Abstract
Studies of mineralization of embryonic spicules and of the sea urchin genome have identified several putative mineralization-related proteins. These predicted proteins have not been isolated or confirmed in mature mineralized tissues. Mature Lytechinus variegatus teeth were demineralized with 0.6 n HCl after prior removal of non-mineralized constituents with 4.0 m guanidinium HCl. The HCl-extracted proteins were fractionated on ceramic hydroxyapatite and separated into bound and unbound pools. Gel electrophoresis compared the protein distributions. The differentially present bands were purified and digested with trypsin, and the tryptic peptides were separated by high pressure liquid chromatography. NH2-terminal sequences were determined by Edman degradation and compared with the genomic sequence bank data. Two of the putative mineralization-related proteins were found. Their complete amino acid sequences were cloned from our L. variegatus cDNA library. Apatite-binding UTMP16 was found to be present in two isoforms; both isoforms had a signal sequence, a Ser-Asp-rich extracellular matrix domain, and a transmembrane and cytosolic insertion sequence. UTMP19, although rich in Glu and Thr did not bind to apatite. It had neither signal peptide nor transmembrane domain but did have typical nuclear localization and nuclear exit signal sequences. Both proteins were phosphorylated and good substrates for phosphatase. Immunolocalization studies with anti-UTMP16 show it to concentrate at the syncytial membranes in contact with the mineral. On the basis of our TOF-SIMS analyses of magnesium ion and Asp mapping of the mineral phase composition, we speculate that UTMP16 may be important in establishing the high magnesium columns that fuse the calcite plates together to enhance the mechanical strength of the mineralized tooth.
The largest currently extant group of sea urchins (phylum Echinodermata, class Echinoidea) is the camarodonts (1), which among other features have a unique mineralized Aristotle's lantern structure housing the five teeth of the urchin. Most studies of mineralization processes within the echinoderms have focused on the mineralized spicules of the larval urchin (2). These give structure to the larvae but are transient and not present in the postlarval maturing animal. The main mineralized structures of the growing urchin, aside from the test and spines, are the lantern stereom and the teeth (3). The teeth are especially interesting, because they grow continuously in a vectorial fashion. The preodontoblasts originating at the aboral plumula arise from a mixed population of coelomocytes. The individual monocytes condense at the outer surface of the plumula (4, 5) just under the epithelial layer, where they fuse and become multinucleated cells, which form sheetlike syncytial layers (6). Calcium carbonate mineral forms at calcification sites between the cellular layers, with the mineral deposition related to the syncytial plasma membranes (7–9). There has been considerable discussion as to whether the chambers in which the mineral forms are within the cells or in the extracellular space between syncytia, but it is clear that the mineralization is related to the membranes enclosing the mineralization space. However, the biogenic urchin tooth calcite crystals themselves contain occluded macromolecules (10).
Thus, our attention has been directed to the questions of which proteins or other macromolecules might be involved in the initiation of mineralization, and specifically where these proteins might be located. We have recently reported on protocols by which the organic components of the urchin tooth can be separated into those readily extracted from the tooth and those intimately related to and protected by the mineralized compartments (6, 11). In the present study, this fractionation scheme has been applied to the isolation of the mineral-related and mineral-bound macromolecular components from the teeth of Lytechinus variegatus, a prominent urchin found in the Gulf of Mexico.
The complete genome of Strongylocentrotus purpuratus, a distantly related urchin, has been reported, and a genome-wide analysis of its putative biomineralization-related proteins has been carried out by Livingston et al. (12). Although some gene and protein sequence differences were anticipated, our approach was to use the derived S. purpuratus protein sequence data base as a guide for cloning any mineral-related and mineral-bound L. variegatus proteins found. The objective of the present work was to move from the putative proteins predicted by the genome sequencing, to examine in detail the proteins actually present in the urchin tooth that might control calcite formation and the process of mineralization. The present paper describes the isolation of two mineral-associated acidic proteins, UTMP16 and UTMP19 from L. variegatus teeth, the cloning and determination of their complete cDNA sequences, and their relation to the mineral phase.
EXPERIMENTAL PROCEDURES
Isolation of the L. variegatus Tooth Proteins
L. variegatus teeth were obtained, and the proteins were isolated as described earlier (6). Briefly, teeth were collected from 100 urchins and immediately submerged in liquid nitrogen. Nine g of teeth were thawed and washed in 100 ml of 0.15 m NaCl containing a freshly prepared protease inhibitor mixture consisting of 5 mm phenylmethylsulfonyl fluoride, 5 mm N-ethylmaleimide, 5 mm EDTA, 50 mm caproic acid, and 2 mm benzamidine, with slow stirring for 2 h at 4 °C. The washed teeth were suspended in 100 ml of 6 m GdnHCl plus a protease inhibitor mixture. The teeth were crushed to a fine powder with pestle and mortar and extracted with gentle stirring for 20 h at 4 °C. The solvent-accessible extracted material (GdnHCl extract) was separated from the insoluble powder by centrifugation. The residual mineralized powder pellet was taken up in 150 ml of 0.6 n HCl plus protease inhibitor mixture, and the contents were stirred for 4 h. Treatment with 0.6 n HCl completely demineralized the powder, leaving a slightly turbid suspension. The mixture was centrifuged, and the supernatant (HCl extract) was collected, dialyzed exhaustively against 0.1 m acetic acid, and then lyophilized.
The lyophilized HCl extract protein was dissolved in 4 ml of 100 mm K2HPO4, pH 7.2, binding buffer. A slurry of 2 g of CHT ceramic hydroxyapatite (Bio-Rad) was prepared in 10 volumes of 200 mm K2HPO4, pH 7.2, stirred for 30 min. After stopping the stirring, the supernatant was decanted, and the slurry was washed two times with the 200 mm buffer and then equilibrated by washing three times with 100 mm K2HPO4, removing the supernatant at each equilibration. The 4 ml of HCl extract in 100 mm K2HPO4 was added to the slurry, and the mix was gently stirred with a slow motion on a platform shaker for 2 h. The mixture was centrifuged, and the supernatant, designated as the unbound fraction, was removed. The hydroxyapatite pellet was washed with 6 ml of 100 mm binding buffer. This process was repeated two more times. The hydroxyapatite-bound proteins were eluted from the final pellet by washing with 6 ml of 400 mm K2HPO4 buffer. The supernatant was separated, and the pellet was extracted again. This elution was repeated two times. The elutes were combined and dialyzed exhaustively against 0.1 m acetic acid using a Spectra Pore 3,500 molecular weight cut-off pore size membrane (Spectra Pore, Rancho Dominques, CA). The acetic acid was removed by lyophilization, and the proteins, representing the initial calcium carbonate-associated proteins also bound by affinity to calcium hydroxyapatite, was saved for further analysis. All procedures were carried out at 4 °C.
Protein Sequencing
The total 0.6 n HCl extract and the hydroxyapatite-eluted fraction were run on SDS-PAGE and stained separately with Coomassie and Stains-All to determine if there were any differential apatite-binding proteins. Typical of acidic proteins, neither fraction stained well with Coomassie, but both were visualized as metachromatic blue-purple bands in the Stains-All stained gels. Unfortunately, Stains-All interferes with sequencing protocols. Therefore, after preliminary gradient gel runs, electrophoresis was carried out on a matched pair of 10–20% gradient gels for a longer time to ensure good band separation. One of the gels was stained with Stains-All. The positions of bands of interest were identified, and the comparable regions corresponding to the stained bands were then cut from the unstained gel. These were sent to the Molecular Structure Facility (Michigan State University, East Lansing, MI) (Dr. Joe Leykam, Director), where the gel slices were digested with trypsin, and the resulting peptides were fractionated by reverse phase HPLC.2 Several prominent peptide peaks were numbered and selected from each chromatogram for sequencing by automated Edman degradation. The proteins were isolated at different times, as were the tryptic peptide digests. In the system used, the peptide chromatograms may shift in overall elution time, but the peak to peak differences stay approximately the same. As described below, the sequence obtained for the selected peptide from the hydroxyapatite-bound protein band was identical to that published for a predicted P16 protein of L. variegatus (13). Another peptide from the mineral-associated but not apatite-bound fraction was found to be similar to a sequence from a putative S. purpuratus protein, P19 (14).
cDNA Isolation and Characterization
On the supposition that the protein translation was likely to be most active in the highly cellular plumula region, and to avoid some problems from the high mineral content of the mature tooth, poly(A)+ mRNA was prepared from the L. variegatus plumula portion of 450 teeth (3.2 g). We had previously determined that the Stains-All staining bands present in the hard part of the tooth were also present in the highly cellular plumula region (results not shown). A custom SMART cDNA library was created in λTriplEX2 to obtain a high representation of full-length mRNAs at Clontech (Mountain View, CA). The cDNA library was constructed into a λTriplEx2 phagemid vector for blue/white screening. Insert sizes ranged from 0.5 to 4.0 kb, with an average insert size of 1.9 kb.
The L. variegatus tooth plumula cDNA library was used to amplify P16 and P19 by PCR, with the primers based on the sequences obtained from the peptides. Since the L. variegatus sequence for P16 was known (13), forward and reverse primers were designed, based on the sequence present in GenBankTM. The PCR was carried out using Platinum Taq Supermix (Invitrogen) to amplify the cDNA. Conditions for PCR were 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 55 °C for 2 min, and 72 °C for 2 min. This was followed by an extension of 72 °C for 10 min. Amplified PCR products were then resolved on a 1% agarose gel, and the bands of interest were excised and cloned into the TA cloning vector (Invitrogen). The resulting plasmids were sequenced at the Biotechnology Center of Northwestern University.
The amino acid peptide sequence obtained for the P19 band showed homology with the putative P19 S. purpuratus sequence (12). Since the L. variegatus sequence for P19 was not known, a forward primer was designed based on the purpuratus gene sequence. This was used in an initial PCR with the TripE Phage 3′ primer, as described above. Following cloning and sequencing, a reverse primer that included the stop codon was designed based on the sequence for the 3′-end obtained and used in a second PCR with the TripE Phage 5′-arm as the forward primer. The PCR product was cloned and sequenced to give the full-length L. variegatus P19 cDNA.
Primer Sequences
The oligonucleotide sequences used for PCR amplification were as follows: P16-Forward, ATGAAGACCATCATCGCTTTATTTGC; P16-Reverse, CTTATGCGTTTGAAAGCATTTGGGCAC; P19-Forward, GGGGCTGAACCTGCAAACAGTTCAGAG; P19-Reverse, TTACTGTTGAAACGCAAACGGCTTGGTTGTCCC; TripE_Phage_5′_arm, TCCGAGATCTGGACGAGC; TripE_Phage_3′_arm, TAATACGACTCACTATAGGG.
Phosphorylation State of the Isolated Proteins
The protein sequences predicted from the cDNA data were analyzed by several programs of the Center for Biological Sequence Analysis of the Technical University of Denmark (available on the World Wide Web). These analyses are discussed below. One set of data, from the NetPhosK 1.0 Server, indicated a high probability that both P16 and P19 were phosphorylated.
Dephosphorylation of the Extracted and Isolated Proteins
The recovered hydroxyapatite bound and non-apatite-bound fractions were concentrated and buffer-exchanged with citrate buffer at pH 4.8, using a 3,000 nominal molecular weight cut-off Amicon Ultra-4 centrifugal filter device (Millipore Corp., Billerica, MA). The concentration and dilution with citrate buffer was done three times to ensure removal of all free phosphate ions. The concentrated fractions were then incubated with 1 unit of potato acid phosphatase (Sigma) in citrate buffer, pH 4.8, for 6 h. After incubation, the reaction was stopped with the addition of an equal amount of 2× Laemmli buffer, boiled for 5 min, and loaded onto a 10–20% gradient gel. After electrophoresis, the gels were stained with Stains-All, followed by destaining and restaining with Coomassie Blue. The gels compared treated and untreated preparations, and as a control, lanes were also loaded with purified (phosphorylated) and phosphatase-treated rat dentin phosphoprotein.
Two-dimensional Gel Electrophoresis and Phosphate Detection
The total HCl extract protein was suspended in two-dimensional cell lysis buffer (30 mm Tris-HCl, pH 8.8, containing 7 m urea, 2 m thiourea, and 4% CHAPS). 50 μg of protein was mixed with 1.0 μl of diluted Cy3 dye and kept in dark on ice for 30 min. The labeling reaction was stopped by adding 1.0 μl of 10 mm lysine to each sample and incubating in the dark on ice for an additional 15 min. The 2× two-dimensional sample buffer (8 m urea, 4% CHAPS, 20 mg/ml dithiothreitol, 2% pharmalytes, and trace amount of bromphenol blue), 100 μl of DeStreak rehydration solution, and buffer (7 m urea, 2 m thiourea, 4% CHAPS, 20 mg/ml dithiothreitol, 1% pharmalytes, and trace amount of bromphenol blue) (GE Healthcare) were added to the labeling mix to make a total volume of 250 μl. These were mixed well and spun before loading the labeled samples into the strip holder.
Isoelectric focusing was run following the Amersham Biosciences protocol. Upon finishing the isoelectric focusing, the immobilized pH gradient strips were incubated in freshly made equilibration buffer-1 (50 mm Tris-HCl, pH 8.8, containing 6 m urea, 30% glycerol, 2% SDS, a trace amount of bromphenol blue, and 10 mg/ml dithiothreitol) for 15 min with gentle shaking. Then the strips were rinsed in freshly made equilibration buffer-2 (50 mm Tris-HCl, pH 8.8, containing 6 m urea, 30% glycerol, 2% SDS, trace amount of bromphenol blue, and 45 mg/ml dithiothreitol) for 10 min with gentle shaking. Next the immobilized pH gradient strips were rinsed in the SDS-gel running buffer before transferring into 13.5% SDS-gels. The SDS-gels were run at 15 °C until the dye front ran out of the gels.
Phosphostaining for Ser, Thr, and Tyr was accomplished using Pro-Q Diamond Phosphoprotein gel stain, as described by the manufacturer (Molecular ProbesTM; Invitrogen). Gel images were scanned immediately following SDS-PAGE using a Typhoon Trio Imager (Amersham Biosciences). The scanned images were then analyzed by ImageQuant software (version 6.0; Amersham Biosciences), followed by in-gel analysis using DeCyder software version 6.0 (Amersham Biosciences).
Production, Purification, and Characterization of Antibodies against L. variegatus P16
To prepare antibodies against P16, a 20-amino acid-long peptide near the amino terminus of the protein shortly following the signal sequence was chosen. The sequence of the peptide was FGNGGGGEIGVNDAYGGGGA. The subsequent peptide synthesis, preparation of the antibodies in rabbits, and affinity purification of the antibody was carried out by Bethyl Laboratories Inc. (Montgomery, TX) according to established procedures. Immunospecificity of the antibody was verified by immunoprecipitation.
Immunoprecipitation
50 μl (corresponding to 40 μg of protein) of the total HCl extract was added to 500 μl of immunoprecipitation buffer (which consisted of 250 mm NaCl, 50 mm Tris, pH 7.5, 0.5% Nonidet P-40). To this was added 25 μg of affinity-purified anti-P16 antibody and 20 μl of protein A-Sepharose. The reaction was rotated (end-to-end) overnight at 4 °C. The next day, the reaction was centrifuged to pellet the complex. The supernatant was removed, and the pellet was washed three times with 500 μl of cold immunoprecipitation buffer. The pellet was then dissolved in 50 μl of Laemmli buffer, boiled for 5 min, and analyzed by SDS-gel electrophoresis.
Immunohistochemistry
L. variegatus teeth were dissected, fixed, and embedded in Epon, and 1-mm serial sections were cut perpendicular to the aboral-adoral axis, as previously described (6). In brief, the best initial fixation was achieved by placing the whole urchin in seawater containing 0.1% glutaraldehyde for 40 min. The teeth were then removed and fixed for another 2 h in the seawater plus 0.1% glutaraldehyde, followed by immersion in 5% glutaraldehyde, 4% paraformaldehyde in 0.08 m Na2HPO4 overnight. After washing with 0.01 m sodium cacodylate, postfixation was with 1% OsO4 before the Epon embedding procedure was initiated. Sections were readily cut without structural disruption from the Epon blocks on a Reichert-Jung Ultracut E microtome with a diamond knife.
The sections were blocked by treatment for 4 h with phosphate-buffered saline containing 3% bovine serum albumin and 1% normal goat serum. Normal goat serum was used, since the secondary antibody was raised in goats. After 4 h, an anti-P16 antibody at a concentration of 1:200 in blocking buffer was added to the experimental slides and reacted at 4 °C overnight. In control slides, the incubation was carried on in blocking buffer without the primary antibody. The next day, sections were washed four times (15 min/wash) with phosphate-buffered saline and reacted for 1 h at room temperature with Alexa Fluor 488 goat anti-rabbit IgG. Following the secondary antibody incubation, sections were once again washed 4 times (15 min/wash) with phosphate-buffered saline. Secondary antibody incubation and subsequent washings were performed in the dark to minimize loss of fluorescent signal due to photobleaching. Immunoreactive areas were visualized on a Nikon C1Si confocal microscope.
RESULTS
Characterization of L. variegatus Tooth Matrix Proteins UTM16 and UTM19
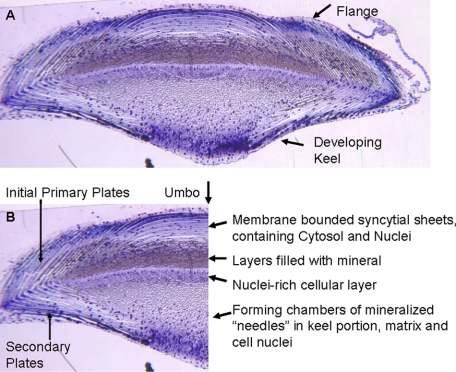
As noted under “Experimental Procedures,” the proteins solubilized during the 0.6 n HCl demineralization of the 6 m GdnHCl-washed teeth include the proteins occluded in the mineral phase as well as in any cellular compartments rendered inaccessible by the surrounding mineral crystals. Histological sections of the mature tooth (9, 16) have shown that there is a complex and intimate distribution of organic and mineralized components throughout the tooth, even in the most highly mineralized portions. This is shown very well in a partially mineralized toluidine blue-stained L. variegatus tooth cross-section cut in the growth region immediately after the tooth keel had begun to develop (Fig. 1A). The left half of the section is printed and labeled (Fig. 1B) to point out the densely stained cell nuclei, the less densely stained syncytial cytosol, and the sharply stained boundaries of the mineral plates. Note that the tooth is surrounded by a cell layer that may be accessible to the initial GdnHCl extraction. The deeper layers of cell syncytia are not equally accessible. The dull reddish-blue along the tooth flange horizontal axis corresponds to the most highly mineralized portion of the densely packed syncytial plates often called the “stone part.” At this point in tooth development, the keel side is less mineralized, but the mineral needles (also called prisms) are beginning to form. They are surrounded by an organic matrix. The sharp boundary between the keel needles and the upper flange of the tooth is composed of an underlying very cellular layer, shown by the band of densely stained cell nuclei. As the tooth matures and the contents of this level are moved toward the incisal edge, where it will ultimately wear away, the mineral fraction increases, and the relative volume occupied by the organic components decreases, but an organic matrix is present at all stages, either between or within the mineral crystals (10). It is easy to see why the 6 m GdnHCl may not be able to gain full access to these densely packed structures although the tooth has been ground to particulate size. The heterogeneity of the tooth mineralization is evident. In the portion of the tooth just under the umbo (Fig. 1B), the central point of the flange where the primary plates and syncytia from each side interdigitate, mineralization is less dense even in the most distal mature part of the tooth.
FIGURE 1.
A, cross-section of developing L. variegatus tooth at an early stage of keel formation. Glutaraldehyde fixed teeth were processed as described by Veis et al. (16). This section was stained with toluidine blue. Cell nuclei stain a deep blue, and the non-mineralized syncytial matrix is a pale blue. The mineral-filled plates are evident as a dull blue-gray. B, the nuclei are seen trapped within the heavily mineralized layer just above the nuclei-rich cellular layer, sometimes referred to as the “stone part” of the tooth. the umbo is at the top center of the flange. The syncytial layers interweave as they grow from each edge. This is the least mineralized portion of the tooth.
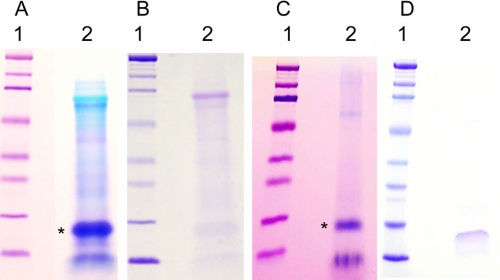
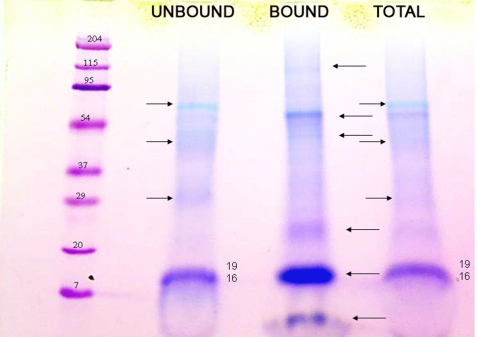
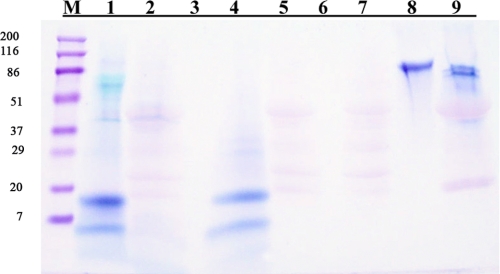
The 0.6 m HCl extraction removed the mineral completely from the pulverized tooth particles and left the resident macromolecular constituents in the soluble extract after dialysis using the molecular weight 3,500 cut-off membrane. Gel electrophoresis of the recovered proteins of the 0.6 m HCl extract in 10–20% gradient gels (Fig. 2, lanes A2 and B2) showed strong polychromatic staining with Stains-All (A2) but very weak staining with Coomassie Blue (B2), typical of strongly acidic proteins. The proteins of the HCl extract were exposed to the ceramic hydroxyapatite at low phosphate concentration, and the particles were washed extensively in the binding buffer. The bound fraction was then eluted with a higher concentration of phosphate, concentrated, and run on a similar 10–20% gradient gel (Fig. 2, lanes C2 and D2). The hydroxyapatite-bound fraction (Fig. 2, lane C2) showed prominent Stains-All-stained bands in the region with apparent molecular weights (Mr) from 15,000 to 20,000. Staining with Coomassie in this Mr region was barely detectable (Fig. 2, lane D2). Compared with the parent total HCl extract, it was evident from the relative intensities and difference in color and broadness of the bands that only a relatively small portion of the total content of the starred band in Fig. 2, lane A2, had been bound to the hydroxyapatite, Fig. 2, lane C2. Reloading the unbound fraction on a fresh hydroxyapatite column did not result in any additional binding, indicating that the unbound fraction does not have an affinity for hydroxyapatite in the presence of the 100 mm K2HPO4, pH 7.2, binding buffer, and is not due to overloading the column. Thus, it appeared likely that some specific component(s) in this low molecular weight range had strong affinity to the hydroxyapatite. The higher Mr components evident in both Stains-All (polychromatic)- and Coomassie-stained bands in lanes A2 and B2 were selectively but weakly bound by the HA. This was confirmed in Fig. 3 by a direct comparison of the apatite bound and unbound fractions. There are obviously several components enhanced in the HA-bound fraction and depleted in the unbound fraction as compared with the total HCl extract. The appearance of the 19/16 kDa band in the total and unbound fractions is quite different from the appearance of the 19/16 kDa band in the bound fraction lane. The expansion and bowed appearance of the 19/16 kDa band in the total and unbound fractions is characteristic of highly acidic Glu-rich proteins, which in Stains-All-stained gels have a pinkish hue in contrast to the deeper purple of the putative 16 kDa band in the “bound” lane. Obviously, the several bands denoted by the left pointing arrows are more concentrated in the BOUND lane than in the TOTAL lane. These data show that the mineral-associated but not apatite-bound (UNBOUND lane, right pointing arrows) components are distinct entities as compared with the apatite-bound components. The closely running P19 and P16 bands were most prominent; thus, those components were examined further.
FIGURE 2.
Gel electrophoresis of the mineral-protected proteins extracted from the previously GdnHCl-extracted mineralized tooth powder with 0.6 n HCl. Gels A and C are stained with Stains-All, and gels B and D are stained with Coomassie. Lanes A1 and B1, the standard proteins; lanes A2 and B2, the total HCl-extracted proteins. After absorption on hydroxyapatite and subsequent high phosphate ion elution, the HA-bound proteins are shown in lanes C2 and D2, stained with Stains-All and Coomassie, respectively. The bands marked with an asterisk were the most prominent, and the proteins recovered from the gels in these bands were subject to trypsin degradation, and certain peptides were then subject to NH2-terminal sequencing.
FIGURE 3.
A direct gel electrophoresis comparison of the initial total HCl extract (TOTAL), the HA-bound fraction (BOUND), and the residual non-HA-bound fraction (UNBOUND). The amount of protein loaded in the BOUND lane was concentrated after elution from the HA; thus, the relative stain intensity is not a quantitative measure of the initial amounts of protein present in that fraction. The bands marked with a left pointing arrow to the right of the BOUND lane are accentuated in that fraction and are not present among the main bands of the UNBOUND lane, marked with a right pointing arrow. The bands visualized in the TOTAL extract (marked with a right pointing arrow) are matched with those in the UNBOUND lane but at lower intensity. The bands in the P19–P16 regions of the BOUND and UNBOUND lanes were the bands used for the sequencing work. The very low mass band (<7 kDa but retained on a 3.5 kDa cut-off membrane during extensive dialysis) in the bound fraction was found consistently but has not yet been analyzed in detail. Similarly, the high mass bands in the bound fraction have not yet been fully analyzed.
Because of the limited quantity of protein within the extracts, the decision was made to move immediately to the preparation of peptides from these two protein fractions for direct sequencing. The region between Mr 15,000 and 20,000 was cut from each of the paired unstained gels (Fig. 2, lanes A2 and C2), eluted, and then digested with trypsin. The digests were fractionated on HPLC under the same conditions. A marked difference in elution patterns (data not shown) confirmed the different compositions of the two peptide digests and that specific proteins had bound selectively to the hydroxyapatite. A sharp peak, Peak 7, from the total HCl extract, and similarly sharp Peak 8 from the apatite bound fraction were NH2-terminal sequenced by Edman degradation.
The most probable amino acid sequence of the Peak 7 peptide was determined to be GAGTGAEPANSSEER. A search of the S. purpuratus sequence data showed this peptide to be homologous to a sequence within the putative protein designated P19, suggested to be the “sea urchin biomineralization protein” (12). Peptide Peak 8 from the digest of the hydroxyapatite-bound protein was also sequenced successfully, and its amino acid sequence was determined to be VPGTGTGGFQGTDGTQF, which proved to be homologous to a sequence within the L. variegatus putative protein designated as P16, found in embryonic L. variegatus spicules (13). P16 had also been proposed to be a “biomineralization-related protein” (12). We have therefore used the designation UTM (urchin tooth matrix) P19 and UTMP16 designations for the two L. variegatus proteins.
Cloning of L. variegatus UTMP16 and UTMP19
The L. variegatus tooth plumula cDNA library (6) was used to amplify UTMP16 and UTMP19 cDNAs by use of PCR. The full sequence for the L. variegatus P19 has not previously been described.
UTMP19
The peptide sequence GAGTGAEPANSSEER, obtained by protein sequencing, showed only partial homology with S. purpuratus P19. However, the underlined sequence portion GAEPANSSEER showed complete identity with the putative purpuratus protein, so a primer based on the nucleotide sequence of the S. purpuratus cDNA was designed (P19-Forward) and used along with the TripE Phage 3′-arm to obtain the 3′-end of the L. variegatus cDNA. This was cloned and sequenced. The L. variegatus sequence obtained was then used to design a reverse primer of 33 nucleotides (P19-Reverse) that included the stop codon at the 3′-end. PCR of the L. variegatus cDNA yielded a single strong amplified band (data not shown), which was cloned in the sequencing vector TOPO 4. The nucleotide sequence of L. variegatus UTMP19 cDNA is shown in Fig. 4A (GenBankTM accession number GQ340974). The complete derived 166-amino acid sequence of the L. variegatus UTMP19, shown in Fig. 4B, was obtained by PCR with the TripE Phage 5′-arm-P19-Reverse primer pair.
FIGURE 4.
The complete nucleotide and amino acid sequences of L. variegatus UTMP19. A, the complete coding region nucleotide sequence of UTMP19. B, the amino acid sequence of L. variegatus UTMP19 compared with the predicted sequences of the two S. purpuratus P19 long and short isoforms. The probability of phosphorylation of threonine (boldface type, red) and serine (boldface type, green) residues at a level of >0.5 is indicated. The underlined residues have phosphorylation probability scores of 0.8–0.94. Note the four highlighted pairs of consecutive glutamic acid residues (boldface type, purple) within the most NH2-terminal portion of the molecule. The dashed line indicates residue identity. There are many more species differences in the COOH-terminal half of the molecule than in the NH2-terminal portions.
The L. variegatus UMTP19 is a highly acidic protein (Fig. 4B), which, without post-translational modification, would have an isoelectric point of 4.72 and a charge of −11.03 at pH 7. The amino acid sequence analyzed by the Net PhosK 1.0 Server (18) suggests that potentially it has 20 threonine phosphorylation sites at threonines 2, 8, 11, 15, 27, 61, 63, 84, 97, and 159 and nine serine phosphorylation sites at serines 31, 39, 52, 60, 62, 78, 91, 92, and 146. All 29 of these have a phosphorylation score greater than the threshold value of 0.5, but Thr61 and Thr63 have scores of 0.88 and 0.94, and Ser62 and Ser78 (boldface type and underlined in Fig. 4B) have scores of 0.93 and 0.80, respectively, suggesting that they represent particularly strategic locations.
As shown in the protein sequence in Fig. 4B, the 19 putative potential phosphorylation sites, of which nine are serine (Thr and Ser highlighted in red and green, respectively), are not distributed evenly throughout the molecule. They are heavily weighted in the amino-terminal half of the molecule along with 22 of the 31 total Glu residues. Four pairs of sequential Glu residues (EE) are prominent in the NH2-terminal portion. Fig. 4B also compares the L. variegatus UTMP19 clone sequence with that of the published predicted S. purpuratus protein from the cDNA. Two forms of S. purpuratus P19 have been predicted. The L. variegatus sequence corresponded in length to the 166-residue S. purpuratus short form (AF516413). Interestingly, only three very conservative substitutions were found in the first 80 NH2-terminal acidic domain portion residues. All of the potential phosphorylated Ser residues were conserved, whereas L. variegatus Thr84 and Thr97 were non-conservatively replaced with Ile and Pro, respectively, in the S. purpuratus sequence. The remaining carboxyl-terminal portion of the molecules had 20 sequence differences. The L. variegatus counterpart of the S. purpuratus long form (AF516414), which has 10 additional residues added directly to the COOH terminus, was not found in the Lytechinus cDNA. It is noteworthy that the L. variegatus UTMP19 has no Cys residues, whereas the S. purpuratus has a single Cys (Cys150) near the COOH terminus.
It is most noteworthy that UTMP19 has neither a predicted signal sequence nor a membrane-spanning sequence; hence, it may be a cytosolic rather than extracellular component. On the other hand, L. variegatus UTMP19, like its S. purpuratus counterpart, has both a nuclear localization signal (NLS), 64RKKK67, and a Leu-Glu-rich nuclear exit signal region, 98EAREIDAELQKRIQDLEQQ116 and 124LQLYRKVNDLE134 (18, 19), respectively, both of which may possibly be α-helical but are immediately preceded by a predicted flexible coil region.
UTMP16
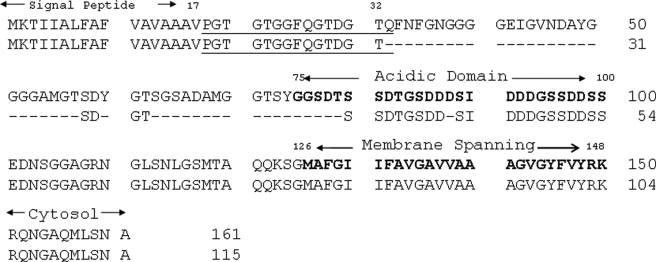
Since the L. variegatus sequence for P16 was known, forward (starting at the ATG start codon) and reverse (ending at the stop codon) primers were designed, based on the sequence present in GenBankTM (accession number AAY59533). PCR of the L. variegatus cDNA yielded a strong amplified band (data not shown), which was then cloned in the sequencing vector TOPO 4. In this case, the cloning yielded two clones with inserts of slightly different sizes. Both clones were sequenced. The obtained nucleotide sequences were passed through a Blast (Basic Alignment Search Tool; available on the World Wide Web) search. The clone with the larger insert was identical to the previously isolated L. variegatus P16. The clone with the smaller size insert was a truncated version of P16 (Fig. 5). P16 has a signal peptide from amino acids 1 to 17. The sequence of the peptide obtained by protein sequencing begins just after the signal peptide cleavage site (underlined in Fig. 5; not a trypsin-sensitive bond), providing the NH2-terminal residue of the secreted protein and demonstrating that the signal peptide had been removed and that P16 was indeed a secreted protein in both long and short forms.
FIGURE 5.
The complete deduced sequences of L. variegatus UTMP16L and UTMP16S. The underlined sequence was that obtained from the amino acid sequence of tryptic peptide 8 (Fig. 4B) and verified by the cloned nucleotide sequences. The signal sequence, central Asp-Ser rich domain, and the transmembrane and cytosolic domains are all indicated. The dashed lines denote the residues deleted in UTMP16S. The high Gly content of the UTMP16L sequence from 32 to 75 suggests that it is a relatively flexible region with no clearly defined structure.
The long form, now designated UTMP16L, has three interesting well defined domains. The sequence 33–74, numbered in Fig. 5 from the start of the signal peptide, has an essentially random conformation with a near zero Kyte-Doolittle hydrophilicity index. In contrast, the following sequence from 75 to 108 is abruptly a strongly acidic and hydrophilic region rich in amino acids Asp and Ser in a sequence pattern similar to those seen in the mammalian protein dentin phosphophoryn (DPP) (20), shown to be involved in mineralization of dentin. This segment is followed shortly from residue 126 to 148, by a hydrophobic sequence with a predicted membrane-spanning domain comprising a major part of the COOH-terminal region. Thus, UTMP16L is likely to be a membrane-associated protein, whereas the central highly acidic region is predicted to be external to the cell membrane. The smaller secreted UMTP16S molecule retains the identical acidic and membrane-spanning carboxyl-terminal sequences but lacks the “neutral, unstructured” NH2-terminal residues 33–74 of UTMP16L noted above. These considerations may explain the appearance of the low molecular mass intensely Stains-All-staining band (presumably UTMP16S) seen in the BOUND lane of the gel in Fig. 3 and the heavily loaded lanes of Fig. 2, lanes A2 and C2, whereas the inclusion of the NH2-terminal segment 33–74 permits the concentrated UTMP16L to stain faintly with Coomassie Blue (Fig. 2, lane D2).
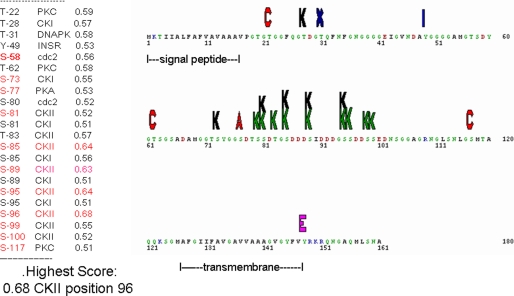
An analysis of the UTMP16L amino acid sequence predicted 12 putative potential serine phosphorylation sites (17), with nine of those in the Asp-rich region and likely to be a substrate for casein kinase II, with some sites good substrates for casein kinase I as well. Thus, it is likely that the external central region has a high probability to be phosphorylated. The potential phosphorylation sites and predicted phosphate landscape map are given in Fig. 6.
FIGURE 6.
Phosphorylation sites and phosphorylation landscape for UTMP16L. The list at the left shows that 22 residues are potentially capable of acting as kinase substrates with a greater than 0.5 probability. The Ser residues at 85, 89, 95, and 96 have the highest phosphorylation probability scores with casein kinase II (CKII) (green K) even without the possibility that phosphorylation would further enhance the potential for phosphorylation of additional Ser or Thr. residues. Thus, this central region exposed at the exterior of the cell syncytial membranes, but anchored by the transmembrane region to the cell interior, may be important in directing mineral localization. Other potential kinases are protein kinase C (red), protein kinase A (red), and casein kinase I (CKI) (black K). Note that several of the Ser residues may be substrates for either casein kinase I or II.
All of the predicted potential UTMP16L serine phosphorylation sites are in the acidic DSS region from residue 73 to 100. The smaller UMTP16S clone retains the identical amino- and carboxyl-terminal sequences and obviously the peptide obtained by protein sequencing but lacks the hydrophobic and Gly-rich central sequence. However, the UTMP16S retains the potentially phosphorylated acidic Asp/Ser-rich region as well as the membrane-spanning region and cytosolic zones. The long and short forms of UTMP16 are acidic proteins with isoelectric points of 3.72 and 3.62 and a charge of −11.08 and −7.09 at pH 7, respectively, even without the effect of the probable phosphorylations. The predicted S. purpuratus P16 protein (14) is 11 residues longer than the UTMP16L, but the COOH-terminal portion, residues 90–172 is highly homologous to residues 80–161 of L. variegatus UTMP16L.
Apatite Binding and Phosphorylation
The gels in Fig. 3 showed that there was differential binding of UTMP19 and UTMP16 fractions to ceramic HA. The HA-bound and unbound fractions were collected and subject to digestion with acid phosphatase and rerun on SDS-gels as before. Fig. 7, lanes 1 and 4, shows the Stains-All stain patterns before digestion for the unbound and HA-bound fractions, respectively. The corresponding lanes 2 and 5 represent the same fractions after digestion, showing that the phosphatase had indeed removed phosphate groups from the proteins of both fractions, resulting in a loss of Stains-All staining. The majority of the very pale staining pink staining in lanes 2 and 5 were identical to the control with phosphatase alone (lane 7). Lanes 3 and 6 were blank, to clearly distinguish the fractions. A second control was the dephosphorylation of rat incisor DPP, shown in lanes 8 and 9, with a decrease in mass of about 6000, corresponding to loss of about 90% of the potential phosphorylated serine residues.
FIGURE 7.
The effect of potato acid phosphatase dephosphorylation on the apatite-bound and unbound fractions of the HCl-extracted proteins, as determined by the Stains-All staining of SDS-gel electrophoresis patterns before and after phosphatase digestion. M, molecular mass markers. Lane 1, unbound, undigested; lane 2, unbound, digested; lanes 3 and 6, blank; lane 4, bound, undigested; lane 5, bound, digested; lane 7, phosphatase plus buffer blank; lane 8, rat DPP (similar phosphorylated sequence control), undigested. Lane 9, phosphophoryn, digested. The faint pink in lane 7 is the control appearance of the acid phosphatase. The enhanced pink bands at ∼45 and 20 kDa suggest that some proteins of DPP may interact stably with components in the phosphatase. Nevertheless, these data show clearly that the majority of the proteins in the mineral-associated preparations had been phosphorylated in vivo, indicating that the NetPhos predictions of phosphorylation were substantiated.
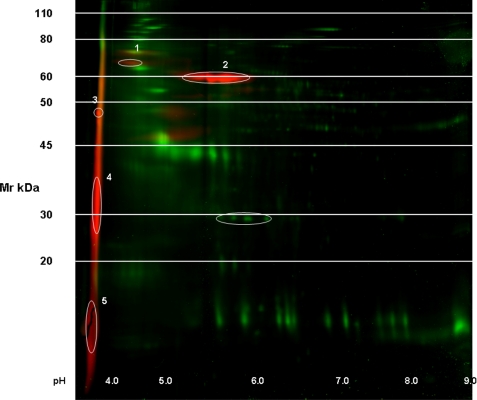
The presence of phosphoryl groups in the initial HCl-soluble extract was demonstrated directly by running two-dimensional gel electrophoresis with phosphoprofiling carried out by Applied Biomics (Hayward, CA) as described. In the gel (Fig. 8), all proteins are shown as labeled with green fluorescence, whereas the phosphorylated proteins are shown by the red fluorescence overlay. Five phosphorylated proteins are denoted and numbered. Of these, proteins numbered 3, 4, and 5 all have strongly acidic isoelectric points, pI < 4 with apparent Mr values ranging from ∼48,000 to ∼15,000. The most striking feature is that the phosphorylated proteins of interest here, UTMP19 and UTMP16, are present in the phosphorylated streak at Mr < 20,000 at such relatively low concentration in the total extract that they barely show up along the pH 3 border of the protein stain gel, whereas they are readily seen as phosphorylated proteins at that locus. P19 is clearly present at higher protein concentration as shown by the Stains-All gels of the HCl extract in Fig. 2 but is less densely phosphorylated. This is probably the basis for the weaker affinity of P19 for binding to the apatite column. The natures of other higher Mr phosphorylated proteins identified as spots 1 and 2 in the two-dimensional gel of Fig. 8 are the object of current work.
FIGURE 8.
Two-dimensional gel electrophoresis protein and phosphorylation analysis of the HCl-extracted mineral-related protein fraction. Green fluorescence, protein; red fluorescence, phosphorylated proteins. The numbered regions were selected by the image analysis software as phosphorylated. Bands 3, 4, and 5 all have isoelectric points of pH <4.0. Band 5 contains UTMP16 and UTMP19. Identification of the components of bands 1–4 is the object of the present studies.
Immunolocalization and Immunoprecipitation of P16
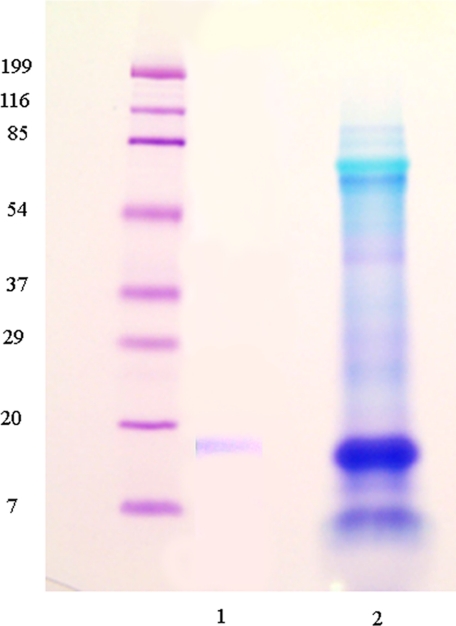
Anti-P16 antibody was used to immunoprecipitate the P16 from the total HCl extract. As shown in Fig. 9, the anti-P16 prepared against the amino-terminal sequence of UTMP16 captured a single protein corresponding to the expected position of UTMP16. Therefore, the focus shifted to immunolocalization. There was a very important problem to consider relative to examination of the tooth structure, since the relationships between mineral and cell syncytium change all along the tooth at each different stage of development. Detailed studies are under way and will be presented elsewhere, but here, to be consistent with the detailed toluidine blue cross-section shown in Fig. 1, at the point where mineralization of the flange region primary plates is taking place, a comparable region is shown in Fig. 10. Three important points can be seen. In the predominant syncytial cell layers, the labeling is both on the layer membranes and widely seen in the cytosol background. In the areas of dense mineral, there is essentially no fluorescence. In the intervening region of mineral deposition, the syncytial membranes are strongly labeled, whereas the cytosol labeling is decreased. Thus, the UTMP16 appears to be localized to the syncytial membrane, as predicted from the membrane-spanning nature of the P16.
FIGURE 9.
The specificity of anti-UTMP16. Gel electrophoresis of the total HCl-extracted protein. Immunoprecipitation of this preparation with anti-UTMP16 yielded the single UTMP16 band shown in lane 1. Lane 2, the expected metachromatically blue stained bands.
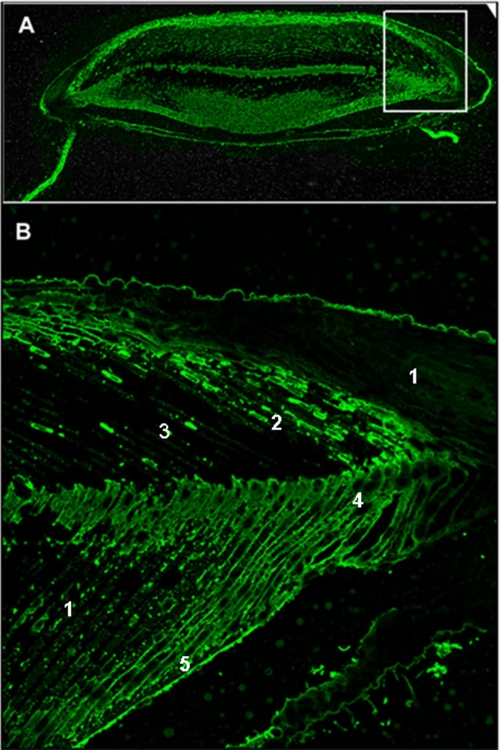
FIGURE 10.
Immunolocalization of UTMP16 within a tooth cross-section early in the tooth development, where the primary and secondary plate mineralization has begun, and the keel has begun to develop. A, a total tooth cross-section (section 1090, ∼10 mm from the origin of the plumula). B, the boxed region of A shown at higher resolution. The mineralized primary plates, developing secondary plates, and cellular parts of the cell syncytia are labeled. The UTMP16 fluorescence diffusely labels the syncytia in the less mineralized or unmineralized regions but concentrates brightly at the cell membrane boundaries of the calcite plates. This localization suggests that the UTMP16 moves to the syncytial membrane when mineralization is initiated. 1, cell syncytia; 2, forming primary plates, membrane labeling; 3, primary plates, mineralized; 4, forming secondary plates, membrane labeling; 5, mineralized secondary plates.
DISCUSSION
Our first studies of the urchin tooth proteins (7, 9) had shown that the proteins of urchin teeth and vertebrate teeth shared several properties, including the fact that many were highly acidic and contained sequences that had common antigenic properties. There was a strong cross-reactivity of the urchin tooth mineralized tissue proteins with antibodies made to purified vertebrate tooth dentin matrix proteins (7). The objective of this study was to characterize the proteins of the mineralized teeth of the sea urchin to determine if they had any relation to proteins involved in the mineralization of vertebrate teeth. The approach was to isolate the proteins from the heavily mineralized portions of the teeth, fractionate them on hydroxyapatite, sequence them, and relate them to functions within the tooth structure. We report here the isolation of two mineral phase-associated acidic proteins with different affinities for hydroxyapatite from the calcite mineral-associated phase of the mature tooth of L. variegatus. They correspond to two predicted proteins, P16 and P19, previously detected by screening of a primary mesenchymal cell library and shown by in situ hybridization to be present in the spicules of the sea urchin S. purpuratus (13, 14). To our knowledge, this is the first report showing that proteins present in the embryonic spicules, mineralized tissues arising transiently during embryogenesis, are also present in the adult mineralized tooth.
Both UTMP16 and UTMP19 are highly acidic proteins with a number of predicted serine phosphorylation sites and, in the case of UTMP19, also numerous threonine residues. Both proteins were obtained from the mineralized portion of the tooth after thorough extraction and washing of the pulverized tooth powder with the lyotropic, dissociative 6.0 m GdnHCl reagent to remove all of the exposed organic components. The residual mineralized particulates were then dissolved in cold 0.6 n HCl. This seemingly harsh solvent completely dissolved the mineral phase, but, as demonstrated in a number of earlier studies, did not degrade or hydrolyze the mineral-trapped proteins, leaving them in essentially their intact form in the HCl extract. The key factor in the isolation and separation of these in vivo calcium carbonate mineral phase sequestered proteins was their differential binding to a ceramic hydroxyapatite. UTMP19 was readily soluble in the HCl and, in the presence of 100 mm KH2PO4, pH 7.2, did not bind to the apatite ceramic. However, the smaller UTMP16L, also soluble in the 0.6 n HCl, bound more avidly to the apatite and needed 400 mm KH2PO4 to be extracted from the apatite ceramic at neutral pH. The principal differences in composition are the ratios of Asp to Glu and Ser to Thr: P16L (Asp/Glu = 15:2, Ser/Thr = 18:11), P19 (Asp/Glu = 8:31, Ser/Thr = 11:13). Without consideration of the potential phosphorylations, P19 has a total of 39 acidic residues, whereas P16L has only 17. Based on studies of the binding of poly-Glu and poly-Asp polymers to apatite (20), both polymers bind to hydroxyapatite with similar efficiency and can displace many acidic proteins. However, poly-Asp has a Kd value about twice that (weaker binding) of poly-Glu; thus, P19 should have had a stronger binding to apatite than the P16. It appears that this general electrostatic interaction is counterbalanced by the 78SDTSSDTGSDDDSIDDDGSSDDSSED103 sequence in central P16L, reminiscent of both the extensive (SSD/DSS)n repeats and the COOH-terminal sequence SDSDSDSDSDSEGSDS of the rat tooth protein DSPP (21), similar in all vertebrate mineralized dentins. In the vertebrate tooth, the Ser residues are usually phosphorylated. As noted earlier, the NetPhos program (available on the World Wide Web) predicts 10 potential phosphorylation sites in the UTMP16L 78–103 region (Fig. 7). This sequence is probably the basis for the enhanced apatite binding for P16L as compared with the Glu-rich P19.
The two-dimensional gel electrophoresis and phospholabeling gels showed that the UTMP16 and UTMP19 were indeed phosphorylated and had isoelectric points around the predicted pH < 3.5 values (Fig. 9). The phosphorylation of both proteins was confirmed qualitatively by the decrease in Stains-All staining after digestion with acid phosphatase (Fig. 8). The acidic and possibly phosphorylated stretches may have a special function or relevance in all mineralized tissues independent of whether the mineral phase is carbonate or apatite. Other invertebrate mineral-related proteins (11, 22, 23) are rich in Asp and Ser and have extensive SSD and SDSD motifs.
The sequence property predictions for the UTMP16L (residues 126–148) and UTMP16S (residues 61–101) predict that both forms are probably anchored via their COOH-terminal domain to the syncytial membranes, with their most COOH-terminal segment (149–161 and 102–116, respectively) in the syncytial cytosol. The hydrophilic and acidic SSD and SD motifs would be exposed on the outside of the syncytial membranes in direct interaction with Ca2+ carbonate on the surface of the mineral phase and possibly embedded within the calcite. Morpholino knockdown studies have shown that SpP16 plays an essential role in embryonic spicule growth and acts at a late step in this process (i.e. downstream of primary mesenchymal cell specification, migration, and fusion) (13). Immunolocalization studies with the anti-P16 antibody, made to the common NH2-terminal sequence of both UTMP16L and UTMP16S, confirmed the concentration of the UTMP16 at the syncytial membranes bounding the mineralized primary plates (Fig. 10). A surprising observation was that the anti-UTMP16 stained the cell cytosol quite generally and was most intense in the non-mineralized regions of the tooth. We observed, however, that the cytosol was less intensely stained in the regions becoming mineralized (i.e. the P16 moved from cytosol to membrane as if in response to a “mineralization signal”). Much more detailed studies are in progress as a function of the changing organization of the tooth structure.
The short form, UTMP16S, described here as 45 residues shorter than the UTMP16L form, with a mass of ∼11 kDa, was not shown to be present in the embryonic spicules, but the deleted sequence, 31TQFNFGNGGGGEIGVNDAYGGGGAMGTSDYGTSGSADAMG70, is essentially structure-neutral, with no preference for extracellular or cytosolic environments. The UTMP16S retains the signal peptide, the central membrane-external Asp and Ser-rich sequence, and the carboxyl-terminal transmembrane domain that may anchor the protein to the syncytial membrane. It is likely that the UTMP16S is a specific alternatively spliced form of UTMP16L, with similar function, although we could not discern classical splice sites within the nucleotide sequence on the UTMP16L mRNA. Similarly, rat tooth phosphophoryn, with its extensive Asp-Ser domains, also has been shown to have a number of splice variants, probably created by cryptic or non-classical splice sites (24).
The putative protein, sea urchin P19, whose message was shown by Illies et al. (14) to be one of the most abundant mRNA transcripts in urchin primary mesenchymal cells based on a qualitative impression of the relative signal strength in whole mount in situ hybridization studies, had not been found in mature urchin tissues. We describe for the first time its presence as a protein abundant in the L. variegatus tooth. It is evidently an intracellular rather than extracellular protein, with neither a signal nor transmembrane sequence. Since the protein was collected from the mineral-protected fraction, it must be in the cells occluded in the mineral phase. Its function remains to be established. Nevertheless, its properties are reminiscent of the vertebrate mineral-related protein DMP1, which upon synthesis enters the nucleus. The DMP1 nuclear form is not phosphorylated and does not bind to hydroxyapatite (25). In the nucleus, DMP1 acts as a transcription factor. In response to an efflux of calcium into the nucleus, DMP1 is then phosphorylated and exits the nucleus, whereupon it is secreted into the extracellular matrix to possibly play a more direct role in mineralization (26). If this scenario applies to UTMP19, the protein may act as a nuclear transcription factor important for mineralization, or in its phosphorylated form, it may be involved in some aspect of calcium transport within the cell, thus indirectly rather than directly functioning as a mineralization-related protein. DMP1 bears a signal peptide and is secreted into the extracellular matrix after phosphorylation, whereas it appears that the UTMP19 is retained within the cell syncytial cytosol without the possibility of entering into the usual cell secretory pathway. Although prior to this report the UTMP19 was found only in the primary mesenchymal cells of the embryonic spicules as one of the most abundant transcripts (12, 14), it is clearly also a major protein component in the urchin tooth, a constantly growing and developing organ in the urchin. If the P19 protein is directly functional in mineralization, that aspect of the process must take place intracellularly. The term “biomineralization-related” protein (12–14) thus has to be more specifically narrowed in terms of function, and this is the approach we are following. The work on the definition of the complete S. purpuratus genome has been indispensable in this study, but we clearly need to delve more deeply into the protein functions and mechanisms of action of the specific proteins with regard to mineralization. There are several questions that stream naturally from this work. Is it necessary or beneficial to have phosphorylated proteins actively participating in the biogenic development of carbonate minerals? Where, with respect to the syncytial membranes, are these proteins localized? Currently, we are making the recombinant UTMP16L, UTMP16S, and UTMP19 proteins so that we can carry out direct studies of the binding of the proteins to calcite and to make specific antibodies that can be used to localize the proteins in situ.
One of the most intriguing properties of the urchin tooth is the well known presence of high magnesium calcite and its uneven distribution within the tooth. We have examined the distribution of tooth components at high resolution using secondary ion mass spectroscopy of intact polished tooth surfaces (15). These data showed that the very high magnesium calcite was present in the most mature part of the tooth in the columns of calcite that fuse the lower magnesium calcite plates and prisms together, presumably to enhance mechanical strength in the adoral region. The interesting fact was that there was a direct correlation between the high magnesium calcite and a high content of Asp-rich protein (i.e. Asp and Mg content were tightly correlated). We had postulated that since the high magnesium columns were a late development during tooth maturation, an Asp-rich protein might be specifically also expressed concomitantly, later rather than early in tooth maturation when the primary lower magnesium calcite plates were formed. It is interesting that we found the P16 proteins retained within the most mature part of the mineralized tooth. In future studies, we intend to focus on the high magnesium structures with regard to mapping UTMP16L and S protein concentrations and in the development of the keel prism and needle regions.
This work was supported, in whole or in part, by National Institutes of Health Grant DE 001374 (to A. V.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) GQ340974.
- HPLC
- high pressure liquid chromatography
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- DPP
- dentin phosphophoryn
- HA
- hydroxyapatite.
REFERENCES
- 1.Smith A. B. (1988) Mol. Biol. Evol. 5, 345–365 [Google Scholar]
- 2.Wilt F. H. (1999) J. Struct. Biol. 126, 216–226 [DOI] [PubMed] [Google Scholar]
- 3.Wang R. Z., Addadi L., Weiner S. (1997) Philos. Trans. R. Soc. Lond. B Biol. Sci. 352, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kniprath E. (1974) Calcif. Tissue Res. 14, 211–228 [DOI] [PubMed] [Google Scholar]
- 5.Markel K., Roser U., Mackendstedt U., Klosterman M. (1986) Zoomorphology 106, 232–243 [Google Scholar]
- 6.Alvares K., Dixit S. N., Lux E., Barss J., Veis A. (2007) J. Exp. Zool. B Mol. Dev. Evol. 308, 357–370 [DOI] [PubMed] [Google Scholar]
- 7.Veis D. J., Albinger T. M., Clohisy J., Rahima M., Sabsay B., Veis A. (1986) J. Exp. Zool. 240, 35–46 [DOI] [PubMed] [Google Scholar]
- 8.Ameye L., Hermann R., Killian C., Wilt F., Dubois P. (1999) J. Histochem. Cytochem. 47, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 9.Veis A., Barss J., Dahl T., Rahima M., Stock S. (2002) Microsc. Res. Tech. 59, 342–351 [DOI] [PubMed] [Google Scholar]
- 10.Robach J. S., Stock S. R., Veis A. (2005) J. Struct. Biol. 151, 18–29 [DOI] [PubMed] [Google Scholar]
- 11.Veis A., Dixit S., Barss J. T., Robach J. S., Alvares K., Malone J. P. (2007) Procedings of the 9th International Symposium on Biomineralization ( Arias J. L., Fernández M. S. eds) pp. 329–342, Editorial Universitaria, Santiago, Chile [Google Scholar]
- 12.Livingston B. T., Killian C. E., Wilt F., Cameron A., Landrum M. J., Ermolaeva O., Sapojnikov V., Maglott D. R., Buchanan A. M., Ettensohn C. A. (2006) Dev. Biol. 300, 335–348 [DOI] [PubMed] [Google Scholar]
- 13.Cheers M. S., Ettensohn C. A. (2005) Dev. Biol. 283, 384–396 [DOI] [PubMed] [Google Scholar]
- 14.Illies M. R., Peeler M. T., Dechtiaruk A. M., Ettensohn C. A. (2002) Dev. Genes Evol. 212, 419–431 [DOI] [PubMed] [Google Scholar]
- 15.Robach J. S., Stock S. R., Veis A. (2006) J. Struct. Biol. 155, 87–95 [DOI] [PubMed] [Google Scholar]
- 16.Veis A., Dahl T., Barss J. (2004) Echinoderms: Munchen, IEC2003 ( Heinzeller T., Niebelsick J. H. eds) pp. 365–370, Taylor and Francis Group, London [Google Scholar]
- 17.Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 18.Begitt A., Meyer T., van Rossum M., Vinkemeier U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10418–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 20.Goldberg H. A., Warner K. J., Li M. C., Hunter G. K. (2001) Connect. Tissue Res. 42, 25–37 [DOI] [PubMed] [Google Scholar]
- 21.George A., Bannon L., Sabsay B., Dillon J. W., Malone J., Veis A., Jenkins N. A., Gilbert D. J., Copeland N. G. (1996) J. Biol. Chem. 271, 32869–32873 [DOI] [PubMed] [Google Scholar]
- 22.Gotliv B. A., Kessler N., Sumerel J. L., Morse D. E., Tuross N., Addadi L., Weiner S. (2005) ChemBioChem 6, 304–314 [DOI] [PubMed] [Google Scholar]
- 23.Tsukamoto D., Sarashina I., Endo K. (2004) Biochem. Biophys. Res. Commun. 320, 1175–1180 [DOI] [PubMed] [Google Scholar]
- 24.Ritchie H. H., Wang L. (2000) Biochim. Biophys. Acta. 1493, 27–32 [DOI] [PubMed] [Google Scholar]
- 25.Massa L. F., Ramachandran A., George A., Arana-Chavez V. E. (2005) Histochem. Cell Biol. 124, 197–205 [DOI] [PubMed] [Google Scholar]
- 26.Narayanan K., Ramachandran A., Hao J., He G., Park K. W., Cho M., George A. (2003) J. Biol. Chem. 278, 17500–17508 [DOI] [PubMed] [Google Scholar]