Abstract
Since interleukin (IL)-18 is a proinflammatory cytokine, mice lacking IL-18 or its ligand-binding receptor (IL-18R) should exhibit decreased cytokine and chemokine production. Indeed, production of IL-1α, IL-6, and MIP-1α was reduced in IL-18 knock-out (ko) mouse embryonic fibroblast (MEF)-like cells. Unexpectedly, we observed a paradoxical 10-fold increase in IL-1β-induced IL-6 production in MEF cells from mice deficient in the IL-18R α-chain (IL-18Rα) compared with wild type MEF. Similar increases were observed for IL-1α, MIP-1α, and prostaglandin E2. Likewise, coincubation with a specific IL-18Rα-blocking antibody augmented IL-1β-induced cytokines in wild type and IL-18 ko MEF. Stable lines of IL-18Rα-depleted human A549 cells were generated using shRNA, resulting in an increase of IL-1β-induced IL-1α, IL-6, and IL-8 compared to scrambled small hairpin RNA. In addition, we silenced IL-18Rα with small interfering RNA in primary human blood cells and observed up to 4-fold increases in the secretion of lipopolysaccharide- and IL-12/IL-18-induced IL-1β, IL-6, interferon-γ, and CD40L. Mechanistically, despite increases in Stat1 and IL-6, induction of SOCS1 and -3 (suppressor of cytokine signaling 1 and 3) was markedly reduced in the absence of IL-18Rα. Consistent with these observations, activation of the p38α/β and ERK1/2 MAPKs and of protein kinase B/Akt increased in IL-18Rα ko MEF, whereas the negative feedback kinase MSK2 was more active in IL-18 ko cells. These data reveal a role for SOCS1 and -3 in the seemingly paradoxical hyperresponsive state in cells deficient in IL-18Rα, supporting the concept that IL-18Rα participates in both pro- and anti-inflammatory responses and that an endogenous ligand engages IL-18Rα to deliver an inhibitory signal.
Often shown to function as a proinflammatory cytokine, structurally related to IL-1β,3 and requiring caspase-1 for processing of its inactive precursor into an active cytokine (1–3), IL-18 is a unique member of the IL-1 family. For example, IL-18 and IL-18 receptor α-chain (IL-18Rα) knock-out (ko) mice unexpectedly overeat and spontaneously become obese, developing insulin resistance and atherosclerosis (4, 5). This phenotype does not occur in mice deficient in other members of the IL-1 family. In the absence of IL-12 and similar co-stimulatory cytokines, IL-18 can act as a typical Th2 cytokine in murine models (6, 7). The affinity of the naturally occurring IL-18-binding protein (IL-18BP) for IL-18 is higher than that of IL-18 for its cognate receptor; thus, low levels of this naturally occurring antagonist effectively neutralize the activity of IL-18 (8–11). In some studies, IL-18 opposes the proinflammatory properties of IL-1β (12). In dextran sodium sulfate-induced colitis, neutralizing antibodies to IL-18 or IL-18BP ameliorate the disease (13, 14), whereas in other studies, mice deficient in IL-18Rα exhibit worsening of the disease (15).
IL-18 Induces Several Proinflammatory Cytokines, Such as IL-1β and TNFα, as well as chemokines, nitric oxide, and vascular adhesion molecules (reviewed in Ref. 16). Using mice deficient in IL-18 or neutralization of IL-18, the cytokine appears to play an important role in models of rheumatoid arthritis (17), lupus-like autoimmune disease (18), metastatic melanoma (19), graft versus host disease (20), and myocardial suppression (21, 22). Unlike IL-1, IL-18 also induces Fas ligand and has been proposed as a key mediator of macrophage activation syndrome (23).
We have previously reported that whereas deficiency in IL-18 attenuated inflammatory responses to various exogenous stimuli, these responses paradoxically were exaggerated in IL-18Rα ko mice (24). In addition to rejecting insulin-producing islet allografts, splenocytes and peritoneal macrophages from IL-18Rα ko mice produced significantly greater amounts of several proinflammatory cytokines upon stimulation with concanavalin A, TLR2 agonist heat-killed Staphylococcus epidermidis, or anti-CD3 antibodies (24).
In the present study, we set out to investigate the fundamental differences in cytokine production between IL-18 ko and IL-18Rα ko mice using mouse embryonic fibroblasts (MEF), which are highly responsive to IL-1 and TNFα stimulation. We also studied the role of IL-18Rα in human cells. IL-18Rα was silenced in human A549 epithelial cells using small hairpin RNA (shRNA) to the IL-18R (shIL-18R) as well as in freshly obtained human peripheral blood mononuclear cells (PBMC). Furthermore, using inhibitors as well as kinase activation studies, real time PCR, and Western blotting, we shed light on the IL-18Rα ko-mediated differences in expression and activation of signaling mediators, such as the suppressors of cytokine signaling (SOCS), MAPKs, protein kinase B/Akt, NF-κB, MSK2/RSKβ, and p70 S6 kinase. The mechanisms underlying the disinhibition of inflammatory responses in IL-18Rα-deficient cells appear to be due to a yet unidentified anti-inflammatory ligand of the IL-18 receptor.
EXPERIMENTAL PROCEDURES
Reagents
S. epidermidis was obtained from the American Tissue Culture Collection (strain 49134), grown overnight in suspension cultures in LB medium (Difco), centrifuged, and washed in pyrogen-free saline, and a small sample was removed for determination of number of organisms by pour plate cultures. The suspension was boiled for 30 min and then remained at room temperature for 24 h. The boiled suspension was diluted in pyrogen-free saline to 10 million organisms/ml and frozen in small aliquots at −70 °C.
Antibodies for the mouse cytokine assays MIP-1α, MIP-2, TNFα, and IL-1α were obtained from R&D Systems (Minneapolis, MN). Recombinant human IL-1β was as previously described (25), and murine TNFα was obtained from PeproTech (Rocky Hill, NJ). Anti-mouse IL-18Rα was a kind gift from Dirk Smith (Amgen, Seattle, WA). Anti-mouse IL-18Rα was also obtained from R&D Systems. The murine IL-6 ELISA and goat biotinylated IgG were from R&D Systems. Alexa Fluor 488 wheat germ agglutinin was purchased from Invitrogen. Bisbenzimide was from Sigma, and ibuprofen was provided by The Upjohn Co. Cell culture media were obtained from Cellgro (Herndon, VA). For extracting total RNA, the RNeasyPlus Mini Kit was used (Qiagen, Hilden, Germany). GeneAmp® RNA PCR kit, core kit, and GeneAmp® Fast PCR Master Mix were purchased from Applied Biosystems (Foster City, CA).
Animals
IL-18 ko and IL-18Rα ko mice (both C57BL/6 background) and wild type C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME), housed five per cage, and kept on a 12-h light-dark cycle. All experiments were approved by the University of Colorado Institutional Animal Care and Use Committee.
Preparation of MEF
Mice of each strain were anesthetized with isoflurane on days 13–16 of pregnancy and sacrificed by rapid cervical dislocation. Embryos were removed, separated from the uterine horns, and washed three times with 10 ml of PBS without calcium/magnesium. After removal of the head and organs, the embryos were washed three times with 10 ml of PBS without calcium or magnesium. The embryos were then minced, and 2 ml of trypsin/EDTA with 100 Kunitz units of DNase I per embryo was added (Sigma). The mixture was incubated, gently shaking at room temperature for 15 min together with 5-mm glass beads to create a single cell suspension.
The trypsin digestion was stopped by adding 20 ml of MEF medium (Dulbecco's modified Eagle's medium, 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 1% non-essential amino acids). The undigested tissue pieces settled to the bottom, and the supernatant was removed and subjected to low speed centrifugation for 5 min at 300 × g. The resulting pellet was resuspended in 20 ml of MEF medium and plated in a culture flask. The medium was changed the following day. Examination on day 3 revealed a homogenous layer of fibroblast-like cells. For the experiments in this study, MEF passages 1–5 were used.
MEF
Cells were detached with trypsin/EDTA (Sigma) and plated in flat-bottom wells. After 24–48 h of growth in Dulbecco's modified Eagle's medium containing 2% fetal calf serum, penicillin/streptomycin, 1% non-essential amino acids, cells were stimulated with the cytokines or inhibitors for the indicated time periods. After 24 h, the supernatant was removed for measuring secreted cytokines. The cells in the wells were then washed and lysed as described previously (26). After 20 min, the wells were scraped with a rubber policeman. The homogenate was centrifuged for 10 min at 14,000 × g, and the supernatant was removed for protein determination and IL-1α determination by the ECL assay.
Generation of Stable A549 Clones with Reduced Levels of IL-18Rα by shRNA
Human lung carcinoma A549 cells (3 × 105 cells/well) were seeded in a 6-well plate a day before transfection. Cells were washed with 2 ml of TOM medium (Welgene, Dae-gu, Korea) and incubated for 30 min in 1 ml of the same medium prior to transfection. In the meantime, the transfection mixture for each well was prepared as follows. 5 μl of Lipofectamine (Invitrogen) was suspended in 100 μl of TOM medium. After a 15-min incubation at room temperature, 1 μg of plasmid DNA comprising equal parts of four sequences of pGeneClip-IL-18Rα shRNA (SABiosciences, Frederick, MD) was added, followed by another 15-min incubation. The transfection mixture was then added to the wells, and the plate was incubated for 5 h at 5% CO2 and 37 °C. Thereafter, 1 ml of cell culture medium containing 20% fetal bovine serum was added. On the next day, cells were trypsinized and transferred into a 9-cm2 plate. When the cells had adhered to the plate, the culture medium was replaced with fresh medium containing 1 μg/ml of puromycin dihydrochloride (Cellgro, Manassas, VA). The selection medium was exchanged every 3 days with fresh selection medium until individual colonies appeared, which took place between 10 and 14 days. Small pieces of circular 3 MM papers were sterilized, wet with trypsin, and then used for picking individual colonies. Colonies were transferred into 24-well plates containing 1 ml of selection medium in each well. An individual clone was grown until the cells reached a population of 106, and each clone was screened by Western blot analysis.
Culture and Transfection of Human PBMC
PBMC were isolated from freshly obtained blood from healthy volunteers by density gradient centrifugation. Transfection with the Amaxa (Cologne, Germany) Nucleofector device was performed as described previously (27). A pool of four OnTargetplus antisense RNA sequences from Thermo Fisher Scientific (Lafayette, CO) at 62.5 nm each (250 nm total) was used for silencing of IL-18Rα. For control, PBMC were transfected with identical concentrations of OnTargetplus non-targeting pools. After overnight recovery, 106 PBMC were transferred into 48-well plates containing 400 μl of RPMI with 1% pooled human serum and primocin (1:500, Amaxa) and then stimulated for 4 or 20 h. Supernatants from these cultures were assayed for cytokines; lysates were subjected to Western blot or real time PCR analysis.
Western Blot
To analyze A549 lysates, 5 × 105 cells were harvested and lysed in 100 μl of sample buffer containing β-mercaptoethanol and then subjected to 10% SDS-PAGE. The samples were transferred to nitrocellulose membranes, which were then blocked in 5% skim milk. The membranes were probed with mouse anti-IL-18Rα (R&D Systems) and normalized with goat anti-actin (Zymed Laboratories Inc., San Francisco, CA). For SOCS1 and SOCS3 analysis in PBMC and MEF, lysates were subjected to 12% SDS-PAGE, blotting, and staining with rabbit anti-mouse SOCS1 or rabbit anti-human/mouse SOCS3.
Cytokine and Chemokine Measurements
Murine MIP-1α, MIP-2, and IL-1α as well as human IL-1α, IL-6, and IL-8 were measured by an ECL assay as described previously (28) using the Origen Analyzer (BioVeris, Gaithersburg, MD). Murine IL-6 and human TGF-β1 were determined by ELISA (R&D Systems). Supernatants from the PBMC experiments were subjected to multiplex analysis using microtiter plates spotted with antibodies to eight cytokines from R&D Systems (MosaicTM ELISA human cytokine panel 1) and then analyzed in the Q-ViewTM Imager from Quansys Biosciences (Logan, UT) according the manufacturer's protocols. In addition, the samples were diluted and measured by specific ELISA (R&D Systems).
Confocal Microscopy
Isolated MEF were grown in culture slides, washed with PBS, and fixed with a 70%/30% mixture of acetone/methanol. After fixation for 15 min at room temperature, the cells were blocked for 1 h in PBS, 1% bovine serum albumin (Sigma) containing 10% donkey serum (Sigma) at room temperature. Staining was performed overnight at 4 °C using a biotinylated goat anti-mouse IL-18Rα antibody (2.5 μg/ml; R&D Systems) in PBS containing 1% BSA. The corresponding concentration of nonimmune goat IgG was used as a negative control. Streptavidin conjugated to Cy3 (dilution 1:1000) was used for detections. All membranes were stained with Alexa Fluor 488® conjugate of wheat germ agglutinin; nuclei were stained with bisbenzimide (Sigma) in a concentration of 1 μg/100 ml. Digital confocal imaging was performed using a Leica DM RXA microscope equipped with SlideBook software for MacIntosh (Intelligent Imaging Innovations, Denver, CO).
Real Time Quantitative PCR
First strand cDNA was synthesized from random hexamer-primed RNA, isolated from MEF samples, using TaqMan® reverse transcription reagents (Applied Biosystems). The first strand cDNA template synthesized from each MEF RNA sample was then used for the amplification of target genes in a quantitative PCR assay using TaqMan® Universal PCR Master Mix (Applied Biosystems) and Source Molecular Diagnostics (Boulder, CO) primer-probe sets (29). Each target gene assay was multiplexed with an 18 S rRNA endogenous control and performed in triplicate on the ABI Prism® 7900HT Sequence Detection System (Applied Biosystems). Normalized gene expression values, termed ΔCT values, for each amplified gene were calculated by taking the difference between the PCR cycle threshold (CT) value of the target gene and its own endogenous 18 S rRNA or glyceraldehyde-3-phosphate dehydrogenase control. The difference in normalized gene expression values between samples (ΔΔCT value) was calculated to determine the relative difference in transcript levels between sample types. For example, a ΔΔCT value of 2 is equivalent to 4-fold change in the amount of the transcript between sample types.
Statistical Analyses
Paired and unpaired t tests were used for statistical analysis. Treatments that differed from controls by more than 2 S.D. values were considered as significant. Individual p values are reported.
RESULTS
Characterization of MEF Cell Lines
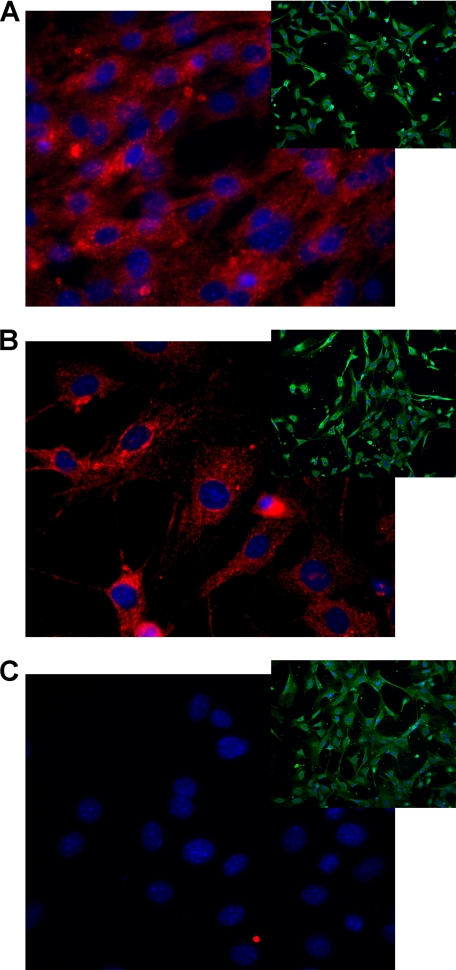
To investigate the differences in cytokine production between wild type (WT), IL-18 ko, and IL-18Rα ko mice in a single cell type, embryonic fibroblasts of each mouse strain were generated. As shown in Fig. 1, immunofluorescence was used to detect expression of IL-18Rα; IL-18Rα was absent in IL-18Rα ko MEF cells (C). An IgG control antibody was used as the control staining in the Cy3 channel (insets). In addition, MEF cells derived from IL-18 ko mice appear slender and to have at least two pseudopods. By comparison, WT cells appeared as normal spindle-shaped fibroblasts, whereas IL-18Rα ko MEF cells have a prominent cytosol and are round.
FIGURE 1.
IL-18Rα in the three MEF strains. MEF cells grown for 2 days were stained against mouse IL-18Rα chain and visualized using confocal digital microscopy. Red dye, anti-IL-18Rα; blue dye, nuclei; green dye, cell membranes (the last only shown in insets). A, WT; B, IL-18 ko; C, IL-18Rα ko. The insets show each strain stained with an IgG control instead of the anti-IL-18Rα antibody.
During the course of these studies, we generated several MEF lines of WT, IL-18 ko, and IL-18Rα ko, each from a different mother. In Table 1, examples of four MEF lines from each strain are presented. The differences between the three MEF cell types were remarkably consistent, despite each being derived from a different mother. In some experiments, more than six lines were studied and exhibited similar results. Replicate experiments, therefore, do not represent MEF cells derived from a single mother but rather data from MEF cells derived from at least four different mothers.
TABLE 1.
MIP-1α protein in supernatants from individual MEF isolations
Each value indicates a MIP-1α measurement (given in pg/mg of total protein) in the supernatant of MEF isolated from embryos of one individual mother exposed for 20 h to IL-1β (1 ng/ml).
| Number | Mouse strain | MIP-1α |
|---|---|---|
| pg/mg | ||
| 1 | Wild type | 74 |
| 2 | Wild type | 67 |
| 3 | Wild type | 60 |
| 4 | Wild type | 62 |
| 1 | IL-18 ko | 20 |
| 2 | IL-18 ko | 8 |
| 3 | IL-18 ko | 74 |
| 4 | IL-18 ko | 46 |
| 1 | IL-18Rα ko | 168 |
| 2 | IL-18Rα ko | 157 |
| 3 | IL-18Rα ko | 200 |
| 4 | IL-18Rα ko | 154 |
MIP-1a Production in MEF Cells Lacking the IL-18Rα Chain
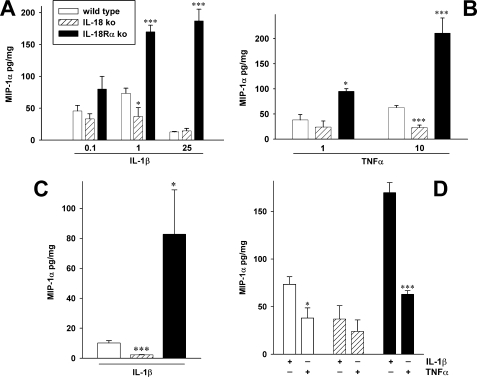
Fig. 2A shows that compared with WT MEF, IL-18Rα ko MEF cells stimulated with different concentrations of IL-1β secreted considerably more MIP-1α per mg of total protein, whereas MEF cells deficient in IL-18 secreted less of the chemokine. These differences in the production of MIP-1α between the three cell types were apparent using concentrations of IL-1β as low as 100 pg/ml (Fig. 2A, left). When the concentration of IL-1β was increased to 25 ng/ml, there was a marked decrease in the amount of MIP-1α released from the WT as well as the IL-18-deficient MEF cells. In contrast to WT and IL-18-deficient cells exposed to higher concentrations of IL-1β, IL-18Rα ko MEF cells produced more than 10-fold higher levels of MIP-1α.
FIGURE 2.
Production of MIP-1α in IL-1β- and TNFα-stimulated MEF cells. WT, IL-18 ko, and IL-18Rα ko MEF cells were stimulated with the indicated concentrations of either IL-1β or TNFα, and after 20 h (A, B, and D) or 6 h (C; 25 ng/ml IL-1β), the levels of MIP-1α were measured in the supernatants. Results represent MIP-1α in pg/mg total protein in IL-1β-exposed (A, C, and D) or TNFα-exposed (B and D) cells. The data are means ± S.E. of four independent experiments, each experiment performed on a separate cell line derived from a different mother (see Table 1). A–C, *, p < 0.05; ***, p < 0.001 compared with the values in WT cells. D, comparison of IL-1β (1 ng/ml) with TNFα (10 ng/ml) as stimuli of MEF cells. Data are means ± S.E. (n = 4); *, p < 0.05; ***, p < 0.001 for IL-1β- compared with TNFα-stimulated MEF.
The three MEF cell lines were then studied for their responses to TNFα. As shown in Fig. 2B, the differences were similar to those observed in response to IL-1β. In IL-18Rα ko MEF cells stimulated with TNFα, 2-fold higher levels of MIP-1α were observed compared with WT, but MIP-1α was 10-fold lower in cells deficient in IL-18. These differences were observed at either 1 or 10 ng/ml of TNFα. Fig. 2D illustrates that 1 ng/ml IL-1β is more effective at inducing MIP-1α in each MEF strain than 10 ng/ml TNFα.
We next compared the responses in MEF cells after 6 h of stimulation with IL-1β. Under these conditions, cells deficient in IL-18Rα produced 10-fold more MIP-1α (Fig. 2C).
Effect of IL-18Rα Deficiency on IL-1α, IL-6, and PGE2
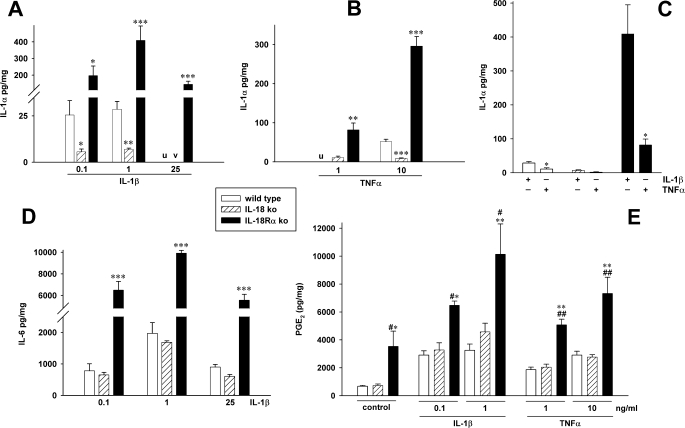
Whereas chemokines, such as MIP-1α, are readily secreted, IL-1α is primarily an intracellular cytokine. Therefore, we measured the intracellular levels of IL-1α in the three MEF cell lines from the same cultures depicted in Fig. 2. When stimulated with IL-1β, IL-18Rα ko MEF cells contained 20-fold higher levels of IL-1α compared with wild type cells and 40-fold more IL-1α than IL-18-deficient MEF cells. Using a lower concentration of IL-1β (100 pg/ml), nearly the same differences were observed (Fig. 3A). Using 25 ng/ml IL-1β, the levels of IL-1α in WT and IL-18-deficient cells were below detection (<20 pg/mg total protein), whereas in IL-18Rα ko MEF (Fig. 3A), the levels were impressively greater. Upon TNFα stimulation, similar differences were obtained (Fig. 3B). The comparison of IL-1β and TNFα as inducers of IL-1α in the different MEF strains (Fig. 3C) revealed similar regulation of MIP-1α, in that 1 ng/ml IL-1β stimulated more IL-1α release than 10 ng/ml TNFα.
FIGURE 3.
Differential induction of IL-1α, IL-6, and PGE2 in the MEF cell strains. WT, IL-18 ko, and IL-18Rα ko were stimulated with IL-1β or TNFα. After 20 h, supernatants were removed and used to generate D and E and the data shown in Fig. 2. The cells were lysed, and levels of IL-1α were measured. The graphs show means of normalized cytokine or prostaglandin concentrations ± S.E. (n = 4). A, B, D, and E, *, p < 0.05; **, p < 0.01; ***, p < 0.001 comparing the ko strains with WT cells. u and v, values in wild type (u) and/or IL-18 ko MEF (v) were below the detection levels of the ECL. A and D, note the breaks in IL-18Rα bars. E, #, p < 0.05; ##, p < 0.01 for IL-18 ko versus IL-18Rα ko. C, 1 ng/ml IL-1β versus 10 ng/ml TNFα. The data are mean ± S.E. of the same four independent experiments as in Fig. 2D. *, p < 0.05 for IL-1β-stimulated compared with TNFα-stimulated cells.
We next examined IL-1β-induced IL-6 in the three different MEF strains. As shown in Fig. 3D, IL-1β at 100 pg/ml induced more than 6,000 pg of IL-6/mg of total protein in IL-18Rα ko cells, whereas one-tenth this amount was present in the supernatants of WT or IL-18 ko cells. Unlike production of IL-1α, IL-6, and MIP-1α, we observed no differences in the production of PGE2 between WT and IL-18 ko MEF. However, IL-18Rα-deficient cells secreted 2–3-fold more PGE2 than the other two strains under unstimulated conditions as well as after stimulation with IL-1β or TNFα (Fig. 3E).
Blocking the IL-18Rα Chain Increases Spontaneous and IL-1β-induced Cytokines
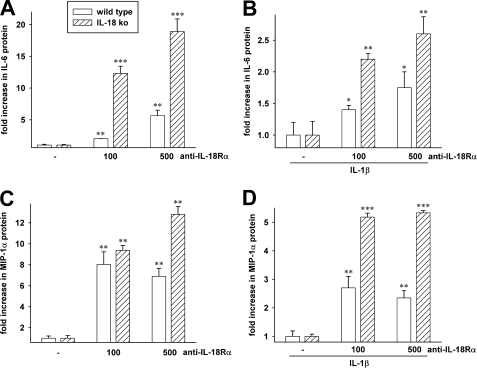
In order to examine decreased function of IL-18Rα in WT MEF cells, we used an anti-murine IL-18Rα-blocking antibody. After 20 h of incubation with the antibody, there was a dose-dependent increase in spontaneous IL-6 production in WT (2.5-fold at 100 ng/ml antibody and 6.5-fold at 500 ng/ml) compared with the level of IL-6 in WT MEF cells incubated without the antibody (Fig. 4A). We next stimulated WT MEF cells with IL-1β in the presence of the blocking antibody. IL-1β-stimulated IL-6 production was set at 1.0 in WT MEF cells incubated without the antibody; in the presence of the antibody, IL-6 increased (1.4-fold at 100 ng/ml and 1.75-fold at 500 ng/ml). As shown in Fig. 4C, the spontaneous production of MIP-1α was also increased in the presence of the antibody. At a 100 ng/ml concentration of the blocking antibody, spontaneous MIP-1α increased 8-fold. In WT cells stimulated with IL-1β, the increase was 2.5-fold (Fig. 4D).
FIGURE 4.
Effect of an anti-IL-18Rα blocking antibody on cytokine production. Increasing concentrations of an anti-IL-18Rα monoclonal blocking antibody (shown below the horizontal axis in ng/ml) were added to WT or IL-18 ko MEF cells. After 20 h of incubation, the levels of spontaneous IL-6 were measured in the supernatant. A, the level of IL-6 in the absence of anti-IL-18Rα was set at 1.0 for each of three independent experiments, and the -fold increase was calculated. The data are mean ± S.E. -fold increase. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with in the absence of the antibody. B, an experiment similar to A, except IL-1β (1 ng/ml) was added to the cells 20 min after the anti-IL-18Rα antibody. The data in C and D are from the same cultures as shown in A and B. In C and D, MIP-1α was measured in the supernatants. **, p < 0.01; ***, p < 0.001 compared with no antibody.
The effect of blocking IL-18Rα in MEF cells deficient in IL-18 itself was also studied. The rationale for this experiment was to evaluate blocking IL-18Rα in the complete absence of IL-18 in these cells. As such, there is no endogenous IL-18 to compete with a putative other ligand utilizing IL-18Rα. As shown in Fig. 4A, spontaneous IL-6 was markedly increased in IL-18 ko cells in the presence of the blocking antibody (12- and 18-fold at antibody concentrations of 100 and 500 ng/ml, respectively). These increases are significantly greater than the increases in WT MEF cells under the same conditions. In IL-18 ko MEF cells stimulated with IL-1β, the increases were again greater compared with the increases in WT cells. Fig. 4, C and D, depicts the data on MIP-1α production in the same cultures. In MEF cells deficient in IL-18 and stimulated with IL-1β, the increases in MIP-1α are 2-fold greater when the IL-18Rα is blocked compared with the WT cells.
In these experiments, we also examined an antibody to murine IL-18Rα obtained from another source (R&D Systems) and observed similar data (not shown). For each antibody tested, an appropriate isotype control antibody was used. Applied at the same concentrations, there were no significant increases in IL-6 or MIP-1α production. Each antibody was tested for contaminations with LPS, and levels were less than 1 enzyme unit/mg.
Silencing the IL-18 Receptor in Human Epithelial A549 Cells
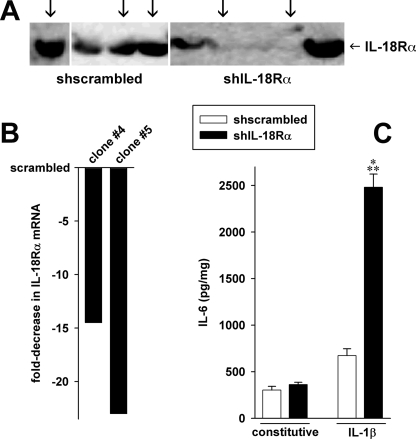
To demonstrate that the hyperinflammatory responses associated with the absence of IL-18Rα are not limited to murine fibroblast-like cells, we generated several stable cell lines derived from human lung epithelial A549 cells in which expression of IL-18Rα was reduced by specific shRNA. Stable clones of scrambled shRNA were also generated. As shown in Fig. 5B, shIL-18Rα reduced steady-state IL-18Rα mRNA up to 23-fold compared with scrambled-transfected control cells. The reduction in mRNA resulted in a marked but not complete inhibition of IL-18Rα protein levels (Fig. 5A). Nevertheless, upon stimulation with IL-1β, this reduction of IL-18Rα was accompanied by an increase in the levels of IL-6 (Fig. 5C). However, in unstimulated cells, there was no difference in cytokine production. We also determined the levels of IL-1α, IL-8, and TGF-β1 in these cultures. Consistent with the MEF cells, the knockdown of IL-18Rα in A549 cells resulted in 2-fold increases in IL-1α and IL-8.
FIGURE 5.
Silencing of the IL-18R in A549 lung epithelial cells. Using shRNA, stable A549 cell lines depleted of the IL-18R α-chain were created. For control, A549 cells were transfected with scrambled shRNA. The knockdown of the IL-18Rα chain was assessed by Western blotting (A) and real time PCR (B). A, the arrows indicate the clones used in the experiments. C, the three scrambled-transfected and two shIL-18Rα-transfected cell lines shown in A and B were stimulated with 10 ng/ml IL-1β or were left untreated for 20 h. Thereafter, IL-6 levels were determined. The panel depicts means of normalized cytokine concentrations ± S.E.; n = 6 clones times 4 independent experiments; ***, p < 0.001 for shIL-18R versus small hairpin scrambled (shscrambled).
Role of Cyclooxygenase and Endogenous IL-1 in IL-6 Production
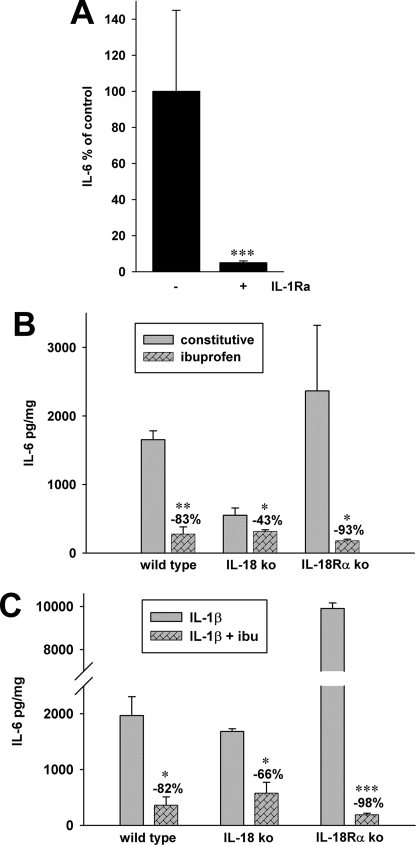
We consistently observed that constitutive IL-6 production was elevated in each of the three MEF lines without exogenous cytokine stimulation (Fig. 6B). However, even in the absence of an exogenous stimulant, MEF cells deficient in IL-18 produced 66% less IL-6 compared with wild type cells (p = 0.04) and 77% less compared with MEF deficient in IL-18Rα (p = 0.04). Also shown in Fig. 6B is the marked decrease in spontaneous IL-6 production induced by inhibition of cyclooxygenase with ibuprofen. A reduction of 93% was observed in MEF deficient in the IL-18Rα, although the other MEF lines also exhibited statistically significant ibuprofen-induced reductions in spontaneous IL-6 production. Stimulating each of the MEF lines with IL-1β (1 ng/ml) in the absence or presence of ibuprofen showed nearly the same reductions in IL-6 levels (Fig. 6C). These data are consistent with the observation that IL-1β-induced IL-6 is cyclooxygenase-sensitive (30).
FIGURE 6.
Effects of ibuprofen and IL-1Ra on the production of IL-6 in MEF cells. A, constitutive secretion of IL-6 was measured in the absence or presence of the IL-1Ra. IL-1Ra was added to the cultures of IL-18Rα ko cells at 10 μg/ml, and after 20 h, levels of IL-6 were measured. In cultures without IL-1Ra (−), the level of IL-6 was set at 100%, and the mean percentage change of parallel cultures with IL-1Ra was calculated. Data are mean ± S.E. percentage change from three independent experiments. ***, p < 0.001 compared with cultures without IL-1Ra. B, constitutive production of IL-6 in WT, IL-18 ko, and IL-18Rα ko MEF cells in the absence and presence of ibuprofen (10 μm) after 20 h of incubation. C, each of the MEF strains were stimulated with IL-1β (1 ng/ml) in the absence or presence of ibuprofen (ibu; 10 μm). After 20 h, levels of IL-6 were measured in the supernatants. Note the break in the IL-18Rα bar. B and C, the data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 comparing cultures with ibuprofen with those without.
In fibroblasts derived from the dermis or the kidney, endogenous IL-1α mediates proliferation, PGE2, and IL-6 synthesis (30–33). As shown in Fig. 3, A–C, IL-1α levels are increased in IL-18Rα ko MEF compared with WT, whereas IL-18 ko MEF cells have lower IL-1α levels. To examine the role of IL-1α in spontaneous as well as IL-1β-induced IL-6 production, we used the IL-1 receptor antagonist (IL-1Ra) to block the IL-1 receptor. In the presence of IL-1Ra, background IL-6 production was decreased in IL-18Rα ko (92.5%; Fig. 6A). We also observed a similar reduction in spontaneous IL-6 production in WT (90%) and IL-18 ko (90.5%) MEF cells. Thus, elevated constitutive levels of cytokines in IL-18Rα-deficient MEF cells are due to endogenous IL-1.
Kinase Profiles of the Three MEF Strains
To further investigate the mechanisms underlying the hyperresponsiveness of cells lacking the IL-18Rα, we subjected lysates from IL-1β-stimulated MEF to a phosphokinase array analysis. As shown in Table 2, we observed differential regulation of several kinases. For example, activity of protein kinase B/Akt was increased in IL-18Rα ko compared with WT MEF, whereas in IL-18 ko cells, the activity of this kinase was reduced (α and γ isoforms) or unchanged (β). ERK1 and -2, p38α and -β, GSK-3α and -β, and p70 S6 kinase exhibited a similar pattern (Table 2, top). HSP27 and RSK1 and -2 were largely unaffected by the knockouts (Table 2, bottom). Table 2 (middle) shows that in the case of p38δ and RSKβ, the pattern was reversed (i.e. activity of these kinases was increased in IL-18 ko MEF and unchanged or slightly reduced in IL-18Rα ko cells). Regulation of the JNK group was unique in that both knockouts consistently increased activation of these kinases.
TABLE 2.
Phosphokinase profile of the three MEF strains
MEF were stimulated with 1 ng/ml IL-1β for 30 min, and then the lysates were subjected to protein array analysis and densitometry. The table shows mean ± S.E. -fold changes in kinase activity in the two knockout strains compared with WT MEF, which is set at 1; n = 2. The top comprises kinases that exhibit increased activation in IL-18Rα ko and/or decreased activation in IL-18 ko compared with WT. The middle shows kinases the activity of which is increased in IL-18 ko MEF, and the bottom shows pathways with little change. Underlining, kinase activity decreased by more than −33%; normal type, change between −33 and +33%; italic type, increase between 33 and 67%; boldface type, increase of >67%.
| IL-18 ko | IL-18Rα ko | |
|---|---|---|
| Activation of IL-18Rα ko > IL-18 ko | ||
| Akt1/PKBα | −36.5 ± 3.3 | 122.6 ± 28.4 |
| Akt2/PKBβ | 2.1 ± 13.7 | 52.6 ± 5.3 |
| Akt pan | −23.7 ± 6.7 | 89.7 ± 21.7 |
| ERK1-p44 | −0.9 ± 15.4 | 68.7 ± 17.7 |
| ERK2-p42 | −5.8 ± 18.1 | 60.1 ± 1.7 |
| p38α | 15.5 ± 17.3 | 76.3 ± 20.4 |
| p38β | 1.2 ± 28.1 | 88.1 ± 19.3 |
| p38γ | −12 ± 3 | 31.1 ± 17.1 |
| GSK3α and -β | −31.5 ± 12.3 | 19.9 ± 4.3 |
| GSK3β | −46.3 ± 9.4 | 6.2 ± 5.8 |
| p70 S6 kinase | −9.2 ± 2.1 | 36.3 ± 10.5 |
| Activation of IL-18Rα ko < IL-18 ko | ||
| p38δ | 56.0 ± 15.4 | −5.4 ± 3.8 |
| MSK2/RSKβ | 40.6 ± 10.3 | −10.6 ± 9 |
| Little change in activation | ||
| Akt3/PKBγ | −11.7 ± 20.5 | 31 ± 23.4 |
| JNK1 | 11.6 ± 4 | 13.3 ± 13.2 |
| JNK2 | 0.5 ± 3.7 | 32.6 ± 4.4 |
| JNK3 | 16.3 ± 6 | 38.0 ± 14.4 |
| JNK pan | 12.3 ± 9.9 | 29.7 ± 22 |
| HSP27 | −3.1 ± 3.6 | 6.1 ± 1.3 |
| RSK1 | −6.3 ± 12.4 | 28.2 ± 0.4 |
| RSK2 | 4.2 ± 21.5 | −0.6 ± 16.5 |
Regulation of SOCS1, SOCS3, and Other Genes in the MEF Strains
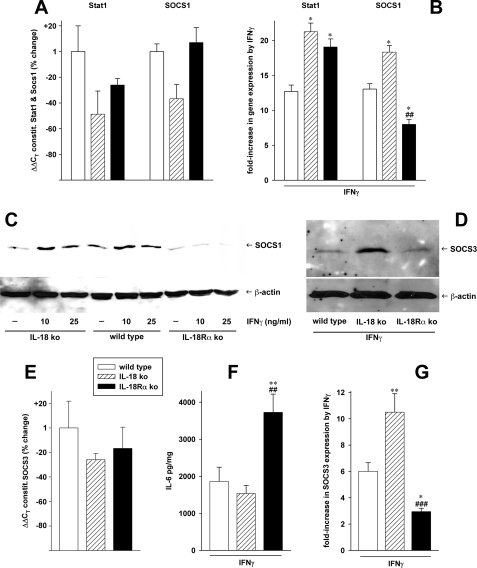
Total RNA was obtained from the MEF cells 4 h after stimulation with IL-1β, IFNγ, or vehicle and subjected to quantitative PCR as described previously (29). The results are shown in Fig. 7 and Table 3. SOCS are important negative feedback inhibitors of cytokine signaling; therefore, we hypothesized that a dysregulation of their expression may play a role in the hyperinflammation in IL-18Rα-deficient cells. Indeed, real time PCR analysis revealed that whereas Stat1 (the target of the negative-feedback inhibition of SOCS1) was increased almost 20-fold by IFNγ in IL-18Rα ko MEF, SOCS1 expression was only 8-fold higher (Fig. 7B). In contrast, the increases in Stat1 and SOCS1 were almost equal in WT and IL-18 ko MEF. Although under constitutive conditions there was a trend toward lower Stat1 expression, especially in IL-18 ko cells, none of the differences between the three strains in Stat1 and SOCS1 were significant in unstimulated cells (Fig. 7A). As shown in Fig. 7E, the same is valid for constitutive levels of SOCS3. Upon stimulation with IFNγ, however, mRNA levels of SOCS3 exhibited large increases in IL-18 ko cells (10.5-fold) (Fig. 7G) and WT MEF (6-fold). In IL-18Rα ko cells, SOCS3 was only 2.9-fold higher than without IFNγ stimulation. This regulatory pattern contrasted with release of IL-6, which is a major inducer of Stat3 and SOCS3 activation; as shown in Fig. 7F, IL-18Rα ko MEF produced about 2-fold more IL-6 than WT and IL-18 ko cells following incubation with IFNγ. Western blot analysis revealed that the regulation of mRNA was translated at the protein level. Induction of both SOCS1 (Fig. 7C) and SOCS3 (Fig. 7D) by IFNγ was markedly reduced in IL-18Rα ko MEF. Background production of SOCS3 was low in each of the MEF strains (not shown).
FIGURE 7.
Expression of SOCS1, Stat1, and SOCS3 in the three MEF strains. mRNA and protein levels of SOCS1, Stat1 (A–C), and SOCS3 (D, E, and G) in WT, IL-18 ko, and IL-18Rα ko MEF were assayed by real time PCR (A, B, E, and G; differences in gene expression were calculated using the ΔΔCT method) or Western blotting (C and D). A, B, E, and G, the panels comprise three independent experiments, which are shown as mean changes ± S.E. 4 h past stimulation. *, p < 0.05; **, p < 0.01; ***, p < 0.001 for both ko strains versus WT. ##, p < 0.01; ###, p < 0.001 for IL-18 ko versus IL-18Rα ko. A and E, constitutive levels of the target genes. mRNA quantity in WT MEF is defined as 1; the bars show the changes conferred by the knockouts in percentage, none of which are significant. B and G, the graphs represent induction in SOCS1, Stat1, and SOCS3 mRNA by 25 ng/ml IFNγ as -fold increase over base lines. F, normalized IL-6 levels in the supernatants from the same IFNγ-stimulated MEF cultures ± S.E. are depicted. C and D, MEF cells from the three strains were stimulated with the indicated concentrations of IFNγ (C) or 25 ng/ml IFNγ (D) for 20 h. Lysates were then analyzed by Western blotting with staining for SOCS1, SOCS3, or β-actin. One blot representative of four others with similar results is shown.
TABLE 3.
-Fold changes in mRNA transcripts due to IL-1β stimulation in MEF cells
MEF were stimulated with 1 ng/ml IL-1β for 4 h; thereafter, changes in gene expression were determined by quantitative real time PCR and calculated using the ΔΔCT method. Values are from individual experiments and represent the -fold change over non-IL-1β-stimulated MEF cells, set at 1.0.
| Gene | Wild type | IL-18Rα KO |
|---|---|---|
| -fold | -fold | |
| NF-κB p50 | 2.18; 2.13 | 3.79; 3.84; 5.74 |
| TGF-β | 1.05; 1.34 | 1.11; 1.16; 0.31 |
| Caspase-3 | 0.94; 1.05 | 0.91; 1.07; 0.86 |
| IL-1RAcP | 1.03; 1.26 | 2.03; 1.93; 3.09 |
| ICAM-1 | 2.57; 2.68 | 9.31; 7.16; 17.58 |
| IL-1Ra | 0.46; 0.41 | 0.51; 0.40; 0.25 |
| IL-18 | 0.66; 0.71 | 0.46; 0.93; 1.39 |
| Vascular endothelial growth factor | 1.30; 1.37 | 2.93; 2.17; 4.48 |
Furthermore, we examined IL-1β-induced mRNA levels of caspase-3, IL-18, IL-1Ra, IL-1 receptor accessory protein, ICAM-1, NF-κB p50, TGF-β1, and vascular endothelial growth factor in WT and IL-18Rα ko MEF. Control cultures incubated without IL-1β were set at 1.0, and the -fold change in the number of transcripts due to IL-1β stimulation was calculated (Table 3). The largest changes in transcript numbers were observed for ICAM-1. In WT MEF cells, IL-1β induced a 2.5-fold increase, whereas in IL-18Rα ko MEF cells, the mean increase was 11-fold. Transcripts for NF-κB p50 in IL-1b-stimulated IL-18Rα ko MEF cells were approximately doubled compared with levels in WT cells stimulated with IL-1β. 2-fold increases were also observed for vascular endothelial growth factor and IL-1 receptor accessory protein in IL-18Rα ko MEF cells when compared with WT controls. There were no differences between IL-1β-induced TGF-β1, caspase-3, IL-1Ra, or IL-18 in IL-18Rα ko MEF cells compared with IL-1β-stimulated WT MEF cells.
Knockdown of IL-18Rα in Human PBMC
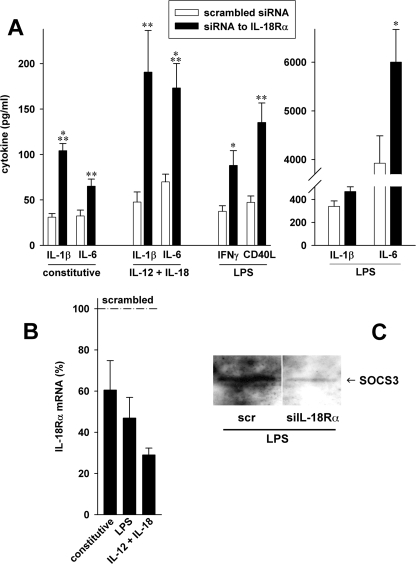
Next, we sought to confirm the observations in MEF and A549 in human primary cells. Transfection with specific siRNA reduced IL-18Rα mRNA up to 3.5-fold (Fig. 8B) compared with scrambled siRNA-transfected controls. In 7 of 10 volunteers, this reduction in IL-18Rα expression was accompanied by up to 4-fold increases in constitutive as well as LPS- and IL-12/IL-18-induced IL-1β and IL-6 production (Fig. 8A). After stimulation with LPS, IFNγ and CD40L also increased up to 2.9-fold. Investigation of SOCS3 by Western blotting revealed that, similar to IL-18Rα ko MEF cells, silencing of IL-18Rα was associated with a decrease in SOCS3 protein levels (Fig. 8C).
FIGURE 8.
Reduction of IL-18Rα in human PBMC. PBMC from 10 healthy volunteers were transfected with concentration-matched pairs of 250 nm siRNA to IL-18Rα or scrambled siRNA. A, cells were stimulated with 1 μg/ml LPS or 25 ng/ml IL-12 and 50 ng/ml IL-18. Supernatants were harvested after 20 h, and cytokine levels were determined by multiplex or regular ELISA. Means ± S.E. of absolute cytokine concentrations from 7 of the 10 donors are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001 for siIL-18Rα versus scrambled. B, cells from four donors were lysed 2 and 20 h after stimulation. RNA was isolated and subjected to reverse transcription and quantitative real time PCR. Changes in mRNA expression were calculated using the ΔΔCT method. Scrambled controls were set at 100% for each condition; percentage changes in IL-18Rα mRNA levels normalized to β-actin and glyceraldehyde-3-phosphate dehydrogenase are shown. C, lysates from the same experiment were subjected to Western blot analysis with staining for SOCS3. One representative of three blots giving similar results is shown. The bands depicted are from one gel; between the pair, one lane containing a condition not pertaining to this study was cut out.
DISCUSSION
In the initial study of IL-18Rα deficiency, murine T-cells cells lacking IL-18Rα exhibited impaired anti-CD3-induced production of IFNγ (34). Later, animal studies indicated a role for IL-18 in host responses to infection and inflammation (16); e.g. mice deficient in IL-18Rα showed an attenuated response to cigarette smoke (35). However, using the same IL-18Rα-deficient mice, dextran sodium sulfate colitis was worse (15). Injection of kainate into the brain of mice induces both IL-1β and IL-18, resulting in cerebellar ataxia (12). In mice deficient in IL-1 receptor type I or MyD88, an accelerated recovery from the ataxia was reported, consistent with proepileptic properties of kainate-induced IL-1 (12). Unexpectedly, mice deficient in IL-18Rα showed a significant delay in recovering from the self-limiting effects of kainate on locomotion. Although complete recovery was observed after 90 min in WT mice, in IL-18Rα ko mice, a return to base-line performance did not take place for 6 h. Therefore, deficiency of IL-18Rα worsened the course of disease.
The MRL/lpr mouse develops an autoimmune lupus-like disease in which a destructive nephritic process can be lethal. In MRL/lpr mice cross-bred with mice deficient in IL-18Rα, a reduction in autoantibodies, nephritis, and death was reported (36). However, Lin and Peng (37) noted that the IL-18Rα ko strain used in backcrossing with the MLR/lpr strain was itself only backcrossed three generations and probably contained genes of the 129 genetic background. Loci in the 129 strain, particularly on chromosome 1, where the IL-18Rα is located, are thought to be a source of genes protective in models of autoimmune disease. Using over eight generations of backcrossing, these investigators created a strain of mice that were homozygous for 24 disease-associated loci on chromosome 1. When crossed with the IL-18Rα ko mouse, MLR/lpr mice homozygous for the IL-18Rα deficiency state developed full-blown lupus. In fact, these mice deficient in IL-18Rα developed disease manifestations more severe than MLR/lpr mice with intact IL-18Rα. The authors concluded that IL-18 is not needed for lupus in the MLR/lpr model (37). However, this interpretation is inconsistent with the large body of evidence on neutralization of IL-18 and on increases in the expression of the IL-18Rβ (18). The alternative explanation for this observation is that IL-18Rα delivers an anti-inflammatory signal triggered by an unknown ligand. The concept of another ligand for IL-18Rα is consistent with the report comparing IL-18Rα ko mice with IL-18 ko in a model of experimental allergic encephalomyelitis (38). Moreover, antibodies to IL-18Rβ are more effective in reducing IL-18 activity than antibodies to the IL-18Rα (39).
Nonetheless, the observation that pancreatic islet allografts derived from mice deficient in the IL-18Rα exhibited a more rapid rejection compared with allografts from WT mice was unexpected (24). In contrast, allografts from mice deficient in IL-18 exhibited a statistically significant delay in rejection; moreover, islets in transgenic mice overexpressing IL-18BP were protected from toxic damage. Cultured spleen cells from IL-18Rα-deficient mice stimulated with concanavalin A produced significantly more IFNγ, TNFα, MIP-1α, and MIP-2 compared with splenocytes from IL-18 ko or WT mice (24).
To investigate these seemingly paradoxical findings, we generated fibroblast-like MEF cells from the three mouse strains in order to avoid interactions between multiple cell types (splenocytes contain macrophages and B and T cells as well as fibroblasts and other bystander cells). In vitro stimulation of these MEF cells with IL-1β and TNFα resulted in a pattern similar to splenocytes with a high level of production of IL-6, IL-1α, MIP-1α, and PGE2 in IL-18Rα ko compared with WT and IL-18 ko cells. This pattern was not only consistent in several MEF lines derived from different mothers but was also reproducible by blockade of IL-18Rα with specific antibodies or specific siRNA.
To extend these paradoxical findings beyond fibroblasts and the murine system, we created several cell lines derived from human lung epithelial A549 cells in which the IL-18Rα was stably silenced by shRNA. Moreover, we silenced IL-18Rα in primary human blood cells. A high degree of knockdown was confirmed by real time PCR and Western blotting. Consistent with MEF cells, silencing of the IL-18Rα in human A549 cell lines resulted in increased IL-1β-induced IL-6, IL-1α, and IL-8. In primary human PBMC, reduction of IL-18Rα increased the levels of LPS- and IL-12/IL-18-induced IL-1β, IL-6, IFNγ, and CD40L. Together with the other reports (12, 15, 37, 38), these data support the hypothesis that both pro- and anti-inflammatory signals converge on the IL-18Rα. In this regard, it is noteworthy that the proinflammatory effect of the anti-IL-18Rα antibodies was more pronounced in the absence of IL-18. One may speculate that this could be due to the role of IL-18 as a competitor of a putative anti-inflammatory ligand, the effect of which is more pronounced in the absence of its competitor IL-18 (hence also the reduced cytokine levels in IL-18 ko MEF).
Inhibition of cytokine signaling by the SOCS family constitutes a major negative feedback mechanism to prevent runaway inflammation (reviewed in Refs. 40 and 41). The transcription of SOCS proteins is rapidly up-regulated in cells stimulated with cytokines; the SOCS then reduce the impact of cytokines by interacting with Jaks and by other mechanisms (42). In the present report, SOCS1 and SOCS3 mRNA expression and protein production were greatly reduced in MEF lacking IL-18Rα compared with IL-18 ko and WT cells (8-fold versus up to 18-fold and 3-fold versus up to 11-fold, respectively). Importantly, this was also the case in primary PBMC in which IL-18Rα was reduced by siRNA. In MEF, the targets or inducers of these SOCS, Stat1 (target of SOCS1) and IL-6 (inducer of SOCS3), were regulated in the opposite way (i.e. exhibited higher levels) in the same cultures. We conclude that this dysregulation of SOCS1 and -3 probably contributes to the events leading to hyperinflammation in cells deficient in IL-18Rα.
The role of SOCS1 and -3 is underscored by higher levels of the proinflammatory p38α and -β MAPKs in IL-18Rα ko compared with WT and IL-18 ko MEF. Since both SOCS1 (43–45) and SOCS3 (46, 47) interfere with IL-1/TLR-MAPK/NF-κB pathway signaling, the increase in activity of these kinases in IL-18Rα ko cells may be due to less inhibition resulting from reduced levels of both SOCS. The kinase profiling experiments furthermore revealed a similar pattern in the activation of ERK1 and -2, protein kinase B/Akt, and p70 S6 kinase. These changes may also be partly caused by upstream differences in SOCS expression, since ERK and/or protein kinase B/Akt are known to be affected by SOCS1 (48) and SOCS3 (49). It is possible that such regulation of ERK, protein kinase B, and p70 S6 kinase plays a role in the hyperresponsiveness in IL-18Rα ko cells by stimulating the cellular protein synthesis machinery and by promoting survival and growth (50).
Interestingly, p38δ and MSK2/RSKβ were regulated the opposite way (i.e. increased in IL-18 ko and slightly reduced in IL-18Rα ko). Although there are hardly any published data on a possible specific role of p38δ in inflammation, MSK2 was reported to act as a negative feedback inhibitor of TLR-triggered inflammation (51) and may thus contribute to the reduction of cytokine levels in IL-18 ko cells. Of note, the facts that there was no change in the activation of HSP27 and RSK2 as well as that the entire JNK group was moderately up-regulated in both knock-out strains provide additional evidence for the specificity of the regulation of kinases and cytokines conferred by the knockouts.
We also assessed changes in IL-1β-induced gene expression of eight additional mediators. We observed a 2-fold increase in steady-state levels of NF-κB p50 in IL-18Rα-deficient MEF cells compared with WT cells 4 h following stimulation with IL-1β. Given the substantial increases in NF-κB-dependent cytokines (IL-1α, IL-6, and MIP-1α) in these cells, it seems surprising that the difference in NF-κB expression is not larger. These data thus suggest that knock-out of the IL-18Rα either affects expression of components NF-κB other than p50 (e.g. p65, as this subunit is affected by SOCS3 (45)); that NF-κB activity is regulated at another level (IKK, NIK, etc.); or, less likely, that NF-κB plays a minor role in IL-18Rα ko-induced hyperresponsiveness. Unlike NF-κB p50, ICAM-1 transcripts were over 16-fold higher than those in WT cells. ICAM-1 does participate in signal transduction with LFA-1 on T-cells, but MEF cells do not express LFA. Therefore, the elevated levels of ICAM-1 mRNA may reflect the hyperresponsiveness to IL-1β. There was little difference in the expression levels of IL-1Ra, TGF-β, and caspase-3; it thus seems unlikely that these mediators are involved in cytokine dysregulation in IL-18Rα ko cells.
In summary, the present study illustrates the paradoxical findings of exaggerated cytokine and prostaglandin responses in murine and human single cell type cultures as well as in primary human PBMC deficient in IL-18Rα compared with WT and IL-18-deficient cells. We speculate that another ligand, most likely from the IL-1 family, uses the IL-18Rα to deliver an inhibitory signal in which SOCS1 and SOCS3 play a prominent role. In the absence of the IL-18Rα, this ligand is unable to trigger its inhibitory mechanisms, such as SOCS1 and SOCS3; the result is a hyperinflammatory state with possible effector functions for the p38 and ERK MAP kinases and the protein kinase B/Akt pathway.
Acknowledgments
We thank Fabia Gamboni-Robertson and Tania Azam for excellent technical assistance. We also thank Dirk Smith of Amgen Inflammation Research (Seattle, WA) for providing the blocking anti-mouse IL-18Rα antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 15614 and CA-04 6934 (Colorado Cancer Center) (to C. A. D.). This work was also supported by Deutsche Forschungsgemeinschaft Grant 747/1-1 (to M. F. N.).
- IL
- interleukin
- IL-18BP
- IL-18-binding protein
- IL-18Rα
- IL-18 receptor α-chain
- IL-1Ra
- IL-1 receptor antagonist
- ko
- knockout
- MEF
- mouse embryonic fibroblast(s)
- PBMC
- peripheral blood mononuclear cell(s)
- shIL-18R
- shRNA to the IL-18R
- shRNA
- small hairpin RNA
- SOCS
- suppressor(s) of cytokine signaling
- WT
- wild type
- TNFα
- tumor necrosis factor α
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- PGE2
- prostaglandin E2
- IFN
- interferon
- ICAM
- intercellular cell adhesion molecule
- MAPK
- mitogen-activated protein kinase
- LPS
- lipopolysaccharide
- siRNA
- small interfering RNA.
REFERENCES
- 1.Ghayur T., Banerjee S., Hugunin M., Butler D., Herzog L., Carter A., Quintal L., Sekut L., Talanian R., Paskind M., Wong W., Kamen R., Tracey D., Allen H. (1997) Nature 386, 619–623 [DOI] [PubMed] [Google Scholar]
- 2.Gu Y., Kuida K., Tsutsui H., Ku G., Hsiao K., Fleming M. A., Hayashi N., Higashino K., Okamura H., Nakanishi K., Kurimoto M., Tanimoto T., Flavell R. A., Sato V., Harding M. W., Livingston D. J., Su M. S. (1997) Science 275, 206–209 [DOI] [PubMed] [Google Scholar]
- 3.Fantuzzi G., Puren A. J., Harding M. W., Livingston D. J., Dinarello C. A. (1998) Blood 91, 2118–2125 [PubMed] [Google Scholar]
- 4.Netea M. G., Joosten L. A., Lewis E., Jensen D. R., Voshol P. J., Kullberg B. J., Tack C. J., van Krieken H., Kim S. H., Stalenhoef A. F., van de Loo F. A., Verschueren I., Pulawa L., Akira S., Eckel R. H., Dinarello C. A., van den Berg W., van der Meer J. W. (2006) Nat. Med. 12, 650–656 [DOI] [PubMed] [Google Scholar]
- 5.Zorrilla E. P., Sanchez-Alavez M., Sugama S., Brennan M., Fernandez R., Bartfai T., Conti B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11097–11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konishi H., Tsutsui H., Murakami T., Yumikura-Futatsugi S., Yamanaka K., Tanaka M., Iwakura Y., Suzuki N., Takeda K., Akira S., Nakanishi K., Mizutani H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11340–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. (2001) Annu. Rev. Immunol. 19, 423–474 [DOI] [PubMed] [Google Scholar]
- 8.Novick D., Kim S. H., Fantuzzi G., Reznikov L. L., Dinarello C. A., Rubinstein M. (1999) Immunity 10, 127–136 [DOI] [PubMed] [Google Scholar]
- 9.Stuyt R. J., Kim S. H., Reznikov L. L., Fantuzzi G., Novick D., Rubinstein M., Kullberg B. J., van der Meer J. W., Dinarello C. A., Netea M. G. (2003) Cytokine 21, 65–73 [DOI] [PubMed] [Google Scholar]
- 10.Reznikov L. L., Kim S. H., Zhou L., Bufler P., Goncharov I., Tsang M., Dinarello C. A. (2002) J. Interferon Cytokine Res. 22, 593–601 [DOI] [PubMed] [Google Scholar]
- 11.Netea M. G., Stuyt R. J., Kim S. H., Van der Meer J. W., Kullberg B. J., Dinarello C. A. (2002) J. Infect. Dis. 185, 963–970 [DOI] [PubMed] [Google Scholar]
- 12.Andoh T., Kishi H., Motoki K., Nakanishi K., Kuraishi Y., Muraguchi A. (2008) J. Immunol. 180, 2322–2328 [DOI] [PubMed] [Google Scholar]
- 13.Sivakumar P. V., Westrich G. M., Kanaly S., Garka K., Born T. L., Derry J. M., Viney J. L. (2002) Gut 50, 812–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegmund B., Fantuzzi G., Rieder F., Gamboni-Robertson F., Lehr H. A., Hartmann G., Dinarello C. A., Endres S., Eigler A. (2001) Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1264–R1273 [DOI] [PubMed] [Google Scholar]
- 15.Takagi H., Kanai T., Okazawa A., Kishi Y., Sato T., Takaishi H., Inoue N., Ogata H., Iwao Y., Hoshino K., Takeda K., Akira S., Watanabe M., Ishii H., Hibi T. (2003) Scand. J. Gastroenterol. 38, 837–844 [DOI] [PubMed] [Google Scholar]
- 16.Boraschi D., Dinarello C. A. (2006) Eur. Cytokine Netw. 17, 224–252 [PubMed] [Google Scholar]
- 17.Gracie J. A., Forsey R. J., Chan W. L., Gilmour A., Leung B. P., Greer M. R., Kennedy K., Carter R., Wei X. Q., Xu D., Field M., Foulis A., Liew F. Y., McInnes I. B. (1999) J. Clin. Invest. 104, 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bossù P., Neumann D., Del Giudice E., Ciaramella A., Gloaguen I., Fantuzzi G., Dinarello C. A., Di Carlo E., Musiani P., Meroni P. L., Caselli G., Ruggiero P., Boraschi D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14181–14186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidal-Vanaclocha F., Fantuzzi G., Mendoza L., Fuentes A. M., Anasagasti M. J., Martín J., Carrascal T., Walsh P., Reznikov L. L., Kim S. H., Novick D., Rubinstein M., Dinarello C. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 734–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy P., Ferrara J. L. (2003) J. Lab. Clin. Med. 141, 365–371 [DOI] [PubMed] [Google Scholar]
- 21.Raeburn C. D., Dinarello C. A., Zimmerman M. A., Calkins C. M., Pomerantz B. J., McIntyre R. C., Jr., Harken A. H., Meng X. (2002) Am. J. Physiol. Heart Circ. Physiol. 283, H650–H657 [DOI] [PubMed] [Google Scholar]
- 22.Pomerantz B. J., Reznikov L. L., Harken A. H., Dinarello C. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2871–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazodier K., Marin V., Novick D., Farnarier C., Robitail S., Schleinitz N., Veit V., Paul P., Rubinstein M., Dinarello C. A., Harlé J. R., Kaplanski G. (2005) Blood 106, 3483–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis E. C., Dinarello C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16852–16857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinarello C. A., Cannon J. G., Mancilla J., Bishai I., Lees J., Coceani F. (1991) Brain Res. 562, 199–206 [DOI] [PubMed] [Google Scholar]
- 26.Petry C., Fritz G., Pfeilschifter J., Huwiler A. (2004) Biochim. Biophys. Acta 1636, 108–118 [DOI] [PubMed] [Google Scholar]
- 27.Nold M. F., Nold-Petry C. A., Pott G. B., Zepp J. A., Saavedra M. T., Kim S. H., Dinarello C. A. (2008) J. Immunol. 181, 557–565 [DOI] [PubMed] [Google Scholar]
- 28.Fantuzzi G., Reed D., Qi M., Scully S., Dinarello C. A., Senaldi G. (2001) Eur. J. Immunol. 31, 369–375 [DOI] [PubMed] [Google Scholar]
- 29.McLoughlin K., Turteltaub K., Bankaitis-Davis D., Gerren R., Siconolfi L., Storm K., Cheronis J., Trollinger D., Macejak D., Tryon V., Bevilacqua M. (2006) Mol. Med. 12, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon D. Y., Dinarello C. A. (2007) J. Biochem. Mol. Biol. 40, 562–570 [DOI] [PubMed] [Google Scholar]
- 31.Pap T., Müller-Ladner U., Gay R. E., Gay S. (2000) Arthritis Res. 2, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lonnemann G., Engler-Blum G., Müller G. A., Koch K. M., Dinarello C. A. (1995) Kidney Int. 47, 845–854 [DOI] [PubMed] [Google Scholar]
- 33.Lonnemann G., Shapiro L., Engler-Blum G., Müller G. A., Koch K. M., Dinarello C. A. (1995) Kidney Int. 47, 837–844 [DOI] [PubMed] [Google Scholar]
- 34.Hoshino K., Tsutsui H., Kawai T., Takeda K., Nakanishi K., Takeda Y., Akira S. (1999) J. Immunol. 162, 5041–5044 [PubMed] [Google Scholar]
- 35.Kang M. J., Homer R. J., Gallo A., Lee C. G., Crothers K. A., Cho S. J., Rochester C., Cain H., Chupp G., Yoon H. J., Elias J. A. (2007) J. Immunol. 178, 1948–1959 [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita K., Yamagata T., Nozaki Y., Sugiyama M., Ikoma S., Funauchi M., Kanamaru A. (2004) J. Immunol. 173, 5312–5318 [DOI] [PubMed] [Google Scholar]
- 37.Lin L., Peng S. L. (2005) Arthritis Rheum. 52, 984–986 [DOI] [PubMed] [Google Scholar]
- 38.Gutcher I., Urich E., Wolter K., Prinz M., Becher B. (2006) Nat. Immunol. 7, 946–953 [DOI] [PubMed] [Google Scholar]
- 39.Debets R., Timans J. C., Churakowa T., Zurawski S., de Waal Malefyt R., Moore K. W., Abrams J. S., O'Garra A., Bazan J. F., Kastelein R. A. (2000) J. Immunol. 165, 4950–4956 [DOI] [PubMed] [Google Scholar]
- 40.Croker B. A., Kiu H., Nicholson S. E. (2008) Semin Cell Dev. Biol. 19, 414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Shea J. J., Murray P. J. (2008) Immunity 28, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimura A., Naka T., Kubo M. (2007) Nat. Rev. Immunol. 7, 454–465 [DOI] [PubMed] [Google Scholar]
- 43.Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., Yoshimura A. (2002) Immunity 17, 583–591 [DOI] [PubMed] [Google Scholar]
- 44.Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 45.Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 46.Frobøse H., Rønn S. G., Heding P. E., Mendoza H., Cohen P., Mandrup-Poulsen T., Billestrup N. (2006) Mol. Endocrinol. 20, 1587–1596 [DOI] [PubMed] [Google Scholar]
- 47.Karlsen A. E., Rønn S. G., Lindberg K., Johannesen J., Galsgaard E. D., Pociot F., Nielsen J. H., Mandrup-Poulsen T., Nerup J., Billestrup N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12191–12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inaba M., Saito H., Fujimoto M., Sumitani S., Ohkawara T., Tanaka T., Kouhara H., Kasayama S., Kawase I., Kishimoto T., Naka T. (2005) Biochem. Biophys. Res. Commun. 328, 953–961 [DOI] [PubMed] [Google Scholar]
- 49.Zanin-Zhorov A., Tal G., Shivtiel S., Cohen M., Lapidot T., Nussbaum G., Margalit R., Cohen I. R., Lider O. (2005) J. Immunol. 175, 276–285 [DOI] [PubMed] [Google Scholar]
- 50.Buchkovich N. J., Yu Y., Zampieri C. A., Alwine J. C. (2008) Nat. Rev. Microbiol. 6, 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ananieva O., Darragh J., Johansen C., Carr J. M., McIlrath J., Park J. M., Wingate A., Monk C. E., Toth R., Santos S. G., Iversen L., Arthur J. S. (2008) Nat. Immunol. 9, 1028–1036 [DOI] [PubMed] [Google Scholar]