Abstract
Glycosaminoglycan (GAG) biosynthesis requires numerous biosynthetic enzymes and activated sulfate and sugar donors. Although the sequence of biosynthetic events is resolved using reconstituted systems, little is known about the emergence of cell-specific GAG chains (heparan sulfate, chondroitin sulfate, and dermatan sulfate) with distinct sulfation patterns. We have utilized a library of click-xylosides that have various aglycones to decipher the mechanism of GAG biosynthesis in a cellular system. Earlier studies have shown that both the concentration of the primers and the structure of the aglycone moieties can affect the composition of the newly synthesized GAG chains. However, it is largely unknown whether structural features of aglycone affect the extent of sulfation, sulfation pattern, disaccharide composition, and chain length of GAG chains. In this study, we show that aglycones can switch not only the type of GAG chains, but also their fine structures. Our findings provide suggestive evidence for the presence of GAGOSOMES that have different combinations of enzymes and their isoforms regulating the synthesis of cell-specific combinatorial structures. We surmise that click-xylosides are differentially recognized by the GAGOSOMES to generate distinct GAG structures as observed in this study. These novel click-xylosides offer new avenues to profile the cell-specific GAG chains, elucidate the mechanism of GAG biosynthesis, and to decipher the biological actions of GAG chains in model organisms.
Proteoglycans play a major role in various cellular/physiological processes, including blood clotting, growth factor signaling, embryogenesis, axon growth and guidance, angiogenesis, and others (1–4). Proteoglycans consists of a core protein and glycosaminoglycan (GAG)2 chains. GAG chains account for >50% of the total molecular weight and are primarily responsible for physiological activity of the proteoglycans (5, 6). GAG chains are composed of repeating disaccharide units of a hexosamine residue and a hexuronic acid residue. The three major types of GAG chains found in the proteoglycans are heparan sulfate (HS), chondroitin sulfate (CS) and dermatan sulfate (DS). These GAG chains are differentiated by the type of hexosamine (glucosamine/galactosamine), the percentage of uronic acid epimers (glucuronic/iduronic acid), the extent of sulfation, and the nature of glycosidic linkage (α-/β-). One of the key steps in the proteoglycan biosynthesis is the xylosylation of certain specific serine residues of the core protein (7–10), which occurs in the late endoplasmic reticulum and/or cis-Golgi compartments (11–13). This key event is an essential prelude for the construction of the proteoglycan linkage region (14) that is followed by sequence of events resulting in the assembly of mature GAG chains by alternative addition of hexosamine and glucuronic acid residues. The maturation of GAG chains occurs in the medial and trans-Golgi compartments and involves the following events: N-sulfation of glucosamine units by N-deacetylase-N-sulfotransferases (for HS only), epimerization of glucuronic acids to iduronic acids by C-5 epimerase, and sulfation of the repeating disaccharide units by a variety of sulfotransferases and their isoforms.
The position, extent, and pattern of sulfation attribute enormous diversity to GAG chains, which confer specificity in binding to a vast array of proteins. These diverse structural features are very tightly regulated in a spatio-temporal manner during and beyond the development of an organism, and these features dictate differential interactions with various growth factors and receptors, and numerous protein targets leading to an array of physiological functions (15, 16).
The presence of free GAG chains has been known to disrupt the interaction of endogenous GAG components of proteoglycans with protein ligands thereby altering the physiological activities. Consequently, they have been used as molecular tools in the elucidation of the role of GAG chains in the activation of cellular events (17–19). Free GAG chains can be synthesized in vitro in cell culture by providing exogenous xylosides containing various hydrophobic aglycone moieties. Thus, the xylosides can act as false acceptors for initiation of linkage region and the subsequent elongation of GAG chains. Xylosides have been used for over three decades both in vitro (20–28) and in vivo (25, 29–31) to probe the functional significance of GAG chains in various dynamic systems under different conditions. The quantity and type of GAG chains synthesized depends on the system where it was tested and on the structure of the aglycone moiety of the xylosides (32–34). Most of these studies have utilized a few O-xylosides that are inherently less stable. Furthermore, synthesis of O-xylosides requires very stringent reaction conditions, toxic Lewis acids, and at times leads to inseparable α and β mixtures with unpredictable yields. As a result, it is tedious to generate diverse xylosides in a rapid fashion and utilize them in biological systems. We envisioned that synthesis of metabolically stable xylosides will advance our knowledge of glycosaminoglycan biosynthesis and how they regulate various pathophysiological processes.
In our earlier communication, we outlined a simple strategy, utilizing click chemical methodology that addresses the above limitations of O-xylosides, to generate a library of xylosides in a robust manner (35). Several studies have shown that the concentration of the primers and the aglycone moieties influence the composition of GAG chains produced (32). In the current study, we show that the aglycone moieties of click-xylosides may not only influence the composition and quantity of GAG chains but also the extent of sulfation, sulfation pattern, disaccharide composition, and chain length using pgsA-745 Chinese hamster ovary (CHO) cell line as a model cellular system. Our findings provide new insights in to the mechanism of GAG biosynthesis and offer new avenues to decipher the biological actions of GAG chains in model organisms.
EXPERIMENTAL PROCEDURES
Materials and Methods
The mutant CHO cell line (pgsA-745) was obtained from American Type Culture Collection (36). All cell culture reagents were obtained from HyClone. The radiochemical, [35S]Na2SO4 was purchased from MP Biomedicals, and d-[-6-3H]glucosamine and flow scintillation mixture, Ultima-FloAP, for Flow radiometric analysis were obtained from PerkinElmer Life Sciences. All other chemicals and biochemicals were obtained from Sigma. The click-xylosides, reported here, were synthesized as described previously (35). The 6× Pronase solution was prepared using Streptomyces griseus protease type XIV (1 mg/ml). The DEAE-Sepharose gel was purchased from Amersham Biosciences. The HPLC columns utilized were DEAE-3SW (7.5 mm × 7.5 cm, 10-μm particle size) for anion-exchange chromatography and G3000SWXL (7.8 mm × 30 cm, 5-μm particle size) for size-exclusion chromatography column, purchased from Tosoh Bioscience LLC. Commercially available unsaturated HS and CS standards were used to determine migration positions and elucidate disaccharide composition of primed GAG chains. Following standard unsaturated heparan sulfate disaccharides were purchased from Sigma-Aldrich: 2-acetamido-2-deoxy-4-O-(4-deoxy-α-l-threo-hexenepyranosyluronic acid)-d-glucose (ΔUA-GlcNAc), 2-deoxy-2-sulfamido-4-O-(4-deoxy-α-l-threo-hexenepyranosyluronic acid)-d-glucose (ΔUA-GlcNS), 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo-α- l-threo-hexenepyranosyluronic acid)-6-O-sulfo-d-glucose (ΔUA-GlcNS6S), 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo-α-l-threo-hexenepyranosyluronic acid)-d-glucose (ΔUA2S-GlcNS), and 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo- α-l-threo-hexenepyranosyluronic acid)-6-O-sulfo-d-glucose (ΔUA2S-GlcNS6S). CS disaccharide standards were also purchased for calibration. GAG disaccharide standards and 3H-labeled lyase disaccharide products were detected at 232 nm using inline UV detector and flow scintillation analyzer, respectively.
Screening of Click-xylosides in Cell Culture
The priming of the xylosides in xylosyl transferase-deficient CHO cell line pgsA-745 was performed as described in our earlier communication (35). Briefly, 1 × 105 cells were plated per well, containing the appropriate complete growth medium, in a 24-well plate and incubated at 37 °C in a humidified incubator for 24 h to reach a confluency of about 50%. The cells were then washed with sterile PBS and replaced with 450 μl appropriate medium containing 10% dialyzed FBS. A serial dilution of the primers at 100X the final concentration was prepared and 5 μl of appropriate 100X primer was added to various wells to yield a final concentration of 0.1 1, 10, 100 and 1000 μm respectively. 50 μCi of [35S]Na2SO4 or d-[6-3H]glucosamine was then added to each well for radiolabeling of the GAG chains. The 24-well plates were placed in the incubator for 24 h before the addition of 6X Pronase solution (100 μl) followed by incubation at 37 °C overnight.
Purification and Quantification of GAG Chains
After treating each well with Pronase solution overnight, the entire contents of the wells were transferred to a microcentrifuge tube and subjected to centrifugation at 16,000 × g for 5 min. The supernatant were transferred to a fresh tube and half-a-volume of 0.016% Triton X-100 was added. The diluted supernatant was loaded on to a DEAE-Sepharose column (0.2 ml) pre-equilibrated with 10 column volumes of wash buffer (20 mm NaOAc buffer, pH 6.0), containing 0.1 m NaCl and 0.01% Triton X-100) and the column was washed with 20 column volumes of wash buffer. The bound GAG was eluted with 6 column volumes of elution buffer (20 mm NaOAc (pH 6.0) containing 1 m NaCl). The amount of GAG chains primed by various click-xylosides was determined by quantifying the 35S- or 3H-radioactivity incorporated in the purified GAG eluate. 50 μl of the various eluates was diluted with 5 ml of scintillation mixture and triplicate samples were measured using a scintillation counter for total radioactivity.
Analysis of GAG Chains by Anion-exchange Chromatography
The homogeneity and extent of sulfation of the GAG chains synthesized on the various primers were determined by measuring migration time on anion-exchange column using high-pressure liquid chromatography (HPLC) with inline flow scintillation analyzer. 25 μl of eluate was diluted 10-fold with 10 mm KH2PO4 (pH 6.0) containing 0.2% CHAPS, and loaded on to a HPLC-DEAE column and eluted with a linear NaCl gradient of 0.2 m to 1 m over 80 min.
Extent of GAG Sulfation in the Presence of Chlorate
Confluent 745-CHO cell cultures containing [3H]GlcNH2 were treated with xyloside 5 (100 μm) in the presence sodium chlorate at various concentrations. GAG chains were recovered from the conditioned media by DEAE-Sepharose chromatography, followed by isolation of [3H]labeled GAG as described in the previous section. To monitor the effect of chlorate concentration on GAG sulfation, samples of [3H]GAG from control and chlorate-treated cells were analyzed on an anion-exchange HPLC column as described in the previous section.
Co-priming of Two Different Xylosides in Cell Culture
The co-priming of the xylosides was performed as described above in the “screening of click-xylosides section” by adding two different xylosides to xylosyl transferase-deficient CHO cells in the same well of a 24-well plate. Briefly, a solution containing two xylosides, 22 and 24, at 100× the final concentration was prepared, and 5 μl of the premixed xylosides was added to yield a final concentration of 100 μm for each xyloside. 50 μCi of [35S]Na2SO4 or d-[6-3H]glucosamine was then added to each well for radiolabeling the GAG chains synthesized. The 24-well plates were placed in the incubator for 24 h before the addition of 6X Pronase solution (100 μl) followed by incubation at 37 °C overnight. The GAG chains were then purified from the solution as described in the earlier section.
Analysis of GAG Chains by Size-exclusion Chromatography
The chain length of the GAG chains synthesized on various primers was determined by measuring their migration time on size exclusion column using HPLC with inline flow scintillation analyzer. The GAG chains were loaded on to two G3000SWXL columns (Tosoh, 7.8 mm × 30 cm), which were connected in tandem, and eluted over 60 min with phosphate buffer (100 mm KH2PO4, 100 mm NaCl, pH 6). The migration time of the GAG chains was determined by measuring the average of peak-width at half maximum, and the average molecular weight was calculated based on the migration time of polystyrene sulfonate standards.
Analysis of GAG Disaccharide Composition
To determine the disaccharide composition of GAG, the total GAG chains were fractionated using size exclusion chromatography as described above and the fractions containing the GAG chains were pooled and dialyzed using a benzoylated dialysis membrane with molecular mass 2000 Da in 10 mm ammonium bicarbonate. To concentrate and remove volatile ammonium bicarbonate, dialyzed GAG samples were placed in a speed-vac concentration system under reduced pressure. The HS in the total GAG was digested with heparitinases I, II, and III overnight at 37 °C and analyzed using strong anion-exchange HPLC using a Phenosphere SAX column (Phenomenex, 250 × 4.6 mm) with inline radiodetection. Solvent A was double-distilled water (pH 3.5). Solvent B was 2 m NaCl (pH 3.5). The disaccharides were eluted at a flow rate of 1 ml/min with the following profile: 100% solvent A for 5 min; linear gradient of 0 to 40% solvent B for 35 min; 100% solvent B for 10 min and finally 100% solvent A for 25 min to equilibrate the column. In a similar fashion, to determine the disaccharide composition of xyloside-primed CS and DS chains, the GAG chains were digested with chondroitinase ABC (ACCI) overnight at 37 °C, and the resulting disaccharides were analyzed as described above.
RESULTS
Priming Activity of Click-xylosides
We have previously reported the synthesis of the click-xylosides and have shown that these molecules prime GAG chains in a cell line that lacks production of endogenous GAG chains due to deficiency of a key enzyme, xylosyl transferase, involved in the biosynthesis of proteoglycans (35). This synthetic methodology allowed us to construct a library of novel xylosides with a wide variety of aglycone moieties in a rapid manner. In this study, we have synthesized additional compounds and expanded the xyloside library to probe the effect of aglycones on the priming ability (Table 1). At the outset, we have examined whether the concentration of xylosides would affect the priming ability and determined the optimal primer concentration of each xyloside for the maximal production of GAG chains.
TABLE 1.
Structures of click-xylosides
A library of metabolically stable xylosides containing a variety of hydrophobic groups was synthesized using click-chemistry (Ref. 35 for details). This library was employed in the current study to elucidate the mechanism of glycosaminoglycan biosynthesis.
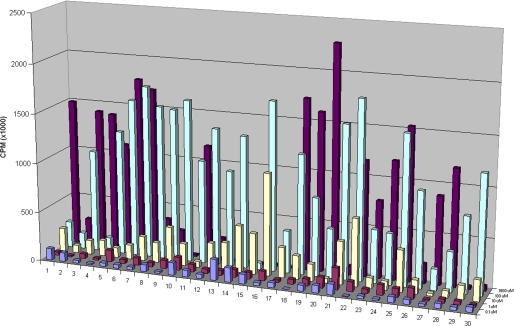
The priming ability of the xylosides was determined as described under “Experimental Procedures” utilizing the mutant CHO cell line (pgsA-745) at the following five concentrations: 0.1 μm, 1 μm, 10 μm, 100 μm, and 1 mm. The results indicate that the priming is concentration-dependent, and significant priming for most xylosides was observed at a concentration of 100 μm or higher (Fig. 1). From the screening experiments with the above concentrations, we were able to determine the optimal concentration range for each xyloside for its maximal priming activity. For a selected group of xylosides, experiments were performed at intermediate concentrations, which showed that click-xylosides can act as acceptors effectively at concentrations as low as 30 μm (data not shown). At 1 mm concentration, the triazolyl xyloside 1, which has no hydrophobic aglycone attached to the triazole group, was able to participate in GAG synthesis, whereas the addition of a hydrophilic -CH2OH group, as shown in xyloside 2, leads to dramatic decrease in the priming activity (Fig. 1). However, the priming activity of hydrophobic xylosides 12, 13, 14, 15, 16, and 17 was higher at 100 μm concentration than at 1 mm concentration (Fig. 1).
FIGURE 1.
Priming activity of various xylosides. The novel xylosides were examined for their priming ability using xylosyl transferase-deficient CHO cells (pgsA-745). 100,000 cells were seeded per well of 24-well plates and treated with various xylosides at 0.1, 1, 10, and 100 μm and 1 mm concentrations for 24 h in the presence of 50 μCi of [35S]O42− or d-[6-3H]glucosamine. The GAG chains were then purified and quantified as described under “Experimental Procedures.” Xylosides with different aglycones primed different amounts of GAG chains at different concentrations suggesting the decisive role played by aglycones. The data indicate the average of three independent experiments.
Effect of Substituents on Priming
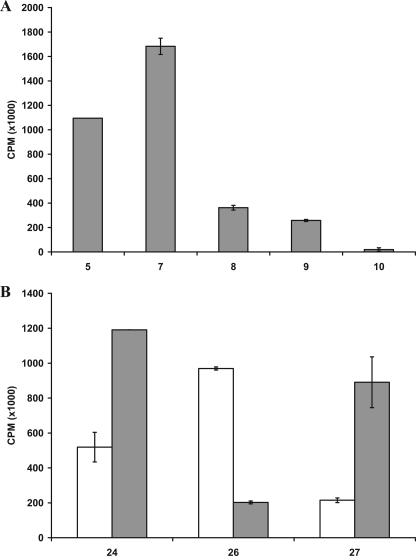
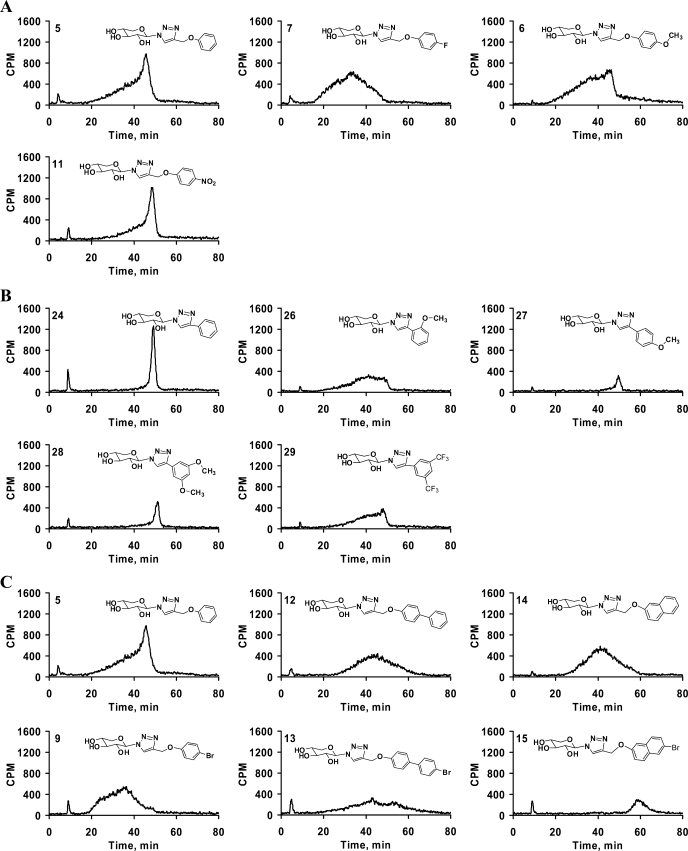
The role of various substituent groups attached to the phenyl aglycone moiety of the xyloside modulating GAG biosynthesis was assessed. Substituents such as -OCH3 group in 6 and the -NO2 group in 11 have a marginal effect on the priming activity at 100 μm concentration. We also found that these substitutions do not have any influence at higher concentrations (Fig. 1). The analysis of the halogen substitutions, fluoro-, chloro-, bromo-, and iodo-substituents in 7, 8, 9, and 10, respectively, show no tangible effect in the amount of GAG chains produced when examined at 100 μm concentration (Fig. 1). Interestingly, the amount of GAG chains primed decreases with increasing atomic weight of the halogen substitutions at 1 mm concentration (Fig. 2A).
FIGURE 2.
Effect of substituents and their positions on priming activity. Cells were treated with various xylosides for 24 h in the presence of 50 μCi of [35S]O42− or d-[6-3H]glucosamine as described under “Experimental Procedures.” Primed GAG chains were then purified from the supernatant using anion-exchange column chromatography and quantified using liquid scintillation. A, effect of various halogen substituents on the priming activity was examined. Fluoro-, chloro-, bromo-, and iodo-substituted xylosides (7, 8, 9, and 10, respectively) were compared against the unsubstituted xyloside 5 at 1 mm concentration. B, effect of methoxy-substituent and its position on the phenyl ring was examined for priming activity. Unsubstituted xyloside 24 was compared with ortho-substituted xyloside 26 and para-substituted xyloside 27 at 100 μm (unshaded) and 1 mm (shaded) concentrations. The data indicate the average of three independent experiments.
Effect of Substituent Positions on Priming Activity
Three xylosides, 24, 26, and 27, were synthesized to investigate the effect of the position of -OCH3 substituent on GAG biosynthesis. At 100 μm, xyloside 26 (ortho) primed nearly two times the GAG chains compared with the unsubstituted xyloside 24, whereas xyloside 27 (para) primed 50% to that of unsubstituted xyloside 24. Interestingly, the para-substituted xyloside primed three times more GAG chains than the ortho-substituted xyloside at 1 mm concentration. Thus, the position of substituent has a dramatic influence on the priming ability of the given xyloside even though the activity trend is different at different concentrations (Fig. 2B).
Extent of Sulfation and Migration Time
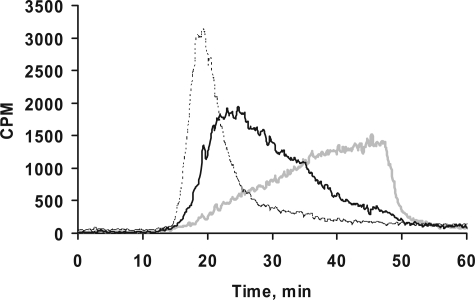
We utilized DEAE anion-exchange chromatography to compare the extent of sulfation of primed GAG chains. However, there are no commercially available GAG standards with various extent of sulfation to correlate the migration time and extent of sulfation. Therefore, we generated GAG chains with different extent of sulfation by incubating xyloside 5 with cells in the presence of chlorate, an inhibitor of 3′-phosphoadenosine 5′-phosphosulfate synthetase that affects GAG sulfation, at various concentrations (37, 38). pgsA-745-CHO cells were treated with 100 μm xyloside 5 and metabolically radiolabeled with [3H]GlcNH2 in the presence of various sodium chlorate concentrations, 1 mm, 5 mm, and 25 mm. GAG chains were recovered from the conditioned media by DEAE-Sepharose chromatography, followed by isolation of [3H]GAG chains as described under “Experimental Procedures.” To monitor the effect of chlorate concentration on GAG sulfation, samples of [3H]GAG from control and chlorate-treated cells were analyzed by an anion-exchange HPLC column chromatography and subsequently analyzed for radioactivity with the aid of inline flow scintillation analyzer. The GAG chains were quantitatively retained by the column, but they required different concentrations of NaCl for their elution (Fig. 3). These findings served as a basis to attribute the differences in the elution profile/migration time of various GAG chains on the HPLC anion-exchange column to the extent of sulfation of GAG chains primed by various xylosides as described. The chain length of the GAG chains can also affect the migration time albeit to a lesser extent than sulfation.
FIGURE 3.
Correlation between extent of sulfation and migration time on HPLC anion-exchange chromatography. Cells containing d-[6-3H]glucosamine were treated with 100 μm xyloside 5 in the presence of chlorate at various concentrations. GAG chains were then isolated and quantified as described under “Experimental Procedures.” The chlorate treatment did not affect the amount of GAG chains produced. 1,000,000 cpm were applied to HPLC DEAE anion-exchange chromatography and eluted with a linear NaCl gradient as described under “Experimental Procedures.” GAG species from chlorate-treated cells were eluted earlier than those from control cells. The elution profiles of GAG chains from control cells (gray tracer), cells treated with 5 mm chlorate (dark tracer), and cells treated with 25 mm chlorate (broken tracer) are shown. The elution profiles are representative of at least two independent experiments.
Comparison of Elution Profiles of GAG Chains
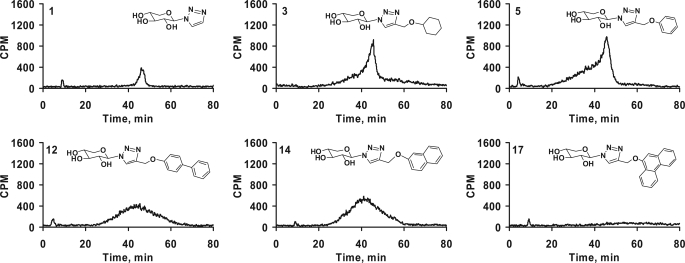
We focused our investigation on HPLC analysis of the GAG chains primed by the exogenous xylosides at the optimal concentration (100 μm). Using anion-exchange HPLC, we compared the elution profile of xyloside primed GAG chains, which is an indicator of extent of sulfation. GAG chains that have less sulfation elute at lower ionic strength, whereas heavily sulfated GAG chains elute at higher ionic strength. First, we compared the HPLC profiles of GAG chains primed by xylosides of the simplest form, xylose-linked to triazole only (1), cyclohexyl (3), or phenyl (5) as shown in Fig. 5. The GAG chains synthesized from the above compounds showed a very similar profile although the quantities of the GAG chains were different for each xyloside. They showed a predominantly homogenous peak along with a nonseparable highly heterogeneous minor portion eluting at lower salt concentration. Xylosides that differ in their aglycone aromatic structure, 5 (phenyl), 12 (biphenyl), 14 (naphthyl), and 17 (phenanthryl), were analyzed for the extent of sulfation by using the DEAE column. GAG chains primed by xyloside 5 eluted from 20 to 52 min with a peak maximum at 45 min, whereas GAG chains primed by 12, 14, and 17 were eluted with an even distribution from 20 to 60 min and with a peak maximum of ∼42–44 min (Fig. 4). The differences in the elution profiles of the primed GAG chains indicate differences in the extent of sulfation among these GAG chains. The phenanthrene containing xyloside 17 primed fewer GAG chains. It may possibly be due to the inability of the enzymes to recognize a highly hydrophobic xyloside even though it may cross cell and Golgi membranes more effectively, or alternatively this may reflect the inability of the xyloside 17 to reach the enzyme active site.
FIGURE 5.
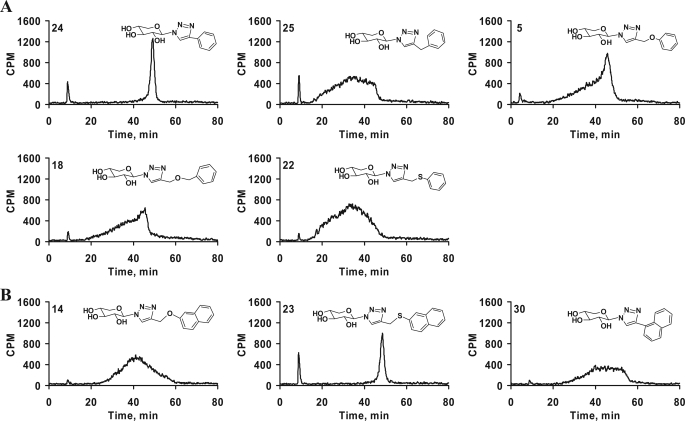
Spacers between the triazole and aromatic ring in the aglycone moiety alter the sulfation pattern and extent of sulfation. Purified GAG chains were diluted 5-fold and analyzed by anion-exchange HPLC column as described under “Experimental Procedures.” A gradient of 0.2 to 1.0 m NaCl for 80 min was used for elution of the GAG chains. The differential elution profile and migration time indicates variations in sulfation pattern and extent of sulfation of GAG chains primed by phenyl group containing xylosides 5, 18, 22, 24, and 25 (A) and naphthyl group containing xylosides 14, 23, and 30 having various spacers were examined (B). The elution profiles are representative of at least two independent experiments.
FIGURE 4.
Effect of various hydrophobic moieties on the DEAE elution profile of GAG chains. GAG chains primed by various xylosides (1, 3, 5, 12, 14, and 17) eluted from small anion-exchange column with 1 m NaCl, were diluted 5-fold and analyzed by anion-exchange HPLC column as described under “Experimental Procedures.” The variations in the elution profiles and migration times indicate differences in both sulfation pattern and extent of sulfation of the primed GAG chains. The elution profiles are representative of at least two independent experiments.
Effect of Spacer on Sulfation Patterns of GAG Chains
The xylosides were attached to the phenyl moiety with many different spacers to confer conformational flexibility and were then examined for their influence on priming activity. The first set of compounds that we compared differs by spacer length. The phenyl ring was linked directly to the triazole in 24 or through the spacers such as -CH2- in 25, -CH2-O- in 5, -CH2-O-CH2- in 18, or -CH2-S- in 22. The elution profiles of the GAG chains primed by these xylosides are shown in Fig. 5A. It was interesting to find that the direct linking of a phenyl group in xyloside 24 led to the synthesis of GAG chains that were highly homogenous and eluted as a very narrow single peak at 48 min. Introducing one carbon unit (-CH2-) between the phenyl and triazole groups in 25 led to loss of the homogeneity of GAG chains synthesized, and these eluted between 18 and 45 min. Introduction of various other spacers such as -CH2-O- in 5, -CH2-O-CH2- in 18, and -CH2-S- in 22 resulted in the production of GAG chains with different elution profiles and failed to re-establish the narrow peak that was observed for the xyloside 24 (Fig. 5A). In contrast to xyloside 24, xyloside 30 that has a naphthyl group directly attached to triazole produced GAG chains with broad elution profile. When the naphthyl group was separated from the triazole group by a -CH2-S- spacer (23) narrow GAG chains production was restored. However, xyloside 14 that has the -CH2-O- spacer produced heterogeneous GAG chains with a broad elution profile (Fig. 5B). This study has revealed that conformational flexibility likely influences the biosynthesis of GAG chains with differential sulfation pattern.
The GAG chains primed by compounds 5, 6 (-OCH3), 7 (fluoro), 8 (chloro), 9 (bromo), 10 (iodo), and 11 (-NO2) were analyzed by HPLC to determine the effect of various substituents on the extent of GAG sulfation. The halogen substituents significantly altered the extent of sulfation causing an increase in the heterogeneity of the GAG chains with little difference among the halogen containing xylosides (Fig. 6A; data not shown for xylosides 8–10). It was interesting to note that halogen substitution resulted in the production of GAG chains that elute earlier from the DEAE column than those from the xyloside 5. The xylosides containing -OCH3 group in 6 and -NO2 group in 11 primed GAG chains that had different elution profiles and reaffirmed that the type of substituent influences the GAG biosynthetic pathways (Fig. 6A). All of the above substituents were located at the para position of the phenyl ring. We then examined whether the position of substituent influences the sulfation patterns of the GAG chains. We examined the sulfation patterns of the GAG chains produced by xylosides that were substituted at various positions on the phenyl ring with the -OCH3 group. The xylosides with a single substituent at the ortho position in 26 and at the para position in 27 resulted in different elution profiles for the primed GAG chains (Fig. 6B). The para-substituted xyloside 27 primed homogenous GAG chains that eluted at 50 min, albeit in lower quantity. On the other hand, the GAG chains primed by the ortho-substituted xyloside 26 eluted between 20 and 50 min indicating these chains have various sulfation patterns (Fig. 6B). The highly electron withdrawing substituent in the place of -OCF3 29 also resulted in broad GAG chains. In addition, we investigated the effect of the same substituent on different aglycone moieties by comparing the bromo substitution of phenyl moiety in 9, biphenyl moiety in 13, and naphthyl moiety in 15. We had observed that a bromo substitution on the phenyl ring led to a decrease in the extent of sulfation of GAG chains, whereas bromo substitution on biphenyl and naphthyl groups in the aglycone moieties led to a significant increase in the extent of sulfation (Fig. 6C).
FIGURE 6.
Effect of substituents and their positions on GAG fine structures. Purified GAG chains were diluted 5-fold and analyzed by HPLC DEAE anion-exchange chromatography as described under “Experimental Procedures.” The differential elution profiles and migration times indicate variations in sulfation pattern and extent of sulfation. A, effect of various substituents at the para position of the phenyl ring was examined. The elution suggests that different substituents on xylosides, 5, 6, 7, and 11 affect the elution profile of the GAG chains suggesting changes in the fine structures. B, influence of position of the substituent on the fine structures was examined. Unsubstituted xyloside 24 primed GAG chains that elute as a narrow peak, whereas ortho methoxy-substituted xyloside 26 produced GAG chains that elute as a broad peak. On the other hand, para-substituted xyloside 27 and dimethoxy-substituted xyloside 28 produce GAG chains that elute as narrow peaks. C, bromo-substitution on phenyl, biphenyl, and naphthyl moieties was found to affect the extent of sulfation and sulfation pattern. Unsubstituted xylosides 5, 12, and 14 were compared with their corresponding bromo-substituted xylosides 9, 13, and 15 for the change in the extent of sulfation and sulfation pattern. The elution profiles are representative of at least two independent experiments.
Co-priming Experiment
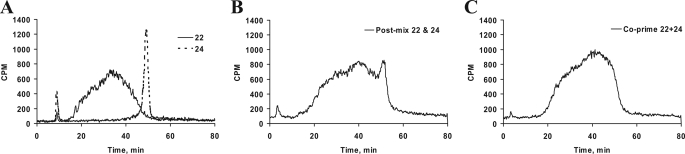
Investigation of the extent of sulfation and the sulfation pattern of the GAG chains that were synthesized by various xylosides showed distinct elution profile on anion-exchange column. These differences among GAG chains primed by xylosides are attributed to the presence of different aglycones on these xylosides. Hence, it is imperative that we understand the molecular mechanisms that can generate such differential priming of GAG chains. Xylosides 22 and 24 primed GAG chains with different profiles in anion-exchange chromatography. Xyloside 22 stimulates GAG chains that elute as a narrow peak with high sulfate density, whereas xyloside 24 produces GAG chains that elute as a broad peak with less sulfate density. To determine whether distinct GAG chains can be primed by cells at the same time, xylosides 22 and 24, were co-primed by addition to the same well, and their anion-exchange profiles were compared with individually primed GAG chains. The resulting GAG chains were isolated and analyzed by anion-exchange chromatography (Fig. 7). The individually primed GAG chains have distinctive elution profiles due to differences in their extent of sulfation (Fig. 7A). Also, an analysis was performed using GAG chains that were primed individually but mixed together before the anion-exchange chromatography. The mixing of individually primed GAG chains results in less resolution of the peaks that have otherwise very distinct profile (Fig. 7B). On the other hand, GAG chains that were co-primed in the presence of 22 and 24 show a very broad peak due to overlap of the individual peaks. The slope of the front of peak increases gradually similar to that observed for GAG chains from 22 while the tail of the peak drops off rather sharply comparable to that of GAG chains primed by 24 (Fig. 7C). This suggests that the GAG chains with different extent of sulfation were primed by two xylosides that were co-primed even though the peaks of the different GAG chains cannot be completely resolved using anion-exchange chromatography.
FIGURE 7.
Co-priming experiment with xylosides that prime distinct GAG chains. A, xylosides 22 and 24 were primed at 100 μm concentration in pgsA-745 cells for 24 h individually, and GAG chains were purified. Individually primed GAG chains were then analyzed on HPLC DEAE anion-exchange chromatography under similar conditions in separate runs. The solid tracer indicates GAG chains primed by xyloside 22, and the broken tracer indicates the narrow elution profiles of the GAG chains primed by xyloside 24. B, individually primed GAG chains from xylosides 22 and 24 were mixed together and analyzed under similar conditions as described in A. C, 100 μm each of xylosides 22 and 24 were primed together for 24 h in cell culture experiments. The resulting GAG chains were purified and analyzed by DEAE anion-exchange chromatography as described earlier. The elution profile suggests further loss of resolution of two distinct GAG chains when xylosides 22 and 24 are co-primed. The elution profiles are representative of at least two independent experiments.
Chain Length Analysis of GAG Chains
To examine the effect of various aglycones on GAG chain length, primed GAG chains were analyzed by size-exclusion chromatography as described under “Experimental Procedures.” GAG chains had an average molecular weight in the range of 6,000 to 34,000 (Table 2). A significant number of xylosides in this study primed GAG chains that have molecular weight greater than 10,000, whereas some generated GAG chains with lower molecular weight. Only a few studies report chain length analysis for xyloside-primed GAG chains (39, 40). In our study, most of the click-xylosides prime significantly long GAG chains. Xylosides that predominantly prime HS chains, 5 and 22, have shorter chain lengths in the range of 6,000 to 12,000. Also, heterogeneous GAG chains with different extent of sulfation observed from the DEAE elution profile have shorter chain lengths. Significantly, GAG chains that have narrow elution profile with high extent of sulfation, primed by 19, 20, 23, and 24, have higher molecular weights (Table 2).
TABLE 2.
The molecular mass of GAG chains primed by various xylosides
The molecular mass of the GAG chains synthesized on various primers was determined by measuring their migration time on size-exclusion column as described under “Experimental Procedures.” The average migration time was determined using peak-width at half-maximum. The average molecular mass (MM) was calculated using the migration time in comparison to that of polystyrene sulfonate standards performed under similar conditions.
| Xyloside | Average MM | Kav |
|---|---|---|
| Da | ||
| 3 | 21,600 | 0.33 |
| 5 | 10,500 | 0.50 |
| 6 | 10,500 | 0.50 |
| 7 | 7,800 | 0.57 |
| 8 | 8,700 | 0.54 |
| 11 | 31,000 | 0.25 |
| 18 | 15,000 | 0.42 |
| 19 | 33,900 | 0.23 |
| 20 | 33,900 | 0.23 |
| 22 | 5,800 | 0.65 |
| 23 | 31,000 | 0.25 |
| 24 | 25,800 | 0.29 |
| 25 | 11,100 | 0.49 |
| 27 | 37,100 | 0.21 |
| 29 | 23,600 | 0.31 |
Disaccharide Composition of GAG Chains
The influence of various aglycones on the disaccharide compositions of GAG chains was not rigorously examined in earlier studies. To determine the GAG disaccharide composition, we chose various xylosides that prime HS (5 and 22), DS (19 and 20), and CS (7 and 14). To determine disaccharide composition, the purified GAG chains were digested with heparitinases I, II, and III, or chondroitinase ABC lyase, and analyzed using a strong anion-exchange HPLC column as described under “Experimental Procedures.” Radiolabeled disaccharides were identified by comparison of their elution positions relative to those of disaccharide standards. Disaccharide analysis revealed the following HS disaccharides in HS chains obtained from xylosides: ΔUA-GlcNAc, ΔUA-GlcNS, ΔUA-GlcNS(6S), ΔUA(2S)-GlcNS, ΔUA(2S)-GlcNAc(6S), and ΔUA(2S)-GlcNS(6S), see supplemental Figs. S3A and S3B. Disaccharide analysis of xyloside-primed DS and CS showed two disaccharides, ΔUA-GalNAc and ΔUA-GalNAc(6S). Xyloside-primed CS chains contained ∼50% unsulfated disaccharides, whereas xyloside-primed DS chains contained >75% 6-O-sulfated disaccharide (see supplemental Figs. S4A, S4B, S5A, and S5B). These results suggest that aglycones may aid in the selective transport of xylosides to different Golgi compartments that have different combinations of biosynthetic enzymes and isoforms resulting in the generation of GAG chains with distinct sulfation patterns.
DISCUSSION
Most of the previous investigations used O-xylosides and a few S-xylosides, which are susceptible to degradation, to determine their ability to prime GAG chains with the exception of two studies that have examined stable C-xylosides (29, 41). We have synthesized metabolically stable click-xylosides, using simple click chemistry. We have also determined that these click-xylosides can continuously prime GAG chains for at least 5 days suggesting their stability (see supplemental Fig. S6).
The priming activity of the xylosides in this study shows that most of the primers generate significant quantity of GAG chains, although a few are not effective primers. One possible explanation is that the diffusion rates of the primers depend on the aglycone and lead to differential biosynthesis of the GAG chains. The biosynthesis of GAG chains is a very complex process as the priming activity depends on many enzymatic reactions in the synthesis of the linkage region as well as polymerization. Therefore, we carefully examined the priming activity of various xylosides at different concentrations. It is interesting to note that the xyloside 15, containing bromonaphthyl aglycone moiety, primed more effectively at 10 μm than it did at 100 μm. If diffusion is the only factor that governs priming activity, the priming activity would be higher at 100 μm. In a similar manner, the xylosides 26 and 29 primed very effectively at 100 μm but primed few GAG chains at 1 mm. We also found that naphthyl xyloside 30 primed better at 100 μm than it did at the 1 mm concentration. This inhibition of priming at higher concentrations of xylosides might possibly be due to substrate level inhibition of enzymes involved assembly of linkage region that would lead to reduction in the amount of GAG chains primed. These data clearly suggest that differential priming activity among various xylosides does not solely depend on the diffusion but also on various factors that are influenced by the aglycones.
Several in vitro studies earlier examined the various characteristics of the GAG chains primed by xylosides such as extent of sulfation and the sulfation pattern using anion-exchange chromatography (39, 42), the chain lengths (39), HS/CS composition (32), and disaccharide composition (32). All of these studies have utilized a single xyloside and compared the xyloside-primed GAG chains to that of endogenous GAG chains. It is known that xylosides tend to make 5–20 times more GAG chains than endogenous core proteins and, therefore, are expected to see the differences between the endogenous and xyloside-primed GAG chains (43, 44). On the other hand, in this study we have exhaustively analyzed the GAG chains stimulated by the click-xylosides for their priming activity, sulfation pattern, extent of sulfation, chain lengths, and disaccharide profile and compared the above characteristics as a function of the aglycone moieties.
Influence of Aglycone on Fine Structures
Our experimental data have shown that most xylosides with aromatic aglycone moieties are good primers of GAG chains. The quantity of total GAG chains, HS/CS/DS composition, extent of sulfation, and sulfation pattern were distinct for each xyloside in our study. The priming ability of the synthetic xylosides depends not only on the aglycone (Fig. 4), type of aromatic ring (Fig. 4), and substitutions (Figs. 6), but also on the distance between the xylose and aglycone (Fig. 5) in addition to the hydrophilic nature of the spacer (Fig. 5). It is intriguing that the wide variety of xylosides primed GAG chains with enormous diversity in their HS/CS/DS composition and the chain length. It is also notable that xyloside-primed HS chains have subtle differences in their disaccharide composition. We plan to examine the fine structure of these HS chains using state-of-the-art techniques, nuclear magnetic resonance and mass spectrometry, to further decipher the difference between the HS chains stimulated by the corresponding xyloside.
Existence of GAGOSOMES
GAG biosynthesis is a multistep process, and the sequence of biosynthetic events is somewhat resolved using reconstituted systems such as recombinant enzymes and microsomal fractions (45–48). However, the factors that regulate the emergence of cell-specific GAG chains with distinct sulfation patterns remain largely unknown. It has been proposed that glycosaminoglycans are synthesized in the Golgi by two different widely debated mechanisms (6, 49, 50). In one mechanism, most enzymes are anchored to the Golgi membrane at different locations and randomly modify the nascent chains resulting in diverse structures. In the second proposed mechanism, enzymes are predicted to be physically interacting within a macromolecular complex called GAGOSOME and concurrently coordinate elongation, epimerization, and sulfation (6). The second mechanism requires channeling of substrates so that the chains are assembled with specific sulfation patterns. Evidence for the first model comes from the fact that GAG chains isolated from a specific cell type or a specific tissue are always found to be polydisperse in nature. This is a more relaxed model that can account for emergence of diverse structures from a given cell. There are various evidences available that also suggest the existence of GAGOSOMES as proposed in the second model. For example, ext1 and ext2 are found to be more active when they are co-expressed than either is expressed alone (51–54). Recent studies have also shown that the relative concentrations of ext1, ext2, and NDST1 influence the sulfation pattern of HS (55). In addition, HS C-5 epimerase and 2-OST are shown to be co-localized in the medial Golgi (56). However, there is no direct evidence for interaction among all of these enzymes.
In this study, we observed a wide variation in the sulfation patterns of GAG chains primed by various xylosides. These variations in the sulfation patterns should be attributed to the presence of discrete enzyme complexes in different Golgi sub-compartments that may differentially regulate the biosynthesis of GAG chains. Several proteoglycans have been found to harbor a conserved motif. A chimeric core protein containing this conserved motif was able to regulate the level of the epimerization of d-glucuronosyl residues to l-iduronosyl residues (57). Thus, this conserved motif is suggested to direct the chimeric protein to a specific sub-cellular compartment, enriched with specific enzymes that can lead to the differential modification of the GAG chains. In a similar fashion, we anticipate that the characteristics of aglycone moieties may dictate the localization of different primers to specific sub-compartments, binding differentially to the biosynthetic enzymes, to generate GAG chains with distinct sulfation patterns or specific type of GAG chain (HS, CS, or DS). In our study, some click-xylosides, 5 and 22, stimulated predominantly HS (>90%) as determined by their susceptibility to digestion with heparitinases I, II, and III. This is the first report to shows that HS can be synthesized exclusively on an exogenous GAG acceptor. In contrast, xylosides 7, 8, 14, and 25 synthesized up to 80% CS, whereas xylosides 19, 20, 23, and 24, exclusively synthesized DS chains that elute as sharp narrow peaks in anion-exchange column. We have analyzed these GAG chains that elute as narrow peaks by digestion of GAG chains with chondroitinase B enzyme that selectively cleaves GalNAcβ(1–4)IdoA linkage and determined these narrow peaks are predominantly DS (Fig. 8). Interestingly, the DS chains that elute as narrow peaks have fewer unsulfated disaccharides. In contrast, CS chains that elute as broad peaks have a significant amount of unsulfated disaccharides (see supplemental Figs. S4 and S5). We surmise that these xylosides may be preferentially partitioned into a specific sub-compartment that results in the synthesis of DS. Thus, selective compartmentalization of certain xylosides would likely result in the synthesis of homogenous and distinct populations of the GAG chains. However, other xylosides may be targeted to more than one sub-cellular compartment resulting in the production of heterogeneous GAG chains with different composition of HS/CS/DS, chain length, and disaccharide composition. A number of modifications have been identified in the linkage region of the proteoglycans that may determine the type of GAG chain synthesized (58–63). These modifications may be selectively made on certain xylosides, which have different aglycones, leading to the dramatic change in the synthesis of specific GAG chains. However, the influence of these modifications in the selective transport of linkage oligosaccharides to specific compartments that may result in unique sulfation pattern is not known. We plan to examine in detail the type of modifications found in the linkage regions of GAG chains primed by these xylosides and their effect on the class switching in addition to specific sulfation patterns.
FIGURE 8.
Chondroitinase B treatment of GAG chains primed by xyloside 20. Xyloside 20 was primed at 100 μm concentration in pgsA-745 cells for 24 h. The GAG chains were then purified as described under “Experimental Procedures.” The purified GAG chains were digested with chondroitinase B, analyzed by HPLC DEAE anion-exchange chromatography, and compared with the undigested GAG chains. The elution profiles of the undigested (broken tracer) and digested (solid tracer) GAG chains are shown.
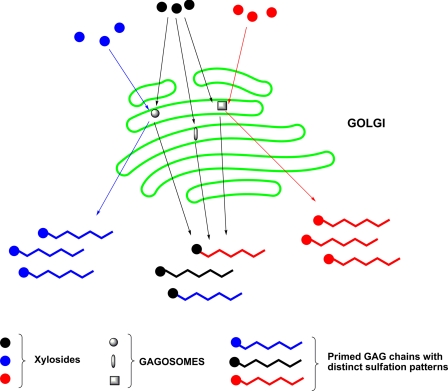
Our findings provide further suggestive evidence for presence of GAGOSOMES that regulate the generation of cell-specific combinatorial structures that are often erroneously referred to as heterogeneous structures. We surmise that these xylosides with different aglycones can either selectively partition into a specific GAGOSOME or be recognized differentially by various GAGOSOMES to generate distinct GAG structures as observed in this study (Fig. 9). Furthermore, the extent of diversity of GAG structures synthesized in a given cell is likely influenced by the number and composition of GAGOSOMES. These enzyme complexes themselves may also be spatio-temporally regulated both at the transcriptional and translational level and are thus expected to dynamically synthesize various unique GAG chains throughout the lifetime of the cell.
FIGURE 9.
Regulation of GAG biosynthesis by GAGOSOME model. Each GAGOSOME can have different combination of enzymes that generate cell-specific combinatorial GAG structures with differential sulfation pattern required for binding to diverse proteins. While some xylosides (blue and red) are selectively primed by a specific GAGOSOME to generate distinct fine structures, other xylosides (black) are promiscuously primed by more than one GAGOSOME resulting in heterogeneous GAG chains.
In conclusion, we have demonstrated for the first time that the aglycone moieties of the xylosides influence sulfation pattern, extent of sulfation, chain length, disaccharide composition, and type of GAG chains. These findings compel us to propose a GAGOSOME model for the GAG biosynthesis. Therefore, we predict that these xylosides would become an important glycobiological tool to diligently probe how the status of the cell would affect the biosynthetic machinery, GAGOSOMES, and also to probe cell-specific dynamic changes both in fine structures and amounts of GAG chains produced during and beyond the developmental stages of an organism.
Supplementary Material
Acknowledgment
We thank Prof. Darrell R. Davis for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM075168 (to B. K.). This work was also supported by Human Frontiers Science Program (to B. K.) and the Vietnam Education Fund (to T. K. N. N. and K. V. N.).
This work is dedicated to Prof. Robert D. Rosenberg.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- CHO
- Chinese hamster ovary
- HPLC
- high-pressure liquid chromatography.
REFERENCES
- 1.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 2.Lander A. D. (1993) Curr. Opin. Neurobiol. 3, 716–723 [DOI] [PubMed] [Google Scholar]
- 3.Capila I., Linhardt R. J. (2002) Angew. Chem. Int. Ed. Engl. 41, 391–412 [DOI] [PubMed] [Google Scholar]
- 4.Powell A. K., Yates E. A., Fernig D. G., Turnbull J. E. (2004) Glycobiology 14, 17R–30R [DOI] [PubMed] [Google Scholar]
- 5.Salmivirta M., Lidholt K., Lindahl U. (1996) FASEB J. 10, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 6.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 7.Lindahl U., Cifonelli J. A., Lindahl B., Roden L. (1965) J. Biol. Chem. 240, 2817–2820 [PubMed] [Google Scholar]
- 8.Bourdon M. A., Krusius T., Campbell S., Schwartz N. B., Ruoslahti E. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3194–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muir H. (1958) Biochem. J. 69, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory J. D., Laurent T. C., Roden L. (1964) J. Biol. Chem. 239, 3312–3320 [PubMed] [Google Scholar]
- 11.Vertel B. M., Walters L. M., Flay N., Kearns A. E., Schwartz N. B. (1993) J. Biol. Chem. 268, 11105–11112 [PubMed] [Google Scholar]
- 12.Lohmander L. S., Hascall V. C., Yanagishita M., Kuettner K. E., Kimura J. H. (1986) Arch. Biochem. Biophys. 250, 211–227 [DOI] [PubMed] [Google Scholar]
- 13.Lohmander L. S., Shinomura T., Hascall V. C., Kimura J. H. (1989) J. Biol. Chem. 264, 18775–18780 [PubMed] [Google Scholar]
- 14.Lindahl U., Roden L. (1965) J. Biol. Chem. 240, 2821–2826 [PubMed] [Google Scholar]
- 15.Kuberan B., Lech M., Borjigin J., Rosenberg R. D. (2004) J. Biol. Chem. 279, 5053–5054 [DOI] [PubMed] [Google Scholar]
- 16.Sugumaran G., Katsman M., Silbert J. E. (1998) Biochem. J. 329, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt J. L., Brown D. M., Granlund K., Oegema T. R., Klein D. J. (1987) Dev. Biol. 123, 293–306 [DOI] [PubMed] [Google Scholar]
- 18.Yost H. J. (1990) Development 110, 865–874 [DOI] [PubMed] [Google Scholar]
- 19.Rapraeger A. (1989) J. Cell Biol. 109, 2509–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanwar Y. S., Hascall V. C., Jakubowski M. L., Gibbons J. T. (1984) J. Cell Biol. 99, 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopwood J. J., Dorfman A. (1977) J. Biol. Chem. 252, 4777–4785 [PubMed] [Google Scholar]
- 22.Okayama M., Kimata K., Suzuki S. (1973) J. Biochem. 74, 1069–1073 [PubMed] [Google Scholar]
- 23.Levitt D., Dorfman A. (1973) Proc. Natl. Acad. Sci. U.S.A. 70, 2201–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galligani L., Hopwood J., Schwartz N. B., Dorfman A. (1975) J. Biol. Chem. 250, 5400–5406 [PubMed] [Google Scholar]
- 25.Gibson K. D., Segen B. J., Audhya T. K. (1977) Biochem. J. 162, 217–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handley C. J., Lowther D. A. (1977) Biochim. Biophys. Acta 500, 132–139 [DOI] [PubMed] [Google Scholar]
- 27.Sandy J. D., Brown H. L., Lowther D. A. (1980) Biochem. J. 188, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morriss-Kay G. M., Crutch B. (1982) J. Anat. 134, 491–506 [PMC free article] [PubMed] [Google Scholar]
- 29.Sobue M., Habuchi H., Ito K., Yonekura H., Oguri K., Sakurai K., Kamohara S., Ueno Y., Noyori R., Suzuki S. (1987) Biochem. J. 241, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson K. D., Segen B. J. (1977) Biochem. J. 168, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hjelle J. T., Gibson K. D. (1979) J. Embryol. Exp. Morphol. 53, 179–202 [PubMed] [Google Scholar]
- 32.Fritz T. A., Lugemwa F. N., Sarkar A. K., Esko J. D. (1994) J. Biol. Chem. 269, 300–307 [PubMed] [Google Scholar]
- 33.Lugemwa F. N., Sarkar A. K., Esko J. D. (1996) J. Biol. Chem. 271, 19159–19165 [DOI] [PubMed] [Google Scholar]
- 34.Mani K., Belting M., Ellervik U., Falk N., Svensson G., Sandgren S., Cheng F., Fransson L. A. (2004) Glycobiology 14, 387–397 [DOI] [PubMed] [Google Scholar]
- 35.Kuberan B., Ethirajan M., Victor X. V., Tran V., Nguyen K., Do A. (2008) Chembiochem 9, 198–200 [DOI] [PubMed] [Google Scholar]
- 36.Esko J. D., Stewart T. E., Taylor W. H. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safaiyan F., Kolset S. O., Prydz K., Gottfridsson E., Lindahl U., Salmivirta M. (1999) J. Biol. Chem. 274, 36267–36273 [DOI] [PubMed] [Google Scholar]
- 38.Greve H., Cully Z., Blumberg P., Kresse H. (1988) J. Biol. Chem. 263, 12886–12892 [PubMed] [Google Scholar]
- 39.Silbert J. E., Sugumaran G., Cogburn J. N. (1993) Biochem. J. 296, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrino D. A., Caplan A. I. (1994) Matrix Biol. 14, 121–133 [DOI] [PubMed] [Google Scholar]
- 41.Malmberg J., Mani K., Säwén E., Wirén A., Ellervik U. (2006) Bioorg. Med. Chem. 14, 6659–6665 [DOI] [PubMed] [Google Scholar]
- 42.Nishimoto S. K., Kajiwara T., Ledger P. W., Tanzer M. L. (1982) J. Biol. Chem. 257, 11712–11716 [PubMed] [Google Scholar]
- 43.Cheng F., Havsmark B., Sakurai K., Habuchi H., Suzuki S., Yoshida K., Fransson L. A. (1997) Glycoconj. J. 14, 297–305 [DOI] [PubMed] [Google Scholar]
- 44.Cöster L., Hernnäs J., Malmström A. (1991) Biochem. J. 276, 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuberan B., Lech M. Z., Beeler D. L., Wu Z. L., Rosenberg R. D. (2003) Nat. Biotechnol. 21, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 46.Kuberan B., Beeler D. L., Lawrence R., Lech M., Rosenberg R. D. (2003) J. Am. Chem. Soc. 125, 12424–12425 [DOI] [PubMed] [Google Scholar]
- 47.Lidholt K., Riesenfeld J., Jacobsson K. G., Feingold D. S., Lindahl U. (1988) Biochem. J. 254, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobsson I., Lindahl U., Jensen J. W., Rodén L., Prihar H., Feingold D. S. (1984) J. Biol. Chem. 259, 1056–1063 [PubMed] [Google Scholar]
- 49.Sasisekharan R., Venkataraman G. (2000) Curr. Opin. Chem. Biol. 4, 626–631 [DOI] [PubMed] [Google Scholar]
- 50.Ledin J., Ringvall M., Thuveson M., Eriksson I., Wilén M., Kusche-Gullberg M., Forsberg E., Kjellén L. (2006) J. Biol. Chem. 281, 35727–35734 [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi S., Morimoto K., Shimizu T., Takahashi M., Kurosawa H., Shirasawa T. (2000) Biochem. Biophys. Res. Commun. 268, 860–867 [DOI] [PubMed] [Google Scholar]
- 52.McCormick C., Duncan G., Goutsos K. T., Tufaro F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim B. T., Kitagawa H., Tanaka J., Tamura J., Sugahara K. (2003) J. Biol. Chem. 278, 41618–41623 [DOI] [PubMed] [Google Scholar]
- 54.Senay C., Lind T., Muguruma K., Tone Y., Kitagawa H., Sugahara K., Lidholt K., Lindahl U., Kusche-Gullberg M. (2000) EMBO Rep. 1, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seidler D. G., Breuer E., Grande-Allen K. J., Hascall V. C., Kresse H. (2002) J. Biol. Chem. 277, 42409–42416 [DOI] [PubMed] [Google Scholar]
- 58.Sugahara K., Ohi Y., Harada T., de Waard P., Vliegenthart J. F. (1992) J. Biol. Chem. 267, 6027–6035 [PubMed] [Google Scholar]
- 59.Oegema T. R., Jr., Kraft E. L., Jourdian G. W., Van Valen T. R. (1984) J. Biol. Chem. 259, 1720–1726 [PubMed] [Google Scholar]
- 60.Sugahara K., Mizuno N., Okumura Y., Kawasaki T. (1992) Eur. J. Biochem. 204, 401–406 [DOI] [PubMed] [Google Scholar]
- 61.Sugahara K., Yamada S., Yoshida K., de Waard P., Vliegenthart J. F. (1992) J. Biol. Chem. 267, 1528–1533 [PubMed] [Google Scholar]
- 62.Fransson L. A., Silverberg I., Carlstedt I. (1985) J. Biol. Chem. 260, 14722–14726 [PubMed] [Google Scholar]
- 63.Shibata S., Midura R. J., Hascall V. C. (1992) J. Biol. Chem. 267, 6548–6555 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.