Abstract
Endonuclease VIII (Nei) is a DNA glycosylase of the base excision repair pathway that recognizes and excises oxidized pyrimidines. We determined the crystal structures of a NEIL1 ortholog from the giant Mimivirus (MvNei1) unliganded and bound to DNA containing tetrahydrofuran (THF), which is the first structure of any Nei with an abasic site analog. The MvNei1 structures exhibit the same overall architecture as other enzymes of the Fpg/Nei family, which consists of two globular domains joined by a linker region. MvNei1 harbors a zincless finger, first described in human NEIL1, rather than the signature zinc finger generally found in the Fpg/Nei family. In contrast to Escherichia coli Nei, where a dramatic conformational change was observed upon binding DNA, the structure of MvNei1 bound to DNA does not reveal any substantial movement compared with the unliganded enzyme. A protein segment encompassing residues 217–245 in MvNei1 corresponds to the “missing loop” in E. coli Nei and the “αF–β10 loop” in E. coli Fpg, which has been reported to be involved in lesion recognition. Interestingly, the corresponding loop in MvNei1 is ordered in both the unliganded and furan-bound structures, unlike other Fpg/Nei enzymes where the loop is generally ordered in the unliganded enzyme or in complexes with a lesion, and disordered otherwise. In the MvNei1·tetrahydrofuran complex a tyrosine located at the tip of the putative lesion recognition loop stacks against the furan ring; the tyrosine is predicted to adopt a different conformation to accommodate a modified base.
All organisms must cope with the generation of potentially lethal or mutagenic oxidative DNA base damage produced by endogenous free radicals. The enzymes that recognize and initiate the repair of these lesions are the DNA glycosylases, which are found ubiquitously in all three kingdoms of life (for reviews see Refs. 1–4). Some of these enzymes are bifunctional, i.e. they catalyze the hydrolysis of the N-glycosyl bond linking a base to a deoxyribose (glycosylase activity) and subsequently cleave the DNA 3′ to the apurinic/apyrimidinic site (lyase activity), whereas others are monofunctional and only carry out the glycosylase reaction, generating abasic sites as products. Structural studies indicate that the DNA glycosylases that recognize oxidative DNA damages fall into two family groups: the helix-hairpin-helix superfamily and the Fpg/Nei family (for reviews see Refs. 1, 5, 6). The helix-hairpin-helix superfamily includes a diverse group of enzymes with varying substrate specificities which nonetheless share a helix-hairpin-helix motif that consists of two α-helices connected by a hairpin loop, followed by a Gly/Pro-rich loop and a conserved catalytic aspartate residue (7). Glycosylase members of the second family, the Fpg/Nei family, share a two-domain architecture: The N-terminal domain consists of a two-layered β-sandwich with two α-helices, whereas the C-terminal domain contains four α-helices, of which two are involved in a conserved helix-two-turn-helix (H2TH)3 motif, and two β-strands that make up the zinc finger motif (2, 6). Both these motifs are involved in DNA binding. Escherichia coli formamidopyrimidine DNA glycosylase (Fpg) was the first identified member of this family and was characterized by its ability to recognize and remove formamidopyrimidines (8); however, the principal biological substrate for Fpg is 8-oxoguanine (8-oxoG) (9, 10).
More than a decade ago a second DNA glycosylase, which recognizes oxidized pyrimidines, Nei (endonuclease VIII), was identified in E. coli and found to be similar in sequence to Fpg (11, 12). In contrast to Fpg, Nei is only sparsely distributed among prokaryotes (2). Several years ago homologs of this enzyme were identified and characterized in vertebrates, the so-called Nei-like or Neil proteins (13–17). The evolutionary origin of the Neil proteins in vertebrates is unclear (2), and this issue was further confounded several years ago when two Nei-like proteins were identified in the giant Mimivirus (mimicking microbe) (18). This is the largest virus characterized to date with a genomic size of 1.2 Mb, larger than many bacteria (19) and with more than 900 protein coding genes (20). The host for Mimivirus is Acanthamoeba polyphaga, a soil and freshwater protozoan (18).
We have recently cloned and characterized the two Nei glycosylases from Mimivirus, MvNei1 (L315) and MvNei2 (L720) (21). Sequence analysis suggests that Mimivirus Nei1 possesses a zincless finger β-hairpin motif first described in human NEIL1, rather than the signature zinc finger (22–24) characteristic of the Fpg/Nei family. Furthermore, we have shown that MvNei1 and MvNei2 have enzymatic properties very similar to their human homologs, NEIL1 and NEIL2 (21). MvNei1 and NEIL1 share substrate preferences for oxidized pyrimidines in duplex DNA. Although MvNei1 and NEIL1 do not recognize 8-oxoguanine, both enzymes cleave its further oxidation products (25, 26), guanidinohydantoin (Gh) and spiroiminodihydantoin, when paired with C (21, 27). Single stranded DNA with the same base lesions, as well as bubble structures, are also substrates for both enzymes (15, 21, 28).
Here we report the crystal structures of MvNei1 unliganded and in complex with DNA containing tetrahydrofuran (THF), a structural analog of the cyclic hemiacetal form of an abasic site. Human NEIL1 and MvNei1 bind DNA containing THF but do not cleave the DNA backbone. The MvNei1·THF complex is the first structure of an Nei with an abasic site analog. MvNei1 shares significant structural similarity with human NEIL1 (29). In contrast to E. coli Nei where a dramatic conformational change was observed upon binding DNA (30), the structure of MvNei1 bound to furan containing DNA does not reveal any substantial conformational change compared with the unliganded protein. The MvNei1 loop corresponding to the αF-β9/10 lesion recognition loop of Fpg is ordered in both the unliganded and furan-bound structures, unlike what was reported for other Fpg/Nei enzymes where the loop is generally ordered when the enzyme is unliganded or bound to a lesion, and disordered otherwise. In the MvNei1·THF complex a tyrosine located at the tip of the putative lesion recognition loop stacks against the furan ring; the tyrosine and/or the tip of the loop are predicted to adopt a different conformation in the presence of a modified base.
EXPERIMENTAL PROCEDURES
Cloning, Overexpression, and Purification
Cloning, expression, and purification of MvNei1 have been described previously (21). After purification, MvNei1 was dialyzed into crystallization buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 10% (v/v) glycerol, and 1 mm dithiothreitol) and concentrated to 10 mg/ml. For the determination of the apparent dissociation constants, MvNei1 and EcoFpg were prepared as described previously (21, 31). The active fraction of each enzyme was determined as reported previously (32) and found to be 15.8% and 21.7% for MvNei1 and EcoFpg, respectively.
DNA Preparation and Complex Formation
The DNA oligonucleotide containing THF was purchased from Midland Certified Reagent Co. The 13-mer oligonucleotides 5′-CGTCCAXGTCTAC-3′ (X = THF) and 5′-GTAGACCTGGACG-3′ were annealed in a 1:1.2 ratio after PAGE purification. The protein·DNA complex was prepared by adding 1.2 m excess of DNA to MvNei1 at 5 mg/ml in the crystallization buffer. After buffer exchange into 20 mm HEPES, pH 7.5, 50 mm NaCl, 10% glycerol, and 5 mm β-mercaptoethanol, the protein·DNA complex was concentrated to 5.5–6 mg/ml as measured by Bradford assay. For the determination of the apparent dissociation constants, a 23-mer 5′-CTCTCCCTTCXTCCTTTCCTCT-3′ (X = THF) oligonucleotide and its complementary strand were used (33). The modified strand was labeled at the 5′-end using [γ-32P]ATP with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and annealed to the complementary strand in a 1:2 ratio.
Determination of the Apparent Dissociation Constants
Electrophoretic mobility shift assays were performed to determine the apparent dissociation constant of MvNei1 and EcoFpg bound to THF-containing DNA. Reaction volumes contained 30 pm of 32P-labeled oligonucleotide duplex, 20 mm Hepes-KOH, pH 7.8, 75 mm KCl, 1 mm EDTA, 1 mm MgCl2, 0.1 mg/ml bovine serum albumin, 1 mm dithiothreitol, 5% (v/v) glycerol, and varying amounts of MvNei1. All reaction samples of the protein·DNA mixture were incubated at 18 °C for 5 min. For the EcoFpg experiments we followed the protocol described by Grollman and co-workers (33). The protein·DNA mixture samples were loaded onto a 6% non-denaturing polyacrylamide gel (29:1 acrylamide/Bis) and run at 4 °C in 0.5× Tris borate EDTA buffer at 10 mA for 2 h. The gel was pre-run at 4 °C at 10 mA for 2 h before loading the samples. Gels were dried and exposed to an Imaging screen K (Bio-Rad) overnight. Binding constants were obtained from three independent experiments and fitted using Prism (GraphPad Software, La Jolla, CA).
Crystallization and Data Collection
Crystals of unliganded MvNei1 were obtained in two different space groups under similar crystallization conditions. Hexagonal (P61 or P65) crystals (Form I) were obtained at 12 °C by the hanging drop, vapor-diffusion method by mixing 1 μl of protein solution in a 1:1 ratio with a reservoir solution comprising 0.25 m ammonium sulfate, 23–25% (w/v) polyethylene glycol 2000 monomethyl ether, and 0.1 m Bis-Tris-HCl, pH 6.3, then transferred into a solution consisting of the mother liquor supplemented with 25% (v/v) glycerol. Triclinic (P1) crystals (Form II) of MvNei1 were obtained at 12 °C by the hanging drop, vapor-diffusion method by mixing 1 μl of protein solution in a 1:1 ratio with a reservoir solution containing 0.2 m ammonium sulfate, 23% (w/v) polyethylene glycol 4000, and 0.1 m Bis-Tris-HCl, pH 6.0. Crystals were mounted in a cryo-loop and flash cooled directly into liquid nitrogen. The crystals in complex with THF-containing DNA were obtained at 12 °C by the hanging drop, vapor-diffusion method by mixing 0.5 μl of DNA·protein solution in a 1:1 ratio with a reservoir solution made of 0.15 m magnesium nitrate, 10% polyethylene glycol 3350 (w/v), and 100 mm Bis-Tris-HCl, pH 6.0–6.3. The crystals of MvNei1 in complex with THF were transferred to a solution of mother liquor supplemented with 30% (v/v) glycerol, followed by flash cooling. The hexagonal and triclinic unliganded MvNei1 crystals diffract to a resolution of 2.6 and 2.3 Å, respectively, by using a rotating anode x-ray generator and MAR image plate detector (MarResearch, Hamburg, Germany). The hexagonal data set was processed with Denzo and Scalepack (34), whereas the triclinic data set was processed with Mosflm (35) and reduced with SCALA (36). A 2.2-Å data set of MvNei1 in complex with THF was collected at a wavelength of 1.08 Å at beamline X29 (National Synchrotron Light Source) and processed with Mosflm (35). The data were reduced with SCALA (36). Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics for MvNei1
Rmerge = Σ|I − 〈I〉|/ΣI, where 〈I〉 is the average intensity from multiple observations of symmetry-related reflections. Phasing power = ΣhklFH/Σ|FPH − FPH, calc|. Rwork and Rfree = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 10% of the reflections not used in refinement. MIR, multiple isomorphous replacement.
| Form-I |
Form-II |
Form-III |
||||
|---|---|---|---|---|---|---|
| Native | Iodide | Lead | Platinum | Native | MvNei1·THF complex | |
| X-rays | Home source | Home source | NSLS X29 | |||
| Space group | P65 | P1 | P21 | |||
| Unit-cell parameters (Å, °) | a = b = 123.24, c = 44.85 | a = 38.67, b = 54.42, c = 87.31 | a = 39.51, b = 121.46, c = 80.51 | |||
| α = β = 90, γ = 120 | α = 107.00, β = 99.86, γ = 93.42 | β = 95.54 | ||||
| Molecules per asymmetric unit | 1 | 2 | 2 | |||
| Data collection statistics | ||||||
| Resolution (Å) | 15-2.60 (2.69-2.60) | 15-2.70 (2.70-2.60) | 15-2.70 (2.70-2.60) | 15-3.20 (2.70-2.60) | 15-2.30 (2.42-2.30) | 35-2.2 (2.32-2.20) |
| Unique reflections | 12,173 (1,209) | 10,804 (1,085) | 10,703 (1,048) | 6,513 (654) | 27,932 (4,034) | 38,069 (5461) |
| Redundancy | 11.8 | 11.0 | 11.1 | 5.4 | 2.0 | 3.9 |
| Rmergea | 0.059 (0.270) | 0.088 (0.436) | 0.066 (0.406) | 0.183 (0.557) | 0.043 (0.146) | 0.080 (0.652) |
| Completenessa (%) | 100 (100) | 100 (100) | 98.5 (95.5) | 99.5 (100) | 94.4 (93.2) | 99.4 (97.9) |
| Overall I/σ(I)a | 44.4 (4.8) | 30.8 (5.0) | 45.0 (5.9) | 9.72 (2.22) | 17.1 (8.1) | 14.6 (2.2) |
| MIR phasing statistics | ||||||
| No. of sites | 2 | 1 | 1 | |||
| Figure of merit (solve) | 0.359 | |||||
| Figure of merit (resolve) | 0.619 | |||||
| Refinement statistics | ||||||
| Rwork (%) | 20.9 | 20.3 | 21.3 | |||
| Rfree (%) | 26.8 | 26.5 | 26.0 | |||
| r.m.s.d. values | ||||||
| Bond length (Å) | 0.0071 | 0.0057 | 0.0058 | |||
| Bond angles (°) | 1.33 | 1.22 | 1.24 | |||
| B-factor (Å2) | ||||||
| Protein | 55 | 27 | 28 | |||
| Water | 53 | 29 | 36 | |||
| DNA | 43 | |||||
| Ramachandran plot | ||||||
| Most favored (%) | 85.7 | 86.6 | 87.9 | |||
| Additional allowed (%) | 14.3 | 13.4 | 12.1 | |||
| Generously allowed (%) | 0.0 | 0.0 | 0.0 | |||
| Disallowed (%) | 0.0 | 0.0 | 0.0 | |||
| r.m.s.d. versus native (Form-I) (Å) | 0.6 (A)/0.8 (B) | 0.7 (A)/0.8 (B) | ||||
a Values for the highest resolution shell are shown in parentheses.
Structure Determination and Refinement
The structure of MvNei1 in hexagonal space group (P61 or P65) was solved by multiple isomorphous replacement by using a native and three derivative data sets (sodium iodide, trimethyl lead acetate, and platinum chloride). The absence of cysteine residues in the MvNei1 sequence prompted us to use heavy atoms other than mercury or gold. Crystals were soaked in 200 mm NaI for 5 min, 20 mm trimethyl lead acetate for 1 week or 0.1 mm K2PtCl4 for 12 h. Two iodide sites were located by SHELXD (37) and one lead site with a difference Patterson map. The initial phases calculated using the iodide and lead sites were used to locate one platinum site by isomorphous difference Fourier methods. The heavy atom sites were refined by SOLVE (38), and the initial model was built by RESOLVE (39). The Z-score of the initial phase calculation by SOLVE (38) indicated that the hexagonal space group was P65. Phase improvement was performed using SHARP (40), which confirmed the choice of the P65 enantiomorph. About 35% of the model was built by RESOLVE (39). The rest of the model was built into the electron density map using O (41) and Coot (42) and refined to 2.6-Å resolution, using simulated annealing, bulk solvent correction, and grouped B-factor and B-individual refinement with CNS (43). Water molecules were picked automatically from residual electron density maps using CNS (43) and manually checked.
The structure of MvNei1 in the triclinic space group was solved by Phaser (44) using the hexagonal crystal structure as the starting model. The model was refined to 2.3-Å resolution, using rigid body minimization, simulated annealing, and grouped B-factor and B-individual refinement as implemented in CNS (43). Final rounds of refinement using the TLS option were performed using REFMAC5 (45). Water molecules were picked automatically from Fo − Fc map using CNS (43) and manually checked.
The structure of the complex with furan containing DNA was solved by molecular replacement with MolRep (46) using the triclinic structure as a starting model. All refinement was performed by CNS (43) in a fashion similar to that described for the triclinic crystal structure. The final models for all three structures were found to exhibit good geometry, as determined using Procheck (47). There are no residues in the disallowed region of the Ramachandran plot. Refinement statistics are shown in Table 1. All structure figures were prepared using PyMOL (DeLano Scientific).
Protein Data Bank Accession Codes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank and are available under the following accession codes: 3A42 (form I) and 3A45 (form II) for unliganded MvNei1 and 3A46 for MvNei1 in complex THF containing DNA (form III).
RESULTS
Structure Determination
The unliganded MvNei1 protein was crystallized in two forms (forms I and II). Form I crystals belong to hexagonal space group P65, and form II crystals to triclinic P1. MvNei1 in complex with DNA containing an AP site analog, THF, was co-crystallized in a third form, which belongs to monoclinic space group P21 (Form III). We determined the MvNei1 structure at 2.6-Å resolution by multiple isomorphous replacement method using form I crystals with three different heavy atom derivatives. The model of unliganded form I crystal was refined to an R-factor of 20.9% and an Rfree of 26.8% (Table 1). The form I crystals contain one MvNei1 monomer including residues 2–270 and 277–289 per asymmetric unit (residues 271–276 of the zincless finger tip are missing in the final model). We then determined the structure of form II crystals by molecular replacement method using the form I structure as a starting model. Form II crystals contain two MvNei1 monomers (residues 2–289) per asymmetric unit. The structure of form III crystals was determined by molecular replacement method using the form II structure as a starting model. Form III crystals contain two MvNei1 monomers (residues 2–289) each bound to a 13-mer THF DNA duplex per asymmetric unit. The models of forms II and III were refined to an R-factor of 20.3% and Rfree of 26.5% at 2.3-Å resolution and R-factor of 21.3% and Rfree of 26.0% at 2.2-Å resolution, respectively (Table 1).
MvNei1 Protein Structure
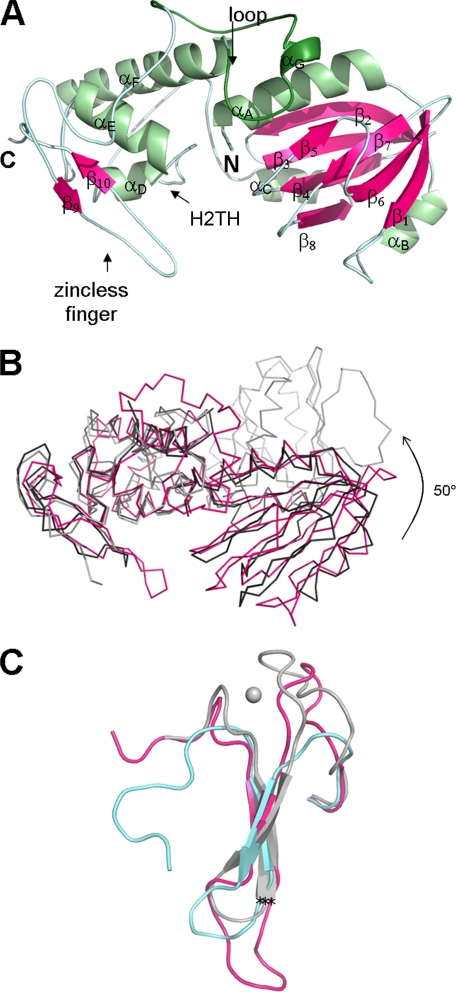
The current study provides three independent structures of unliganded MvNei1 protein observed in two different space groups (forms I and II). All three protein models are nearly identical: the root mean square deviation values calculated for the polypeptide chain Cα atoms are 0.6 and 0.8 Å (Table 1). The approximate dimensions of the MvNei1 monomer are 30 × 65 × 35 Å. MvNei1 is composed of two globular domains connected by a linker (Fig. 1A). The N-terminal domain comprises three α-helices and seven β-strands, which form a two-layer β-sandwich. The C-terminal domain contains four α-helices, of which two form the helix-two turn-helix (H2TH) motif, and two β-strands that form a structural motif mimicking an antiparallel β-hairpin zinc finger, a motif first described in the human NEIL1 (hNEIL1) structure (29).
FIGURE 1.
A, ribbon diagram of unliganded MvNei1. The model from form II crystals comprises residues 2–289. The secondary structure elements were defined by DSSP (59) and are as follows: αA (4–18), β1 (22–27), αB (41–45), β2 (50–58), β3 (61–67), β4 (75–81), β5 (87–89), β6 (96–102), β7 (107–111), β8 (118–122), αC (125–133), αD (157–162), αE (173–183), αF (196–215), αG (226–229), β9 (265–267), and β10 (279–281). Helices are shown in light green and β-strands in magenta. The putative lesion binding loop is highlighted in a darker shade of green. B, comparison of MvNei1 with E. coli Nei (EcoNei). Superposition of MvNei1 (pink) with unliganded EcoNei (gray; PDB code 1Q3B (30) and the EcoNei trapped DNA complex (black; PDB code 1K3W (23)). C, close-up of the zinc-finger motif. Shown are residues 230–262 for EcoNei, 263–290 for hNEIL1, and 253–289 for MvNei1. The asterisks indicate the position of the Cα of the conserved arginine (Arg-252 in EcoNei and Arg-277 in hNEIL1 and MvNei1).
Although MvNei1 shares substantial structure similarity with EcoNei, there is one significant difference. EcoNei has been reported to adopt two different conformations: an elongated “open” conformation in unliganded EcoNei (30) and a more compact “closed” conformation in a DNA-bound complex (23). A superposition of the unliganded MvNei1 structure onto the two EcoNei structures (unliganded and in complex with DNA) shows that MvNei1 is in a closed conformation, even in the absence of a DNA substrate (Fig. 1B). In the unliganded EcoNei structure, the conformational change occurs via the flexible hinge region (Leu-122 to Pro-127), which connects the two Nei domains. The open conformation is stabilized via hydrogen bonds between the hinge region residues Leu-122 and Gln-123 and residues Arg-50 and Arg-147 (30). These interactions are lost in the EcoNei·DNA complex (23). A similar situation is observed in MvNei1: The residue corresponding to EcoNei Arg-147 is Lys-151 in MvNei1 (supplemental Fig. 1). In the unliganded MvNei1 structures this lysine residue does not interact with the hinge region; it participates instead in a salt bridge with Asp-196, a residue found in helix αF of the C-terminal domain. MvNei1 adopts a closed conformation in the absence of DNA, and this conformation is independent of crystal packing, because the same closed conformation was observed in three independent molecules obtained from two crystals (two molecules per asymmetric unit in P1 crystals, and one molecule in P65 crystals). A closed conformation was similarly observed in the unliganded structure of hNEIL1 (29).
Because there are no cysteine residues in the MvNei1 amino acid sequence, MvNei1 was predicted to lack the C-terminal zinc finger motif (21). Our structure showed that indeed MvNei1 contains a motif mimicking an antiparallel β-hairpin zinc finger that follows the H2TH motif (29). The zincless finger motif superimposes quite well onto the corresponding segment in EcoNei and hNEIL1, including the conserved Arg-277 (Arg-252 in EcoNei and Arg-277 in hNEIL1), which is known to interact with DNA (23) (Fig. 1C). The superposition reveals that the loop connecting the two β-strands of the zincless finger in MvNei1 is longer than in its bacterial and human counterparts (Fig. 1C). In addition the two loops immediately preceding β strand 9 (residues 254–264) and following β strand 10 (residues 282–289) are not truncated as in hNEIL1: they are similar in length to the loops in EcoNei, with the difference that in MvNei1 the loops lack the four cysteines and therefore do not coordinate a zinc ion. What, then, stabilizes the loops in the absence of a metal ion? There are four main-chain H-bond interactions in the loop preceding β strand 9 involving residues 256, 258, 260–262, and 264. The C-terminal loop is stabilized by an H-bond involving main-chain atoms from residues 282 and 286.
A crystal structure of an Nei in complex with lesion-containing DNA is not yet available. On the other hand, the crystal structures of Bacillus stearothermophilus Fpg (BstFpg) and Lactobacillus lactis Fpg (LlaFpg) in complex with DNA oxidative damage were published recently (48–50). The segment encompassing residues 217–245 in MvNei1 corresponds to the “missing loop” in the borohydride trapped covalent complex of EcoNei·DNA (23) and the “αF–β9/10 loop” in Fpg structures (22, 51, 52). This loop was recently shown to play a part in the recognition of the lesion in Fpg complexes with 8-oxoG and 5,6-dihydrouracil (DHU) (48) and carbocyclic FapydG (cFapydG) (50). In trapped Fpg·DNA complexes (22, 51) and Fpg complexes with DNA containing an AP site (51–53) the lesion binding loop was observed to be disordered. In contrast, this region is ordered in unliganded Thermus thermophilus Fpg (TthFpg) (24), EcoNei (30), and hNEIL1 (29). It is also ordered in complexes with DNA containing a lesion (48). Unexpectedly the corresponding loop is ordered in all of the MvNei1 structures, unliganded, and in the complex with furan-containing DNA (Figs. 1A and 2A).
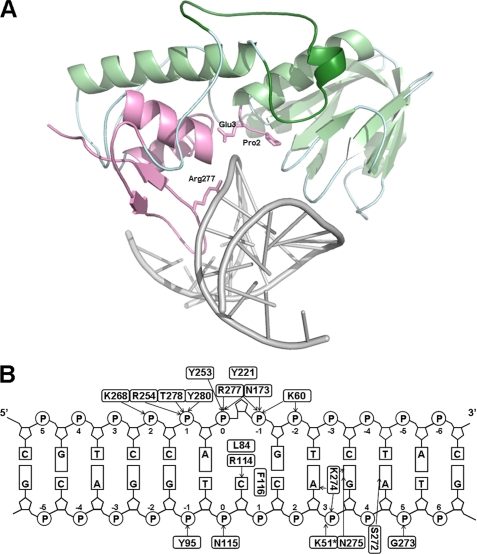
FIGURE 2.
A, MvNei1·THF complex structure. The zincless finger, H2TH, catalytic proline and glutamic acid, and conserved arginine are highlighted in pink. The DNA is shown in gray. The putative lesion binding loop is shown in dark green. B, schematics of MvNei1·DNA interactions. Nucleotides are numbered beginning from THF0, with positive numbers toward the 5′-end. Hydrogen bonds are represented with black arrows pointing toward the acceptors. Tyr-221 stacks with THF.
Overall Structure of the MvNei1·THF Complex
The MvNei1 complex crystallized in a P21 space group with two protein·DNA complexes per asymmetric unit. The two complexes are nearly identical, and we describe below the structure of complex A. The 13-mer duplex is bound, as expected, in the DNA binding cleft between the two domains and lies perpendicularly to the long axis of MvNei1 (Fig. 2A). The overall DNA axis bend measured by CURVES (54) is ∼40°, a number comparable to that reported for EcoNei (45°) (23). Although the DNA is distorted in the vicinity of the abasic site analog, it is essentially a B-form away from the lesion (54).
As mentioned earlier, a significant conformational change was reported in EcoNei between the free and liganded forms of the enzyme, which adopted an open and closed conformation (30). In contrast, superposition of the MvNei1·THF complex structure onto unliganded MvNei1 structures (Form II crystal) showed that the protein structures are identical except the zincless finger tip region and a few amino acid side chains (root mean square deviation of Cα atoms are 0.7 and 0.8 Å, see Table 1). The orientation of Pro-2 differs from that seen in unliganded MvNei1: The amino group of Pro-2 moves by ∼1.0 Å to accommodate the DNA, and the zincless finger tip moves by ∼4 Å upon binding DNA. A conformational change of the same magnitude was observed in the EcoNei zinc finger (55). The formation of a MvNei1 complex, however, is not accompanied by any hinge movement between two domains, which makes this Nei enzyme more similar to an Fpg, because no conformational change was observed in Fpg upon binding its substrate (51).
The H2TH and zincless finger motifs in the C-terminal domain play a key role in DNA binding. These DNA binding motifs are mostly conserved within the Fpg/Nei family (23, 53). Asn-173 in the H2TH motif hydrogen bonds to the backbone phosphates P0 and P−1 via main-chain and side-chain interactions (Fig. 2B). Asn-173 is conserved within the Fpg/Nei family, and residues in its vicinity are also involved in binding DNA in the EcoNei·DNA and Fpg·DNA complexes (22, 23, 51, 53). Adjacent to the H2TH motif, the conserved Lys-60 interacts with P−2 and P−1 (Fig. 2B). Lys-60 is known to play a critical role in the excision of both 8-oxoG by EcoFpg (56) and 5-OHU by NEIL1 (28). The zincless finger motif is also involved in binding DNA, especially the conserved Arg-277 (Figs. 1C and 2B). Mutating this arginine in EcoNei and hNEIL1 results in a protein variant with a marked decrease in glycosylase activity; the lyase activity, on the other hand, is mostly unaffected (23, 29). The side chain of Arg-277 hydrogen bonds to phosphates P0 and P−1, and neighboring residues Thr-278 and Tyr-280 interact with phosphate P1. As mentioned earlier, the loop connecting the two β-strands of the zincless finger in MvNei1 is longer (11 residues) than the corresponding loops in EcoNei (2 residues) or hNEIL1 (5 residues). A direct consequence is that the DNA is pushed further from the H2TH motif, and fewer residues in that motif partake in the interaction with the DNA: Asn-173 is the only residue in the H2TH motif to take part in binding DNA. In contrast, two amino acid residues (Gln-160 and Asn-168) interact with DNA directly in EcoNei·DNA (23). Another consequence of the loop being longer is that residues located at the tip of the loop interact with the strand opposite the lesion (Fig. 2B). As a result, there are more interactions to the complementary strand in MvNei1 than in EcoNei or EcoFpg (22, 23).
Interactions with THF and Opposite Base
MvNei1 interacts with the THF strand via several hydrogen bonding interactions to phosphates P−2, P−1, P0, and P1. THF is extruded from the DNA and lies in a pocket that is mostly hydrophobic. The furan moiety is surrounded by four aromatic residues, Tyr-174, Tyr-221, Phe-250, and Tyr-253, and two catalytic residues, Glu-3 and Pro-2. Tyr-174 and Tyr-253 are within van der Waals distance of THF, and Tyr-221 stacks on the furan moiety (Fig. 3A). A molecular dynamic simulation of EcoNei predicted that its binding pocket for thymine glycol would be built by Tyr-169, Phe-227, and Phe-230, three aromatic residues that correspond to MvNei1 Tyr-174, Phe-250, and Tyr-253 (23). More recently, homology modeling and molecular dynamics simulations were also used to generate a model of hNEIL1 bound to thymine glycol or spiroiminodihydantoin; in the resulting model Pro-2, Glu-3, Glu-6, Tyr-177, and Tyr-263 are predicted to participate in binding the non-planar oxidative DNA lesions (57). In this case again, the two tyrosines correspond to the tyrosines in MvNei1 (Tyr-174 and Tyr-253) that are within van der Waals distance of THF.
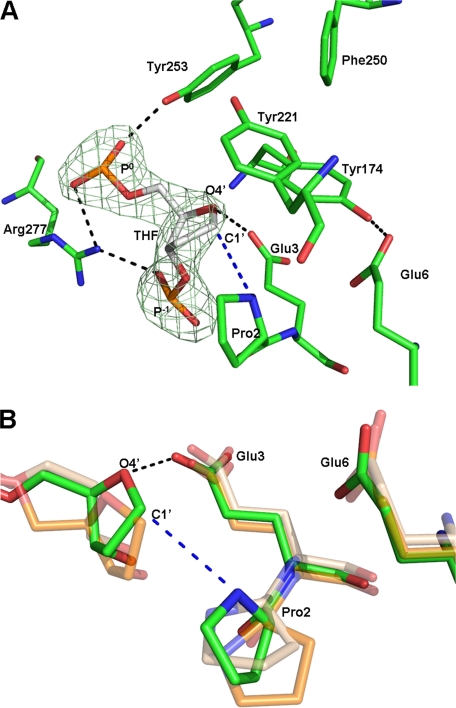
FIGURE 3.
A, close-up view of THF and interacting residues with overlaid simulated annealing omit map contoured at 5 σ (lime green). MvNei1 is shown in green and DNA in gray. Hydrogen bonds are represented by black dashed lines. B, superposition of the MvNei1·THF complex onto LlaFpg·AP site complexes (THF and Pr). The MvNei1·THF complex is shown in green, LlaFpg/Pr complex (PDB code 1PJI (52)) in beige, and the LlaFpg·THF complex (PDB code 1PM5 (52)) in orange. The hydrogen bond is shown as a black dashed line. The blue dashed line indicates the distance between the amino group of Pro-2 and C1′ of THF.
The distance between the α-nitrogen atom of the catalytic residue Pro-2 and the C1′ atom of THF is 3.7 Å. Although the α-amino group of the N-terminal proline seems well aligned for a nucleophilic attack at C1′ (if furan were replaced by a damaged nucleoside), we note that it is still unclear whether Fpg/Nei enzymes employ an SN2- or SN1-type mechanism (48). The distance between the carboxyl group of Glu-3 and O4′ of THF is 3.1 Å: too short for a van der Waals interaction but compatible with a hydrogen bond, which would imply that this glutamic acid is protonated. The possibility that Glu-3 may be protonated has been suggested by others (23, 48). Glu-3 also engages in H-bond interactions with the backbone amides of three residues of the H2TH motif (Gly-172, Tyr-174, and Leu-175), which could help orient the glutamate's carboxylate group for protonation of O4′ (23).
The only other available structure of an Fpg/Nei enzyme with furan is that of LlaFpg (52). In comparison, the distances between THF and the residues described above are longer than in MvNei1: The C1′-α-N distance is 4.3 Å in the LlaFpg·THF complex, and the distance between the carboxyl group of Glu-3 and O4′ of THF is 3.7 Å. In addition the αF–β9 loop is partly disordered in the LlaFpg·THF complex, and the tyrosine corresponding to Tyr-221 (LlaFpg Tyr-222) is not seen in the structure. Moreover the αF–β9 loop is in the open conformation and would have to adopt the closed conformation to interact with the lesion (see below) (50). The weaker interactions observed in the LlaFpg structure are consistent with the weaker binding affinity of this enzyme for furan, as compared with MvNei1.
A comparison of the MvNei1·THF complex with unliganded MvNei1 shows that the side chains of several amino acid residues, Pro-2, Arg-114, Phe-116, and Arg-227, had to shift to accommodate the DNA. Leu-84 (located in the loop between β4 and β5), Arg-114, and Phe-116 (in the loop joining β6 and β7) are the residues that fill the void created by the eversion of the damaged base (supplemental Fig. 2). These residues correspond to the consecutive residues Gln-69, Leu-70, and Tyr-71 in EcoNei (23) and Met-75, Arg-109, and Phe-111 in LlaFpg (53). The non-contiguous arrangement and the nature (Leu, Arg, and Phe) of these residues suggest that MvNei1 may be closer to Fpg than Nei when it comes to opposite base recognition. Leu-84 takes the place of the excised damaged base, and Phe-116 is wedged between the orphaned base (cytosine) and its 5′ neighbor. Arg-114 interacts with the orphaned cytosine via two hydrogen bonds, as described in the BstFpg·DNA (51) and LlFpg·DNA complexes (53). This arginine was shown to contribute to the observed specificity of Fpg for the base opposite the lesion (51, 52). We showed that MvNei1 prefers thymine glycol and 5-hydroxyuracil and cleaves these lesions with little or no specificity for the base opposite the damage. In the case of guanidinohydantoin, however, MvNei1 cleaves the damage with a marked preference for C opposite the lesion (Km = 1.3 nm for C and 1.1 μm for A) (21). A similar disparity in the preference for the opposite base depending on the nature of the oxidative lesion was also reported for EcoNei (55).
MvNei1 Exhibits a High Affinity for DNA Containing THF
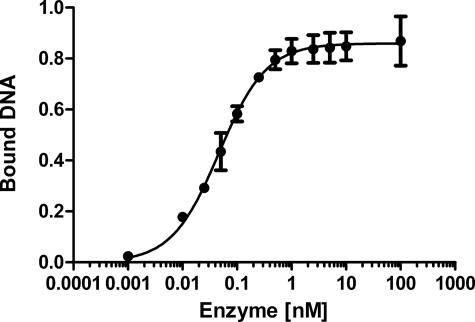
In the MvNei1·DNA complex, the DNA is kinked at the THF site, and the abasic site analog is flipped out of the DNA helix. Although several structures of Fpg and Nei in complex with DNA have been reported (22, 23, 48, 50–53) only one structure in complex with DNA containing THF is available for comparison, that of L. lactis Fpg (52). The superposition of MvNei1·THF complex onto LlaFpg·THF revealed two significant differences: First, the DNA backbone torsion angles of the MvNei1·THF complex indicate that the conformation at the THF site is more extrahelical than in the LlaFpg·THF complex. The MvNei1·THF complex is in fact more similar to the complex of LlaFpg with propanediol (52) (Table 2). The difference in the DNA backbone conformation might explain the different orientation of Pro-2 in the MvNei1·THF and LlaFpg·THF complexes (Fig. 3B). The differences observed in the phosphodiester backbone in the structures of LlaFpg in complex with various abasic site analogs were found to be consistent with the measured binding affinity of this enzyme to the AP site analogs (58), that is, THF was the least extrahelical and the least efficiently recognized by LlaFpg among the AP site analogues tested (52). Measurements of dissociation constants using electrophoretic mobility shift assay of EcoNei with THF (55) showed that EcoNei exhibits a high affinity for THF (KDapp = 0.21- 0.47 nm) (55) compared with EcoFpg (9.5–90 nm) (33, 58). These differences led us to measure the KDapp of MvNei1 bound to THF-containing DNA. A representative electrophoretic mobility shift assay plot is shown in Fig. 4. The apparent dissociation constant of MvNei1 for THF is 0.047 nm, comparable to the KD of EcoNei for THF opposite C (KD = 0.21 nm) and much tighter than that of Fpg in the same DNA sequence context (KD = 5.3–9.0 nm) (Table 3).
TABLE 2.
Backbone torsion angles α, γ, and ζ of DNA in MvNei1 and LlaFpg DNA complexes containing the AP site analogs THF and Pr
| Backbone torsion angles (°) |
PDB ID code | Ref. | |||
|---|---|---|---|---|---|
| α | γ | ζ | |||
| MvNei1·THFa | −64.86/−53.34 | −55.47/−56.99 | −71.69/−62.42 | 3A46 | This work |
| LlaFpg·Pr | −66.66 | −49.59 | −68.15 | 1PJI | 52 |
| LlaFpg·THF | 63.71 | −174.63 | −56.76 | 1PM5 | 52 |
a Each number is given for molecules A and B.
FIGURE 4.
Determination of the apparent dissociation constant of MvNei1 bound to DNA containing THF. Plot of percent MvNei1 bound to a 23-mer 5′-32P-labeled duplex containing THF as a function of enzyme concentration. Error bars represent the standard error on the average of three separate experiments.
TABLE 3.
Dissociation constants of MvNei1, EcoNei, and EcoFpg bound to furan-containing DNA
These experiments were performed in triplicate with the same 23-mer THF:C oligomer under similar conditions.
DISCUSSION
Lesion Accommodation and Recognition
After solving the unliganded crystal structure of an active C-terminal deletion variant of hNEIL1, we attempted to capture the enzyme in complex with DNA (29). Despite multiple attempts we were unable to crystallize a complex. We then searched for a suitable homolog of hNEIL1 and chose to focus on one of the two NEIL orthologs from the giant Mimivirus, MvNei1. MvNei1 harbors a zincless finger in the C-terminal domain similar to that of hNEIL1, as predicted by sequence alignments. Although the MvNei1 structure revealed some Fpg-like traits, such as the nature and non-contiguous location of the void filling residues and the absence of conformational change upon binding DNA, MvNei1 exhibits a substrate preference that is similar to that of EcoNei and hNEIL1: MvNei1 prefers oxidized pyrimidines and the further oxidation products of 8-oxoG, Gh, or spiroiminodihydantoin, whereas 8-oxoG is removed very inefficiently (21).
As mentioned above, recent crystal structures of BstFpg in complex with DNA containing either 8-oxoG or DHU (48), and LlaFpg in complex with cFapydG and N7-benzyl-FapydG (49, 50) revealed that the oxidative lesion is recognized by residues located in the αF–β9/10 loop. A structural comparison of LlaFpg bound to cFapydG or THF showed that the αF–β9 loop can adopt two conformations, open and closed (50, 52). The closed loop conformation appears to play a key role in the lesion recognition by assembling the substrate binding pocket. The same loop was also described as being flexible in the structures of BstFpg in complex with a reduced abasic site (51) and 8-oxoG (48). Interestingly, the corresponding loop in the unliganded and MvNei1·THF complex structures adopts what appears to be a closed loop conformation. Castaing and coworkers reported that in the closed conformation the tip of the loop contacts cFapydG and observed that the conserved aromatic residue Tyr-222 wraps over the damaged base (50). Tyr-221, the corresponding residue in MvNei1, stacks against the THF moiety (Fig. 3A).
How Nei enzymes recognize a vast repertoire of DNA lesions remains an open question. The αF–β9/10 loop of Fpg was shown to play a part in lesion recognition (48, 50). The αF–β9/10 loops of bacterial Fpgs have very similar size and shape (root mean square deviation is 0.4–0.6Å for a segment of the loop in the closed conformation comprising ∼27 residues: residues 200–228 in TthFpg (PDB ID code 1EE8), 213–239 in BstFpg (PDB ID code 1R2Y), and 210–235 in LlaFpg (1XC8)). In contrast, the loop in Nei enzymes varies in length and secondary structure elements (Fig. 5A). The Nei loop is generally shorter than in Fpg, with the notable exception of plant and fungal Fpgs, which harbor a truncated loop (supplemental Fig. 1). Plant and fungal Fpg have in common with Nei that they excise oxidized pyrimidines and recognize 8-oxoguanine very poorly or not at all. One hypothesis is that the long αF–β10 loop of bacterial Fpg is designed to recognize 8-oxoG; Nei, plant, and fungal Fpg do not and their loop has evolved a shorter segment able to recognize oxidized pyrimidines. In the BstFpg-8-oxoG complex four consecutive main-chain amides (residues 222–225) at the tip of the loop contact the exocyclic O6 of the oxidized guanine (48). Four consecutive residues (219–222) at the tip of the LlaFpg loop also interact with the O6 position of another damaged guanine, cFapydG (50). The first three of these residues are missing in the Nei loop (supplemental Fig. 1 and Fig. 5B).
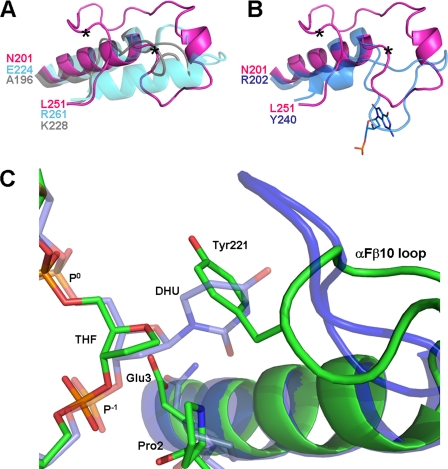
FIGURE 5.
A, superposition of the loop region of the three Nei enzymes with known structures: MvNei1 (pink), EcoNei (gray; PDB code 1Q3B (30)), and hNEIL1 (cyan; PDB code 1TDH (29)). Two asterisks indicate the start and end of the putative lesion binding loop in MvNei1. B, superposition of the MvNei1 loop (pink) with the αF–β10 loop of the BstFpg·8-oxoG complex (blue; PDB code 1R2Y (48)). C, superposition of the MvNei1·THF complex with the BstFpg·DHU complex (PDB code 1R2Z (48)). The MvNei1 complex is shown in green and the BstFpg complex in blue.
In the absence of a structure of MvNei1 with an oxidatively damaged pyrimidine we have modeled DHU in the binding site of MvNei1 based on the structure of the BstFpg·DHU complex (48). The superposition indicates that DHU, or any damaged base in the same conformation, would clash with the aromatic ring of Tyr-221 (Fig. 5C), indicating that the loop would have to adopt a different conformation in the presence of a damaged base. The corresponding loop in Fpg has been shown to undergo different conformations depending on the nature of the AP site analog or lesion at the center of the DNA duplex (48, 50, 52). A model for how the loop of Nei interacts with a damaged base will have to await the structure of an Nei in complex with DNA containing an oxidized pyrimidine.
In conclusion, we present here two new crystal structures of a viral NEIL1 ortholog, MvNei1, unliganded and in complex with furan containing DNA. The study illustrates that (i) the protein conformation does not change upon binding DNA, (ii) the putative lesion binding loop is well ordered in the free and DNA-bound states, (iii) MvNei1 has a higher affinity for THF than bacterial Fpg, and (iv) the flexible segment of the tip of the putative lesion binding loop in MvNei1 would have to adopt a different conformation to accommodate a damaged base.
Supplementary Material
Acknowledgments
We thank Dr. Alexei Soares for data collection at beamline X29 at National Synchrotron Light Source, Alicia Holmes for technical assistance, and Minmin Liu and Drs. Scott Kathe and Joyce E. Heckman for help with the enzyme activity assays and measurements of apparent dissociation constants.
This work was supported, in whole or in part, by National Institutes of Health Grant P01CA098993 awarded by the NCI. Financial support of National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U. S. Department of Energy, and from the National Center for Research Resources of NIH.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (codes 3A42, 3A45, and 3A46) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- H2TH
- helix-two-turn-helix
- Fpg
- formamidopyrimidine DNA glycosylase
- 8-oxoG
- 8-oxoguanine
- Nei
- endonuclease VIII
- Gh
- guanidinohydantoin
- DHU
- 5,6-dihydrouracil
- BstFpg
- B. stearothermophilus Fpg
- LlaFpg
- L. lactis Fpg
- cFapyFpg
- carbocyclic FapydG
- THF
- tetrahydrofuran
- MvNei1
- NEIL1 ortholog from the giant Mimivirus
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TthFpg
- T. thermophilus Fpg
- EcoFpg
- E. coli Fpg.
REFERENCES
- 1.Krokan H. E., Standal R., Slupphaug G. (1997) Biochem. J. 325, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace S. S., Bandaru V., Kathe S. D., Bond J. P. (2003) DNA Repair 2, 441–453 [DOI] [PubMed] [Google Scholar]
- 3.David S. S., O'Shea V. L., Kundu S. (2007) Nature 447, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denver D. R., Swenson S. L., Lynch M. (2003) Mol. Biol. Evol. 20, 1603–1611 [DOI] [PubMed] [Google Scholar]
- 5.Fromme J. C., Banerjee A., Verdine G. L. (2004) Curr. Opin Struct. Biol. 14, 43–49 [DOI] [PubMed] [Google Scholar]
- 6.Zharkov D. O., Shoham G., Grollman A. P. (2003) DNA Repair 2, 839–862 [DOI] [PubMed] [Google Scholar]
- 7.Thayer M. M., Ahern H., Xing D., Cunningham R. P., Tainer J. A. (1995) EMBO J. 14, 4108–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chetsanga C. J., Lozon M., Makaroff C., Savage L. (1981) Biochemistry 20, 5201–5207 [DOI] [PubMed] [Google Scholar]
- 9.Chung M. H., Kasai H., Jones D. S., Inoue H., Ishikawa H., Ohtsuka E., Nishimura S. (1991) Mutat. Res. 254, 1–12 [DOI] [PubMed] [Google Scholar]
- 10.Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4690–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melamede R. J., Hatahet Z., Kow Y. W., Ide H., Wallace S. S. (1994) Biochemistry 33, 1255–1264 [DOI] [PubMed] [Google Scholar]
- 12.Jiang D., Hatahet Z., Melamede R. J., Kow Y. W., Wallace S. S. (1997) J. Biol. Chem. 272, 32230–32239 [DOI] [PubMed] [Google Scholar]
- 13.Bandaru V., Sunkara S., Wallace S. S., Bond J. P. (2002) DNA Repair 1, 517–529 [DOI] [PubMed] [Google Scholar]
- 14.Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., Izumi T. (2002) J. Biol. Chem. 277, 30417–30420 [DOI] [PubMed] [Google Scholar]
- 16.Morland I., Rolseth V., Luna L., Rognes T., Bjørås M., Seeberg E. (2002) Nucleic Acids Res. 30, 4926–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takao M., Kanno S., Kobayashi K., Zhang Q. M., Yonei S., van der Horst G. T., Yasui A. (2002) J. Biol. Chem. 277, 42205–42213 [DOI] [PubMed] [Google Scholar]
- 18.Suzan-Monti M., La Scola B., Raoult D. (2006) Virus Res. 117, 145–155 [DOI] [PubMed] [Google Scholar]
- 19.Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J. M. (2004) Science 306, 1344–1350 [DOI] [PubMed] [Google Scholar]
- 20.Ghedin E., Claverie J. M. (2005) Virol J. 2, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandaru V., Zhao X., Newton M. R., Burrows C. J., Wallace S. S. (2007) DNA Repair 6, 1629–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilboa R., Zharkov D. O., Golan G., Fernandes A. S., Gerchman S. E., Matz E., Kycia J. H., Grollman A. P., Shoham G. (2002) J. Biol. Chem. 277, 19811–19816 [DOI] [PubMed] [Google Scholar]
- 23.Zharkov D. O., Golan G., Gilboa R., Fernandes A. S., Gerchman S. E., Kycia J. H., Rieger R. A., Grollman A. P., Shoham G. (2002) EMBO J. 21, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugahara M., Mikawa T., Kumasaka T., Yamamoto M., Kato R., Fukuyama K., Inoue Y., Kuramitsu S. (2000) EMBO J. 19, 3857–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goyal R. N., Dryhurst G. (1982) J. Electroanal. Chem. 135, 75–91 [Google Scholar]
- 26.Luo W., Muller J. G., Rachlin E. M., Burrows C. J. (2000) Org. Lett. 2, 613–616 [DOI] [PubMed] [Google Scholar]
- 27.Hailer M. K., Slade P. G., Martin B. D., Rosenquist T. A., Sugden K. D. (2005) DNA Repair 4, 41–50 [DOI] [PubMed] [Google Scholar]
- 28.Dou H., Mitra S., Hazra T. K. (2003) J. Biol. Chem. 278, 49679–49684 [DOI] [PubMed] [Google Scholar]
- 29.Doublié S., Bandaru V., Bond J. P., Wallace S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golan G., Zharkov D. O., Feinberg H., Fernandes A. S., Zaika E. I., Kycia J. H., Grollman A. P., Shoham G. (2005) Nucleic Acids Res. 33, 5006–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandaru V., Blaisdell J. O., Wallace S. S. (2006) Methods Enzymol. 408, 15–33 [DOI] [PubMed] [Google Scholar]
- 32.Blaisdell J. O., Wallace S. S. (2007) Nucleic Acids Res. 35, 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tchou J., Michaels M. L., Miller J. H., Grollman A. P. (1993) J. Biol. Chem. 268, 26738–26744 [PubMed] [Google Scholar]
- 34.Otwinowski Z., Minor W. (1997) Meth. Enzymol. 276, 307–325 [DOI] [PubMed] [Google Scholar]
- 35.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 36.Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 40.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 41.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43.Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 44.McCoy A. J., Storoni L. C., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1220–1228 [DOI] [PubMed] [Google Scholar]
- 45.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 46.Vagin A., Teplyakov A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 47.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 48.Fromme J. C., Verdine G. L. (2003) J. Biol. Chem. 278, 51543–51548 [DOI] [PubMed] [Google Scholar]
- 49.Coste F., Ober M., Carell T., Boiteux S., Zelwer C., Castaing B. (2004) J. Biol. Chem. 279, 44074–44083 [DOI] [PubMed] [Google Scholar]
- 50.Coste F., Ober M., Le Bihan Y. V., Izquierdo M. A., Hervouet N., Mueller H., Carell T., Castaing B. (2008) Chem. Biol. 15, 706–717 [DOI] [PubMed] [Google Scholar]
- 51.Fromme J. C., Verdine G. L. (2002) Nat. Struct. Biol. 9, 544–552 [DOI] [PubMed] [Google Scholar]
- 52.Pereira de Jésus K., Serre L., Zelwer C., Castaing B. (2005) Nucleic Acids Res. 33, 5936–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serre L., Pereira de Jésus K., Boiteux S., Zelwer C., Castaing B. (2002) EMBO J. 21, 2854–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavery R., Sklenar H. (1989) J. Biomol. Struct. Dyn. 6, 655–667 [DOI] [PubMed] [Google Scholar]
- 55.Kropachev K. Y., Zharkov D. O., Grollman A. P. (2006) Biochemistry 45, 12039–12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sidorkina O. M., Laval J. (1998) Nucleic Acids Res. 26, 5351–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia L., Shafirovich V., Geacintov N. E., Broyde S. (2007) Biochemistry 46, 5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castaing B., Fourrey J. L., Hervouet N., Thomas M., Boiteux S., Zelwer C. (1999) Nucleic Acids Res. 27, 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.