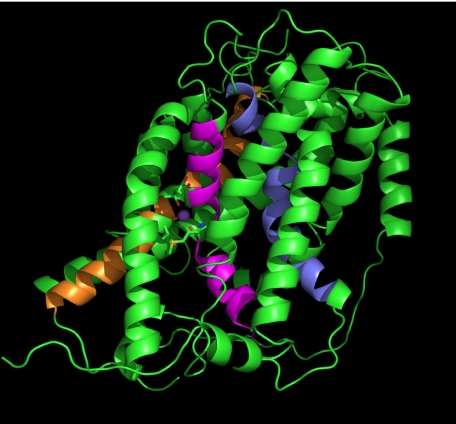
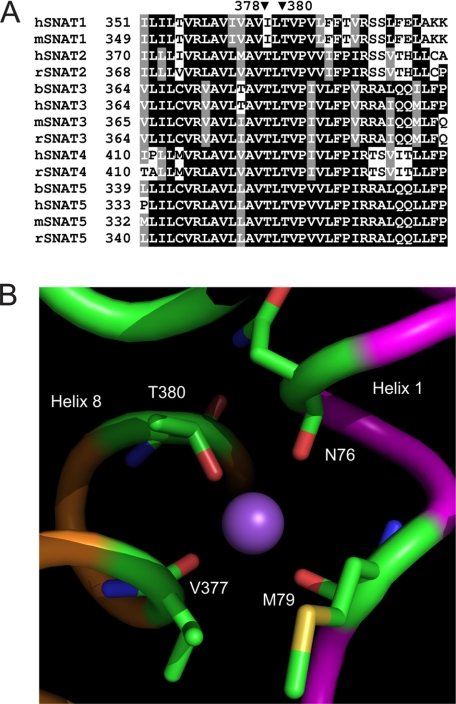
The effect of Asn-76 mutations on coupled and uncoupled ion transport suggested that Asn-76 is located in the translocation pore of the transporter. High resolution structures of membrane transporters have so far revealed only a limited number of different folds with 12 transmembrane spanning regions.3 Widespread among mammalian transporters appears to be the fold of the major facilitator superfamily of transporters exemplified by lactose and glycerol permeases (32, 33), a fold adopted by Na+/H+ exchangers (34), and a fold common to a large number of different transporter families, such as the sodium-dependent neurotransmitter transporters (35), the hydantoin permease (Mhp1) (36), the Na+-dependent glucose transporters (vSGLT1) (37), and the betain transporter BetP (38). We, thus, hypothesized that SNAT3 might adopt one of these folds. BLAST or PSI-BLAST searches did not reveal significant homology of SNAT3 to any of the transport proteins for which high resolution structures are available. However, using HHpred, a homology detection server based on a pair wise alignment of profile hidden Markov models (26), we could identify the hydantoin permease Mhp1 as a protein related to SNAT3 (E-value, 1.6 × 10−6), other proteins being significantly less or not related. The resulting sequence alignment is shown as supplemental Fig. 1. To confirm that the proteins are structurally related, we aligned the hydropathy plots, a method previously used to determine successfully structural similarity between proteins with little or no sequence homology (39). A comparison of two topology-predicting plots as provided by the Topcons server indeed showed a remarkable alignment between SNAT3 and Mhp1 (Fig. 6, A and B). Using this information we could develop a homology model for SNAT3 that is homologous to that of Mhp1 (Fig. 7). In this model helices 1 and 6 line the translocation pore as typical in this fold. In the Mhp1 structure, a potential cation binding site was predicted in the center of transmembrane helix 1, also involving residues in transmembrane helix 8 (36). The site is equivalent to that identified in the related structures of LeuT (35) and vSGLT1 (37). A Na+ ion inserted into this position in an initial SNAT3 homology model (not shown) interacted with Asn-76 (backbone) and Met-79 (backbone), Ile-375 (backbone), and Thr-378 (side chain). Analysis of the sequence alignment of SLC38 family transporters, however, showed that Thr-378 is not conserved in the Na+-dependent transporter SNAT1, whereas Thr-380 is highly conserved in all members (Fig. 8A). To incorporate this information, part of the alignment was moved by two positions to place Thr-380 in the same spatial position as Thr-378 in the initial model. The revised putative SNAT3 Na+-binding site would, thus, involve Asn-76, Met-79 in helix 1, and Val-377 (backbone) and Thr-380 (side-chain) in helix 8 in Na+ binding (Fig. 8B). The equivalent residues in Mhp1 are Ala-38 (backbone), Ile-41 (backbone) in helix 1, and Ser-312 (side chain) and Thr-313 (side chain) in helix 8. The precise orientation of residues around this putative Na+-binding site of SNAT3 is hypothetical at this stage, but the sensitivity of residue Asn-76 to mutation is consistent with its suggested central position in the protein.
FIGURE 6.
SNAT3 is structurally related to Mhp1. The peptide sequence of rat SNAT3 (red) and of the hydantoin permease Mhp1 (black) were both analyzed using the Topcons topology prediction program. Subsequently, the absolute value of the Z-coordinate (A) and the free energy of insertion into the Sec61 translocon (B) were aligned according to the profile alignment as provided by the HHpred server. The optimally aligned sequence (supplemental Fig. 1) was then used to build a homology model along the structure of the hydantoin permease.
FIGURE 7.
Homology model of SNAT3. The homology model was generated using Modeler with the optimized alignment between Mhp1 and rSNAT3 (supplemental Fig. 1). The structure was visualized using PyMol (DeLano Scientific). Helix 1 is shown in purple, helix 6 is in blue, and helix 8 is in orange.
FIGURE 8.
Sequence alignment of members of the SNAT (SLC38) family and putative SNAT3 Na+-binding site. A, reference sequences for SNAT1–5 were downloaded and aligned using ClustalW. The alignment around rat SNAT3 residue Thr-380 is shown. The corresponding positions in other family members are indicated. Human, mouse, rat, and bovine sequences are indicated by prefix h, m, r, and b, respectively. B, close view of the proposed Na+-binding site of SNAT3. Helix 1 is shown in purple, and helix 8 is in orange. Residues potentially involved in Na+ binding are indicated.
DISCUSSION
Several lines of evidence suggest that asparagine 76 is a crucial residue for ion translocation in the SNAT3 transporter. First, mutation of Asn-76 to any other residue abolishes substrate-induced conductances. Second, mutation of Asn-76 to glutamate or aspartate introduces a large chloride conductance and renders the transporter Na+-independent. Third, all mutants increase the alkali-induced substrate-independent conductance. The equivalent residue asparagine 82 in rat SNAT2 has recently been suggested to be involved in Na+ binding (40), supporting this conclusion. Furthermore, a histidine residue in helix 1 that is located one turn of a helix before asparagine 76 is thought to be involved in proton co-transport in the SLC36 family of proton amino acid co-transporters (41). Thus, a single residue in SNAT3 affects strongly both coupled and uncoupled ion movements in SNAT3.
Hydropathy plots suggested that asparagine 76 is located in the first transmembrane spanning helix but did not provide more detailed structural information. High resolution structures have greatly increased our understanding of carrier-mediated solute transport. Thus far only prokaryotic transporter structures have been resolved, but functional studies have shown that reliable homology models can be generated for related eukaryotic transporters. These have been very informative, for example, in the elucidation of ion binding (42), substrate binding (43), and of conformational changes (44). Less than 10 different folds have been identified thus far for secondary active transporters (for an up-to-date list see the Stephen White Laboratory at UC Irvine, online). Some of these protein folds appear to be particularly widespread among mammalian membrane metabolite transporters, such as the protein fold exemplified by the structures of the major facilitator superfamily (45), the Na+/H+ exchanger fold (34), and another fold represented by the structures of LeuT (35), vSGLT1 (37), Mhp1 (36), and BetP (38). The presence of only a limited number of folds among transporters in the data base has also been suggested as a result of a comparative analysis of hydropathy plots (46). Using an advanced homology search based on profile alignment and secondary structure prediction, we could identify Mhp1 as being related to SNAT3. The relation was further confirmed by alignment of hydropathy plots. During the revision of this manuscript the structure of the arginine/agmatine antiporter AdiC from two different microorganisms has been published (47, 48). Its structure is closely related to that of LeuT, vSGLT, MhP1, and BetP. The transporter belongs to the amino acid/polyamine/organocation superfamily, which in turn is distantly related to the amino acid and auxin permease superfamily (49). Thus, it appears likely that the overall folding pattern of the amino acid and auxin permease superfamily member SNAT3 will be close to the general structure proposed here.
The homology model predicts asparagine 76 to be located in the center of the translocation pore, consistent with its role in ion binding and translocation. In addition to residues in helix 1, the initial homology model of SNAT3 predicted Ile-375 and Thr-378 in helix 8 to be involved in Na+ binding. We have previously mutated Thr-380 to alanine and noticed that this mutation removed the cation conductance at pH 7.4 and strongly reduced the ability of Li+ to replace Na+ as the cotransported ion (17). In addition, Thr-380 is highly conserved, whereas Thr-378 is not. As a result we adjusted the model by sliding the alignment of two residues to involve Thr-380 in the Na+-binding site. The current model is consistent with the data, but further studies will be necessary to adjust and confirm the model particularly with regard to the precise arrangement of helix 8.
The most surprising result was the appearance of a large substrate-dependent chloride conductance at alkaline pH in SNAT3 (N76D and N76E). The conductance was only observed in the absence of Na+ and, thus, is unlikely to play a physiological role. However, the ease by which an anion can pass through SNAT3 suggests that anion permeation could be a normal function of SNAT3. Although we do not have direct experimental evidence, this finding could suggest that instead of a Na+-glutamine cotransport-H+-antiport, SNAT3 may in fact carry out a Na+-OH−-glutamine cotransport. This would also explain why, quite counterintuitive, we have observed a proton conductance at alkaline pH (17). Experiments discriminating between OH− symport and H+ antiport are, however, difficult to conceive. An anion pathway might indeed be conserved inside the SLC38 family. In the related transporter SNAT2, a tonic anion leak current was observed, which could be suppressed by substrate binding (50). However, substrate transport via SNAT1 or SNAT2 is not accompanied by changes of cytosolic pH (51, 52).
The cation conductance of SNAT3 could have an important physiological function. Glutamine transport in the liver and in pancreatic β-cells is mediated by SNAT3 (12, 53). Exposure to glutamine causes hepatocytes to swell (54) and β-cells to depolarize (55). In both cases this initiates signal transduction pathways resulting in metabolic changes (liver) or insulin release (pancreas). The SNAT3-associated ion conductance could cause significant depolarization and would increase the osmotic effect of Na+-glutamine cotransport.
Supplementary Material
Acknowledgment
We are indebted to Tancred Frickey (Australian National University) for help with bioinformatics in this project.
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft Grant De 231/16-2 and a grant from the Landesschwerpunkt “Membrantransport” (to J. W. D.), by Australian Research Council Grant LX0881952 (to J. W. D. and S. B.), and by National Health and Medical Research Council Grant 366713 (to S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The term “fold” refers to a typical protein folding pattern observed in a group of proteins.
REFERENCES
- 1.Sundberg B. E., Wååg E., Jacobsson J. A., Stephansson O., Rumaks J., Svirskis S., Alsiö J., Roman E., Ebendal T., Klusa V., Fredriksson R. (2008) J. Mol. Neurosci. 35, 179–193 [DOI] [PubMed] [Google Scholar]
- 2.Gasnier B. (2004) Pflugers Arch. 447, 756–759 [DOI] [PubMed] [Google Scholar]
- 3.Boll M., Daniel H., Gasnier B. (2004) Pflugers Arch. 447, 776–779 [DOI] [PubMed] [Google Scholar]
- 4.Bröer S., Bailey C. G., Kowalczuk S., Ng C., Vanslambrouck J. M., Rodgers H., Auray-Blais C., Cavanaugh J. A., Bröer A., Rasko J. E. (2008) J. Clin. Invest. 118, 3881–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackenzie B., Erickson J. D. (2004) Pflugers Arch. 447, 784–795 [DOI] [PubMed] [Google Scholar]
- 6.Sonders M. S., Amara S. G. (1996) Curr. Opin. Neurobiol. 6, 294–302 [DOI] [PubMed] [Google Scholar]
- 7.Blakely R. D., Defelice L. J., Galli A. (2005) Physiology 20, 225–231 [DOI] [PubMed] [Google Scholar]
- 8.Mager S., Min C., Henry D. J., Chavkin C., Hoffman B. J., Davidson N., Lester H. A. (1994) Neuron 12, 845–859 [DOI] [PubMed] [Google Scholar]
- 9.Galli A., Blakely R. D., DeFelice L. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8671–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan R. M., Vandenberg R. J. (2002) J. Biol. Chem. 277, 13494–13500 [DOI] [PubMed] [Google Scholar]
- 11.Wadiche J. I., Kavanaugh M. P. (1998) J. Neurosci. 18, 7650–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry F. A., Krizaj D., Larsson P., Reimer R. J., Wreden C., Storm-Mathisen J., Copenhagen D., Kavanaugh M., Edwards R. H. (2001) EMBO J. 20, 7041–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bröer A., Albers A., Setiawan I., Edwards R. H., Chaudhry F. A., Lang F., Wagner C. A., Bröer S. (2002) J. Physiol. 539, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerangue N., Kavanaugh M. P. (1996) J. Biol. Chem. 271, 27991–27994 [DOI] [PubMed] [Google Scholar]
- 15.Bröer A., Wagner C., Lang F., Bröer S. (2000) Biochem. J. 346, 705–710 [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhry F. A., Reimer R. J., Krizaj D., Barber D., Storm-Mathisen J., Copenhagen D. R., Edwards R. H. (1999) Cell 99, 769–780 [DOI] [PubMed] [Google Scholar]
- 17.Schneider H. P., Bröer S., Bröer A., Deitmer J. W. (2007) J. Biol. Chem. 282, 3788–3798 [DOI] [PubMed] [Google Scholar]
- 18.Wadiche J. I., Amara S. G., Kavanaugh M. P. (1995) Neuron 15, 721–728 [DOI] [PubMed] [Google Scholar]
- 19.Ryan R. M., Mitrovic A. D., Vandenberg R. J. (2004) J. Biol. Chem. 279, 20742–20751 [DOI] [PubMed] [Google Scholar]
- 20.Borre L., Kanner B. I. (2001) J. Biol. Chem. 276, 40396–40401 [DOI] [PubMed] [Google Scholar]
- 21.Leary G. P., Stone E. F., Holley D. C., Kavanaugh M. P. (2007) J. Neurosci. 27, 2938–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bröer S. (2003) Methods Mol. Biol. 227, 245–258 [DOI] [PubMed] [Google Scholar]
- 23.Deitmer J. W. (1991) J. Gen. Physiol. 98, 637–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munsch T., Deitmer J. W. (1994) J. Physiol. 474, 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bröer S., Schneider H. P., Bröer A., Rahman B., Hamprecht B., Deitmer J. W. (1998) Biochem. J. 333, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söding J. (2005) Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 27.Söding J., Biegert A., Lupas A. N. (2005) Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granseth E., Viklund H., Elofsson A. (2006) Bioinformatics 22, e191–e196 [DOI] [PubMed] [Google Scholar]
- 29.Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 30.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 31.Gouaux E., Mackinnon R. (2005) Science 310, 1461–1465 [DOI] [PubMed] [Google Scholar]
- 32.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 33.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 34.Padan E. (2008) Trends Biochem. Sci. 33, 435–443 [DOI] [PubMed] [Google Scholar]
- 35.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 36.Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ressl S., Terwisscha van Scheltinga A. C., Vonrhein C., Ott V., Ziegler C. (2009) Nature 458, 47–52 [DOI] [PubMed] [Google Scholar]
- 39.Lolkema J. S., Slotboom D. J. (1998) FEMS Microbiol. Rev. 22, 305–322 [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z., Gameiro A., Grewer C. (2008) J. Biol. Chem. 283, 12284–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metzner L., Natho K., Zebisch K., Dorn M., Bosse-Doenecke E., Ganapathy V., Brandsch M. (2008) Biochim. Biophys. Acta 1778, 1042–1050 [DOI] [PubMed] [Google Scholar]
- 42.Zomot E., Bendahan A., Quick M., Zhao Y., Javitch J. A., Kanner B. I. (2007) Nature 449, 726–730 [DOI] [PubMed] [Google Scholar]
- 43.Vandenberg R. J., Shaddick K., Ju P. (2007) J. Biol. Chem. 282, 14447–14453 [DOI] [PubMed] [Google Scholar]
- 44.Forrest L. R., Zhang Y. W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abramson J., Kaback H. R., Iwata S. (2004) Curr. Opin. Struct. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 46.Lolkema J. S., Slotboom D. J. (2008) Mol. Memb. Biol. 25, 567–570 [DOI] [PubMed] [Google Scholar]
- 47.Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009) Science 324, 1565–1568 [DOI] [PubMed] [Google Scholar]
- 48.Fang Y., Jayaram H., Shane T., Kolmakova-Partensky L., Wu F., Williams C., Xiong Y., Miller C. (2009) Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jack D. L., Paulsen I. T., Saier M. H. (2000) Microbiology 146, 1797–1814 [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z., Grewer C. (2007) Biophys. J. 92, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albers A., Bröer A., Wagner C. A., Setiawan I., Lang P. A., Kranz E. U., Lang F., Bröer S. (2001) Pflugers Arch. 443, 92–101 [DOI] [PubMed] [Google Scholar]
- 52.Chaudhry F. A., Schmitz D., Reimer R. J., Larsson P., Gray A. T., Nicoll R., Kavanaugh M., Edwards R. H. (2002) J. Neurosci. 22, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gammelsaeter R., Jenstad M., Bredahl M. K., Gundersen V., Chaudhry F. A. (2009) Biochem. Biophys. Res. Commun. 381, 378–382 [DOI] [PubMed] [Google Scholar]
- 54.Haussinger D., Graf D., Weiergraber O. H. (2001) J. Nutr. 131, Suppl. 9, 2509–2514 and 2523–2524 [DOI] [PubMed] [Google Scholar]
- 55.Liu Y. J., Cheng H., Drought H., MacDonald M. J., Sharp G. W., Straub S. G. (2003) Am. J. Physiol. Endocrinol. Metab. 285, E380–E389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.