Abstract
The endonuclease Argonaute2 (Ago2) mediates the degradation of the target mRNA within the RNA-induced silencing complex. We determined the binding and cleavage properties of recombinant human Ago2. Human Ago2 was unable to cleave preformed RNA duplexes and exhibited weaker binding affinity for RNA duplexes compared with the single strand RNA. The enzyme exhibited greater RNase H activity in the presence of Mn2+ compared with Mg2+. Human Ago2 exhibited weaker binding affinities and reduced cleavage activities for antisense RNAs with either a 5′-terminal hydroxyl or abasic nucleotide. Binding kinetics suggest that the 5′-terminal heterocycle base nucleates the interaction between the enzyme and the antisense RNA, and the 5′-phosphate stabilizes the interaction. Mn2+ ameliorated the effects of the 5′-terminal hydroxyl or abasic nucleotide on Ago2 cleavage activity and binding affinity. Nucleotide substitutions at the 3′ terminus of the antisense RNA had no effect on human Ago2 cleavage activity, whereas 2′-methoxyethyl substitutions at position 2 reduced binding and cleavage activity and 12–14 reduced the cleavage activity. RNase protection assays indicated that human Ago2 interacts with the first 14 nucleotides at the 5′-pole of the antisense RNA. Human Ago2 preloaded with the antisense RNA exhibited greater binding affinities for longer sense RNAs suggesting that the enzyme interacts with regions in the sense RNA outside the site for antisense hybridization. Finally, transiently expressed human Ago2 immunoprecipitated from HeLa cells contained the double strand RNA-binding protein human immunodeficiency virus, type 1, trans-activating response RNA-binding protein, and deletion mutants of Ago2 showed that trans-activating response RNA-binding protein interacts with the PIWI domain of the enzyme.
RNA interference is a mechanism by which double-stranded RNA triggers the loss of RNA of homologous sequence (1). Long double strand RNAs are processed by the double strand endonuclease Dicer into short RNA duplexes (siRNA)2 ranging from 21 to 23 nucleotides in length (2). The double strand RNA-binding proteins Dicer and human immunodeficiency virus, type 1, trans-activating response RNA-binding protein (TRBP) transfer the siRNAs to the RNA-induced silencing complex (RISC) (3). The antisense strand of the siRNA binds to the RISC endonuclease Argonaute 2 (Ago2), which then cleaves the target mRNA at a single phosphodiester bond bridging the ribonucleotides opposing the 10th and 11th nucleotide from the 5′ terminus of the antisense strand (4–11).
The structure-activity relationships of siRNAs in human cultured cells have been studied extensively, but these types of studies offer few insights into the underlying mechanisms contributing to the observed activities of the siRNA and, in particular, their interaction with the RISC endonuclease human Ago2. Surprisingly, the little that is known about the interaction between human Ago2 and the substrate comes from a single report describing the preliminary characterization of recombinant human Ago2 (11). Specifically, human Ago2 cleavage activity was magnesium-dependent, and the antisense RNA containing a phosphate at the 5′ terminus exhibited greater cleavage activity compared with the antisense RNA with a 5′-hydroxyl. The enzyme was unable to cleave a DNA target or use a DNA antisense strand to trigger the cleavage of a complementary RNA (11). In addition, UV cross-linking experiments showed that single strand but not double strand RNA was able to cross-link with the recombinant enzyme. Finally, unlike RISC activity from cellular extracts, which has been shown to catalyze multiple rounds of cleavage, recombinant Ago2 exhibited single-turnover kinetics (11, 12).
The architecture of the human Ago2 protein consists of a PIWI domain at the amino terminus, a centrally located Mid domain and a PAZ domain at the carboxyl terminus (13–17). The PIWI domain constitutes the catalytic domain of the enzyme and exhibits a three-dimensional structure similar to RNase H, sharing the same aspartic acid-aspartic acid-glutamic acid (DDE) catalytic triad and metal cofactor requirements (10, 16, 17). Recently, the structures of argonaute from Thermus thermophilus and Archaeoglobus fulgidus bound to the antisense strand have been solved (15, 18). The structures show that the PAZ, Mid, and PIWI domains form an extended nucleic acid binding surface for the antisense strand. In addition, a basic binding pocket positioned within the Mid domain and a basic cleft in the PIWI domain were shown to bind, respectively, the 5′-terminal phosphate and the backbone at the 5′-pole of the antisense strand (15, 18). Aside from the two 3′-terminal nucleotides of the antisense strand, which were shown to bind a hydrophobic pocket within the PAZ domain, no interactions were observed between the enzyme and the 3′-pole of the antisense strand. An important difference between the structures of the two prokaryotic proteins was that the A. fulgidus protein contained a tyrosine residue positioned in the basic binding pocket, which formed a stacking interaction with the heterocycle base of the 5′-terminal nucleotide in the antisense strand. The human Ago2 protein appears to differ significantly from the prokaryotic argonaute proteins in that the key amino acids that make up the nucleic acid binding surface of the prokaryotic proteins are not conserved in the human enzyme. Consequently, the structures of the prokaryotic proteins appear to offer limited insights into the interaction between the human enzyme and the antisense strand of the siRNA.
Given that Ago2 is responsible for the siRNA-mediated cleavage of the target RNA, understanding the properties important for the interaction between the antisense strand and Ago2 could lead to the identification of siRNA configurations with improved potency. To better understand the substrate specificity of human Ago2, we determined the cleavage activities, binding affinities, and binding kinetics of human Ago2 for various antisense oligonucleotides. The antisense oligonucleotides were designed to evaluate the interaction between human Ago2 and various regions in the antisense RNA, including the 5′ and 3′ termini and 2′-hydroxyl. The activities and binding affinities were compared for two different preparations of the enzyme as follows: a human Ago2 protein containing a glutathione S-transferase tag (GST-Ago2) that was expressed in insect cells and purified to homogeneity and an HA-tagged protein that was expressed in HeLa cells and immunoprecipitated with HA antibody (HA-Ago2). In addition, we evaluated the effects of divalent cation metals on the substrate specificity of human Ago2. Finally, we identified endogenous TRBP in the immunoprecipitated HA-Ago2 preparation and demonstrated using deletion mutants that the PIWI domain of Ago2 interacts with TRBP.
MATERIALS AND METHODS
Preparation of Oligonucleotides and 32P-Labeled Substrate
Synthetic ribonucleotide, deoxyribonucleotide, and 2-methoxy-modified oligonucleotides were manufactured by Dharmacon Research, Inc. (Lafayette, CO). Antisense oligoribonucleotides containing abasic and 2′-methoxyethyl substitutions were synthesized as described previously (19–21). The oligonucleotides were 5′-end-labeled with 32P using 10 units of T4 polynucleotide kinase (Promega, WI), 200 pmol (5000 Ci/mmol) [α-32P]ATP (GE Healthcare), 40 pmol of oligonucleotide, 40 mm Tris, pH 7.5, 10 mm MgCl2, and 5 mm DTT. The kinase reaction was incubated at 37 °C for 30 min. The oligonucleotides were 3′-end-labeled using [32P]cytidine bisphosphate and RNA ligase as described previously (22). The labeled oligonucleotide was purified by electrophoresis on a 12% denaturing polyacrylamide gel (23). The specific activity of the labeled oligonucleotide is ∼3000–8000 cpm/fmol.
Preparation of Human HA-Ago2 Expression Vector
cDNA encoding full-length human Ago2 was generated by reverse transcription-PCR from HeLa-extracted total RNA. Human Ago2 with an amino-terminal HA epitope was generated by PCR with the following primers: phCMV2–3′_E2 (5′-TAA CAA TGT ACC CAT ACG ATG TTC CGG ATT ACG CTT ACT CGG GAG CCG GCC CCG CAC TTG-3′) and phCMV2–3′_E2 (5′-CCC GGG CCC GCG GTA CCG TCG ACT GCA GAA TTA AGC AAA GTA CAT GGT GCG CAG AGT GT-3′). Plasmid pE2_N-HA was constructed by homologous recombination of the human Ago2 PCR product generated from the HeLa cDNA with a linear phCMV-2/Xi vector according to the manufacturer's protocol (Gene Therapy Systems).
Preparation of Recombinant Human GST-Ago2
cDNA from the previously described amino-terminal hAgo2 plasmid construct was subcloned into the pGEX-3x vector (GE Healthcare) using the 5′ BamHI and 3′ EcoRI sites. DNA from the PCR amplification of the GST-Ago2 fusion DNA using 5′ EcoRI and 3′ NotI primers was subcloned into the EcoRI and NotI sites of the baculovirus shuttle vector pENTR2B (Invitrogen). The shuttle vector was subsequently transferred into baculovirus using BaculoDirect baculovirus expression system (Invitrogen) for generation of high viral titers in Sf9 insect cells. Sf9 cells were plated at 107 cells per 3 ml and infected with three viruses per cell. After 72 h the infected cells were harvested using cell scrapers and gently lysed in lysis buffer (Dulbecco's phosphate-buffered saline solution, 0.5% Nonidet P-40, 1 mm DTT, protease inhibitors). Lysate was then centrifuged at 10,000 × g for 30 min after which time the pellet was resuspended and subjected to sonication. Both supernatant and sonicated pellet fractions were run over a GST affinity purification column on a AKTA basic high pressure liquid chromatograph (Amersham Biosciences). Human GST-Ago2 protein recovered by this method yielded purities greater than 95% (supplemental Fig. 1).
Cleavage Activity and Binding Assays for Recombinant Human GST-Ago2
For human GST-Ago2 cleavage activity, 1 ng of recombinant Ago2 was incubated with 10 nm antisense oligoribonucleotide in cleavage buffer (10 mm Tris, pH 7.5, 100 mm KCl, 2 mm MgCl2, protease inhibitor, and 0.5 mm DTT) for 2 h at 37 °C. 0.1 nm 32P-labeled target RNA was added, and cleavage reactions were quenched at 30 min in gel loading buffer (Ambion, TX). Cleavage products were resolved by denaturing PAGE and quantitated with Storm 850 PhosphorImager (GE Healthcare). The errors reported for the cleavage activities are based on three trials.
The binding affinity for recombinant human GST-Ago2 was determined using homologous and competitive saturation. Briefly, 32P-labeled oligoribonucleotide was incubated with 1 ng of GST-Ago2 enzyme in the presence of increasing concentrations of unlabeled homologous oligoribonucleotide or competitor oligoribonucleotide. The enzyme was added to 50 μl of Miltenyi Biotec μMACS Anti-GST Microbeads (Auburn, CA) and allowed to bind for 2 h. The beads were then loaded onto paramagnetic columns supplied by Miltenyi, and unbound 32P-labeled oligoribonucleotide was washed away with 1 ml of cleavage buffer. The remaining, i.e. bound, radioactivity was counted in a scintillation counter, and the bound counts were plotted as a function of the concentration of the unlabeled oligoribonucleotide. The dissociation constants (Kd) were calculated from nonlinear least squares fit of the data to the equation for the one-site binding hyperbola (Y = Bmax·X/Kd + X, where Bmax is maximum binding and Kd is the concentration of unlabeled oligoribonucleotide at half-maximal binding). The errors reported for the binding affinities are based on three trials.
The association (k1) and dissociation (k−1) rates for the binding of the oligoribonucleotide to Ago2 were determined by adding 32P-labeled oligoribonucleotide to the enzyme coupled to the GST beads and quenching the reactions at various time points by washing the unbound oligoribonucleotide off the beads. The bound counts were plotted as a function of time, and the observed association rate (kobs) was calculated from nonlinear least squares fit of the data to the equation for the one-site binding hyperbola (Y = Bmax·X/Kobs + X, where Bmax is maximum binding, and kobs is the amount of time required for the oligoribonucleotide to reach half-maximal binding). The kobs were plotted as a function oligoribonucleotide concentration, and the slope and y intercept from the linear least squares fit of the data were used to calculate, respectively, the k1 and k−1.
The binding affinity of Ago2 containing the 19-nucleotide antisense RNA for various target sense RNAs were determined as described above with the exception that the enzyme coupled to the GST beads was incubated with 1 μm antisense RNA, and the unbound antisense RNA was washed away prior to adding the 32P-labeled sense target RNA. The data were analyzed as described above, and the errors reported for the binding affinities are based on three trials.
Cleavage Activity and Binding Assays for Immunoprecipitated HA-Ago2
The HeLa cell line (CCL-2) used in these experiments was obtained from the American Type Culture Collection (Manassas, VA) and was cultured in Dulbecco's modified Eagle's medium-high glucose (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). HeLa cells were seeded at an initial density of 3,200,000 cells/150-mm plate (BD Biosciences) on the day prior to the transfection and incubated at 37 °C and 10% CO2 in Dulbecco's modified Eagle's medium-high glucose containing 10% fetal bovine serum. The next day, plasmid pE2-N-HA was delivered to cells (typically at 70–75% confluency) using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions and incubated for 24 h at 37 °C. Cells were combined into a 15-ml conical tube upon harvesting with trypsin, washed twice with 1 ml of cold phosphate-buffered saline, and centrifuged at 1000 × g. The cell pellet was resuspended in 500 ml of lysis buffer (150 mm NaCl, 0.5% Nonidet P-40, 2 mm MgCl2, 2 mm CaCl2, 20 mm Tris, pH 7.5) that contained a Complete mini protease inhibitor tablet (Roche Applied Science) and 1 mm DTT, then passed 10 times through a 1-cc U-100 insulin syringe (BD Biosciences). All lysate supernatants were clarified using a 10,000 × g clearing spin for 10 min, and protein concentrations were determined using QuickStart Bradford protein assay (Bio-Rad).
1.8 and 0.18 mg of total protein was added for, respectively, the cleavage activity and binding assays to 50 μl of Miltenyi Biotec μMACS anti-HA microbeads (Auburn, CA) and allowed to bind for 2 h. The beads were then loaded onto paramagnetic columns supplied by Miltenyi and washed with 1 ml of cleavage buffer. The cleavage activities and binding affinities for immunoprecipitated human HA-Ago2 were determined as described above.
RESULTS
Purified (GST-Ago2) and Immunoprecipitated (HA-Ago2) Recombinant Human Ago2 Preparations Exhibit Similar Substrate Specificities
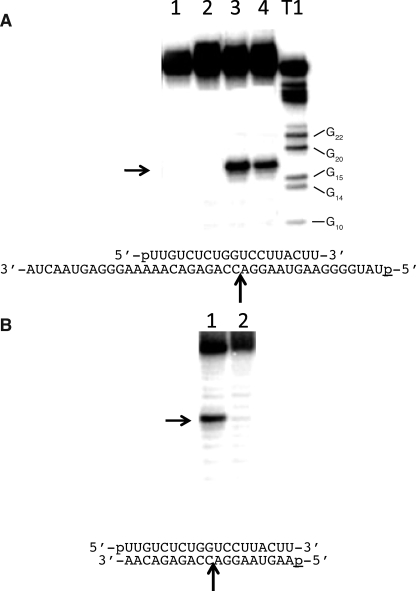
The cleavage activities for human Ago2 were determined for two different preparations of the enzyme as follows: a human Ago2 protein containing a glutathione S-transferase tag (GST-Ago2) that was expressed in insect cells and purified to homogeneity and an HA-tagged protein that was expressed in HeLa cells and immunoprecipitated with an HA antibody (HA-Ago2) (supplemental Fig. 1). Fig. 1 shows the cleavage activities of purified GST-Ago2 and immunoprecipitated HA-Ago2 enzyme preparations in which the enzymes were presented with the 19-nucleotide antisense RNA prior to adding the 40-nucleotide target RNA or with preformed RNA duplexes containing the 19-nucleotide antisense RNA annealed to either a 19- or 40-nucleotide sense RNA. Cleavage of the target RNA was only observed for the reactions in which either the HA-Ago2 or GST-Ago2 enzyme preparations were incubated with the antisense RNA prior to adding the target sense RNA (Fig. 1, A and B). Both enzyme preparations produced the canonical cleavage site for Ago2, i.e. a single cleavage site on the sense RNA opposing the 10th and 11th nucleotides from the 5′ terminus of the antisense RNA (Fig. 1, A and B). No cleavage activity was observed for either the GST-Ago2 or HA-Ago2 preparations containing the preformed 19/19 or 19/40 nucleotide RNA duplexes (Fig. 1, A and B).
FIGURE 1.
Cleavage activity of human Ago2 for preformed RNA duplexes and single strand RNA. A, denaturing PAGE of 40-nucleotide sense RNA digested with human Ago2. Lanes 1 and 2, 19-nucleotide antisense RNA and 40-nucleotide sense RNA were annealed for 1 h prior to incubation with, respectively, GST-Ago2 or HA-Ago2 for 1 h. Lanes 3 and 4, 19-nucleotide antisense RNA was incubated with, respectively, GST-Ago2 or HA-Ago2 for 1 h prior to incubation with the 40-nucleotide sense RNA for an additional 1 h. T1, 40-nucleotide sense RNA digested with RNase T1 for 10 min at 24 °C. The positions of the T1 digestions adjacent to guanidine residues are shown next to the ladder. Arrow indicates the position of the human Ago2 cleavage site. The sequences of the 19-nucleotide antisense RNA and 5′-32P-labeled (p) 40-nucleotide sense RNA are shown below the gel. B, denaturing polyacrylamide gel of 19-nucleotide sense RNA digested with GST-Ago2. Lane 1, 19-nucleotide antisense RNA was incubated with GST-Ago2 for 1 h prior to incubation with the 19-nucleotide sense RNA for an additional 1 h. Lane 2, 19-nucleotide antisense RNA and 19-nucleotide sense RNAs were annealed for 1 h prior to incubation with GST-Ago2 for 1 h. Arrow indicates the position of the human Ago2 cleavage site. The sequences of the 19-nucleotide antisense RNA (top sequence) and 5′-32P-labeled (p) 19-nucleotide sense RNA (bottom sequence) are shown below the gel.
To better understand the lack of cleavage activity observed for the preformed RNA duplexes, we determined the binding affinities of the Ago2 preparations for the single and double strand RNAs tested in Fig. 1. Both the GST-Ago2 and HA-Ago2 enzyme preparations exhibited significantly tighter binding affinities for the single strand RNA compared with the RNA duplexes (Table 1). Specifically, Ago2 bound the single strand RNA (19-as) 70–100-fold tighter than the RNA duplexes (19-as/40-s and 19-as/19-s) (Table 1). The weak binding affinity of human Ago2 for double strand RNA is consistent the lack of Ago2 cleavage activity observed for the RNA duplexes.
TABLE 1.
Dissociation constants for human Ago2 preparations binding to single and double strand RNA
The dissociation constants (Kd) for recombinant (GST-Ago2) and immunoprecipitated (HA-Ago2) Ago2 binding to 19-nucleotide antisense RNA (19-as), 19-nucleotide antisense RNA (19-as) hybridized to 40-nucleotide sense RNA (40-s), and 19-nucleotide antisense RNA (19-as) hybridized to 19-nucleotide sense RNA (19-s) were determined as described under “Materials and Methods.” The errors reported for the Ago2 binding affinities are based on three trials.
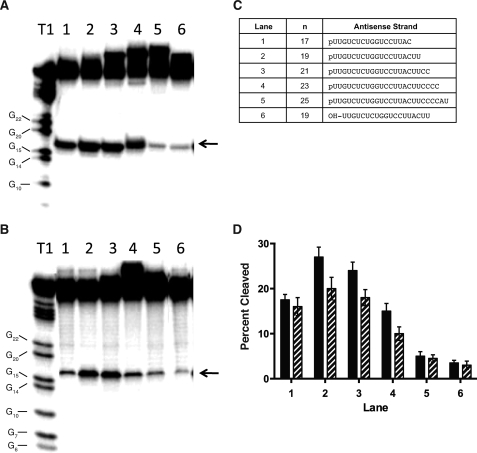
The optimum length of the antisense RNA for human Ago2 cleavage activity was determined for the GST-Ago2 and HA-Ago2 enzyme preparations (Fig. 2). The length of the antisense RNA was increased from 17 to 25 nucleotides in two nucleotide increments by introducing additional ribonucleotides to the 3′ terminus of the RNA, and the resulting antisense RNAs were incubated with the enzyme preparations prior to adding the 32P-labeled target (Fig. 2C). Similar cleavage sites were observed for both the GST-Ago2 and HA-Ago2 preparations containing the various length antisense RNAs (Fig. 2, A and B). The cleavage activities for both enzyme preparations exhibited a bell-shaped response with respect to antisense RNA length, with the 19- and 21-nucleotide antisense RNAs resulting in the greatest cleavage of the sense RNA (Fig. 2D). We also show that the GST-Ago2 and HA-Ago2 enzyme preparations containing the 19-nucleotide antisense RNA with a 5′-terminal hydroxyl exhibited significantly weaker cleavage activity compared with the 19-nucleotide antisense RNA containing a 5′-phosphate (Fig. 2). Taken together, these data suggest that the GST-Ago2 enzyme expressed and purified from insect cells and HA-Ago2 preparation expressed and immunoprecipitated from HeLa cells using HA antibody exhibit similar cleavage and binding specificities for single and double strand RNAs, and the GST tag of the purified enzyme appears to have no effect on the cleavage activity of the enzyme.
FIGURE 2.
Cleavage activities for human Ago2 containing various length antisense RNAs. A, denaturing PAGE of 40-nucleotide sense RNA digested with GST-Ago2 containing the antisense RNAs shown in C, ranging in length from 17 to 25 nucleotides. B, denaturing PAGE of 40-nucleotide sense RNA digested with HA-Ago2 containing the antisense RNAs shown in C. Antisense RNA was incubated with Ago2 for 1 h prior to incubation with the 40-nucleotide sense RNA for an additional 1 h. T1, 40-nucleotide sense RNA digested with RNase T1 for 10 min at 24 °C. The positions of the T1 digestions adjacent to guanidine residues are shown next to the ladder. Arrows indicate the position of the human Ago2 cleavage site. C, sequences of the antisense RNAs with the corresponding lane designations. The 19-nucleotide antisense RNA contained either a 5′-phosphate (p) or 5′-hydroxyl (OH). The 19-nucleotide antisense RNA containing a 5′-phosphate corresponds to the antisense RNA in Fig. 1. D, percent sense RNA cleaved with GST-Ago2 (solid bar) or HA-Ago2 (hatched bar). The standard errors reported for the Ago2 cleavage activities are based on three experiments. The antisense RNA sequences tested and corresponding lane designations are described in C.
Divalent Cations Affect the Binding Specificity and Cleavage Activity of Human Ago2
The crystal structures of Ago2 indicate that the enzyme contains two metal-binding sites as follows: the first is positioned at the DDE catalytic triad in the PIWI domain, and the second is positioned at the basic binding pocket in the Mid domain, which was shown to interact with the 5′ terminus of the antisense RNA (10, 15–18). To better understand the roles of the divalent metal in both binding of the enzyme to the antisense RNA and catalysis of the sense RNA, the cleavage activities of human Ago2 containing various antisense oligonucleotides were determined in the presence of either magnesium or manganese. Additionally, nucleotide substitutions were introduced at the 5′ terminus of the antisense RNA to evaluate the interaction of the antisense RNA with the basic binding pocket of the enzyme. The modifications at the 3′ terminus of the antisense RNA were designed to evaluate the interaction between the antisense RNA and the PAZ domain of Ago2.
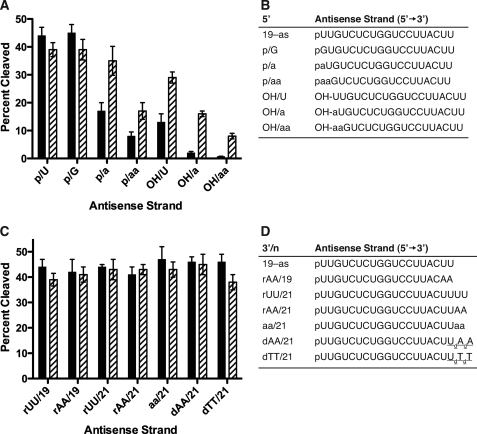
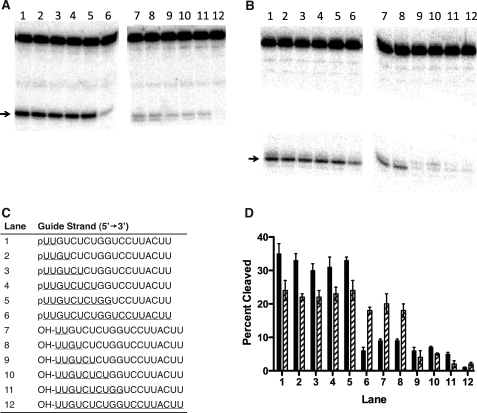
The antisense RNAs containing various 5′-terminal substitutions are shown in Fig. 3B. Similar cleavage activities were observed for human Ago2 containing the unmodified antisense RNA (19-as) with a uridine residue at the 5′ terminus compared with the RNA with a guanidine substitution (p/G) in the presence of either Mg2+ or Mn2+ (Fig. 3, A and B). In the presence of Mg2+, reduced human Ago2 cleavage activities were observed for the enzyme containing the antisense RNAs with abasic substitution (p/a, p/aa) (Fig. 3, A and B). In addition, a significant reduction in Ago2 cleavage activity was observed for the antisense RNAs containing a 5′-hydroxyl (e.g. OH/U, OH/a, and OH/aa) (Fig. 3, A and B).
FIGURE 3.
Influence of divalent cations and composition of the 3′ and 5′ terminus in the antisense RNA on human Ago2 cleavage activity. A, percent cleaved of the 40-nucleotide sense RNA by GST-Ago2 containing antisense sRNAs with various 5′-terminal substitutions (shown in B) in the presence of Mg2+ (solid bar) or Mn2+ (hatched bar). B, sequences of the unmodified 19-nucleotide antisense RNA (19-as) and antisense RNAs containing the following 5′-terminal substitutions: 5′-phosphorylated guanidine (p/G), 5′-phosphorylated abasic substitutions (p/a and p/aa), 5′-dephosphorylated uridine (OH/U), or 5′-dephosphorylated abasic substitutions (OH/a and OH/aa). C, percent cleaved of the 40-nucleotide sense RNA by GST-Ago2 containing antisense sRNAs with various 3′-terminal substitutions (shown in D) in the presence of Mg2+ (solid bar) or Mn2+ (hatched bar). D, sequences of the unmodified 19-nucleotide antisense RNA (19-as) and 21 (/21) nucleotide antisense RNAs containing the following 3′-terminal substitutions: riboadenine (AA), abasic residues (aa), deoxyriboadenine (dAA), or deoxyribothymidine (dTT). The errors reported for the Ago2 cleavage activities are based on three trials.
In contrast, Mn2+ appeared to ameliorate the effects of the abasic and dephosphorylated antisense RNAs on human Ago2 cleavage activity. For example, the Ago2 cleavage activities for the antisense RNAs containing the abasic and 5′-hydroxyl substitutions (p/a, p/aa, and OH/U) were ∼2-fold greater in the presence of Mn2+ compared with the cleavage activities of these antisense RNAs in the presence of Mg2+ (Fig. 3, A and B). Additionally, significantly greater Ago2 cleavage activities were observed for the antisense RNAs containing the abasic substitutions and a 5′-hydroxyl (e.g. OH/a and OH/aa) in the presence of Mn2+ compared with the cleavage activities of these antisense RNAs in the presence of Mg2+ (Fig. 3, A and B). Importantly, in the presence of Mn2+ only a slight reduction in the Ago2 cleavage activity was observed for the antisense RNA containing the 5′-hydroxyl (OH/U) compared with the unmodified antisense RNA (19-as) (Fig. 3, A and B). Again, these data suggest that the enhanced Ago2 cleavage activities observed for the antisense RNAs containing either a 5′-terminal hydroxyl or abasic residue in the presence of Mn2+ compared with Mg2+ are consistent with the metal affecting the interaction between the basic binding pocket of the enzyme and the antisense RNA.
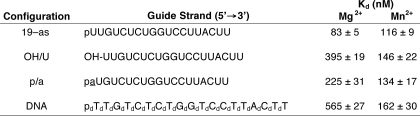
The binding affinities of human Ago2 for the antisense RNAs with the 5′-substitutions correlated with the cleavage activities observed for Ago2 containing these antisense RNAs. In the presence of Mg2+, ∼3-fold lower cleavage activities and 5-fold weaker binding affinities were observed for Ago2 containing either the OH/U or p/a antisense RNAs compared with the unmodified antisense RNA (19-as) (Fig. 3, A and B, and Table 2). In the presence of Mn2+, comparable Ago2 cleavage activities and binding affinities were observed for the p/a and 19-as antisense RNAs, whereas the slight reduction in the Ago2 cleavage activity observed for the OH/U antisense RNA corresponded with a slightly weaker binding affinity of human Ago2 for the this antisense RNA compared with the 19-as RNA (Fig. 3, A and B, and Table 2).
TABLE 2.
Effect of divalent cations on the binding affinity of human Ago2 for various antisense oligonucleotides
The dissociation constants (Kd) for human Ago2 containing the various antisense oligonucleotide configurations in the presence of either Mg2+ or Mn2+ were determined as described under “Materials and Methods.” Sequences of the unmodified 19-nucleotide antisense RNA (19-as) and antisense RNAs containing 5′-hydroxyl (OH), abasic residue (p/a), or deoxyribonucleotides substitutions (dN). The errors reported for the Ago2 binding affinities are based on three trials.
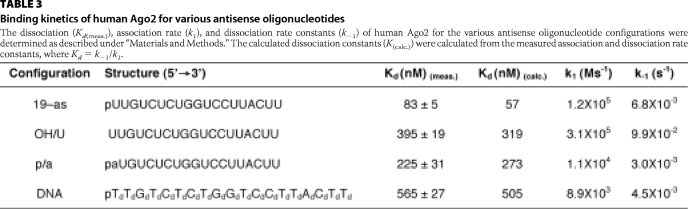
The binding kinetics of Ago2 for the antisense RNAs with the 5′-terminal substitutions are shown in Table 3. The association (k1) and dissociation (k−1) constants observed for Ago2 binding the OH/U antisense RNA were, respectively, 3- and 15-fold faster compared with the 19-as antisense RNA (Table 3). In contrast, a slightly faster k−1 and ∼10-fold slower k1 were observed for Ago2 binding the p/a antisense RNA compared with the 19-as RNA (Table 3). Taken together, these data suggest that the weaker Kd value observed for human Ago2 binding to the single abasic (p/a) antisense RNA was the result of a slower association rate (k1) of the enzyme for the p/a antisense RNA compared with the 19-as RNA (Table 3). In contrast, the weaker binding affinity observed for the OH/U antisense RNA was due to a faster dissociation rate (k−1) of the Ago2 for the OH/U RNA compared with the 19-as RNA.
TABLE 3.
Binding kinetics of human Ago2 for various antisense oligonucleotides
The dissociation (Kd(meas.)), association rate (k1), and dissociation rate constants (k−1) of human Ago2 for the various antisense oligonucleotide configurations were determined as described under “Materials and Methods.” The calculated dissociation constants (K(calc.)) were calculated from the measured association and dissociation rate constants, where Kd = k−1/k1.
Unlike the substitutions at the 5′ terminus of the antisense RNA, substitutions at the 3′ terminus in the presence of either of the divalent metals appeared to have no effect on human Ago2 cleavage activity. For example, the substitution of the 3′-terminal uridine residues of the antisense RNA with adenines appeared to have no effect on the cleavage activity of human Ago2 in the presence of either Mg2+ or Mn2+ (Fig. 3, C and D). Similarly, the antisense RNAs containing additional uridine, abasic, adenine ribose, or deoxyribose dinucleotides exhibited similar cleavage activities in the presence of either Mg2+ or Mn2+ (Fig. 3, C and D).
To further explore the influence of the divalent metals on the interaction between the antisense RNA and the enzyme, successive deoxyribonucleotide substitutions were introduced at the 5′-pole of the antisense oligonucleotide (Fig. 4). In the presence of Mg2+, similar cleavage activities were observed for Ago2 containing the antisense oligonucleotides with the 5′-phosphate and 2–10 DNA residues at the 5′-pole compared with the unmodified antisense RNA with a 5′-phosphate (Fig. 2, A and D, and Fig. 4, A and D). A significant reduction in cleavage activity was observed for Ago2 containing the antisense with all DNA residues and a 5′-phosphate (Fig. 4, A and D). A similar trend was observed for the antisense oligonucleotides containing DNA substitutions and a 5′-hydroxyl, although the cleavage activities for Ago2 containing these antisense oligonucleotides were significantly weaker than the activities observed for the antisense oligonucleotides with a 5′-phosphate (Fig. 4, A and D).
FIGURE 4.
Influence of divalent cations on the cleavage activities of human Ago2 containing antisense RNA with deoxyribonucleotide substitutions. A, denaturing PAGE of 40-nucleotide sense RNA digested with GST-Ago2 containing antisense RNAs with various deoxyribonucleotide substitutions in the presence of Mg2+. B, denaturing PAGE of 40-nucleotide sense RNA digested with GST-Ago2 containing antisense RNAs with various deoxyribonucleotide substitutions in the presence of Mn2+. Antisense RNA was incubated with Ago2 for 1 h prior to incubation with the 40-nucleotide sense RNA for an additional 1 h. Arrows indicate the position of the human Ago2 cleavage site. C, sequences of the antisense oligonucleotides with the corresponding lane designations. The antisense oligonucleotides contained either a 5′-phosphate (p) or 5′-hydroxyl (OH) and DNA substitutions (underlined). D, percent cleaved of the 40-nucleotide sense RNA by GST-Ago2 containing the antisense sequences shown in C in the presence of either Mg2+ (solid bar) or Mn2+ (hatched bar). The errors reported for the Ago2 cleavage activities are based on three trials.
Again, Mn2+ appeared to ameliorate the effects of the 5′-hydroxyl and DNA substitutions on human Ago2 cleavage activity. For example, only a slight reduction in cleavage activity was observed for Ago2 containing the antisense DNA and a 5′-phosphate, suggesting that in the presence of Mn2+ Ago2 exhibits significant RNase H activity (Fig. 4, B and D). In addition, no reduction in the cleavage activities for Ago2 containing the antisense oligonucleotides with a 5′-hydroxyl and 2–4 deoxyribonucleotide substitutions were observed compared with the same antisense oligonucleotides containing a 5′-terminal phosphate (Fig. 4, B and D). The observed RNase H activity was determined by incubating the GST-Ago2 enzyme bound to paramagnetic beads with the antisense DNA and washing the unbound DNA prior to adding the target sense RNA. Given that RNase H has been shown to bind preformed RNA/DNA heteroduplexes but not single strand DNA oligonucleotides, it is unlikely that the RNase H activity observed for human Ago2 is because of RNase H contamination from the host cells (24, 25). To ensure that the GST-Ago2 preparation did not contain an RNase H contaminant, the GST-Ago2 enzyme was incubated with the preformed antisense DNA/sense RNA heteroduplex in the presence of Mn2+. No cleavage activity was observed for the GST-Ago2 enzyme in the presence of the preformed heteroduplex (supplemental Fig. 2). In addition, the GST-Ago2 preparation did not bind the preformed RNA/DNA heteroduplex (supplemental Fig. 2). These data combined with the purity of the GST-Ago2 preparation suggest that human Ago2 is responsible for the observed RNase H activity (supplemental Fig. 1).
The divalent metal-dependent cleavage activities observed for Ago2 containing the antisense DNA were consistent with the metal-dependent binding affinities of the enzyme for the antisense DNA (Table 2). For example, the dissociation constants (Kd) of Ago2 for the antisense DNA and unmodified antisense RNA (19-as) were, respectively, 565 and 83 nm in the presence of Mg2+ (Table 2). In contrast, a smaller difference in the Kd values was observed for human Ago2 binding to the same antisense oligonucleotides in the presence of Mn2+ (e.g. Kd of 162 nm for the DNA compared with 116 nm for the 19-as RNA) (Table 2). Additionally, the antisense DNA exhibited a similar k−1 and ∼10-fold slower k1 compared with the 19-as RNA (Table 3). These data suggest that the weaker Kd value observed for human Ago2 binding to the antisense DNA compared with the antisense RNA was the result of a slower association rate (k1).
Positional Effects of 2′-Ribose Modifications in the Antisense RNA on Human Ago2 Activity
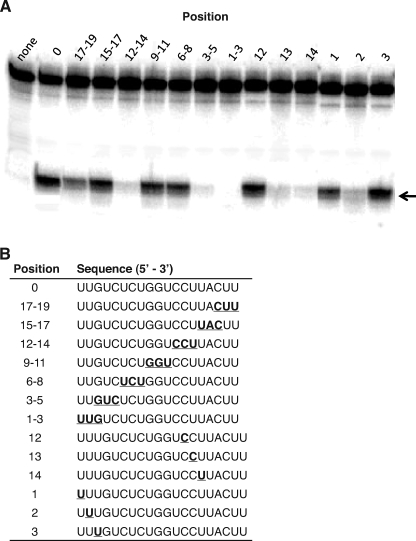
The cleavage activities for human Ago2 containing antisense RNAs with 2′-methoxyethyl (2′-MOE) substitutions are shown in Fig. 5. Similar Ago2 cleavage activities were observed for the antisense RNAs containing three successive 2′-MOE substitutions at positions 6–8, 9–11, 15–17, and 17–19 compared with the unmodified antisense RNA (0) (Fig. 5). In contrast, significantly lower cleavage activities were observed for the antisense RNAs containing 2′-MOE substitutions at positions 3–5 and 12–14 compared with the unmodified antisense RNA. No Ago2 cleavage activity was observed for the antisense RNA containing 2′-MOE substitutions at positions 1–3 (Fig. 5). To identify the specific 2′-MOE substitution responsible for the reduction in Ago2 cleavage activity, single 2′-MOE substitutions were introduced at positions 1–3 and 12–14 (Fig. 5). Significantly lower Ago2 cleavage activities were observed for the antisense RNAs containing a single 2′-MOE substitution at positions 2, 13, and 14, whereas 2′-MOE substitutions at positions 1, 3, and 12 appeared to have no effect on the cleavage activity (Fig. 5).
FIGURE 5.
Effect of 2′-methoxyethyl substitutions in the antisense RNA on human Ago2 cleavage activity. A, denaturing PAGE of 40-nucleotide sense RNA digested with GST-Ago2 in the absence of antisense strand (none) and containing the antisense RNAs with 2′-methoxyethyl substitutions at various positions, counting from the 5′ terminus of the antisense RNA. The unmodified antisense RNA (0) corresponds to the 19-as RNA shown in Figs. 1–3. Arrow indicates the position of the human Ago2 cleavage site. B, sequences of the modified antisense RNAs containing the 2′-methoxyethyl substitutions at various positions (underlined).
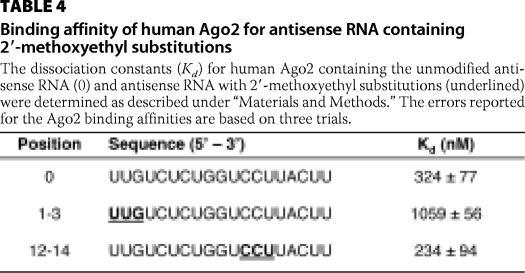
To better understand the effects of 2′-MOE substitutions on the activity of human Ago2, we determined the binding affinity of Ago2 for the antisense RNAs containing 2′-MOE substitutions at positions 1–3 and 12–14 (Table 4). Comparable binding affinities were observed for the antisense RNA containing the 2′-MOE substitutions at positions 12–14 compared with the unmodified antisense RNA. In contrast, the binding affinity of Ago2 for the antisense RNA containing 2′-MOE substitutions at positions 1–3 was ∼3-fold weaker compared with the unmodified antisense RNA (Table 4). Taken together, these data suggest that the loss in Ago2 cleavage activity observed for the antisense RNA containing 2′-MOE substitutions at positions 1–3 appears to be due to the 2′-MOE residues interfering with the binding of antisense RNA to Ago2. In contrast, the 2′-MOE substitutions at positions 12–14 appeared to have no effect on binding of the antisense RNA to the enzyme suggesting that the loss in Ago2 cleavage activity was due to the 2′-MOE residues interfering with catalysis. In addition, positions in the antisense RNA that were sensitive to 2′-MOE substitutions (e.g. positions 1–3 and 12–14) are consistent with an RNase protection assay in which the antisense RNA was treated with single strand-specific endoribonucleases in either the presence or absence of human Ago2 (supplemental Fig. 3). Specifically, only the first 14 nucleotides from the 5′ terminus of the antisense RNA were protected from RNase degradation suggesting that human Ago2 does not appear to interact with the remaining five 3′-most nucleotides.
TABLE 4.
Binding affinity of human Ago2 for antisense RNA containing 2′-methoxyethyl substitutions
The dissociation constants (Kd) for human Ago2 containing the unmodified antisense RNA (0) and antisense RNA with 2′-methoxyethyl substitutions (underlined) were determined as described under “Materials and Methods.” The errors reported for the Ago2 binding affinities are based on three trials.
Human Ago2 Appears to Interact with Regions in the Target RNA Adjacent the Hybridization Site of the Antisense RNA
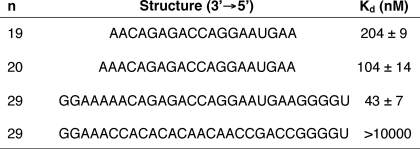
The binding affinity of human Ago2 containing the 19-nucleotide antisense RNA was determined for 19-, 20-, or 29-nucleotide-long sense RNAs (Table 5). The 19-nucleotide sense RNA is length-matched to the antisense RNA, whereas the 20-nucleotide sense RNA contains an additional adenine residue at the 3′ terminus and the 29-nucleotide sense RNA contains five additional residues at both the 3′ and 5′ termini (Table 5). Human Ago2 containing the antisense RNA exhibited a Kd of 204 nm for the length-matched 19-nucleotide sense RNA. A 2- and 5-fold tighter binding affinity was observed for, respectively, the 20 and 29-nucleotide sense RNAs compared with the 19-nucleotide sense RNA (Table 5). The enhanced binding affinities observed for the longer sense RNAs suggest that the enzyme is interacting with regions on the target RNA adjacent to the site of hybridization for the 19-nucleotide antisense RNA. To ensure that the observed binding affinities for the sense RNAs were for Ago2 programmed with the antisense RNA and not Ago2 alone, we determined the binding affinity of Ago2 containing the 19-nucleotide antisense RNA for a 29-nucleotide sense RNA with no sequence complementarity to the antisense RNA (Table 5). Human Ago2 containing the antisense RNA was unable to bind the noncomplementary sense RNA (Table 5).
TABLE 5.
Binding affinity of human Ago2 with the antisense RNA for various length sense RNAs
Human Ago2 was incubated with the 19-nucleotide antisense RNA for 1 h prior to adding the sense RNA and the binding affinities (Kd) of human Ago2 containing the 19-nucleotide antisense RNA for various length complementary target RNAs were determined as described under “Materials and Methods.” The errors reported for the Ago2 binding affinities are based on three trials.
Endogenous TRBP Binds the PIWI Domain of Immunoprecipitated Human Ago2
Denaturing PAGE analysis of the HA-Ago2 preparation that was expressed in HeLa cells and immunoprecipitated using HA antibody revealed several additional protein bands associated with the immunoprecipitated HA-Ago2 (data not shown). Western blot analysis using antibodies against human Ago2, Dicer, and the immunodeficiency virus TRBP identified the Ago2 and TRBP proteins but not the human Dicer protein suggesting that the HA-Ago2 enzyme was binding endogenous TRBP (Fig. 6, B and C). To determine whether the observed binding interaction between human Ago2 and TRBP involves a nucleic acid intermediary (e.g. single or double strand RNA), the immunoprecipitated HA-Ago2 was treated with either a single or double strand specific RNase, washed, and evaluated by Western blot analysis as described under “Materials and Methods.” The immunoprecipitated HA-Ago2 preparations treated with the RNases contained TRBP suggesting that human Ago2 likely binds endogenous TRBP through protein-protein interactions and not through an RNA bridge (supplemental Fig. 4).
FIGURE 6.
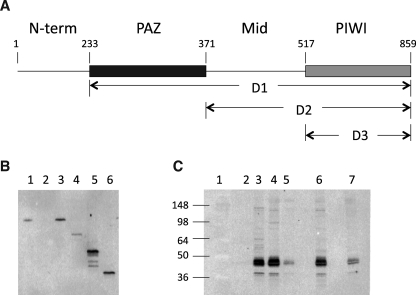
Characterization of the interaction between human Ago2 and TRBP. A, human Ago2 deletion mutants. Amino acid residues are numbered from the amino terminus of the enzyme. Human Ago2 deletion mutant 1 (D1) consists of amino acids 233–859 and contains the PAZ, Mid, and PIWI domains of the enzyme. Human Ago2 deletion mutant 2 (D2) consists of amino acids 371–859 and contains the Mid and PIWI domains. Human Ago2 deletion mutant 3 (D3) consists of amino acids 517–859 and contains the PAZ domain. B, Western blot analysis of HA-Ago2 expressed and immunoprecipitated from HeLa cells using HA antibody and probed with human Ago2 antibody. Lane 1, HeLa cell lysate; lane 2, immunoprecipitate from cells not transfected with HA-Ago2 plasmid; lanes 3–6, immunoprecipitates from cells transfected with, respectively, HA-Ago2, HA-D1, HA-D2, and HA-D3 plasmids. C, Western blot analysis of HA-Ago2 expressed and immunoprecipitated from HeLa cells using HA antibody and probed with human TRBP antibody. Lane 1, molecular weight ladder; lane 2, immunoprecipitate from cells not transfected with HA-Ago2 plasmid; lane 3, purified recombinant TRBP; lanes 4–7, immunoprecipitates from cells transfected with, respectively, Ha-Ago2, HA-D1, HA-D2, and HA-D3 plasmids.
To identify the regions of Ago2 that are interacting with TRBP, deletion mutants of HA-Ago2 were prepared in which either the amino-terminal region of the protein (D1), the amino-terminal region and the PAZ domain (D2), or the amino-terminal region PAZ and Mid domains were deleted (D3) (Fig. 6A). The deletion mutants were cloned into the cytomegalovirus expression vector and then transfected into HeLa cells and immunoprecipitated using HA antibody (Fig. 6B). All three deletion mutants of HA-Ago2 co-immunoprecipitated TRBP suggesting that TRBP was binding to the PIWI domain of Ago2 (Fig. 6C). In addition, the signal observed for the TRBP protein correlated with the level of HA-Ago2 expression (Fig. 6, B and C). For example, the highest TRBP level was observed for the immunoprecipitation of the D2 mutant, which also exhibited the highest expression level of the HA-Ago2 mutants. The D1 mutant, on the other hand, exhibited both the lowest expression level and lowest TRBP signal, whereas the D2 mutant exhibited an intermediate expression level and corresponding intermediate TRBP signal. Finally, TRBP does not appear to influence cleavage activities and binding specificities of human Ago2 given that the GST-Ago2 and HA-Ago2 preparations exhibited similar cleavage activities and binding specificities (Figs. 1 and 2 and Table 1).
DISCUSSION
Substrate Specificity of Human Ago2
We show that recombinant human Ago2 does not cleave preformed RNA duplexes, including a 19-bp siRNA-like duplex and an RNA duplex consisting of a 19-nucleotide antisense RNA hybridized to a 40-nucleotide sense RNA (Fig. 1). The lack of cleavage of the preformed duplexes is because of the fact that human Ago2 selectively binds single strand RNA relative to double strand RNA, exhibiting an ∼2 orders of magnitude tighter binding affinity for the antisense RNA compared with the RNA duplexes (Table 1). These observations are consistent with previous reports in which single strand but not double strand RNA was able to cross-link with human Ago2 (11).
In human cells, the antisense RNA associated with Ago2 is generated by the endonuclease Dicer, which processes long double strand RNAs into short effector double strand RNAs (1, 2). Given the observed binding and cleavage specificity of human Ago2 and that the cleavage products of human Dicer consist of double strand RNAs, our data suggest that the translocation of the Dicer product to Ago2 requires additional factors to either unwind the double strand RNA product or facilitate binding of the double strand RNA product to Ago2, which could then cleave the sense strand of the duplex in a manner similar to that described for Ago2 from Drosophila (26, 27).
The human HA-Ago2 protein expressed and immunoprecipitated from HeLa cells was also unable to cleave the preformed duplexes and exhibited binding specificities similar to the recombinant GST-Ago2 enzyme (Fig. 1 and Table 1). The HA-Ago2 preparation contained the human immunodeficiency virus TRBP but not Dicer suggesting that TRBP alone was insufficient to facilitate the translocation of the RNA duplex to Ago2 (Fig. 6). Previous reports suggest that Dicer and TRBP, which together constitute the minimum RISC loading complex, translocate the Dicer cleavage products to Ago2 (2–4, 28–34). The addition of recombinant human Dicer to the HA-Ago2 preparation had no effect on the ability of the enzyme to cleave a preformed duplex (data not shown). Only when the siRNA duplex was transfected into HeLa cells expressing the HA-Ago2 protein was the immunoprecipitated HA-Ago2 preparation able to cleave the 32P-labeled target RNA (data not shown). Therefore, the endogenous cellular factors required to convert the RNA duplex to the active moiety appear to associate with the transiently expressed HA-Ago2 enzyme in the cell but not with the immunoprecipitated HA-Ago2 preparation. Finally, consistent with a previous report, the HA-Ago2 preparation exhibited single-turnover kinetics similar to that observed for the GST-Ago2 enzyme suggesting that TRBP did not alter the cleavage kinetics of the enzyme (data not shown) (11). This suggests that in the cell other factors must be responsible for removing the cleavage products from Ago2 or that Ago2 cleavage is not necessary to cause degradation of the mRNA.
Nature of the Interaction between the Antisense RNA and Human Ago2
Our data suggest that human Ago2 interacts with the 5′-pole and not the 3′ terminus of the antisense RNA. First, the RNase protection assay showed that human Ago2 protected the 5′-pole and not the 3′ terminus of the antisense RNA from ribonuclease degradation (supplemental Fig. 3). Second, nucleotide substitutions, including abasic nucleotides positioned at the 3′ terminus of the antisense RNA, had no effect on human Ago2 cleavage activity (Fig. 3, C and D). The architecture of the human Ago2 protein consists of a PIWI, Mid, and PAZ domains, which together form an extended nucleic acid binding surface for the antisense RNA (13–18). A basic binding pocket, positioned within the Mid domain, binds the 5′ terminus of the antisense strand, and a hydrophobic pocket within the PAZ domain binds the two 3′-terminal nucleotides of the antisense strand (13–18). Based on these structures, our data suggest that the antisense RNA interacts with the Mid and PIWI domains but not the PAZ domain of the enzyme. Interestingly, Ago2 containing the 25-nucleotide antisense RNA exhibited lower Ago2 cleavage activity compared with the 19–23-nucleotide antisense RNAs suggesting that beyond a certain length, additional nucleotides at the 3′ terminus of the antisense RNA interfere with the interaction between the antisense RNA and the enzyme (Fig. 2).
The observed biochemical properties for the interaction between the 5′-pole of the antisense RNA and human Ago2 indicated that both the 5′-terminal phosphate and heterocycle base of the antisense RNA were important for binding to the enzyme (Fig. 2, Fig. 3, A and B, and Table 3). The tighter binding affinity observed for the antisense RNA with the 5′-phosphate was due to both a slightly slower association rate (k1) and significantly slower dissociation rate (k−1) compared with the antisense RNA with the 5′-hydroxyl (Table 3). In addition, the weaker binding affinities observed for the antisense RNAs containing the abasic residues were due to a slower k1 (Table 3). The structures of the Argonaute proteins show that the 5′-terminal phosphate of the antisense RNA forms electrostatic interactions with a divalent metal and two lysine residues positioned within the basic binding pocket of the enzyme (15, 18). Furthermore, the heterocycle base of the 5′-terminal nucleotide of the antisense RNA formed a stacking interaction with a tyrosine residue positioned in the basic binding pocket (15). This tyrosine residue is also conserved in human Ago2 (15). Consequently, the slower k1 observed for the antisense RNA containing the 5′-phosphate is presumably because of the coordination of the phosphate with the divalent metal and basic amino acids within the binding pocket. In addition, the slower k−1 suggests that once positioned within the basic binding pocket, the 5′-phosphate serves to stabilize the interaction between the antisense RNA and human Ago2. The slower k1 observed for human Ago2 binding the antisense RNA with abasic substitutions suggests that the heterocycle base of the 5′-terminal nucleotide functions as a lure to nucleate the interaction between the 5′ terminus of the antisense RNA and the basic binding pocket of the enzyme. Interestingly, the antisense RNA containing two abasic residues at the 5′ terminus exhibited an ∼2-fold lower Ago2 cleavage activity compared with the antisense RNA with a single abasic substitution (Fig. 3, A and B). Examination of the structure from the A. fulgidus protein indicates that the heterocycle base of the nucleotide at the second position from the 5′ terminus of the antisense RNA does not appear to interact with the protein (15). Thus, the effect of the second abasic substitution on the cleavage activity of human Ago2 suggests that the human enzyme may exhibit a different structure than that observed for the A. fulgidus protein.
The observed effects of the 5′-terminal phosphate and heterocycle base of the antisense RNA on the cleavage activity and binding affinity of human Ago2 were less pronounced in the presence of Mn2+ compared with Mg2+ (Fig. 3A and Fig. 4B and Table 2). The metal-dependent effects on the interaction of the 5′ terminus of the antisense RNA with human Ago2 is likely due to the observed coordination of a divalent metal within the basic binding pocket (15, 18). Although Mn2+ and Mg2+ are similar in size, the metals exhibit different coordination properties. For example, the coordination of these metals with Escherichia coli RNase H shows a single Mg2+ ion at the catalytic site of the enzyme, whereas the same site was able to accommodate two Mn2+ ions (35–37). The Mg2+ ion formed an outer sphere (water-mediated) coordination with the E. coli enzyme, whereas the Mn2+ ions formed an inner sphere (non-water-mediated) coordination, resulting in smaller distances between the Mn2+ ion and the coordinating amino acids relative to Mg2+ (37). In the case of human Ago2, the coordination differences between the metals likely change the spatial orientation of the surrounding amino acids, thereby changing the interaction between the basic binding pocket of the enzyme and the antisense RNA.
The 2′-sugar substitutions also affected the interaction between human Ago2 and the antisense RNA (Fig. 5). In the case of the 2′-MOE substitution at position 2, the reduction in Ago2 cleavage activity was because of a weaker binding affinity of the enzyme for the modified antisense oligonucleotide (Fig. 5 and Table 4). This observation is consistent with the structures of the Argonaute proteins that show that the 2′-hydroxyl of the 5′-terminal nucleotide was positioned toward the solvent and away from the enzyme, and the 2′-hydroxyl of the second nucleotide from the 5′ terminus of the antisense RNA was positioned adjacent to the enzyme (15, 18). Therefore, large 2′-moieties such as 2′-MOE positioned at the 5′-terminal nucleotide should have no effect on the interaction with the enzyme, whereas 2′-substitutions at position 2 would likely cause steric interference with the enzyme. The 2′-MOE substitutions at positions 13 and 14 also inhibited Ago2 cleavage activity, but in this case the 2′-MOE residues appeared to have no effect on binding of human Ago2 for the antisense RNA (Fig. 5 and Table 4). The PIWI domain constitutes the catalytic domain of the enzyme and exhibits a three-dimensional structure similar to RNase H, sharing the same DDE catalytic triad and metal cofactor requirements (10, 16, 17). The co-crystal structure of the PIWI domain with an antisense RNA shows a kinked structure in the backbone of the antisense RNA between the 10th and 11th nucleotides opposing the catalytic site of the enzyme (18). Based on these observations, the 2′-MOE substitutions at positions 12–14 likely effect the interaction of the antisense RNA with the catalytic site of the enzyme.
The observed effects of the divalent metals on the cleavage activities of human Ago2 containing antisense DNA appear to involve the metal coordinated at the catalytic site of the enzyme. For example, human Ago2 exhibited a greater binding affinity for the antisense DNA in the presence of Mn2+ compared with Mg2+ (Table 2). The weaker binding affinity of Ago2 for the antisense DNA in the presence of Mg2+ appeared to be due to a slower k1 suggesting that a structural reorganization of either the antisense DNA or the enzyme may be required for either binding the antisense DNA or cleavage of the target RNA. In fact, the differences in the helical geometries between double strand RNA and a DNA/RNA heteroduplex are predicted to have a profound effect on the precise positioning of the internucleotide phosphates relative to the enzyme. Specifically, the DNA/RNA heteroduplexes exhibit a minor groove width that is ∼2 Å narrower compared with double strand RNA (38–40). In addition, both the axial rise per nucleotide and internucleotide phosphate distance for the DNA/RNA heteroduplex is ∼0.5 Å longer compared with double strand RNA, and given that Ago2 cleaves the target RNA 10 bp from the 5′ terminus of the antisense strand, the increased phosphate distances equate to an ∼5-Å increase in the distance between the 5′ terminus of the antisense strand of the duplex and the cleavage site. Interestingly, human Ago2 containing the antisense DNA cleaved the target RNA at the same position in the presence of Mn2+ (e.g. between the 10th and 11th bp from the 5′ terminus of the antisense RNA), compared with the enzyme containing the antisense RNA (Fig. 4). Presumably, the coordination differences between the Mn2+ and Mg2+ could alter the structure of the catalytic site of human Ago2 sufficiently to accommodate the structural differences between the duplexes and enable cleavage of the sense RNA bound to antisense DNA.
How Is the RNA Translocated from the RISC Loading Complex to Human Ago2?
Our data offer additional insights into how the RNA may be transferred from the RISC loading complex to Ago2. In a previous report we demonstrated that human Dicer is capable of binding short single strand and double strand RNAs with affinities comparable with substrate and that Dicer interacts predominantly with the 3′-overhang of the double strand RNA and 3′ terminus of the single strand RNA, likely involving the PAZ domain of Dicer (41). In addition, the position of the 3′-overhang in the siRNA appeared to dictate which strand of the siRNA was loaded into the human HA-Ago2 enzyme expressed and immunoprecipitated from HeLa cells (41). Here we show that human Ago2 binds to the 5′-pole and not the 3′ terminus of the antisense RNA and that this interaction likely involves the Mid and PIWI domains of the enzyme. Taken together these data suggest several possible routes by which the translocation of the siRNA from Dicer to Ago2 could occur. First, following Dicer cleavage, additional factor(s) promote dissociation of the two strands, and Dicer retains the single strand antisense RNA at its 3′ terminus, transferring the RNA to Ago2 via the 5′ terminus where the heterocycle base of the 5′-terminal nucleotide lures the 5′-terminal phosphate to the basic binding pocket of Ago2. Once introduced to the pocket, the 5′-terminal phosphate then stabilizes the interaction between the antisense RNA and the enzyme. Alternatively, following Dicer cleavage, Dicer retains the RNA duplex via the 3′-overhang and along with other factor(s) loads the double strand RNA onto Ago2, which then cleaves the sense strand promoting its release from the enzyme. In addition, TRBP may function as an intermediary in this process, binding to the PIWI domain of human Ago2 and using one of its three double strand RNA binding domains to position the siRNA in close proximity to the catalytic domain of Ago2 (Fig. 6) (3). Chendrimada et al. (3) showed that TRBP plays an important role in the activities of siRNAs as reduction of TRBP protein expression reduced the activities of siRNAs. In Drosophila, cross-linking studies between Dicer-2 and the TRBP paralog R2D2 bound to the siRNA indicated that Dicer-2 was positioned adjacent to the 5′ terminus of the sense strand, and R2D2 was positioned adjacent to the 5′ terminus of the antisense strand (42). In human cells, TRBP may play a similar role.
Finally, in addition to its role as an endonuclease, Ago2 may serve additional functions, namely facilitating the hybridization of the antisense RNA to the target RNA. For example, human Ago2 containing the 19-nucleotide antisense RNA exhibited greater binding affinities for longer sense RNAs suggesting that the enzyme is forming additional interactions with regions on the sense RNA outside the site of hybridization for the antisense RNA (Table 5). The additional contacts with the target RNA would be particularly advantageous in the case of microRNA-mediated gene silencing, where Ago2 functions primarily as a delivery device and not as an endonuclease for the microRNA to cleave the target mRNA. Specifically, microRNAs exhibit multiple mismatched base pairs with the target RNA at the nucleotides positioned adjacent to the catalytic site of the enzyme and therefore do not trigger Ago2-dependent cleavage of the target RNA. Given that microRNAs form mismatched base pairs with the target RNA, the enhanced binding affinity observed for Ago2 would enhance the affinity of the microRNA for the target mRNA. In the case of siRNAs, the enhanced binding affinity may have a deleterious effect, thus enhancing the potential for greater off-target effects of the siRNA. Clearly more work needs to be done to better understand how microRNAs and siRNA interact with and promote the degradation of the target mRNA and the factors that lead to the degradation of off-target RNAs.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- siRNA
- short interfering RNA
- TRBP
- trans-activating response RNA-binding protein
- Ago2
- Argonaute2
- RISC
- RNA-induced silencing complex
- HA
- hemagglutinin
- 2′-MOE
- 2′-methoxyethyl
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- s
- sense
- as
- antisense.
REFERENCES
- 1.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998) Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. (2001) Nature 409, 363–366 [DOI] [PubMed] [Google Scholar]
- 3.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir S. M., Lendeckel W., Tuschl T. (2001) Genes Dev. 15, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomari Y., Zamore P. D. (2005) Curr. Biol. 15, R61–R64 [DOI] [PubMed] [Google Scholar]
- 6.Nykänen A., Haley B., Zamore P. D. (2001) Cell 107, 309–321 [DOI] [PubMed] [Google Scholar]
- 7.Hammond S. M., Caudy A. A., Hannon G. J. (2001) Nat. Rev. Genet. 2, 110–119 [DOI] [PubMed] [Google Scholar]
- 8.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004) Mol. Cell 15, 185–197 [DOI] [PubMed] [Google Scholar]
- 9.Rand T. A., Ginalski K., Grishin N. V., Wang X. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14385–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L. (2004) Science 305, 1434–1437 [DOI] [PubMed] [Google Scholar]
- 11.Rivas F. V., Tolia N. H., Song J. J., Aragon J. P., Liu J., Hannon G. J., Joshua-Tor L. (2005) Nat. Struct. Mol. Biol. 12, 340–349 [DOI] [PubMed] [Google Scholar]
- 12.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P. D. (2004) Science 306, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 13.Song J. J., Liu J., Tolia N. H., Schneiderman J., Smith S. K., Martienssen R. A., Hannon G. J., Joshua-Tor L. (2003) Nat. Struct. Biol. 10, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 14.Lingel A., Simon B., Izaurralde E., Sattler M. (2004) Nat. Struct. Mol. Biol. 11, 576–577 [DOI] [PubMed] [Google Scholar]
- 15.Ma J. B., Yuan Y. R., Meister G., Pei Y., Tuschl T., Patel D. J. (2005) Nature 434, 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker J. S., Roe S. M., Barford D. (2005) Nature 434, 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. (2004) Science 305, 1437–1441 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D. J. (2008) Nature 456, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima W. F., Nichols J. G., Wu H., Prakash T. P., Migawa M. T., Wyrzykiewicz T. K., Bhat B., Crooke S. T. (2004) J. Biol. Chem. 279, 36317–36326 [DOI] [PubMed] [Google Scholar]
- 20.Baker B. F., Lot S. S., Condon T. P., Cheng-Flournoy S., Lesnik E. A., Sasmor H. M., Bennett C. F. (1997) J. Biol. Chem. 272, 11994–12000 [DOI] [PubMed] [Google Scholar]
- 21.McKay R. A., Miraglia L. J., Cummins L. L., Owens S. R., Sasmor H., Dean N. M. M. (1999) J. Biol. Chem. 274, 1715–1722 [DOI] [PubMed] [Google Scholar]
- 22.Ikehara M., Ohtsuka E., Tokunaga T., Nishikawa S., Uesugi S., Tanaka T., Aoyama Y., Kikyodani S., Fujimoto K., Yanase K. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 4695–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1988) in Current Protocols in Molecular Biology, p. 3.10.3, John Wiley & Sons, Inc., New York [Google Scholar]
- 24.Lima W. F., Crooke S. T. (1997) Biochemistry 36, 390–398 [DOI] [PubMed] [Google Scholar]
- 25.Wu H., Lima W. F., Crooke S. T. (1999) J. Biol. Chem. 274, 28270–28278 [DOI] [PubMed] [Google Scholar]
- 26.Rand T. A., Petersen S., Du F., Wang X. (2005) Cell 123, 621–629 [DOI] [PubMed] [Google Scholar]
- 27.Matranga C., Tomari Y., Shin C., Bartel D. P., Zamore P. D. (2005) Cell 123, 607–620 [DOI] [PubMed] [Google Scholar]
- 28.Zamore P. D., Tuschl T., Sharp P. A., Bartel D. P. (2000) Cell 101, 25–33 [DOI] [PubMed] [Google Scholar]
- 29.Hammond S. M., Bernstein E., Beach D., Hannon G. J. (2000) Nature 404, 293–296 [DOI] [PubMed] [Google Scholar]
- 30.Sontheimer E. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 127–138 [DOI] [PubMed] [Google Scholar]
- 31.Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. (1991) Science 251, 1597–1600 [DOI] [PubMed] [Google Scholar]
- 32.Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., Baillie D. L., Fire A., Ruvkun G., Mello C. C. (2001) Cell 106, 23–34 [DOI] [PubMed] [Google Scholar]
- 33.Hutvágner G., McLachlan J., Pasquinelli A. E., Bálint E., Tuschl T., Zamore P. D. (2001) Science 293, 834–838 [DOI] [PubMed] [Google Scholar]
- 34.Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., Plasterk R. H. (2001) Genes Dev. 15, 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Nakamura H., Ikehara M., Matsuzaki T., Morikawa K. (1992) J. Mol. Biol. 223, 1029–1052 [DOI] [PubMed] [Google Scholar]
- 36.Katayanagi K., Okumura M., Morikawa K. (1993) Proteins Struct. Funct. Genet. 17, 337–346 [DOI] [PubMed] [Google Scholar]
- 37.Goedken E. R., Marqusee S. (2001) J. Biol. Chem. 276, 7266–7271 [DOI] [PubMed] [Google Scholar]
- 38.Fedoroff O. Yu., Salazar M., Reid B. R. (1993) J. Mol. Biol. 233, 509–523 [DOI] [PubMed] [Google Scholar]
- 39.Egli M., Portmann S., Usman N. (1996) Biochemistry 35, 8489–8494 [DOI] [PubMed] [Google Scholar]
- 40.Saenger W. (1984) in Principles of Nucleic Acid Structure, ( Cantor C. R. ed) pp. 24–27, Springer-Verlag Inc., New York [Google Scholar]
- 41.Lima W. F., Murray H., Nichols J. G., Wu H., Sun H., Prakash T. P., Berdeja A. R., Gaus H. J., Crooke S. T. (2009) J. Biol. Chem. 284, 2535–2548 [DOI] [PubMed] [Google Scholar]
- 42.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P. D. (2004) Science 306, 1377–1380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.