Abstract
Metastatic progression of melanoma is associated with overexpression and activity of cAMP-response element-binding protein (CREB). However, the mechanism by which CREB contributes to tumor progression and metastasis remains unclear. Here, we demonstrate that stably silencing CREB expression in two human metastatic melanoma cell lines, A375SM and C8161-c9, suppresses tumor growth and experimental metastasis. Analysis of cDNA microarrays revealed that CREB silencing leads to increased expression of cysteine-rich protein 61 (CCN1/CYR61) known to mediate adhesion, chemostasis, survival, and angiogenesis. Promoter analysis and chromatin immunoprecipitation assays demonstrated that CREB acts as a negative regulator of CCN1/CYR61 transcription by directly binding to its promoter. Re-expression of CREB in CREB-silenced cells rescued the low CCN1/CYR61 expression phenotype. CCN1/CYR61 overexpression resulted in reduced tumor growth and metastasis and inhibited the activity of matrix metalloproteinase-2. Furthermore, its overexpression decreased melanoma cell motility and invasion through Matrigel, which was abrogated by silencing CCN1/CYR61 in low metastatic melanoma cells. Moreover, a significant decrease in angiogenesis as well as an increase in apoptosis was seen in tumors overexpressing CCN1/CYR61. Our results demonstrate that CREB promotes melanoma growth and metastasis by down-regulating CCN1/CYR61 expression, which acts as a suppressor of melanoma cell motility, invasion and angiogenesis.
Cutaneous melanoma is the most aggressive type of skin cancer, and it can metastasize very rapidly (1). An estimated 62,480 new cases of melanoma were diagnosed in the United States during 2008, 8,420 of which resulted in death (2). The transition of melanoma from the radial growth phase to the vertical growth phase to metastasis is accompanied by multiple molecular changes (3–8). We and others have shown that two transcription factors, activating transcription factor-1 (ATF-1)2 and cAMP-response element-binding protein (CREB), are activated and overexpressed in melanoma during its progression toward the malignant phenotype (9–13).
CREB and ATF-1 belong to the leucine zipper class of transcription factors. Stimuli such as growth factors, neurotransmitters, inflammatory biolipids, stress signals, or other factors that elevate intracellular cAMP or Ca2+ levels can activate CREB/ATF-1 through phosphorylation at Ser133 by protein kinase A or mitogen-activated protein kinases (MAPK) (14–17). Following activation, CREB/ATF-1 regulates the expression of genes that suppress apoptosis, induce cell proliferation, and mediate inflammation and tumor metastasis by binding to cAMP-response elements (CREs) within the promoter and enhancer regions of these genes (15, 18–20).
A number of reports have suggested that CREB is involved in melanoma progression We have demonstrated previously that quenching CREB activity in metastatic melanoma cells by means of a dominant-negative form of CREB (KCREB) leads to a decrease in their tumorigenicity and metastatic potential in nude mice (21). In that study, we identified two mechanisms by which overexpression of CREB/ATF-1 contributes to the metastatic phenotype: first, CREB/ATF-1 plays an essential role in cell invasion by regulating the CRE-dependent expression of matrix metalloproteinase-2 (MMP-2) and the adhesion molecule genes MCAM/MUC18 (21); second, CREB and ATF-1 act as survival factors for human melanoma cells. Indeed, expression of a dominant-negative form of CREB (KCREB) in metastatic melanoma cells sensitizes them to thapsigargin-induced apoptosis (12). In an analogous manner, intracellular expression of an inhibitory anti-ATF-1 single chain variable fragment (ScFv) antibody in MeWo melanoma cells suppresses their tumorigenicity and metastatic potential in nude mice (21, 22). Expression of ScFv anti-ATF-1 renders the melanoma cells susceptible to thapsigargin-induced apoptosis in vitro and causes massive apoptosis in tumors transplanted subcutaneously into nude mice (23). Recently, we have demonstrated that phosphorylation of CREB and ATF-1 can be stimulated by a bioactive lipid platelet-activating factor (PAF) in metastatic melanoma cells (17). PAF-induced CREB phosphorylation leads to the overexpression and activation of MMP-2 and membrane type 1-MMP (17). In line with our observations, another study demonstrated that down-regulation of CREB expression with small interfering RNA in non-small cell lung carcinoma (NSCLC) cells suppresses their growth by inducing apoptotic cell death (24).
To better understand the mechanisms of CREB-induced tumor growth and metastasis and identify other proteins/factors involved in CREB-induced tumor growth and metastasis, we silenced CREB expression by stably transfecting the highly metastatic human melanoma cell lines A375SM and C8161-c9 with a lentivirus-based short hairpin RNA (shRNA). We found that CREB silencing resulted in the up-regulation of cysteine-rich protein 61 (CCN1/CYR61) expression. CCN1/CYR61 is a member of the growth factor-inducible immediate-early gene family consisting of CYR61/CCN1, connective tissue growth factor/CCN2, and nephroblastoma overexpressed/CCN3. CCN proteins have been shown to mediate functions as diverse as cell proliferation, migration, adhesion, survival, differentiation, and extracellular matrix formation (25). They also regulate more complex processes, such as angiogenesis and tumorigenesis (26). CCN1/CYR61 was the first cloned member of the CCN family. It is a 40-kDa cysteine-rich heparin-binding protein that either localizes intracellularly or is secreted into the extracellular milieu where it associates with the extracellular matrix and cell surfaces (27, 28).
Here, we demonstrate that CCN1/CYR61 inhibits tumor growth and metastasis, decreases angiogenesis, and induces apoptosis of melanoma cells in vivo. This is the first report to identify CCN1/CYR61 as a tumor suppressor in human melanoma.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
The A375SM human melanoma cell line was established from pooled lung metastases produced by A375-P cells injected intravenously into nude mice (29) and maintained in Eagle's minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) as described previously (30). The aggressive amelanotic human melanoma cell line C8161-c9 was maintained in Dulbecco's modified Eagle's medium-F12 (DMEM-F12) supplemented with 5% FBS as described previously (31). The SB-2 melanoma cell line was isolated from a primary cutaneous lesion as described previously (32) and maintained in MEM supplemented with 10% FBS. The 293FT cells (Invitrogen) used to produce the lentiviral shRNA were maintained in DMEM supplemented with 10% FBS according to the manufacturer's instructions.
Lentiviral shRNA
CREB-targeting shRNA (CREB-shRNA) (target sequence: 5′-GAGAGAGGTCCGTCTAATG-3′), CCN1/CYR61 shRNA (target sequence: 5′-CGCATCCTATACAACCCTTTA-3′), and a nontargeting shRNA (NT-shRNA) (target sequence: 5′-UUCUCCGAACGUGUCACGU-3′) were designed with a hairpin and sticky ends (ClaI and MluI) for use with the lentiviral system developed and kindly provided by Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) (33). The oligonucleotides were annealed into the lentiviral gene transfer vector, pLVTHM, using the ClaI and MluI restriction enzyme sites. Competent Escherichia coli were transformed with the annealed lentiviral vector and grown overnight. Several bacterial colonies were then isolated and grown in Luria broth overnight. DNA was isolated from the bacterial culture using a Maxi Plasmid DNA purification kit (Qiagen). The DNA was sequenced to test for proper insertion and length of the inserts. The lentivirus was then produced by transfecting human embryonic kidney cells (293FT; Invitrogen) with the sequence-verified PLVTHM vector containing either the CREB-shRNA or the CCN1/CYR61 shRNA sequences, the packaging plasmid (MD2G), and the envelope plasmid (PAX2), which are required for viral production. Three days later, the viral supernatant was collected and filtered to remove cellular debris. To silence CREB, highly metastatic A375SM and C8161-c9 cell lines plated at 70% confluency in 6-well plates were transduced with the virus. To silence CCN1/CYR61, SB-2 cells were plated at 60% confluency in 6-well plates and transduced with the virus. After 16 h, the virus-containing medium was removed and replaced with normal growth medium. Transduced cells were sorted by green fluorescent protein.
Quantitative Real-time PCR
RNA (20 ng/μl) from A375SM and C8161-c9 cell lines transduced with CREB-shRNA or nontargeting (NT) shRNA were harvested using the RNAqueous kit (Ambion) according to the manufacturer's instructions. The RNA was then made into cDNA using TaqMan reverse transcriptase reagents (Applied Biosystems). The primers and fluorescence probes were obtained from Applied Biosystems (CCN1/CYR61: Assay ID Hs00155479_m1). Reaction components for reverse transcription-PCR and amplifications were described previously (34). Amplifications were run in triplicates, and averages were obtained after normalization with 18s rRNA (Applied Biosystems). Data were expressed in -fold increase.
CCN1/CYR61 Expression Vector Construct
The total RNA was extracted from A375SM and reverse-transcribed using a commercial kit (Clontech). The ORF of CCN1/CYR61 was PCR-amplified from the reverse transcription product with the following two primers: C61C-Xba-F, 5′-GCTCTAGAATGAGCTCCCGCATCGCCAGGG-3′ and C61C-Cla-H3-R: 5′-CCCAAGCTTATCGATTTAGTCCCTAAATTTGTGAATGTC-3′. The PCR product was digested with XbaI and HindIII and cloned into pcDNA3.1(−) through the same two restriction enzyme sites. The inserted CCN1/CYR61 ORF was confirmed by sequencing. For making the lentiviral construct the DNA fragment containing cytomegalovirus promoter and CCN1/CYR61 ORF was cut out from the above pcDNA3.1- CYR61 vector with MluI and ClaI and cloned into lentiviral vector. The recombinant lentivirus was produced with 293T according to the standard protocol. To make CCN1/CYR61 stably expressing lines, A375SM and C8161-c9 were plated in 6-well plate and transduced with the virus containing the expression vector. After 48 h, the cells were replated in growth medium containing 500 μg/ml puromycin. Two weeks later, colonies were isolated and plated into 6-well plates. The overexpression of CCN1/CYR61 was confirmed by Western blot. As control, cells were transduced with empty vector (EV).
Nontargetable CREB Expression Vector
The lentiviral CREB expression vector was developed as described above. Briefly, total RNA was extracted from the A375SM cells, and the ORF of CREB was amplified by PCR from the reverse transcription product with the following two primers: CreB-XbaF, 5′-GCTCTAGAATGACCATGGAATCTGGAGCCGAG-3′ and CreB-Cla-H3R: 5′-CCCAAGCTTatcgaTTAATCTGATTTGTGGCAGTAAAG-3′. To create a nontargetable CREB expression vector we utilized the following oligonucleotides: CreB1B-ReF, 5′-GAAGCAGCACGAAAGAGAGAAGTGCGACTGATGAAGAACAGGGAAGCAG-3′; and CreB1B-ReR, 5′-CTGCTTCCCTGTTCTTCATCAGTCGCACTTCTCTCTTTCGTGCTGCTTC-3′. To rescue CREB expression in stably CREB-silenced cells, A375SM and C8161-c9 CREB-shRNA or NT-shRNA were plated in 6-well plates and transduced with the virus containing either the nontargetable CREB expression vector or empty vector. After 48 h, the cells were replated and selected as described previously. The CREB expression was confirmed by Western blot.
Western Blot Analysis
CREB, pCREB, CCN1/CYR61, and MMP-2 were detected in total cell extracts (20 μg) by 10% SDS-PAGE and transferred to an Immobilon-P transfer membrane (Millipore). The membranes were washed in Tris-buffered saline with Tween 20 (10 mm Tris-HCl, pH 8, 150 mm NaCl, and 0.05% Tween 20) and blocked with 5% nonfat milk in Tris-buffered saline with Tween 20 overnight at 4 °C. The blots were then probed overnight at 4 °C with primary antibodies at dilutions of 1:2000 (anti-CREB or pCREB, Cell Signaling Technology), 1:1000 (anti-CYR61, Santa Cruz Biotechnology), and 1:1000 (anti-MMP-2, Cell Signaling Technology). After 2 h of incubation with horseradish peroxide-conjugated secondary antibody, immunoreactive proteins were detected by enhanced chemiluminescence using the ECL detection system per the manufacturer's instructions (GE Healthcare). For detecting CCN1/CYR61 in the nuclear extract, A375SM and C8161-c9 nuclear extracts were prepared using a nuclear extraction kit (Panomics) according to the manufacturer's instructions. Conditioned media were collected after culturing cells in serum-free media for 48 h. The methanol precipitation method was performed with 10 ml of each sample. Briefly, 3 volumes of methanol, 1 volume of chloroform, and 4 volumes of H2O were added, mixed well, and centrifuged at maximum speed for 1 min. The upper aqueous phase was removed without disturbing the protein at the interface. Subsequently, 8 volumes of methanol were added, and the sample was vortexed to mix. Following methanol precipitation at −20 °C for 30 min, samples were centrifuged at maximum speed for 15 min to form protein pellets. The pellets were then air-dried and resuspended in phosphate-buffered saline (PBS). Protein concentrations were determined by using the Bradford protein assay (Bio-Rad).
cDNA Microarray
Microarray analysis was performed using a human genome U133 Plus 2.0 array (Affymetrix). The microarrays were produced in the microarray core facility of Codon Bioscience (Houston, TX). Total RNA was isolated from NT-shRNA- and CREB-shRNA-transduced A375SM cells using the Clontech Advantage RT-for-PCR kit according to the manufacturer's instructions. The data were analyzed using the Affymetrix software program as described previously (35).
Reporter Constructs and Luciferase Activity Assays
The CNN1/CYR61 promoter region (nucleotides −960 to +65 from the transcription initiation site) was amplified from A375SM genomic DNA using the following primers: forward, 5′-GAAGATCTGGAGAAGGCGCGGAGGGCGC-3′; reverse, 5′- ggggtacCTCCCCGCGTTCGTTTCCTCTCG-3′. The fragment was digested with KpnI and BglII and ligated into the pGL3-basic vector (Promega). Analysis of transcription factor binding sites was performed using GENOMATIX software. Site-directed mutagenesis of the CRE sites, replacing CG of the GACGTCA CRE site with AT, was performed using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. Transient transfections were performed using Lipofectin or Lipofectamin 2000 (Invitrogen) according to the manufacturer's instructions. In a 24-well plate, a total of 2.5 × 104 cells/well were transfected with 0.5 μg of the empty pGL3 expression vector or with 0.5 μg of the pGL3-CYR61 or pGL3-CYR61-promoter-mutant-containing firefly luciferase expression constructs. For each transfection, 2.5 ng of cytomegalovirus-driven Renilla luciferase reporter construct (pRL-CMV, Promega) was included. After 4 h, the transfection medium was replaced with serum-containing growth medium. After 48 h, the cells were harvested and lysed, and the luciferase activity was assayed utilizing a Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions. The luciferase luminescence (relative light intensity × 106) was measured with a LUMIstar microplate reader (BMG Labtech). The ratio of firefly luciferase activity to cytomegalovirus-driven Renilla luciferase activity was used to normalize any differences in transfection efficiency among samples. All constructs were fully sequenced in both directions before use.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were performed using the ChIP-IT Express kit from Active Motif according to the manufacturer's protocol. Briefly, cells were fixed with 1% formaldehyde, and the cross-linking reaction was stopped with 0.125 m glycine. The cells were pelleted and resuspended in a hypotonic buffer, and cell nuclei were isolated using a Dounce homogenizer. The chromatin was then sheared into 200–1000-bp fragments by adding an enzymatic solution for 10 min at 37 °C. Fractions of the chromatin solutions were incubated overnight at 4 °C with either 3 μg of anti-CREB or IgG control antibodies cross-linked to magnetic beads. The immune complexes were then eluted from the magnetic beads, and proteins were reverse cross-linked at 65 °C for 2.5 h. Proteins were digested with 2 μl of proteinase K at 37 °C for 1 h, extracted in elution buffer, and analyzed by PCR. A 250-bp fragment spanning the −515 to −265 region of the CCN1/CYR61 promoter was amplified by PCR using the following primer sequences: forward, 5′-CAGATAACTTGCCTCTCACC-3′; and reverse, 5′-TACGACTTATGTTGGGAAGG-3′.
Invasion Assay through Matrigel
The invasion assay was performed using BioCoat Matrigel invasion chambers (BD Biosciences) primed according to the manufacturer's directions. A solution of 20% FBS in DMEM-F12 or MEM was placed in the lower well to act as a chemoattractant. For experiments using A375SM or C8161-c9 cell lines, 2.5 × 103 cells in 500 μl of serum-free medium were placed in the upper chamber of the Matrigel plate and incubated at 37 °C for 24 h. Experiments using SB-2 cells were carried out as described previously (36). Cells on the lower surface of the filter were stained with Protocol Hema3 stain set (Fisher Scientific). Each sample was analyzed under a Nikon Microphot-FXA microscope at ×10 magnification. Pictures were taken using a Leica DFC 320 R2, and images were evaluated by Photoshop. The data were expressed as the average number ± S.D. of cells from four fields that migrated to the lower surface of the filter.
Monolayer Wound Healing Assay
A375SM and C8161-c9 cells were seeded in 6-well plates and allowed to reach complete confluence. To make the wound, the growth medium was aspirated and replaced by calcium-free PBS to prevent the killing of cells at the edge of the wound caused by exposure to high calcium concentrations. Subsequently, a blue plastic P1000 pipette tip (Fisher) was used to scratch the cell monolayer to create a cleared area. The wounded cell layer was washed once with fresh medium to remove loose cells and then refed with fresh growth medium. The wounds were observed using phase contrast microscopy on an inverted microscope. Images were taking at regular intervals over the course of 12 to 48 h. Images were analyzed by digitally drawing lines (using Adobe Photoshop) and averaging the positions of the migrating cells at the wound edges. We determined the cell migration distance by measuring the width of the wound, dividing by 2, and subtracting this value from the initial half-width of the wound.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were deparaffinized by sequential washing with xylene, graded ethanol, and PBS. Antigen retrieval was done by heating in a steam cooker in 1× Target Retrieval Solution (Dako) for 30 min. After cooling and washing with PBS, endogenous peroxide was blocked with 3% hydrogen peroxidase inhibitor in PBS for 12 min. Nonspecific proteins were blocked in 5% horse serum and 1% goat serum for 20 min. Slides were incubated with anti-MMP-2 (1:400; Chemicon) or anti-human CCN1/CYR61 (polyclonal, 1:100; Novus Biologicals) antibodies overnight at 4 °C and then with a peroxidase-labeled anti-rabbit antibody (1:500; Jackson ImmunoResearch) for 1 h at room temperature. Signal was detected by staining with 3,3′-diaminobenzidine (DAB; Phoenix Biotechnologies) substrate for 6 min and then counterstaining with Gill's hematoxylin No. 3 (Sigma) for 20 s. For CD31 staining, frozen tissue sections were fixed by incubation with cold acetone for 5 min followed by a 1:1 mixture of acetone/chloroform and again by cold acetone for 5 min each. Nonspecific proteins were blocked in 4% fish gelatin in PBS for 20 min. Overnight incubation at 4 °C with rat anti-mouse CD31 antibody (1:800; Pharmingen) was performed followed by a 1-h incubation with goat anti-rat Alexa 594 (1:500; Invitrogen) at room temperature. After washes in PBS, the samples were counterstained with Hoechst dye (Molecular Probes) for 10 min at room temperature.
In Situ TUNEL Assay
The terminal deoxynucleotidyl transferase-mediated dUTP end labeling (TUNEL) assay was done using a commercial kit (Promega) according to the manufacturer's protocol after CD31 staining. Briefly, tumor sections were rinsed with PBS and incubated in 4% paraformaldehyde in PBS for 10 min at room temperature. After two washes in PBS, the slides were incubated in 0.2% Triton X-100 in PBS for 15 min at room temperature followed by two washes in PBS. Slides were incubated with equilibration buffer for 10 min at room temperature. Tumor sections were then covered with TUNEL incubation buffer (50 μl/slide) and incubated in a humidified chamber at 4 °C overnight. The reaction was terminated by washing the slides with SSC for 15 min at room temperature followed by three washes in PBS to remove unincorporated fluorescein-dUTP. Slides were observed using a fluorescent microscope.
Zymography
MMP-2 activity was determined on substrate-impregnated gels as described previously (17). Approximately 5 × 103 melanoma cells were plated in 6-well dishes and allowed to attach for 24 h. Then the medium (normal growth medium with 10% FBS) was removed and replaced with serum-free medium overnight. The supernatants were collected and their volumes adjusted to the cell number, and a total of 60 μl of supernatant was loaded onto a gelatin-impregnated (1 mg/ml; Sigma) 8% SDS-polyacrylamide gel and separated under nonreducing conditions. As a positive control, 1% FBS in normal growth medium was used. For the negative control, serum-free medium was used. Plates were shaken for 1 h in 2.5% Triton X-100 (Fisher Scientific) to remove all of the SDS from the gels. Plates were then removed, and the gels were incubated for 16 h at 37 °C in 50 mmol/liter Tris, 0.2 m NaCl, 5 mmol/liter CaCl2, and 0.002% Brij 35 (w/v) at pH 7.6. At the end of the incubation, the gels were stained with 0.5% Coomassie G-250 (Bio-Rad) in methanol/acetic acid/H2O (30:10:60). The intensities of the various bands were determined by quantitation of a scanned image.
Animals, Tumor Growth, and Metastasis
Female athymic BALB/c nude mice (National Institutes of Health, NCI, Frederick Cancer Research Institute) were housed in laminar flow cabinets under specific pathogen-free conditions and used at 8 weeks of age. Animals were maintained in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with the current regulations and standards of the U. S. Department of Agriculture, Department of Health and Human Services, National Institutes of Health, and institutional regulations. In accordance with the Institutional Animal Care and Use Committee, when the largest dimension of a subcutaneously injected tumor reached 1.5 cm, the mice were considered moribund and were sacrificed in a CO2 chamber. Subcutaneous tumors were produced by injecting 2.5 × 105 C8161-c9 cells or 5 × 105 A375SM cells (single-cell suspensions, >95% viability by a trypan blue exclusion test) in 0.2 ml of Hanks' buffered salt solution into the right flank of each mouse. Tumor growth was recorded three times weekly with a caliper and calculated as a × b2/2 cm3 (a, long diameter; b, short diameter). Mice were sacrificed 26 or 36 days after injection or when the tumor reached 1.5 cm3 in volume, and tumors were processed for hematoxylin and eosin or immunohistochemical staining. Ten mice were used in each group. To determine metastatic potential, 5 × 105 or 1 × 106 tumor cells, processed as described above, were injected into the tail veins of mice (0.1 ml/mouse). Thirty-five days later, the mice were sacrificed, their lungs harvested, and the number of macroscopic surface tumor nodules counted. Six or seven mice were used in each group.
Statistical Analysis
Student's t test was used to evaluate the statistical significance of the in vitro data. Statistical analysis of the results of the animal studies was performed using the Mann-Whitney U test. Values for tumor growth are given as a mean volume ± S.D., and p values < 0.05 were considered statistically significant.
RESULTS
Silencing CREB Inhibits Tumor Growth and Metastasis of Melanoma
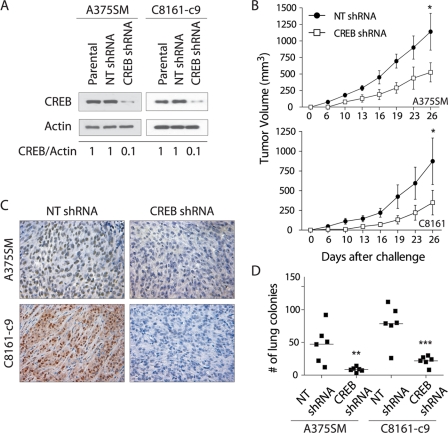
We have shown previously that silencing CREB, using a dominant-negative form of CREB (KCREB) in MeWo metastatic melanoma cells, decreases their tumorigenic and metastatic potentials in nude mice by down-regulating expression of MMP-2 and MCAM/MUC18 and decreasing melanoma cell invasion through the basement membrane (21). To further understand how CREB silencing leads to an increase in melanoma cell tumorigenicity and metastatic potential, here we used a lentiviral construct carrying CREB-shRNA to silence CREB expression in two highly metastatic and aggressive melanoma cell lines, A375SM and C8161-c9. As a control, cells were transduced with a lentiviral construct carrying NT- shRNA with no sequence homology to any known human gene. Cells were transduced with the lentiviral constructs and sorted for green fluorescent protein expression. We then measured CREB expression in the green fluorescent protein-positive cells by using Western blotting. We found a 90% reduction in CREB expression in both the A375SM and C8161-c9 cell lines (Fig. 1A).
FIGURE 1.
Effects of CREB silencing on melanoma growth and metastasis. A, Western blot analysis shows silencing of CREB in the A375SM and C8161-c9 cell lines stably transduced with CREB-shRNA as compared with those transduced with nontargeting control shRNA (NT shRNA). α-Actin was used as a loading control. A 90% reduction in CREB was observed in the CREB-shRNA-transduced cells as compared with the NT-shRNA-transduced cells as measured by densitometry (0.1 and 1, respectively). B, silencing of CREB resulted in a significant inhibition of tumor growth in both the A375SM (CREB-shRNA, 278.1 mm3; NT-shRNA, 1140.0 mm3) and C8161-c9 (CREB-shRNA, 348.4 mm3; NT-shRNA, 873.0 mm3) melanoma cell lines. *, p < 0.05 for both A375SM and C8161-c9 cells. C, immunohistochemical staining for CREB was performed on tumor samples from mice 26 days after injection with CREB-shRNA- or NT-shRNA-transduced A375SM or C8161-c9 cells, demonstrating down-regulation of CREB expression in CREB-shRNA tumors. Images are shown at ×20 magnification. D, effect of CREB silencing on the metastatic potential of cells. There is a significant decrease in the number of lung metastases in CREB-shRNA-transduced A375SM and C8161-c9 cells as compared with the NT-shRNA-transduced cells. Each square represents one mouse (n = 6/group). **, p < 0.01; ***, p < 0.001.
To determine whether CREB silencing affects the tumorigenic and metastatic potential of melanoma cells in vivo, CREB-silenced A375SM and C8161-c9 cells were injected subcutaneously into nude mice. Silencing of CREB resulted in the inhibition of tumor growth in both cell lines. Fig. 1B shows a significant inhibition of tumor growth in mice injected with CREB-silenced A375SM cells as compared with that in mice injected with NT-shRNA A375SM cells (mean tumor volumes, 278.1 and 1140.0 mm3, respectively; p < 0.05). Similar differences were observed between the mice injected with CREB-silenced C8161-c9 cells and mice injected with NT-shRNA C8161-c9 cells (mean tumor volumes, 348.4 and 873.0 mm3, respectively; p < 0.05). Immunohistochemical analysis of CREB expression in tumor samples obtained from mice 26 days after they were injected with either the CREB-shRNA or NT-shRNA melanoma cells revealed very low CREB expression levels in the CREB-shRNA tumors as compared with the NT-shRNA tumors (Fig. 1C).
Next, we sought to determine whether CREB silencing in melanoma cells would affect their ability to metastasize in vivo. To that end, nude mice were injected intravenously with A375SM or C8161-c9 cells transduced with either CREB-shRNA or NT-shRNA. As demonstrated in Fig. 1D, CREB silencing resulted in a significant reduction in the median number of lung metastases in mice injected with either A375SM cells (NT-shRNA, 49; CREB-shRNA, 8; p < 0.01) or C8161-c9 cells (NT-shRNA, 80; CREB-shRNA, 23; p < 0.001). Overall, these results confirm that CREB promotes melanoma tumorigenicity and metastasis.
CREB Acts as a Negative Regulator of CCN1/CYR61 Expression in Melanoma
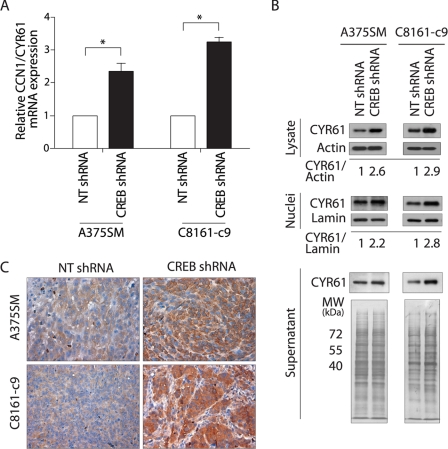
To understand the mechanism by which CREB promotes the tumorigenic and metastatic potentials of melanoma cells, we sought to identify downstream target genes regulated by CREB. To that end, we used cDNA microarrays to measure differences in gene expression between A375SM CREB-silenced and NT-shRNA cells. Among the genes found to be differentially expressed by the cDNA microarray analysis, CCN1/CYR61 was up-regulated by almost 3.0-fold in the CREB-silenced cells. We validated the CCN1/CYR61 up-regulation of both mRNA and protein levels after CREB silencing. Utilizing quantitative real-time PCR, we found that A375SM and C8161-c9 cells transduced with CREB-shRNA had a significant increase of CCN1/CYR61 mRNA levels by 1.4- and 2.3-fold (p < 0.005) respectively, as compared with the NT-shRNA-transduced cells (Fig. 2A). Furthermore, Western blot analysis of the total cell lysates revealed a 2.6- and 2.9-fold increase in CCN1/CYR61 protein levels after CREB silencing in both the A375SM and C8161-c9 cell lines, respectively (Fig. 2B). Cell fractionation revealed that the level of CCN1/CYR61 in the nuclei of CREB-shRNA cells was also increased (Fig. 2B). Because CCN1/CYR61 is a secreted protein, we next analyzed its levels in the supernatants before and after CREB silencing. An increase in CCN1/CYR61 secretion was observed after CREB silencing (Fig. 2B). In addition, immunohistochemical analysis of tumor samples generated in the earlier described experiment (Fig. 1B) demonstrated a strong increase in CCN1/CYR61 expression in the CREB-shRNA tumors 26 days after tumor injections (Fig. 2C).
FIGURE 2.
Validation of CCN1/CYR61 overexpression after CREB silencing in melanoma cells. A, quantitative real-time PCR validation for CCN1/CYR61 gene expression. Expression values shown are -fold change in each CREB-shRNA-transduced cell line relative to the NT-shRNA cells after normalization with 18s RNA. *, p < 0.005. B, Western blot analyses of total cell lysate, nuclei and supernatant show a significant increase in CCN1/CYR61 expression in both A375SM and C8161-c9 cell lines after CREB silencing as compared with NT-shRNA cells. α-Actin, lamin, and whole gel staining were used as indicators of equal sample loading. C, immunohistochemical staining for CCN1/CYR61 was performed on tumor samples from mice 26 days after injection with NT-shRNA- or CREB-shRNA-transduced A375SM or C8161-c9 cells, demonstrating CCN1/CYR61 overexpression in CREB-shRNA-transduced cells in vivo.
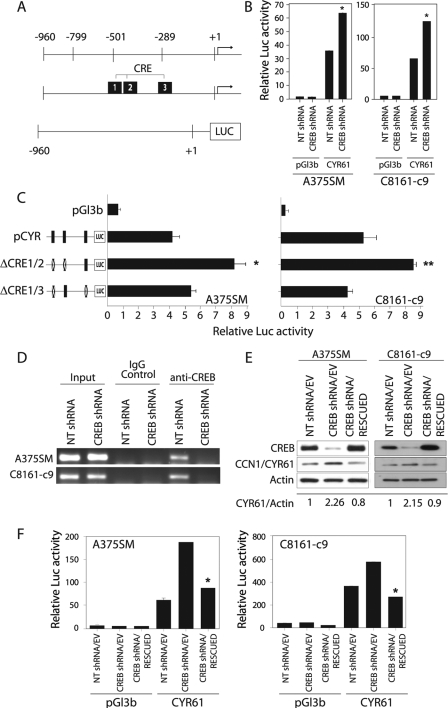
To understand the mechanism of CCN1/CYR61 regulation, we analyzed the promoter sequence of the human CCN1/CYR61 gene (GenBankTM accession number AC092807) and found that it contains three CRE sites. To confirm that CREB regulates CCN1/CYR61 expression at the transcriptional level, we cloned the promoter region (−960 to +65) of CCN1/CYR61 in front of a luciferase reporter gene (Fig. 3A). The luciferase activity driven by the CCN1/CYR61 promoter increased 2-fold (p < 0.001) after CREB silencing in both of the melanoma cell lines tested (Fig. 3B). This result suggests that CREB acts as a negative regulator of CCN1/CYR61 transcription.
FIGURE 3.
Regulation of the CCN1/CYR61 promoter by CREB. A, schematic representation of the CCN1/CYR61 promoter region fused to the luciferase reporter gene and its predicted CRE binding sites. B, the luciferase activity driven by the CCN1/CYR61 promoter increased 2-fold after CREB silencing in both the A375SM and C8161-c9 cell lines as compared with the NT-shRNA-transduced cells. *, p < 0.001. C, a schematic representation of the promoter point mutations is depicted on the left side of the panel. The three identified CRE binding sites were mutated either alone or in combination, as described under “Experimental Procedures.” The luciferase activity driven by the CCN1/CYR61 promoter increased 2.2-fold when both the first and second CRE binding sites were mutated in the A375SM and C8161-c9 cell lines. *, p < 0.001; ** p < 0.01. D, chromatin immunoprecipitation studies showed no binding of CREB to the CCN1/CYR61 promoter in either of the CREB-silenced cell lines (A375SM and C8161-c9). IgG antibodies were used as negative controls. Input DNA was used to ensure an equal amount of chromatin used in each assay. E, rescue of CREB expression in the CREB-silenced cells results in down-regulation of CCN1/CYR61 expression. α-Actin was used as a loading control. F, the luciferase activity driven by the CCN1/CYR61 promoter decreased significantly (*, p < 0.001) after rescue of CREB expression in both the A375SM and C8161-c9 cell lines. NT-shRNA/EV, nontargeting control cells. CREB-shRNA/EV, CREB-silenced control cells. CREB-shRNA/RESCUED, CREB-silenced cells transduced with CREB nontargetable expression vector.
To verify that the CRE sites within the CCN1/CYR61 promoter were involved in the regulation of CCN1/CYR61 transcription, we altered these binding sites by introducing point mutations into the CRE sites. Mutation of single CRE sites did not produce any significant changes in the luciferase reporter activity in the two melanoma cell lines tested (data not shown). However, mutation of the first two CRE elements (Fig. 3A) simultaneously led to a significant increase in the CCN1/CYR61 promoter-driven reporter activity in both the A375SM and C8161-c9 parental cells (Fig. 3C). In fact, the basal luciferase activity of the double-mutant promoter reporter in the parental cells was comparable to the CCN1/CYR61 promoter activity levels seen after CREB silencing. When the assay was repeated with a CCN1/CYR61 promoter in which the first and the third CRE sites were mutated simultaneously, we did not observe any significant differences in the reporter activity (Fig. 3C). Taken together, these site-directed mutation analyses revealed that the first two CRE sites, located at −509 and −445, are important for CCN1/CYR61 transcription regulation by CREB.
To further investigate whether CREB regulates CCN1/CYR61 transcription by directly binding to its promoter, we conducted a chromatin immunoprecipitation assay and found that CREB was bound to the CCN1/CYR61 promoter in the control NT-shRNA cells; this binding was abrogated following CREB silencing in both the A375SM and C8161-c9 cell lines (Fig. 3D). These data confirm that CREB regulates CCN1/CYR61 transcription by directly binding to its promoter.
Finally, to confirm the specificity of the effect of CREB-shRNA on CCN1/CYR61 expression, we were able to restore CREB expression in both CREB-silenced cell lines by overexpressing CREB in which silent mutations were introduced to render it nontargetable by CREB-shRNA. Western blot analysis of these cells demonstrated that this CREB rescue resulted in a restoration of CREB expression to levels comparable with those in control cells (NT-shRNA/EV). Restoring CREB expression in these cells also resulted in a decrease in the expression of CCN1/CYR61 (Fig. 3E) and a decrease in the luciferase activity driven by the CCN1/CYR61 promoter in both melanoma cell lines tested (Fig. 3F). Overall, these experiments confirmed that CREB regulates CCN1/CYR61 transcription in melanoma cells.
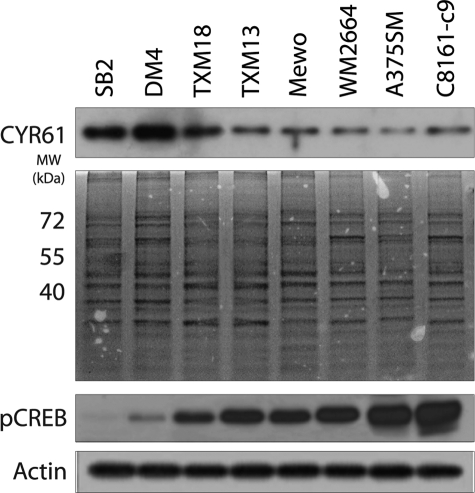
Expression of CCN1/CYR61 Is Inversely Correlated with the Metastatic Potential of Melanoma Cells
To investigate whether CCN1/CYR61 expression correlates with the metastatic potential of melanoma cells, we compared the levels of CCN1/CYR61 secretion in a panel of human melanoma cell lines with low metastatic potential (SB-2, DM4, TXM18, and TXM13) with those in cell lines with high metastatic potential (MeWo, WM2664, A375SM, and C8161-c9). As shown in Fig. 4, higher levels of CCN1/CYR61 were found in the supernatant of cells with low metastatic potential as compared with those in the supernatant of highly metastatic cells. Moreover, analyses of activated CREB measured by its phosphorylation at Ser133 (pCREB) revealed increased levels with melanoma progression and correlated with decreased levels of CCN1/CYR61. Taken together, our data suggest that there is an inverse correlation between CCN1/CYR61 secretion levels and the metastatic potential of human melanoma cell lines. Furthermore, our data confirmed the role of CREB in melanoma progression and metastasis. This result suggests a role for CCN1/CYR61 as a tumor suppressor gene in human melanoma.
FIGURE 4.
There is an inverse correlation between CCN1/CYR61 expression and the metastatic potential of melanoma cells. Western blot analysis of CCN1/CYR61 protein levels in the supernatant of a panel of human melanoma cell lines with either low metastatic potential (SB-2, DM4, TXM18, and TXM13) or high metastatic potential (MeWo, WM2664, A375SM, and C8161-c9). Higher levels of secreted CCN1/CYR61 were found in cell lines with low metastatic potential as compared with levels in the highly metastatic cell lines. Staining of the entire gel is included as an indication of equal sample loading. In contrast, higher levels of phosphorylated CREB (pCREB) were observed in metastatic melanoma cells when compared with low metastic cell lines. α-Actin was used as a loading control.
CCN1/CYR61 Overexpression Suppresses the Tumorigenicity and Metastatic Potential of Melanoma Cells
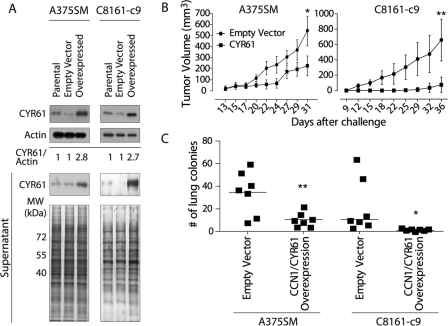
To elucidate the role of CCN1/CYR61 in melanoma, we stably overexpressed CCN1/CYR61 in A375SM and C8161-c9 cells, which are highly metastatic with low levels of CCN1/CYR61, using a lentiviral expression vector. We found a marked increase in CCN1/CYR61 expression in the total cell lysates of both the A375SM and C8161-c9 cells transduced with the CCN1/CYR61-containing vector (2.8- and 2.7-fold increase, respectively) as compared with the cells transduced with the EV control or the parental cells (Fig. 5A, upper panel). In addition, we detected a marked increase in the levels of CCN1/CYR61 secreted from cells after transduction of both cell lines with the CCN1/CYR61-containing vector (Fig. 5A, lower panel). It is noteworthy that the nonmetastatic cell lines expressed 2–10-fold higher levels of CCN1/CYR61 than the highly metastatic A375SM or C8161-c9 cell lines (Fig. 4), suggesting that these overexpression levels are physiologically relevant.
FIGURE 5.
CCN1/CYR61 overexpression inhibits tumor growth and metastasis in vivo. A, Western blot analysis demonstrating overexpression of CCN1/CYR61 in total cell lysates and supernatants of the transduced cells. B, effect of CCN1/CYR61 expression on tumor growth in vivo. CCN1/CYR61 overexpression resulted in a significant inhibition of tumor growth in both the A375SM (CCN1/CYR61 226.4 mm3; EV control, 544.4 mm3) and C8161-c9 (CCN1/CYR61, 75.0 mm3; EV control, 659.6 mm3) melanoma cells. *, p < 0.05; **, p < 0.01. C, effects of CCN1/CYR61 overexpression on the metastatic potential of melanoma cells. We observed a significant decrease in the number of lung metastases in mice injected with cells overexpressing CCN1/CYR61 as compared with the number in mice injected with EV control cells. A375SM (median, EV control, 37; CCN1/CYR61, 10; **, p < 0.01) and C8161-c9 (median, EV control, 10; CCN1/CYR61, 3; *, p < 0.05). Each square represents one mouse (n = 7/group).
Next, we analyzed the CCN1/CYR61-overexpressing cells for their ability to form tumors and metastases in vivo. A375SM and C8161-c9 cells overexpressing CCN1/CYR61 were injected subcutaneously into nude mice. We observed a significant inhibition of tumor growth in mice injected with the A375SM cells overexpressing CCN1/CYR61 as compared with the mice injected with the cells containing the EV control (mean tumor volume at day 31, 226.4 and 544.4 mm3, respectively; p < 0.05). Similar differences were observed between the C8161-c9 cells overexpressing CCN1/CYR61 and the EV control C8161-c9 cells (mean tumor volume at day 36, 75.0 and 659.6 mm3, respectively; p < 0.01) (Fig. 5B). In addition, overexpression of CCN1/CYR61 resulted in a reduction in the median number of lung metastases in both the A375SM-injected (EV control, 37, CCN1/CYR61, 10; p < 0.01) and C8161-c9-injected (EV control, 10, CCN1/CYR61, 3; p < 0.05) mice (Fig. 5C). Taken together, these data show, for the first time, that overexpression of CCN1/CYR61 results in an inhibition of melanoma growth and metastasis. To identify any changes in cell proliferation after CCN1/CYR61 overexpression, we performed 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assays as well as clonogenic assays on both cell lines either overexpressing CCN1/CYR61 or with the EV control. We did not observe any difference in the proliferation rates for any of the cell lines tested (data not shown).
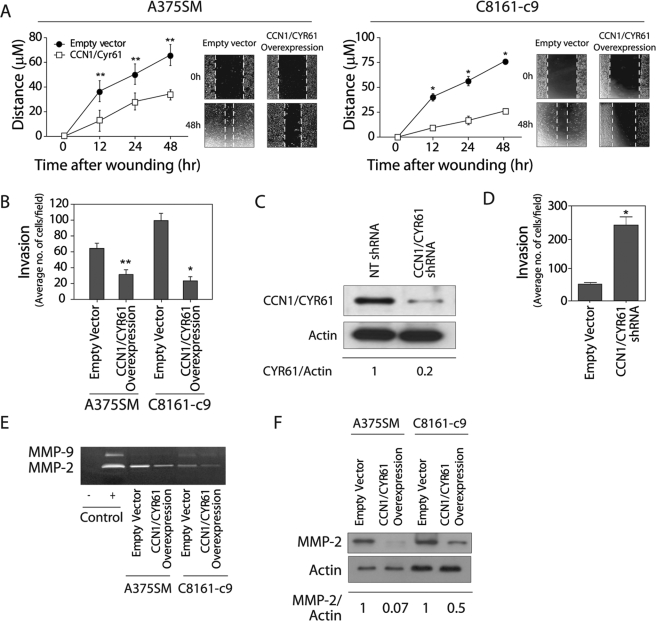
CCN1/CYR61 Inhibits Melanoma Cell Migration, Invasion, and MMP-2 Expression and Activity
To understand the mechanism by which CCN1/CYR61 inhibits the formation of tumors and the metastatic potential of melanoma cells, we further investigated its effect on tumor cell migration and invasion. Cell migration was assessed before and after CCN1/CYR61 overexpression using the wound healing (scratch) assay. As demonstrated in Fig. 6A, 48 h after scratching, cell lines overexpressing CCN1/CYR61 migrated significantly less than in the cells expressing the EV control (A375SM CCN1/CYR61, 34.5 μm, A375SM EV control 66.0 μm; p < 0.01; C8161-c9 CCN1/CYR61, 26.0 μm, C8161-c9 EV control, 75.7; p < 0.001). Furthermore, CCN1/CYR61 overexpression significantly reduced the invasion of A375SM and C8161-c9 cells through Matrigel-coated filters by 2- and 3-fold, respectively (Fig. 6B). To further establish the role of CCN1/CYR61 as a tumor suppressor gene in melanoma, we next attempted to silence the expression of CCN1/CYR61 in low metastatic melanoma cells (SB-2) and subsequently analyzed their invasion potential in vitro. To that end, we stably transduced the SB-2 cell line with CCN1/CYR61 shRNA. Western blot analysis of total protein lysate revealed an 80% decrease in CCN1/CYR61 protein expression levels as compared with NT-shRNA (Fig. 6C). Similar results were found in the supernatant of these cells (data not shown). As seen on Fig. 6D, SB-2 cells transduced with CCN1/CYR61 shRNA significantly increased their invasion ability through Matrigel by 4-fold (p < 0.001). These results suggest that CCN1/CYR61 acts as a negative regulator of melanoma cell migration and invasion.
FIGURE 6.
CCN1/CYR61 decreases cell motility and invasion of melanoma cells by down-regulation of MMP-2. A, overexpression of CCN1/CYR61 in A375SM and C8161-c9 cells inhibits their motility as determined by the scratch wound healing assay (*, p < 0.001; **, p < 0.01). B, CCN1/CYR61 overexpression inhibits the invasive properties of both melanoma cell lines in a Matrigel-coated filter chamber assay (*, p < 0.001; **, p < 0.01). C, Western blot analysis shows silencing of CCN1/CYR61 in the SB-2 cell line stably transduced with CCN1/CYR61 shRNA as compared with NT-shRNA. α-Actin was used as a loading control. 80% reduction in CCN1/CYR61 protein level was observed in the transduced cells as compared with the NT-shRNA-transduced cells as measured by densitometry (0.2 and 1, respectively). D, CCN1/CYR61 silencing increases the invasive ability of SB-2 cells in a Matrigel chamber assay (*, p < 0.001) E, zymography gel analysis of A375SM and C8161-c9 cells overexpressing CCN1/CYR61 demonstrates a significant decrease in the activity of MMP-2 as compared with that in EV control cells. FBS (1%) was loaded and used as a positive control. F, Western blot analysis showing MMP-2 protein levels in both cell lines after CCN1/CYR61 overexpression. α-Actin was used as a loading control. Densitometry analysis reveals that the MMP-2 protein levels decreased by 97 and 50%, respectively, in the A375SM and C8161-c9 cell lines overexpressing CCN1/CYR61.
It is well known that matrix metalloproteinases play a central role in regulating tumor cell invasion and in remodeling the stromal microenvironment of the tumor (37). In melanoma, active MMP-2 can only be observed in highly invasive melanoma cell lines and is absent in non-invasive or poorly invasive cell lines (38). To investigate whether CCN1/CYR61 affects melanoma cell invasion through the activity of MMPs, we performed a zymography assay. In A375SM and C8161-c9 cells, overexpression of CCN1/CYR61 led to a significant decrease in the gelatinase activity of MMP-2, but not that of MMP-9, in the cell's supernatant as compared with the EV control cells (Fig. 6E). Western blot analysis demonstrated that MMP-2 protein levels were also markedly reduced in both cell lines overexpressing CCN1/CYR61 (Fig. 6F). These results suggest that CCN1/CYR61 acts as a negative regulator of melanoma tumor cell motility and invasion, in part by down-regulating the expression and activity of MMP-2. Therefore, the negative effect of CCN1/CYR61 on melanoma tumorigenicity and metastasis could be, at least partly, explained by its ability to control MMP-2 expression.
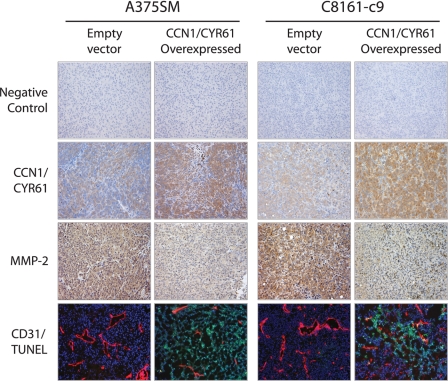
CCN1/CYR61 Overexpression Reduces Angiogenesis and Increases Apoptosis in Vivo
In addition to regulating tumor cell invasion and metastasis, MMP-2 plays an important role in regulating tumor angiogenesis. Therefore, we sought to determine whether overexpression of CCN1/CYR61 could affect MMP-2 expression and angiogenesis in vivo. To that end, we performed immunohistochemical analysis of subcutaneous tumors derived from mice injected with A375SM and C8161-c9 cells with or without overexpression of CCN1/CYR61 (Fig. 5B). We found a significant decrease in the expression of MMP-2 in tumor samples overexpressing CCN1/CYR61 as compared with EV control tumor samples (Fig. 7). Furthermore, we observed marked differences in microvessel density as judged by CD31 staining, i.e. tumor samples overexpressing CCN1/CYR61 showed a dramatic decrease in the number and size of vessels as compared with EV control tumor samples. To monitor whether inhibition of tumor angiogenesis leads to tumor cells apoptosis, we performed a TUNEL assay. The results shown in Fig. 7 (bottom panel) demonstrate that there was a large increase in the number of apoptotic cells in both the A375SM and C8161-c9 tumor samples overexpressing CCN1/CYR61. In total, these experiments established that CCN1/CYR61 acts as a supressor of melanoma tumor growth and metastasis by negatively regulating MMP-2 expression, culminating in a major effect on angiogenesis in vivo. Our results further suggest that CREB acts to augment melanoma tumor growth and metastasis by suppressing CCN1/CYR61 expression.
FIGURE 7.
Effects of CCN1/CYR61 overexpression on MMP-2 expression, angiogenesis, and apoptosis in vivo. Immunohistochemical analyses were performed on tumor samples from mice challenged with A375SM or C8161-c9 cells overexpressing CCN1/CYR61 or EV control. Representative images show that overexpression of CCN1/CYR61 resulted in the down-regulation of MMP-2 expression, a reduction in the number and size of blood vessels (CD31), and an increase in the number of apoptotic cells (TUNEL). Tumor samples were incubated without primary antibody as a negative control. All images are shown at ×10 magnification.
DISCUSSION
We have shown previously that quenching CREB and ATF-1 activities with a dominant-negative form of CREB (KCREB) or with a ScFv anti-ATF-1 antibody inhibits melanoma growth and metastasis, thereby establishing a critical role for these transcription factors in melanoma (13, 21–23). Here, we used a more specific, shRNA-based approach to investigate further the role of CREB in melanoma growth and metastasis. Our results demonstrate that stable CREB silencing with shRNA leads to a decrease in melanoma tumor growth and metastasis, thereby corroborating our previous studies. In addition, using cDNA microarrays, we have identified a novel downstream target, the CCN1/CYR61 gene, which is negatively regulated by CREB in melanoma cells. In subsequent experiments, we confirmed that CREB silencing significantly increases CCN1/CYR61 expression in vitro and in subcutaneously grown melanoma tumors. Promoter analysis and chromatin immunoprecipitation assays demonstrated that CREB negatively regulates CCN1/CYR61 transcription by binding to and inhibiting transcription from the CCN1/CYR61 promoter. Restoring CREB expression after shRNA silencing reduces CCN1/CYR61 protein levels as well as its promoter reporter activity, confirming the specificity of the shRNA for CREB. Although CREB acts usually as transactivator, here we have demonstrated that in the case of CCN1/CYR61, it acts as repressor. This has been shown for other transcriptional factors such as AP-2α (30).
Analysis of the role of CCN1/CYR61 in melanoma demonstrates that it acts as a suppressor of melanoma tumorigenicity and metastasis in vivo. Indeed, when CCN1/CYR61 was overexpressed in A375SM and C8161-c9, we observed a significant decrease in melanoma tumor growth as well as a decrease in the number of lung metastases. Notably, the levels of CCN1/CYR61 after overexpression were similar to those observed in nonmetastatic and low tumorigenic melanoma cell lines such as SB-2 and DM4, suggesting that increased CCN1/CYR61 expression levels in these cells may contribute to their low metastatic and tumorigenic potential. Also, the fact that an inverse correlation of CCN1/CYR61 and CREB activation was found further supports this hypothesis. Overexpression of CCN1/CYR61 also resulted in an inhibition of melanoma cell motility and invasion and MMP-2 expression and activity in vitro. In vivo, overexpression of CCN1/CYR61 was associated with a decrease in MMP-2 expression, inhibition of tumor angiogenesis, and an increase in apoptosis. These results corroborate our previous finding that CREB acts as a positive regulator of MMP-2 expression and melanoma cell invasion, suggesting that these effects can be explained, in part, by the CREB-mediated inhibition of CCN1/CYR61 expression.
However, it is unclear how CCN1/CYR61 may regulate MMP-2 expression in melanoma. Integrins are well known receptors for CCN proteins, and receptor activation may produce a variety of effects (39). Studies using B16F10, a murine melanoma cell line, have shown that ligation of cell surface α5β1 integrin by anti-α5 antibody activates focal adhesion kinase, thereby modulating expression and activation of MMP-2 and MMP-7 (40). Our preliminary data suggest that in human melanoma, CCN1/CYR61 overexpression inhibits phosphorylation of CREB. Thus, whether or not CCN1/CYR61 interacts with integrins in melanoma, and thereby regulates CREB activation, requires further investigation.
Conversely, the role of CCN1/CYR61 in cancer is controversial and clearly tumor-type dependent. For example, down-regulation of CCN1/CYR61 expression has been described in prostate cancer, uterine leiomyoma, rhabdomyosarcoma, embryonic rhabdomyosarcoma, and non-small cell lung carcinoma (41–43). Meanwhile, overexpression of CCN1/CYR61 in lung cancer cells inhibits their tumorigenicity. This inhibitory effect could be attributed in part to CCN1/CYR61-induced cell cycle arrest, up-regulation of both p53 and p21WAF1, and decreased kinase activity of CDK2 (44). CCN1/CYR61 is further reported to inhibit the growth of endometrial cancer and leiomyomas (43, 45). In human gastric carcinomas, CCN1/CYR61 expression also down-regulates MMP-7 expression and inhibits tumor progression (46). On the other hand, earlier reports demonstrated that elevated CCN1/CYR61 levels are associated with advanced breast adenocarcinoma, pancreatic cancer, and gliomas (47–49). CCN1/CYR61 overexpression was found to stimulate the progression of breast cancer (50–52) and increase tumor formation in gastric adenocarcinoma (53).
Even among melanomas, the role of CCN1/CYR61 expression appears contradictory. Previous studies have reported that in uveal melanoma, CCN1/CYR61 expression is increased and that CCN1/CYR61 plays a role in tumor angiogenesis (54, 55). By contrast, our results demonstrate that in cutaneous melanoma, CCN1/CYR61 expression is suppressed by CREB and that CCN1/CYR61 acts as a negative regulator of tumor cell motility, invasion, tumorigenicity, angiogenesis, and metastasis. Furthermore, we found an inverse correlation between CCN1/CYR61 expression and the metastatic potential of melanoma cell lines, which further suggests that CCN1/CYR61 acts as a negative regulator of melanoma progression.
Surprisingly, we detected a considerable amount of CCN1/CYR61 in melanoma cell nuclei. Typically, CCN1/CYR61 either localizes intracellularly or associates with the extracellular matrix and cell surfaces. However, in bladder smooth muscle cells, CCN1/CYR61 has been found to localize in both the cytoplasm and nucleus (56). CCN1/CYR61 does not have any putative nuclear localization sequences; however, a growing list of polypeptides that lack a nuclear localization sequence, including the nephroblastoma overexpressed gene (CCN3), epidermal growth factor, fibroblast growth factor, platelet-derived growth factor, angiogenin, and parathyroid hormone-related peptide, has been reported to localize to the nucleus. Although the nuclear functions of these peptides are not yet completely understood, they might be involved in transcription regulation and/or mRNA transport (56). Thus, the detection of CCN1/CYR61 in the nuclei of human melanoma cells warrants further investigation into its nuclear functions.
Interestingly, we also found a marked decrease in microvessel density and angiogenesis, accompanied by an increase in the number of apoptotic cells, in tumor samples overexpressing CCN1/CYR61. These data are particularly intriguing because CCN1/CYR61 has been described as an angiogenic inducer (25) and has been implicated in the regulation of angiogenesis and matrix remodeling genes, including vascular endothelial growth factor A, vascular endothelial growth factor C, type I collagen, MMP-1, MMP-3, and tissue inhibitors of metalloproteinases (57). However, CCN1/CYR61 can either induce or suppress apoptosis in a cell type-specific manner. Generally, CCN1/CYR61 adhesion to endothelial cells promotes cell survival, whereas CCN1/CYR61 adhesion to fibroblasts induces apoptosis by binding to α6β1 integrin and syndecan-4, leading to the p53-dependent activation of Bax and cytochrome c release (58). In human endometrial cancer cells, overexpression of CCN1/CYR61 results in increased expression of the apoptotic factors Bax and Bad, cytochrome c release, activation of caspase-9 and -3, and apoptosis (58). In our experiments, overexpression of CCN1/CYR61 did not induce melanoma cell death in vitro (data not shown). In vivo, however, it inhibited tumor angiogenesis and induced tumor cell apoptosis.
Taken together, our results demonstrate that CREB contributes to melanoma progression by inhibiting the expression of CCN1/CYR61, which in turn acts as a negative regulator of tumor growth, angiogenesis, and metastasis. In cutaneous melanoma, CCN1/CYR61 inhibits cellular motility and invasion through down-regulation of MMP-2 expression and activity. Overexpression of CCN1/CYR61 in highly metastatic melanoma cells causes a decrease in tumor angiogenesis, which is accompanied by melanoma cell apoptosis. Corroborating these results, we found an inverse correlation between CCN1/CYR61 expression levels and the metastatic potential of melanoma cells. Future experiments will focus on identifying additional molecular mechanisms that drive the tumor- and metastasis-suppressive functions of CCN1/CYR61 in melanoma.
Acknowledgments
We thank Dr. Ruth Lupu from Northwestern University Feinberg School of Medicine for kindly providing the CCN1/CYR61 promoter reporter plasmid and Didier Trono for kindly providing the lentiviral backbone vectors used to incorporate CREB and CCN1/CYR61 shRNA. We also thank Donna Reynolds (Department of Cancer Biology-M. D. Anderson Cancer Center (MDACC)) and Carol M. Johnston (Division of Surgery-MDACC) for sharing their expertise in immunohistochemical techniques.
This work was supported, in whole or in part, by National Institute of Health Grant CA76098 (to M. B.-E.). This work was also supported by a grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES-Brazil) (to A. S. D.).
- ATF-1
- activating transcription factor-1
- CRE
- cAMP-response element
- CREB
- cAMP-response element-binding protein
- EV
- empty vector
- MMP
- matrix metalloproteinase
- NT
- nontargeting
- shRNA
- short hairpin RNA
- MEM
- minimum essential medium
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- ORF
- open reading frame
- PBS
- phosphate-buffered saline
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP end labeling.
REFERENCES
- 1.de Gruijl F. R. (1999) Eur. J. Cancer 35, 2003–2009 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 3.de Braud F., Khayat D., Kroon B. B., Valdagni R., Bruzzi P., Cascinelli N. (2003) Crit. Rev. Oncol. Hematol. 47, 35–63 [DOI] [PubMed] [Google Scholar]
- 4.Smith A. P., Weeraratna A. T., Spears J. R., Meltzer P. S., Becker D. (2004) Cancer Biol. Ther. 3, 104–109 [DOI] [PubMed] [Google Scholar]
- 5.McDonald S. L., Edington H. D., Kirkwood J. M., Becker D. (2004) Cancer Biol. Ther. 3, 110–120 [DOI] [PubMed] [Google Scholar]
- 6.Nyormoi O., Bar-Eli M. (2003) Clin. Exp. Metastasis 20, 251–263 [DOI] [PubMed] [Google Scholar]
- 7.Melnikova V. O., Bar-Eli M. (2006) Pigment Cell Res. 19, 395–405 [DOI] [PubMed] [Google Scholar]
- 8.Leslie M. C., Zhao Y. J., Lachman L. B., Hwu P., Wu G. J., Bar-Eli M. (2007) Gene Ther. 14, 316–323 [DOI] [PubMed] [Google Scholar]
- 9.Rutberg S. E., Goldstein I. M., Yang Y. M., Stackpole C. W., Ronai Z. (1994) Mol. Carcinog. 10, 82–87 [DOI] [PubMed] [Google Scholar]
- 10.Yang Y. M., Dolan L. R., Ronai Z. (1996) Oncogene 12, 2223–2233 [PubMed] [Google Scholar]
- 11.Aucoin R., Reiland J., Roy M., Marchetti D. (2004) J. Cell. Biochem. 93, 215–223 [DOI] [PubMed] [Google Scholar]
- 12.Jean D., Harbison M., McConkey D. J., Ronai Z., Bar-Eli M. (1998) J. Biol. Chem. 273, 24884–24890 [DOI] [PubMed] [Google Scholar]
- 13.Jean D., Bar-Eli M. (2000) Mol. Cell. Biochem. 212, 19–28 [PubMed] [Google Scholar]
- 14.Shaywitz A. J., Greenberg M. E. (1999) Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 15.Mayr B., Montminy M. (2001) Nat. Rev. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 16.Iourgenko V., Zhang W., Mickanin C., Daly I., Jiang C., Hexham J. M., Orth A. P., Miraglia L., Meltzer J., Garza D., Chirn G. W., McWhinnie E., Cohen D., Skelton J., Terry R., Yu Y., Bodian D., Buxton F. P., Zhu J., Song C., Labow M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12147–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnikova V. O., Mourad-Zeidan A. A., Lev D. C., Bar-Eli M. (2006) J. Biol. Chem. 281, 2911–2922 [DOI] [PubMed] [Google Scholar]
- 18.Desdouets C., Matesic G., Molina C. A., Foulkes N. S., Sassone-Corsi P., Brechot C., Sobczak-Thepot J. (1995) Mol. Cell. Biol. 15, 3301–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White P. C., Shore A. M., Clement M., McLaren J., Soeiro I., Lam E. W., Brennan P. (2006) Oncogene 25, 2170–2180 [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie S., Price J. E., Luca M., Jean D., Ronai Z., Bar-Eli M. (1997) Oncogene 15, 2069–2075 [DOI] [PubMed] [Google Scholar]
- 22.Jean D., Bar-Eli M. (2001) Crit. Rev. Immunol. 21, 275–286 [PubMed] [Google Scholar]
- 23.Jean D., Tellez C., Huang S., Davis D. W., Bruns C. J., McConkey D. J., Hinrichs S. H., Bar-Eli M. (2000) Oncogene 19, 2721–2730 [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal S., Kim S. W., Ryu S. H., Chung W. C., Koo J. S. (2008) Cancer Res. 68, 981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brigstock D. R. (2003) J. Endocrinol. 178, 169–175 [DOI] [PubMed] [Google Scholar]
- 26.Perbal B. (2001) Mol. Pathol. 54, 103–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Du X. Y. (2007) J. Cell. Biochem. 100, 1337–1345 [DOI] [PubMed] [Google Scholar]
- 28.Leask A., Abraham D. J. (2006) J. Cell Sci. 119, 4803–4810 [DOI] [PubMed] [Google Scholar]
- 29.Li L., Price J. E., Fan D., Zhang R. D., Bucana C. D., Fidler I. J. (1989) J. Natl. Cancer Inst. 81, 1406–1412 [DOI] [PubMed] [Google Scholar]
- 30.Huang S., Jean D., Luca M., Tainsky M. A., Bar-Eli M. (1998) The EMBO J. 17, 4358–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch D. R., Bisi J. E., Miller B. E., Conaway D., Seftor E. A., Yohem K. H., Gilmore L. B., Seftor R. E., Nakajima M., Hendrix M. J. (1991) Int. J. Cancer 47, 227–237 [DOI] [PubMed] [Google Scholar]
- 32.Verschraegen C. F., Giovanella B. C., Mendoza J. T., Kozielski A. J., Stehlin J. S., Jr. (1991) Anticancer Res. 11, 529–535 [PubMed] [Google Scholar]
- 33.Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villares G. J., Zigler M., Wang H., Melnikova V. O., Wu H., Friedman R., Leslie M. C., Vivas-Mejia P. E., Lopez-Berestein G., Sood A. K., Bar-Eli M. (2008) Cancer Res. 68, 9078–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mourad-Zeidan A. A., Melnikova V. O., Wang H., Raz A., Bar-Eli M. (2008) Am. J. Pathol. 173, 1839–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gershenwald J. E., Sumner W., Calderone T., Wang Z., Huang S., Bar-Eli M. (2001) Oncogene 20, 3363–3375 [DOI] [PubMed] [Google Scholar]
- 37.Hofmann U. B., Eggert A. A., Blass K., Bröcker E. B., Becker J. C. (2003) Cancer Res. 63, 8221–8225 [PubMed] [Google Scholar]
- 38.Kurschat P., Zigrino P., Nischt R., Breitkopf K., Steurer P., Klein C. E., Krieg T., Mauch C. (1999) J. Biol. Chem. 274, 21056–21062 [DOI] [PubMed] [Google Scholar]
- 39.Holbourn K. P., Acharya K. R., Perbal B. (2008) Trends Biochem. Sci. 33, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitra A., Chakrabarti J., Chatterjee A. (2003) J. Environ. Pathol. Toxicol. Oncol. 22, 167–178 [DOI] [PubMed] [Google Scholar]
- 41.Pilarsky C. P., Schmidt U., Eissrich C., Stade J., Froschermaier S. E., Haase M., Faller G., Kirchner T. W., Wirth M. P. (1998) Prostate 36, 85–91 [DOI] [PubMed] [Google Scholar]
- 42.Tong X., Xie D., O'Kelly J., Miller C. W., Muller-Tidow C., Koeffler H. P. (2001) J. Biol. Chem. 276, 47709–47714 [DOI] [PubMed] [Google Scholar]
- 43.Chien W., Kumagai T., Miller C. W., Desmond J. C., Frank J. M., Said J. W., Koeffler H. P. (2004) J. Biol. Chem. 279, 53087–53096 [DOI] [PubMed] [Google Scholar]
- 44.Tong X., O'Kelly J., Xie D., Mori A., Lemp N., McKenna R., Miller C. W., Koeffler H. P. (2004) Oncogene 23, 4847–4855 [DOI] [PubMed] [Google Scholar]
- 45.Sampath D., Zhu Y., Winneker R. C., Zhang Z. (2001) J. Clin. Endocrinol. Metab. 86, 1707–1715 [DOI] [PubMed] [Google Scholar]
- 46.Maeta N., Osaki M., Shomori K., Inaba A., Kidani K., Ikeguchi M., Ito H. (2007) Oncology 73, 118–126 [DOI] [PubMed] [Google Scholar]
- 47.Xie D., Nakachi K., Wang H., Elashoff R., Koeffler H. P. (2001) Cancer Res. 61, 8917–8923 [PubMed] [Google Scholar]
- 48.Tsai M. S., Bogart D. F., Li P., Mehmi I., Lupu R. (2002) Oncogene 21, 964–973 [DOI] [PubMed] [Google Scholar]
- 49.Holloway S. E., Beck A. W., Girard L., Jaber M. R., Barnett C. C., Jr., Brekken R. A., Fleming J. B. (2005) J. Am. Coll. Surg. 200, 371–377 [DOI] [PubMed] [Google Scholar]
- 50.O'Kelly J., Chung A., Lemp N., Chumakova K., Yin D., Wang H. J., Said J., Gui D., Miller C. W., Karlan B. Y., Koeffler H. P. (2008) Int. J. Oncol. 33, 59–67 [PubMed] [Google Scholar]
- 51.Nguyen N., Kuliopulos A., Graham R. A., Covic L. (2006) Cancer Res. 66, 2658–2665 [DOI] [PubMed] [Google Scholar]
- 52.Menéndez J. A., Mehmi I., Griggs D. W., Lupu R. (2003) Endocr. Relat. Cancer 10, 141–152 [DOI] [PubMed] [Google Scholar]
- 53.Lin M. T., Zuon C. Y., Chang C. C., Chen S. T., Chen C. P., Lin B. R., Wang M. Y., Jeng Y. M., Chang K. J., Lee P. H., Chen W. J., Kuo M. L. (2005) Clin Cancer Res. 11, 5809–5820 [DOI] [PubMed] [Google Scholar]
- 54.Walker T. M., Van Ginkel P. R., Gee R. L., Ahmadi H., Subramanian L., Ksander B. R., Meisner L. F., Albert D. M., Polans A. S. (2002) Arch. Ophthalmol. 120, 1719–1725 [DOI] [PubMed] [Google Scholar]
- 55.Kunz M., Moeller S., Koczan D., Lorenz P., Wenger R. H., Glocker M. O., Thiesen H. J., Gross G., Ibrahim S. M. (2003) J. Biol. Chem. 278, 45651–45660 [DOI] [PubMed] [Google Scholar]
- 56.Tamura I., Rosenbloom J., Macarak E., Chaqour B. (2001) Am. J. Physiol. Cell Physiol. 281, C1524–C1532 [DOI] [PubMed] [Google Scholar]
- 57.Chen C. C., Mo F. E., Lau L. F. (2001) J. Biol. Chem. 276, 47329–47337 [DOI] [PubMed] [Google Scholar]
- 58.Todorovicçc V., Chen C. C., Hay N., Lau L. F. (2005) J. Cell Biol. 171, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]