Abstract
The ectodomain of the Ebola virus Gp2 glycoprotein was solubilized with a trimeric, isoleucine zipper derived from GCN4 (pIIGCN4) in place of the hydrophobic fusion peptide at the N terminus. This chimeric molecule forms a trimeric, highly α-helical, and very thermostable molecule, as determined by chemical crosslinking and circular dichroism. Electron microscopy indicates that Gp2 folds into a rod-like structure like influenza HA2 and HIV-1 gp41, providing further evidence that viral fusion proteins from diverse families such as Orthomyxoviridae (Influenza), Retroviridae (HIV-1), and Filoviridae (Ebola) share common structural features, and suggesting a common membrane fusion mechanism.
The filovirus, Ebola virus, has been linked to a number of lethal outbreaks of hemorrhagic fever (1, 2). The virus genome is negative-stranded and encodes for seven structural and regulatory proteins (3, 4), including a surface glycoprotein (Gp) that is synthesized as a precursor molecule and then cleaved into two subunits (5, 6), Gp1 and Gp2, the latter of which is anchored in the membrane. Ebola Gp is encoded in two ORFs (7, 8), which produce a secreted and a membrane-anchored form of Gp, whereas all other filovirus genomes encode only the membrane-anchored Gp. The secreted Ebola Gp dimer interacts with neutrophils through a Fc γ receptor III (CD16b) (9) and the membrane-anchored form binds to a number of target cells, including endothelial cells (9, 10) and liver cells (11), and is thought to mediate viral entry. Infection and replication in endothelial cells was proposed to contribute to the severe hemorrhagic character of the late stages of disease (9, 10).
Amino acid sequences with the potential to form coiled coils have been recognized adjacent to N-terminal fusion peptides in many viral Gps (12–14) and similar α-helical models have been proposed for the HIV-gp41, Avian sarcoma virus, and Ebola virus transmembrane (TM) Gp subunits (12, 14). The x-ray crystal structures of the low-pH induced conformation of influenza virus hemagglutinin (HA) 2 (15) of a protease resistant fragment of HIV-1 env gp41 (16–18), and of a small fragment of the Moloney murine leukemia virus (MoMuLV) TM protein (19) revealed that the central part of all of these molecules is a similar, long, triple-stranded coiled coil, in the former two cases surrounded by an outer layer of antiparallel α-helices. This structural organization that places the membrane anchor in close proximity to the hydrophobic fusion peptide, at the same end of a long rod-shaped molecule, was proposed to facilitate the membrane fusion process (17) and thus viral entry.
Here, we report that the intact extracellular part of the Ebola virus subunit Gp2, lacking the N-terminal fusion peptide, can be expressed and solubilized by adding a trimeric zipper in-frame with the predicted coiled-coil region of Gp2. The oligomeric state of the chimeric molecule was characterized by chemical crosslinking and secondary structure analysis and showed a high α-helical content. Most strikingly, electron micrographs (EM) indicate a long rod-shaped structure similar to EM images observed of the low-pH-induced conformation of influenza virus HA2 (20, 21) and of fragments of HIV-1 env gp41 (22, 23), suggesting a similar role for Ebola Gp2 in membrane fusion.
METHODS
Cloning, Protein Expression, and Purification.
The Ebola gp2 gene sequence encoding residues 552–650 (Zaire subtype) (4) was amplified with synthetic oligonucleotides, and Cys-556 and Cys-609 were mutated to serines by standard PCR methods. DNA encoding GCN4 residues 250–280 with both the a and d positions of the coiled coil mutated to isoleucine (pII) (24) was synthesized as two overlapping oligonucleotides. The DNA fragments encoding pII and Gp2 were subcloned into the expression vector pRSET (Invitrogen) and transformed into Escherichia coli cells BL21 DE3/pUBS (25). The DNA sequence was verified by sequencing. After induction of protein expression by isopropyl β-d-thiogalactoside (Sigma), bacterial pellets were lysed in 50 mM Tris⋅HCl, pH 8.8/100 mM NaCl by sonication, and the supernatant was cleared by centrifugation at 40,000 rpm for 1 h. The soluble fraction was loaded onto a DEAE-Sepharose (Pharmacia) column (5 × 25 cm) and protein was eluted with a 0.1–0.4 M NaCl gradient. Fractions containing pIIgp were identified by SDS/PAGE (26), concentrated in centriprep-30 (Amicon), and further purified by gel filtration chromatography with Superdex 200 (Pharmacia) (20 mM Tris, pH 8.8/100 mM NaCl).
Chemical Crosslinking.
pIIGp(552–650) (2 mg/ml) in 50 mM Hepes, pH 8.3/100 mM NaCl was crosslinked with ethyleneglycol bis(-succinimidylsuccinate) (EGS) (Pierce). The reactions were incubated for 1 h on ice at concentrations of 0.1, 0.5, 2.0, and 5.0 mM EGS and then quenched with 50 mM glycine. Crosslinked products were analyzed under reducing conditions on SDS/PAGE (26).
Circular Dichroism.
CD spectra of pIIGp(552–650) (0.15 mg/ml; 10 mM phosphate, pH 8.0/100 mM NaCl) were recorded at 20°C and 95°C by using a 1-mm cell on an AVIV 62DS spectropolarimeter and averaging five measurements. Thermodynamic stability was measured at 222 nm by monitoring the CD signal between 20°C and 95°C with a scan rate of 1° per min. The protein concentration was calculated by measuring the OD280, with an extinction coefficient of 29,910/M per cm. The percentage of α-helical content was estimated from [Θ]222 by assuming that a value of −33,000 degree cm2 dmol−1 corresponds to 100% α-helix content (27). The baseline value of [Θ]222, equal to −2,500 deg cm2 dmol−1 of unfolded pIIGp2(552–650) was considered to be 0% α-helix content.
To follow the unfolding in guanidine hydrochloride, pH 8.0, pIIGp2(552–650) was incubated in the denaturant at concentrations of 0.5–8.0 M for 30 min at room temperature, and the [Θ]222 was plotted versus the guanidine hydrochloride concentration.
Electron Microscopy.
Samples were adsorbed onto carbon films, negatively stained with 1% sodium silcotungstate (pH 7.0), and examined with a JEOL 1200EX microscope at 100 kV as described previously (28, 29).
RESULTS
Expression and Oligomeric State of an Ebola Virus Gp2 Subunit Chimera.
Residues 552–650 of the extracellular region of Ebola Gp2, without the N-terminal fusion peptide (Fig. 1a), were expressed as a fusion protein in E. coli. The heptad positions a and d in the GCN4 coiled coil were mutated to isoleucine, which has been shown to induce a trimeric coiled-coil conformation (24) and placed in register with a proposed coiled-coil segment in Ebola Gp2 (14) (Fig. 1a). This Gp2-chimera [pIIGp2(552–650)] is highly overexpressed in E. coli and was purified from the soluble fraction of bacterial lysates. Constructs lacking pIIGCN4 were expressed only as insoluble inclusion bodies. In addition, the solubility was dependent on the inclusion of residues 644–650 (data not shown). A difference in mobility of reduced and nonreduced pIIGp2(552–650) (Fig. 1b, lanes 1 and 2) indicated that the disulfide bond between Cys-601 and Cys-608 had formed properly. Cysteine residues 556 and 609 were mutated to Ser to avoid nonspecific disulfide crosslinking. Chemical crosslinking indicates a trimeric oligomeric state. After crosslinking two new bands appear (Fig. 1b, lanes 3–6), migrating at 26 and approximately 44 kDa, corresponding to a dimeric and trimeric form of pIIGp2(552–650).
Figure 1.
(a) Primary structure of the Ebola virus Gp. The numbers indicate amino acid positions (including the signal peptide) for (i) potential carbohydrate sites (4), (ii) the cleavage of Gp into Gp1 and Gp2 occurring at position 501 (5, 6); and (iii) Gp2 anchoring in the membrane by amino acids 651–672. The sequence of the expressed construct pIIGp2(552–650) is shown with the a and d heptad positions of pIIGCN4 in-frame with the predicted a and d positions of the Ebola virus TM protein (14). The cysteines (601 and 608) involved in disulfide formation are indicated and those changed to Ser (556 and 609) are indicated by S. (b) Chemical crosslinking of pIIGp2(552–650). Crosslinked products were separated on 15% SDS/PAGE and bands are stained with Coomassie brilliant blue. Lane 1, not reduced; lane 2, reduced; lanes 3–6 with ethyleneglycol bis(-succinimidylsuccinate) crosslinking concentrations of 0.1, 0.5, 2.0, and 5.0 mM. Molecular weight standards are shown. Bands corresponding to monomer, dimer, and trimer are indicated.
High α-Helical Content and Thermostability.
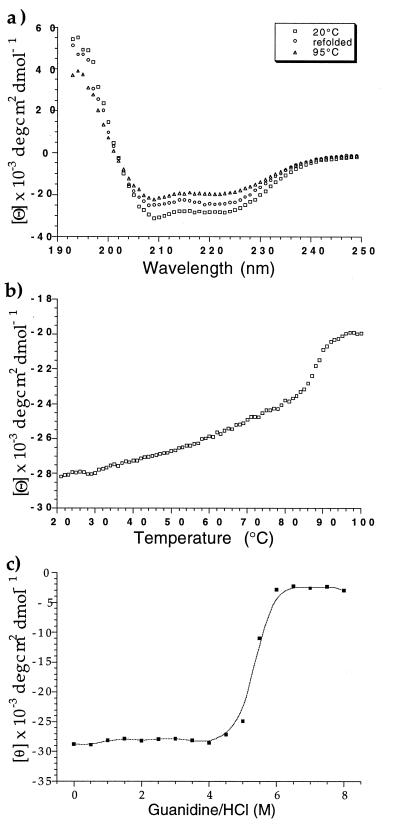
The CD spectrum of pII pIIGp2(552–650) measured from 195 nm to 250 nm had a maximum at 194 nm and minima at 208 and 222 nm characteristic of α-helices (Fig. 2a). The estimated α-helical content is approximately 84%. Thermal unfolding is observed by monitoring the changes in ellipticity at 222 nm. No clear unfolding transition occurs; instead, the increasing temperature slightly, but steadily, reduces the overall helical content and shows some cooperative transition at approximately 90°C. No complete unfolding is observed when the temperature is raised to 98°C (Fig. 2b). This partial unfolding also is shown in a spectrum recorded at 95°C (Fig. 2a), which indicates that the α-helical content is reduced to only 56%. Refolding is not complete as the α-helical content is restored to only 71% (Fig. 2a). Complete unfolding is achieved in guanidine hydrochloride with a cooperative transition at 5.5 M and complete unfolding at 6 M as monitored by CD (Fig. 2c).
Figure 2.
(a) CD spectra of pIIGp2(552–650) recorded at 20°C and 95°C, and after one cycle of heating to 95°C and cooling down to 20°C (refolded). (b) Thermal denaturation curve of pIIGp2(552–650) recorded at 222 nm with a scan rate of 1°C per min. (c) Denaturation of pIIGp2(552–650) in guanidine hydrochloride. The CD signal was monitored at 222 nm.
Rod-Like Structure Revealed by Electron Microscopy.
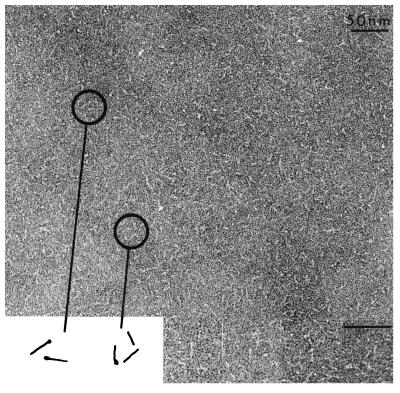
Electron microscopy of pIIGp2(552–650) (Fig. 3) reveals a rod-like shape. Measurement of 70 molecules indicated an average length of about 13 nm. This length is consistent with the presence of an α-helical coiled coil spanning the 72 residues from the N-terminal end of the GCN4 segment to residue 593 of Gp2, where the helix terminates in the MoMuLV TM protein (19). Subtracting the calculated length of the GCN4 coiled-coil domain (32 aa) suggests that the length of Gp2(552–650) contributes about 8 nm to the length of the chimeric molecule. There is a remarkable similarity in the electron micrograph images of the Ebola pIIGp2(552–650) rods and the fragments derived from the low-pH-induced conformation of influenza virus HA2 and HIV-1 gp41 (Fig. 4). Fig. 4 shows rods of influenza virus HA2 generated by lowering the pH of viral HA and proteolysis (TBHA2), which measure about 13.5 nm in length (20). Expression of the same fragment of influenza virus HA2 (TBHA2) in E. coli results in the same image (Fig. 4) (21). In addition, fragments of HIV-1 gp41 produced in insect cells (Fig. 4) results in rods with an average length of 13 nm (22) and those assembled from peptides expressed in E. coli produce rods with an average length of 12 nm (Fig. 4) (23).
Figure 3.
Electron micrographs of pIIGp2(552–650). Diagrammatic representations of the molecules are shown. The average length of the rods is 13 nm.
Figure 4.
Electron micrographs of influenza virus TBHA2 (proteolytic fragment of low pH-treated viral HA, HA2 residues 38–175; (20); influenza virus E.TBHA2 (E. coli-expressed HA2 residues 38–175; (21); HIV-1 Gp41 (residues 21–166, expressed in insect cells) (22); HIV-1 pIIGpPT (proteolytic fragment derived from E. coli; pIIGCN4 and gp41 residues 30–90 and 107- 167/+12; (23); and Ebola pIIGp2(552–650). Diagrammatic representations of the molecules are shown. Micrographs showing fields of particles are found in refs. 20–23.
DISCUSSION
The extracellular part of the Ebola virus TM protein, Gp2, was solubilized by adding a trimeric GCN4 zipper to the N terminus of the molecule in place of the fusion peptide, in-frame with a predicted coiled-coil region (14), using a strategy that was successful with the HIV-1 fusion protein gp41 (17, 23). The Ebola Gp2 chimera forms trimers in solution as detected by chemical crosslinking. A trimeric oligomerization state also has been reported for the closely related Gp from Marburg virus (30), indicating that it was not induced in Ebola Gp2 by the trimeric GCN4 zipper. A number of retroviral membrane-fusion proteins (19, 22, 31–34) as well as the influenza virus HA (35) are also trimers.
The Ebola virus TM ectodomain shares additional structural features with influenza virus HA2, HIV-1 gp41, and MoMuLV TM. Secondary structure analysis by CD revealed a high α-helical content and a high thermostability characteristic of these coiled-coil-containing proteins (20–22, 32, 34, 36). The Ebola pIIGp2(552–650) protein did not unfold completely at high temperature. This partial unfolding might be caused by aggregation at elevated temperatures (W.W., unpublished observations) that stabilizes some of the helices. The irreversibility observed in thermal denaturation and renaturation (Fig. 2a) also suggests aggregation. In contrast to thermal denaturation, guanidine HCl causes a cooperative unfolding of pIIGp2(552–650) as expected for a folded and monodisperse molecule in solution.
The length of the Ebola pIIGp2(552–650) measured by electron microscopy, approximately 13 nm, is that expected for a structure containing an α-helical coiled coil from the N-terminal GCN4 sequence to approximately residue 593. The electron micrograph (EM) images of Ebola pIIGp2(552–650) show thin rods, very similar to EM images of HIV-1 gp41 (22, 23), and the low-pH-induced conformation of the influenza virus HA2 (20, 21, 29). This similarity in shape, the high α-helical content, and the trimeric oligomeric state provide direct evidence for a long α-helical coiled coil as the central structural feature of Ebola Gp2.
Viral envelope Gps, like influenza virus HA and HIV-1 env, undergo a conformational change at an early stage of infection (37–39) that is required for their membrane fusion activities. Influenza virus HA2 folds spontaneously into this fusion-active (low-pH-induced) conformation, when expressed without the receptor binding domain HA1 (21, 40). The structural similarities among influenza virus HA2 (15) without HA1, HIV-1 gp41 without gp120 (16, 17), and MoMuLV TM without the SU (surface subunit) receptor binding domain (19) suggest that viral membrane-fusion proteins, including the Ebola virus Gp2, fold spontaneously into the fusion-active conformation in the absence of the receptor binding domain (21, 22).
The distribution of sequence conservation along the length of Gp2 of Ebola virus and Marburg virus (72% sequence similarity) (4) is consistent with the two-layered α-helical structure observed in gp41 (16–18) and inferred from the structure of a fragment of MoMuLV TM (19). The sequence within the short disulfide-linked loop (Fig. 1a) has the highest sequence conservation, suggesting a conserved function. It has been proposed to be immunosuppressive (41) comparable to sequences found in C- and D-type retroviral TM subunits (42). This similarity to the retrovirus sequences (14) provides evidence that the central coiled coil of Gp2 will end before this disulfide loop and that the path of the polypeptide will reverse to form an outer α-helical layer. As in gp41 sequences, the N-terminal sequences that are proposed to form the central coiled coil in Gp2s (without the fusion peptide) have higher sequence similarity (77%) than the C-terminal sequences (57%) that are proposed to form the antiparallel, outer layer of α-helices after the disulfide loop region.
The crystal structures of influenza virus HA at neutral pH (43) and of the low-pH induced conformation of HA2 (15) define a dramatic conformational refolding in HA2 induced by the pH that triggers viral entry by membrane fusion. Two major rearrangements take place (15): the extension of a triple-stranded coiled coil that moves the fusion peptide over 100 Å, as anticipated by a peptide model (44), and second, a reversal in the polypeptide direction at the C terminus of the coiled coil to allow the formation of an antiparallel outer layer of α-helices packed on the central coiled coil. A similar architecture was found in gp41 (16–18). Based on the crystal structure of a proteolytic fragment of HIV-1 gp41, we proposed that the antiparallel packing of the outer layer of α-helices on the inner triple-stranded coiled coil, also seen in the low pH-induced conformation of influenza virus HA2 and inferred from the structure of a fragment of MoMuLV TM, would place the N-terminal fusion peptide near the C-terminal TM anchor at one end of a long rod, a rearrangement that could allow the cellular and viral membranes to be in close proximity and promote membrane fusion (Fig. 3; ref. 17). The possibility that this architectural feature of viral proteins, a long rod with attachments for two membranes at one end of the rod, was suggested to be shared by the membrane protein complex that directs vesicle fusion in eukaryotes (45). There, the v-SNARE and t-SNARE proteins (46), thought to contain coiled coils (47–49), have been shown to form a parallel rod-like complex, with their membrane anchors at the same end (45, 50).
Acknowledgments
We thank Gary Nabel and Anthony Sanchez for the DNA clone of the Ebola virus Gp. W.W. was supported by the Howard Hughes Medical Institute. This work was supported by a supplement to the National Institute of Allergy and Infectious Diseases grant (5RO1AI13654–20) for Expanded International Research on Emerging and Re-Emerging Diseases, the Medical Research Council (U.K.), and the Howard Hughes Medical Institute. D.C.W. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HA
hemagglutinin
- Gp
glycoprotein
- MoMuLV
Moloney murine leukemia virus
- TM
transmembrane
References
- 1.Bowen E T, Lloyd G, Harris W J, Platt G S, Baskerville A, Vella E E. Lancet. 1977;1:571–573. doi: 10.1016/s0140-6736(77)92001-3. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Morbid Mortal Week Rep. 1995;44:381–384. [Google Scholar]
- 3.Feldmann H, Muhlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H D. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 5.Volchkov, V. E., Volchkova, V. A., Slenczka, W., Klenk, H.-D. & Feldmann, H. (1998) Virology, in press. [DOI] [PubMed]
- 6.Volchkov V E, Feldmann H, Volchkova V A, Klenk D-D. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z-Y, Delgado R, Xu L, Todd R F, Nabel E G, Sanchez A, Nabel G J. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- 10.Schnittler H J, Mahner F, Drenckhahn D, Klenk H D, Feldmann H. J Clin Invest. 1993;91:1301–1309. doi: 10.1172/JCI116329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker S, Spiess M, Klenk H D. J Gen Virol. 1995;76:393–399. doi: 10.1099/0022-1317-76-2-393. [DOI] [PubMed] [Google Scholar]
- 12.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. AIDS Res Hum Retroviruses. 1989;4:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 13.Chambers P, Pringle C R, Easton A J. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 14.Gallaher W R. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 15.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 16.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 17.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 18.Tan K, Liu J, Wang J, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 20.Ruigrok R W, Aitken A, Calder L J, Martin S R, Skehel J J, Wharton S A, Weis W, Wiley D C. J Gen Virol. 1988;69:2785–2795. doi: 10.1099/0022-1317-69-11-2785. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Wharton S A, Weissenhorn W, Calder L J, Hughson F M, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissenhorn W, Wharton S A, Calder L J, Earl P L, Moss B, Aliprandis E, Skehel J J, Wiley D C. EMBO J. 1996;15:1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenhorn W, Calder L J, Dessen A, Laue T, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1997b;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann U, Mattes R E, Buckel P. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.O’Shea E K, Rutkowski R, Kim P S. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 28.Wrigley N G, Brown E, Chillingworth R K. J Microsc (Oxford) 1983;130:225–232. doi: 10.1111/j.1365-2818.1983.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 29.Wharton S A, Calder L J, Ruigrok R W, Skehel J J, Steinhauer D A, Wiley D C. EMBO J. 1995;14:240–246. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldmann H, Will C, Schikore M, Slenczka W, Klenk H D. Virology. 1991;182:353–356. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einfeld D, Hunter E. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 33.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 34.Fass D, Kim P S. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 35.Wiley D C, Skehel J J, Waterfield M. Virology. 1977;79:446–448. doi: 10.1016/0042-6822(77)90371-3. [DOI] [PubMed] [Google Scholar]
- 36.Ruigrok R W, Martin S R, Wharton S A, Skehel J J, Bayley P M, Wiley D C. Virology. 1986;155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 37.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sattentau Q, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones P, Korte T, Blumenthal R. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 40.Carr C M, Chaudhry C, Kim P S. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volchkov V E, Blinov V M, Netesov S V. FEBS Lett. 1992;305:181–184. doi: 10.1016/0014-5793(92)80662-z. [DOI] [PubMed] [Google Scholar]
- 42.Cianciolo G J, Copeland T D, Oroszlan S, Snyderman R. Science. 1985;230:453–455. doi: 10.1126/science.2996136. [DOI] [PubMed] [Google Scholar]
- 43.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 44.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 45.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 46.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof T C, Niemann H. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kee Y, Lin R C, Hsu S C, Scheller R H. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 49.Weimbs T, Low S H, Chapin S J, Mostov K E, Bucher P, Hofmann K. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Söllner T H, Rothman J E. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]