Abstract
H2TF1 is a ubiquitous major histocompatibility complex (MHC) class I-specific transcription factor, which binds to the palindrome κB enhancer site upstream of MHC class I genes. Here we report that H2TF1 consists of a polypeptide with relative molecular mass 110,000, that corresponds to the predicted 100-kDa product (NF-κB2 p100) encoded by the candidate proto-oncogene nfκb2 (lyt-10). H2TF1 was purified by a novel affinity chromatography method and identified as the NF-κB2 p100 polypeptide by peptide sequencing as well as by reactivity with a specific antiserum. Purified H2TF1 binds the MHC κB site with high affinity (KD = 3 × 10−11m), in contrast with previous reports that NF-κB2 p100 did not bind DNA.
The transcription factor H2TF1 has been implicated in the regulation of major histocompatibility complex (MHC)1 class I gene expression (Baldwin and Sharp, 1987). Down-regulation of H2TF1 is correlated with the down-regulation of MHC class I gene expression in the development of certain malignancies (Lenardo et al., 1989; Bernards, 1991). MHC class I proteins function in the presentation of peptide antigens to T cells and are of primary importance for cytotoxic T cell lysis of allogeneic, neoplastic, and virally infected cells. MHC class I expression is down-regulated in disease processes such as viral infection with adenovirus 5 or human immunodeficiency virus-1 (Schrier et al., 1983; Scheppler et al., 1989) and malignancies including neuroblastoma, Burkitt’s lymphoma, small cell lung carcinoma, melanoma, breast carcinoma, embryonal cell carcinoma, and AKR murine leukemia (reviewed by Bernards (1987) and Tenaka et al. (1988)), potentially allowing virally infected or malignant cells to escape cytotoxic T cell lysis.
At least three cis-acting sequences located in the 200-bp region upstream of the initiation site of MHC class I genes have been implicated in the regulation of transcription (reviewed by Singer and Maguire (1990) and David-Watine et al. (1990)). Of particular importance is the palindromic MHC κB enhancer site located at −166 bp relative to the transcription initiation site. This site is an important cis-acting component in both basal and inducible MHC class I expression (Kimura et al., 1986; Israёl et al., 1989). The factors H2TF1, NF-κB (NF-κB1 p50·RelA heterodimer), and KBF1 (NF-κB1 p50 homodimer) all bind to the MHC κB site (Baldwin and Sharp, 1987, 1988, Israёl et al., 1987). NF-κB and KBF1 are members of the Rel family of transcription factors, which have homologous NH2-terminal Rel domains and bind DNA as either homodimers or heterodimers (Kieran et al., 1990; Ghosh et al., 1990; Bours et al., 1990; Meyer et al., 1991; Nolan et al., 1991; reviewed by Blank et al., 1992). The Rel domain contains a DNA binding activity which recognizes a set of binding sites related to the canonical immunoglobulin κB site (Sen and Baltimore, 1986) and a dimerization domain which allows combinatorial dimerization possibilities among the members of the family (reviewed by Blank et al. (1992)). Other members of the human Rel family include Rel (85 kDa; previously named c-Rel) (reviewed by Gilmore (1991)), RelA (65 kDa; previously named p65 NF-κB) (Nolan et al., 1991), RelB (65 kDa) (Ryseck et al., 1992; Ruben et al., 1991), NF-κB2 p100 (Schmid et al., 1991; Neri et al., 1991; Bours et al., 1992), and an alternative splice product related to NF-κB2 p100 called NF-κB p49 (Schmid et al., 1991). Proteolytic processing of NF-κB2 p100 expressed in transfected cells is suspected to be responsible for the presence of a distinct ≈ 52 - kDa species, called NF-κB2 p52, consisting of the NH2-terminal half of p100 (see Fig. 7B). NF-κB2 p49 binds to the MHC κB site with high affinity (Duckett et al., 1993) and p52 is believed to have a similar high binding affinity.
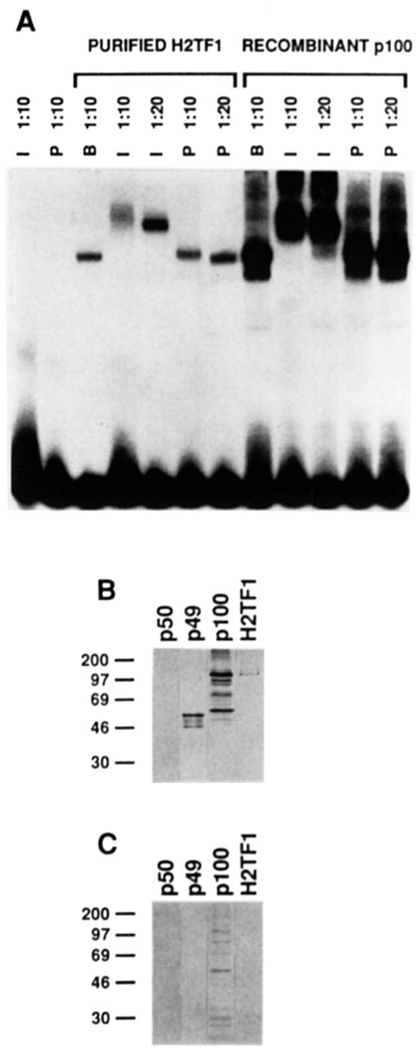
FIG. 7. H2TF1 is recognized by antiserum raised against an 18-amino acid peptide from the NH2 terminus of NF-κB2 p100.
A, supershift of H2TF1 and recombinant NF-κB2 pl00 complexes by an antiserum specific for the NH2-terminal peptide of the NF-κB2 pl00 protein. The gel shift. reactions were assembled and electrophoresed as described under “Materials and Methods.” The H2TF1 tested (Purified H2TF1) was purified by three affinity steps (SDS-PAGE shown in Fig. 2B). The recombinant NF-κB2 pl00 tested (Recombinant p100) was overproduced in 293 cells (immunoblot shown in B and C, lane p100). Both immune (I) and preimmune (P) sera were tested at either 1:10 or 1:20 dilution, as indicated, and antiserum dilution buffer (B) was tested at a 1:l0 dilution. Control reactions with antisera alone are also shown. B, immunoblot of H2TF1 and recombinant Rel family proteins developed with the immune antiserum used in A. The immunoblot procedure was performed as described under “Materials and Methods.” Equivalent amounts (10 ng, estimated by DNA-binding activity) of human NF-κB1 p50, NF-κB2 p49, and NF-κB2 pl00 were tested in lanes p50, p49, and p100, respectively. An equivalent amount of H2TF1 (10 ng, estimated by DNA-binding activity), purified by two affinity steps, was tested in lane H2TF1. C, immunoblot as in B, but the immune antiserum was blocked with the NF-κB2 pl00 NH2-terminal peptide to which it was raised (at 2 mg/ml; sequence presented under “Materials and Methods”) prior to development of the blot.
The importance of Rel family members in basal and inducible MHC class I expression has been tested by expression of a transdominant negative mutant of NF-κB p50, Δsp, which is capable of dimerizing with Rel family members, but incapable of binding DNA (Logeat et al., 1991). This mutant would be expected to inhibit the DNA binding of KBF1, NF-κB, and other Rel family members, by dimerizing with their component subunits. In transfection experiments both basal and cytokine (TNF-α and IL-1)-induced MHC class I expression were inhibited, suggesting that Rel family members are important regulators of MHC class I expression.
H2TF1 was first defined as a factor distinct from NF-κB and KBF1 which binds to the palindromic MHC κB site at −166 bp (Baldwin and Sharp, 1987, 1988). H2TF1 is found ubiquitously in cytoplasmic and nuclear extracts from a wide variety of cell types. The KBF1 factor is similarly widely expressed, but the KBF1·DNA complex has significantly faster mobility on native gel electrophoresis than the NF-κB. DNA complex, which in turn has significantly faster mobility than the H2TF1·DNA complex. The DNA binding specificities of the NF-κB, KBF1, and H2TF1 factors are also distinct. While NF-κB and KBF1 bind to the MHC κB site and the related immunoglobulin κB site with similar high affinities (Baldwin and Sharp, 1987; Fujita et al., 1992), H2TF1 binds the MHC κB site with high affinity, but not the immunoglobulin κB site (Baldwin and Sharp, 1988). Furthermore, methylation interference patterns for the binding of H2TF1 and NF-κB to the MHC κB site are distinctly different (Baldwin and Sharp, 1988).
Several lines of evidence implicate H2TF1 in transcriptional regulation of MHC class I genes. Co-transfection of competitor oligonucleotides corresponding to the MHC κB site resulted in decreased expression of a chloramphenicol acetyltransferase reporter plasmid driven by the MHC class I enhancer while co-transfection of oligonucleotides corresponding to the canonical immunoglobulin κB site did not (Israёl et al., 1989). This result suggested that a factor regulating MHC class I genes had the specificity of H2TF1, rather than that of NF-κB or KBF1. Furthermore, a factor with the DNA binding specificity of H2TF1, rather than NF-κB or KBF1, was found to be diminished in neuroblastoma cells with down-regulated MHC class I expression (Lenardo et al., 1989; Bernards, 1991). A similar correlation has also been observed in the down-regulation of MHC class I protein in AKR leukemia (Henseling et al., 1990) and HLA class I expression in a number of human tumor cell lines (Blanchet et al., 1992).
In the studies reported here, H2TF1 was determined to be a protein of relative molecular weight 110,000 that was able to bind the MHC class I site with high affinity in the absence of other polypeptides. H2TF1 was identified by protein sequencing and immunologic methods as being identical to NF-κB2 p100, a member of the Rel family of factors. NF-κB2 p100 was previously thought to have little or no DNA binding activity until proteolytically processed (Neri et al., 1991; Bours et al., 1992). NF-κB2 p100 has been determined to be the gene product of the candidate proto-oncogene nfkb2 (lyt-10), which is disrupted by gene rearrangements in some human lymphomas (Neri et al., 1991).
MATERIALS AND METHODS
Cell Lines
Washed and frozen HeLa cells, a gift of Ajinomoto Corp., were stored at −135 °C prior to use. The 293 adenovirus 5-transformed primary human embryonal kidney cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum.
Probe and Competitor DNA
Probe and competitor DNA sequences were derived from those used by Baldwin and Sharp (1987). The oligonucleotide GGCTGGGGATTCCCCATCT, containing the MHC κB site, was cloned into the BamHI site of the pUC13 plasmid polylinker. The 68-bp EcoRI-HindIII restriction digest fragment of the pUC13 polylinker containing the MHC κB site, used as a probe and competitor DNA fragment, was designated MHC WT. The oligonucleotide GGCTGCGGATTCCCGATCT, containing a double point mutation of the MHC κB sequence, was cloned into the BamHI site of the pUC13 polylinker. The 68-bp EcoRI-HindIII restriction digest fragment of the pUC13 polylinker containing the mutant MHC κB site, used as a probe and competitor DNA fragment, was designated MHC MT. The oligo AGGGGACTTTCCG, containing the murine immunoglobulin κB site was cloned in the KpnI site of the pUC18 plasmid polylinker. The 72-bp EcoRI-HindIII restriction digest fragment of the pUC18 polylinker containing the immunoglobulin κB site, used as a probe and competitor fragment, was designated NF-κB. The 47-bp BamHI-XhoI restriction digest fragment of the Bluescript IIKS plasmid polylinker (Stratagene), used as an unrelated competitor DNA fragment, was designated BSCRIPT. Unlabeled competitor DNA was isolated either directly from plasmid DNA, or from DNA generated by polymerase chain reaction from these plasmids, by digestion with the appropriate restriction endonucleases. Probe DNA fragments were labeled with [α-32P]dATP by filling in 5′ overhangs with the Klenow fragment of DNA polymerase I.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) conditions were similar to those of Baldwin and Sharp (1987), with some modifications. Binding reactions were done in the presence of 20 mm HEPES KOH (pH 7.5), 10% glycerol, 0.3 mg/ml bovine serum albumin, 50–300 µg/ml poly(dI·dC):poly(dI·dC), 5 fmol of 32P-end-labeled probe (sequence indicated above; specific activity = 4 × 104 cpm/fmol). In some cases, competitor DNA species (indicated below) were added following the probe DNA (fold-excess indicated in the figure legends). The assay sample was added to the binding reaction following the above components. To certain binding reactions sodium deoxycholate was subsequently added to a final concentration of 0.8%, followed by Nonidet P-40 added to a final concentration of 1% (Baeuerle and Baltimore, 1988). The final salt concentration was 60 mm KCl, which was optimal for H2TF1 binding (data not shown). The final volume of each reaction was 10 µl. The reactions were incubated for 15 min at 30 °C and loaded on low salt Tris-HCl/sodium acetate/EDTA (TAE) gels as described by Chodosh (1991). Dried gels were scanned with a Molecular Dynamics PhosphorImager, using ImageQuant(™) software, to quantitate bound and free probe radioactivity. A defined amount of the probe, of known specific activity, was spotted on the gel. The spotted probe served as an internal calibration to correlate integration measurements with fmol of probe. It was thus possible to determine the fmol of bound and free probe in each experiment. Since the free probe was in excess, the amount of shifted (protein-bound) probe was defined as the amount of EMSA DNA-binding activity in the sample. H2TF1 activity was defined as 2 × pmol of shifted MHC κB probe comigrating with a complex having the expected DNA-binding specificity of H2TF1, assuming dimer binding, as is the case for other Rel family members.
Purification of H2TF1
Cytoplasmic S100 and nuclear extracts were prepared from 180 g of HeLa cells essentially as described by Dignam et al. (1983). All chromatography and desalting procedures were done at 4 °C and fractions were stored at −80 °C. The S100 extract was adjusted to 0.42 m NaCl. To remove extraneous nucleic acids, the S100 extract was chromatographed on a 70-ml DEAE-Sephacel (Pharmacia LKB Biotechnology Inc.) column under isocratic conditions in buffer B (0.42 m NaCl,20 mm HEPES KOH (pH 7.9), 20% glycerol, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 1 µm leupeptin, 1 µm pepstatin A, 1 µm chymostatin, and 1 µm antipain). About 1.5 g of protein was chromatographed per column run. The eluted fractions were assayed for H2TF1 activity with the MHC κB site probe. Fractions containing H2TF1 activity were pooled, dialyzed to a 50 mm NaCl concentration, and loaded on a 90-ml heparin-Sepharose (Pharmacia) column at about 15 mg of protein/ml of column matrix, or about 1.4 g of protein/column run. The column was washed with buffer BC 0.05 (50 mm KCl, 20 mm HEPES KOH (pH 7.9), 20% glycerol, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 1 µm leupeptin, 1 µm pepstatin A, 1 µm chymostatin, and 1 µm antipain). The column was developed with a linear gradient from 0.10 to 1.0 m KCl in buffer BC. Representative samples of each fraction were quantitatively assayed by gel shift for H2TF1 activity. H2TF1 DNA-binding specificity of the H2TF1 activity was confirmed by competition assays, with competitor DNA fragments MHC WT, MHC MT, κB, and BSCRIPT. Complexes containing H2TF1 were competed only by MHC WT (data not shown). Fractions containing H2TF1 binding activity were pooled and desalted by dialysis versus 2 changes of 300 volumes of buffer BC containing no KCl, to a final salt concentration of 50 mm, as measured by conductivity.
DNA Affinity Chromatography
Affinity columns of the MHC κB site were made by the method of Larson and Verdine (1992). Columns were synthesized with either monomers of the MHC KB site oligonucleotide or ligated multimers of the oligonucleotide. Both types of column were effective in the purification of H2TF1. These columns had about 200 nmol of binding site/ml of resin and had a bed volume of 1.0 ml. Pooled, desalted eluate containing H2TF1, but depleted of NF-κB, from the heparin-Sepharose column was made 0.5–1.0 mg/ml in salmon sperm DNA and loaded on an MHC κB site column. The column was eluted with a linear gradient from 70 to 400 mm KCl in buffer Y (20 mm HEPES KOH (pH 7.8), 10% glycerol, 1 mm DTT). Fractions containing H2TF1 activity were pooled and desalted by gel filtration on Sephadex PD10 columns. The desalted material was chromatographed a second time on an MHC κB site column under identical conditions. Of note, a single loading of the second affinity step was found to give a higher yield of H2TF1 than reloading the flow-through twice. Optimal yields were obtained with the second affinity step when less than 8 pmol of H2TF1 (by gel shift assay) was loaded per column run. The eluted material from 12 column runs was pooled, and was estimated to contain about 40 pmol of H2TF1 by gel shift assay, assuming dimer binding as is the case for other Rel family members. The amount of double affinity purified H2TF1 was independently estimated to be 80 pmol by densitometry of silver-stained H2TF1 electrophoresed on an SDS-polyacrylamide gel, compared with a purified NF-κB1 p105 standard (gift of Takashi Fujita and David Baltimore).
UV Cross-linking of H2TF1 to the MHC κB Site
H2TF1, purified by three affinity steps (see Fig. 2B), was bound and UV cross-linked to a bromodeoxyuridine (BrdUrd)-substituted probes synthesized as in the legend of Fig. 3. The BrdUrd-substituted probe formed a specific gel shift complex with mobility and competition specificity identical to unsubstituted MHC WT DNA (data not shown). Specific competitor DNA species were all enzymatically synthesized by polymerase chain reaction, because they competed with probe more effectively per fmol than synthetic oligonucleotide competitors. Binding reactions were incubated under standard conditions with 2 fmol of probe at 30 °C for 15 min and subsequently irradiated for 15 min at 30 °C with a UV Products (San Gabriel, CA) UVM-57 Chromato-VUE® midrange wavelength lamp, which had a spectrum centered at 302 nm. The lamp filter was at a distance of 4 cm above the reaction liquid, which was at the bottom of an open 1.5-ml microcentrifuge tube, Following irradiation, CaCl2 was added to a concentration of 8 mm. The reaction was then incubated in the presence of 10 units of micrococcal nuclease for 6 min at 37 °C, made 1 × in SDS-PAGE loading buffer, boiled for 3 min, and subjected to SDS-PAGE on a 10% polyacrylamide gel.
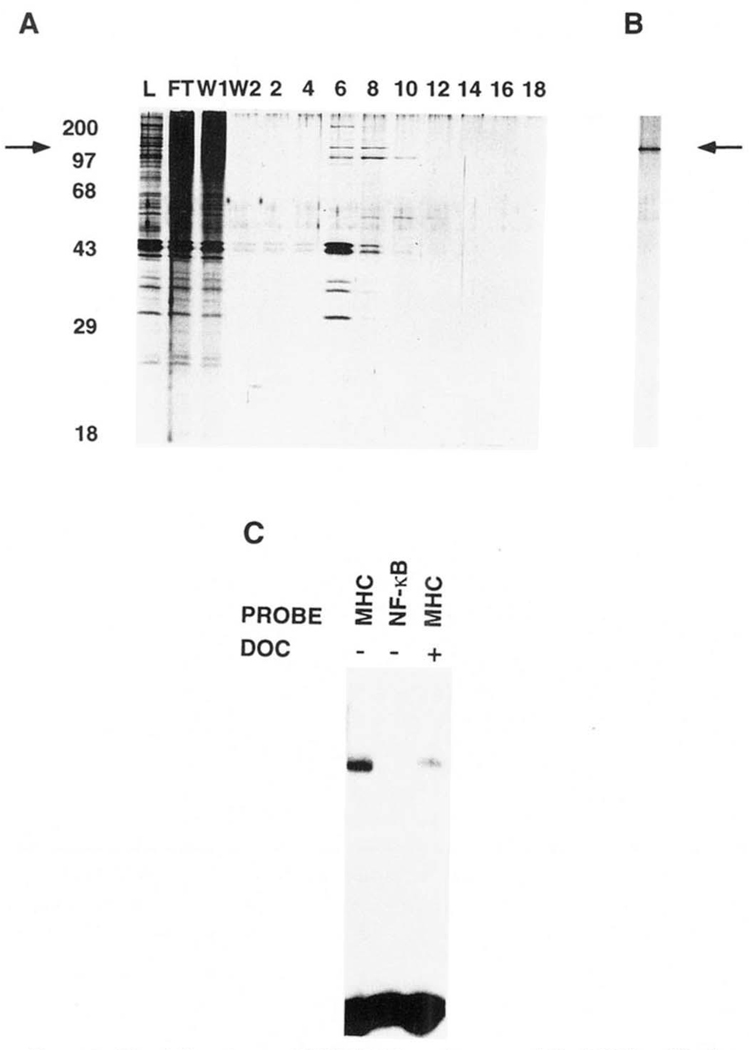
FIG. 2. Purification of H2TF1 by site-specific DNA affinity chromatography.
H2TF1-containing fractions eluted from the heparin-Sepharose column were pooled and desalted on Sephadex PD-10 columns to a final salt concentration of 50 mm KCl. Highly purified sonicated salmon sperm DNA was added to the desalted pool, to a final concentration of 500 mg/ml. The pool was then loaded on a 1.0-ml MHC κB site column, constructed as described under “Materials and Methods.” In some cases the loading was done more than once, but multiple loadings were later shown to be unnecessary for efficient binding (defined as >50% of loaded activity bound, data not shown). The loaded column was washed with 100 mm KCl and then developed with a 100–500 mm KCl gradient, all in buffer Y. The fractions containing H2TF1 activity were then pooled and desalted on Sephadex PD-10 gel filtration columns to a final salt concentration of 50 mm KCl. Sonicated salmon sperm DNA was again added to the pool, to a final concentration of 500 mg/ml. Chromatographic separation on the MHC κB site column was then repeated as above. A, SDS-PAGE of proteins eluted by a linear KCl gradient on the second round of MHC κB site affinity chromatography. The gel was run by the method of Laemmli (1970) and stained with silver by the method of Blum et al. (1987). Representative samples of loaded material (lane L), flow-through (lane FT),the first half of the wash (lane W1), the second half of the wash (lane W2),and fractions eluted by the linear KCl gradient from 190 to 580 mm KCl (lanes 2–18) were electrophoresed. Molecular weight standards (Mr) were myosin (200,000), phosphorylase b (97, 400), BSA (68,000), ovalbumin (43,000), carbonic anhydrase (29,000) and β-lactoglobulin (18,400). The arrow indicates the mobility of a 110,000 Mr polypeptide which further co-purified with H2TF1 activity and cross-linked specifically to the MHC κB site. B, SDS-PAGE of the fraction containing the peak H2TF1 activity, eluted from the third round of MHC κB site affinity chromatography. The protein species indicated by the arrow at 110,000 Mr co-eluted with the H2TF1 activity. The molecular weight standards for this gel were essentially the same as for A. C, gel shift assay of the fraction from the third affinity purification with peak H2TF1 activity, shown in B. The probes are as indicated. DOC +/− indicates the presence or absence of deoxycholate. The complex observed with the MHC WT probe was competed by a 40-fold excess of unlabeled MHC WT competitor, but not MHC MT, κB, or BSCRIPT (data not shown).
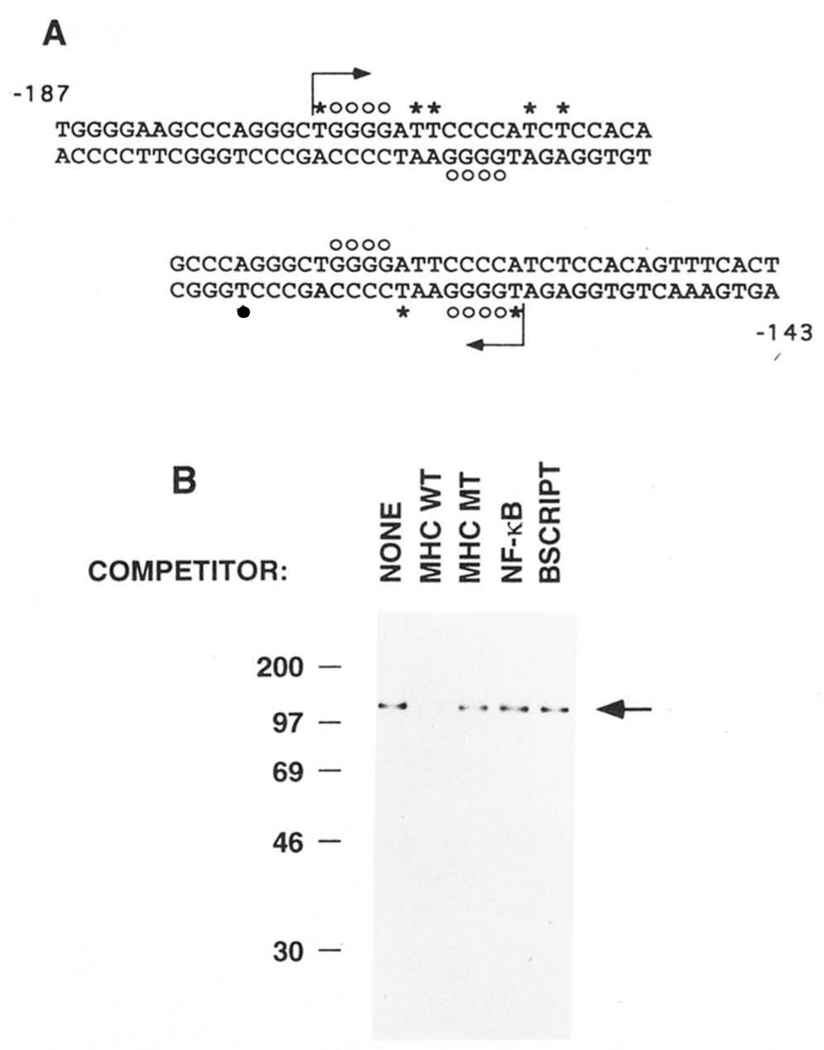
FIG. 3. UV cross-linking of purified H2TF1 to the MHC class I site.
H2TF1, purified by three affinity steps (SDS-PAGE shown in Fig. 2B), was cross-linked by UV irradiation to MHC κB site probes substituted with BrdUrd and 5′ 32P-dC as indicated below. Competitor DNA species, where indicated, were present in 60-fold excess. The binding reactions were incubated and cross-linked as described under “Materials and Methods.” A, DNA sequences of the MHC κB site probes used in the UV cross-linking experiments. The sequences span from −187 to −143 bp upstream from the MHC H2-Kb transcription start site and include the MHC κB site centered at −166 bp. The probe beginning at −187 bp was designated “−187” and the probe ending at −143 bp was designated “−143.” The −187 probe was made by hybridizing a 16-base oligonucleotide primer to the 37-base oligonucleotide bottom strand and then filling in the single stranded gap by synthesis with DNA polymerase I Klenow fragment, as indicated by the starting position and direction of the arrow, allowing substitution of the probe with BrdUrd and 32P-dC. The −143 probe was similarly made. The asterisks mark the dT nucleotides replaced with BrdUrd. The dC nucleotides added in the direction of the arrow were labeled with 32P on their 5′ side. The open circles mark the dG nucleotides which, when methylated, interfere with the binding of H2TF1 (Baldwin and Sharp, 1988). B, SDS-PAGE of the complexes obtained by UV cross-linking purified H2TF1 (Fig. 2B) to the −143 probe. The cross-linking reactions were done in the absence and presence of the indicated competitor DNA species (see “Materials and Methods” for sequences). The arrow indicates the position of a protein of 110,000 Mr, specifically cross-linking to the −143 probe, competed by wild type (MHC WT) competitor, but not by double point mutant (MHC MT), NF-κB site (NF-κB), or Bluescript poly-linker (BSCRIPT) competitors. A similar result was obtained with the −187 probe and H2TF1 purified by two affinity steps (data not shown). The molecular weight standards, labeled with 14C methylation (Amersham C626), were myosin (200,000), phosphorylase b (97,400), BSA (69,000), ovalbumin (46,000), and carbonic anhydrase (30,000).
Renaturation of H2TF1 from an SDS-PAGE Gel
The renaturation was done according to a protocol modified from those of Baeuerle and Baltimore (1988) and Hager and Burgess (1980). A 9% SDS-polyacrylamide gel was loaded with 1.5 pmol of H2TF1 purified by two affinity steps (essentially as the protein loaded on Fig. 2A, lanes 6, 8, and 10). Molecular weight standards were loaded on both sides of the H2TF1 lane. Following electrophoresis, the H2TF1 lane was cut into 15 horizontal sections spanning between 200,000 and 30,000 Mr. The sections were cut with marks extending to the lanes containing the molecular weight markers. The lanes containing the molecular weight markers were stained with silver, allowing assignment of a range of molecular weight to each section. Each gel section was shredded and eluted overnight in 400 ml of buffer EB (50 mm Tris-HCl (pH 7.9), 0.1% SDS, 0.1 mg/ml BSA, 0.2 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 2.5% glycerol) at 4 °C. The eluate was centrifuged to remove gel fragments. The protein in the eluates was precipitated with 4 volumes of acetone at −20 °C in siliconized microcentrifuge tubes. The tubes were centrifuged at 14,000 × g for 30 min. The supernatant was removed by pipetting. The sample was washed with methanol and again microfuged as above. The supernatant was again removed by pipetting. The pellets were immediately dissolved in 6 m guanidine HCl in buffer DB (25 mm HEPES KOH (pH 7.7), 25 rnm KCI, 2 mm DTT), and allowed to sit for 1 h at 4 °C. The dissolved samples were dialyzed in a Pierce microdialyzer at 4 °C against 5 changes of buffer DB, each lasting 35 min. The final volume of the sample corresponding to each gel section was 50 µl. Glycerol was added to each sample to 9% and they were then stored at −80 °C. In each EMSA assay, an 8-µl sample was assayed, or about 15% of the material obtained from each gel section. The EMSA conditions were essentially as described above, except that the reaction volumes were 15 µl.
Protein Sequencing of Purified H2TF1
The pooled H2TF1, twice purified by DNA affinity chromatography on an MHC κB site column was precipitated by addition of one-sixth volume of 90% trichloroacetic acid and centrifuged in an SW41 rotor (Beckman) at 40,000 rpm for 3 h at 4 °C. A pellet was obtained and held at 4 °C. The supernatant was recentrifuged for 3 h and an additional, smaller pellet was obtained, and was similarly processed and pooled. The pellets were suspended by gentle vortexing in cold (−20 °C) acetone and then centrifuged in a siliconized microcentrifuge tube in an Eppendorf Microfuge at 14,000 × g for 30 min. The pellet was washed twice in cold acetone, which removed the trichloroacetic acid. The pellet was taken up in 1 × SDS-PAGE loading buffer (60 mm Tris-HCl (pH 6.8), 1% SDS, 10% glycerol, 140 mm β-mercaptoethanol, and 0.005% bromphenol blue), boiled for 3 min, and subjected to SDS-PAGE on a single lane of a 9% polyacrylamide gel of 1-mm thickness. Following electrophoresis, the gel was equilibrated in transfer buffer (25 mm Tris base, 172 mm glycine, 20% methanol, 0.01% SDS) and transferred in the same buffer to a 0.2-µm nitrocellulose filter (Schleicher & Schuell BA83) in a Bio-Rad Trans-Blot cell for 16 h at 4 °C at a voltage of 50 V. The filter was stained with Ponceau red (0.5%) in 10% acetic acid The expected band ran as a doublet at 110,000 and 105,000 Mr. Both bands were cut out and saved.
Internal sequence analysis of H2TF1 was obtained according to the methods of Tempst et al. (1990). Both the 110,000 and 105,000 Mr bands were digested with trypsin and the proteolytic products were eluted. The peptides were chromatographed by HPLC on a C18 silica column spectroscopically monitored for 277- and 210-nm absorption. The fractions corresponding to the 277-nm absorption peaks were collected and stored at −20 °C. A control consisting of a nonstaining piece of the nitrocellulose filter was also digested with trypsin and the resulting peptides were separated by HPLC to determine which absorption peaks corresponded to trypsin tryptic peptides. Following subtraction of the trypsin peptide peaks, the peptide maps of the 110,000 and 105,000 Mr proteins matched. Three matching nontrypsin peptide peaks were pooled and sequenced. Three peptides were sequenced by Edman stepwise degradation on an Applied Bio-systems model 477/A pulse liquid protein sequenator.
Generation of a NF-κB pl00-specific Antiserum
A rabbit antiserum was raised against a synthetic peptide, MESCYNPGLD-GIIEYDDFC, corresponding to the NH2-terminal 18-amino acid sequence of NF-κB2 p100, with an additional COOH-terminal cysteine residue added for purification purposes. The peptide was purified by HPLC and the molecular weight confirmed by mass spectroscopy, prior to generation of the antiserum. The antiserum was useful in distinguishing p100-related polypeptides from those related to NF-κB1 p105, because the NH2-terminal peptide sequence to which it was raised is p100-specific.
Preparation of Cell Extracts Containing the NF-κB2 P100, NF-κB2 p49, and NF-κB1 p50 Proteins
293 human embryonal kidney cells (see above) were transfected with eukaryotic expression vectors encoding NF-κB2 pl00(human). The cells were lysed in a buffer containing 1% Nonidet P-40. The NF-κB2 p49 (human) was expressed in Escherichia coli and prepared as described earlier (Schmid et al., 1991). Purified NF-κB1 p50 (human, greater than 90% pure), prepared from an E. coli overexpression system, was a gift of Kenneth LeClair.
Supershift of the H2TF1 and Recombinant NF-κB2 pl00 Protein-DNA Complexes
Gel shift protein-DNA binding reactions were assembled as above. Polyclonal antiserum raised to the NH2-terminal peptide of NF-κB2 pl00 was diluted in a buffer containing 150 mm KCl, 20 mm HEPES KOH (pH 7.8), and 1% BSA. Preimmune serum was similarly prepared. The final KCl concentration in the reaction mixture after the addition of antibody was 50 mm. H2TF1 purified to apparent homogeneity or recombinant NF-κB2 pl00 expressed in a cellular extract from 293 cells was then added to the reaction mixture. The reactions were incubated under standard conditions, at 30 °C for 15 min, loaded on a nondenaturing 4% polyacrylamide gel, and electrophoresed as described above.
Immunoblot Procedure
H2TF1 purified by two affinity step was acetone precipitated and then solubilized in SDS-PAGE loading buffer. The amount of H2TF1 protein loaded was estimated to be 10 ng by quantitative gel shift. Equivalent amounts of the NF-κB2 p100, NF-κB1 p50, and NF-κB2 p49 proteins, as determined by EMSA activity, were loaded as controls. Following SDS-PAGE, the gel was equilibrated in transfer buffer (as above) and proteins were electro-phoretically transferred to a 0.2-µm nitrocellulose filter (Schleicher & Schuell BA83) in a Bio-Rad Mini Trans-Blot electrophoretic transfer cell for 2 h at 4 °C. Following transfer, the blot was blocked with 5% Blotto (5% dry milk, 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 1 mm EDTA, and 1 mm DTT) overnight. The next day, the blot was rinsed with TBST (150 mm NaCl, 10 mm Tris-HCl (pH 8), and 0.05% Tween 20) for 10 s. Following the rinse, the primary antibody or preimmune serum, at a dilution of 1:250, was bound for 30 min at room temperature. Following the binding, the blot was washed with TBST containing 0.2% Tween 20. The blot was then incubated for 30 min with the secondary antibody, goat anti-rabbit IgG-alkaline phosphatase (Promega), at a dilution of 1:7500 in TBST. The blot was subsequently washed three times with TBST containing 0.2% Tween 20. The blot was developed for 30 s with a mixture of nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Pro-mega) in alkaline phosphatase buffer (100 mm NaCl, 100 mm Tris-HCl (pH 9.5), 15 mm MgCl2). To end development the blot was washed with two changes of deionized H2O.
RESULTS
H2TF1 Is Found Predominantly in HeLa Cytoplasmic Extracts and Is Separable from NF-κB by Heparin Chromtography
Previous studies have demonstrated the presence of H2TF1 activity in whole cell extracts of a wide variety of cell types including BALB/c 3T3 cells, murine erythroleukemia cells, and HeLa cells (Baldwin and Sharp, 1987). HeLa cell S100 cytoplasmic and nuclear extracts were tested for H2TF1 DNA-binding activity, defined by a complex with slower mobility than that of the NF-κB complex and with a binding preference for the MHC κB site as compared to the canonical immunoglobulin κB site (Baldwin and Sharp, 1988). Such a complex was identified in both Sl00 cytoplasmic and nuclear extracts. Approximately 90% of the H2TF1 activity within the HeLa cell was present in the former (data not shown). S100 cytoplasmic extract of HeLa cells was therefore used as the source for the purification of H2TF1.
The H2TF1 activity could be separated from the NF-κB activities by heparin-Sepharose chromatography (Fig. 1). Elution of the heparin-Sepharose column with a linear KCl gradient yielded H2TF1 in the 230–310 mm KCl fractions (Fig. lB), while the majority of the NF-κB binding activity, defined by activation by the ionic detergent sodium deoxycholate (Baeuerle and Baltimore, 1988) and a distinct DNA binding specificity, eluted between either 120 and 220 or 350 and 450 mm KCl (Fig. 1A). The basis of the separation of these two NF-κB activities is unknown. The H2TF1 activity was also distinguishable from the NF-κB activity in that it was inhibited by detergent treatment with sodium deoxycholate. The heparin-Sepharose chromatography step resulted in a 4-fold purification of H2TF1 with 60% recovery (Table I) and separated this factor from other proteins which bind the MHC KB site, allowing application of DNA affinity chromatography in the next step.
FIG. 1. Fractionation of HeLa S100 extract by heparin-Sepharose chromatography.
HeLa S100 cytoplasmic extract protein, prepared as described under “Materials and Methods,” was separated from extraneous nucleic acid species by DEAE-Sephacel chromatography. The eluted protein was then dialyzed and loaded onto a heparin-Sepharose column at 50 mm NaCl. The heparin-Separose column was washed with 50 mm KCl and then developed with a linear gradient between 100 and 600 mm KCl in buffer BC. A, gel shift assay of the heparin column fractions for NF-κB. done in the presence of deoxycholate, which eliminates the H2TF1 protein. DNA complex (MHC WT probe). B, gel shift assay of the heparin column fractions for H2TF1. done in the absence of deoxycholate (MHC WT probe). The arrow indicates the complex with H2TF1 DNA-binding specificity (data not shown).
Table I.
Purification of H2TF1
| Fraction | Total proteina | H2TF1b | Specific activity | Purification | Yield |
|---|---|---|---|---|---|
| mg | pmol | pmol/mg | -fold | % | |
| I. HeLa S100 extractc | 4500 | 800 | 0.18 | – | – |
| II. DEAE/heparin | 720 | 500 | 0.69 | 4 | 60 |
| III. MHC κB affinity I | 6.5 | 140 | 22 | 130 | 18 |
| IV. MHC κB site affinity II | 0.067 | 40 | 600 | 3300 | 5 |
I–II, protein determined by Bradford assay; III–IV, protein determined by micro-Bradford assay or SDS-PAGE and comparison with standards.
I–IV, H2TF1 determined by EMSA; SDS-PAGE analysis with standards suggested that the amount of H2TF1 was 1.5–2-fold underestimated by EMSA in step IV.
From 180 g of HeLa cells.
H2TF1 Was Purified to Apparent Homogeneity Using an Improved DNA Affinity Methodology
Attempts to further purify H2TF1 by conventional DNA affinity chromatography methods from enriched heparin-Sepharose fractions were unsuccessful. Most of the H2TF1 activity failed to bind a conventional ligated MHC κB site oligonucleotide matrix (Kadonaga and Tjian, 1986) during the loading process. A small amount of H2TF1 was eluted by a linear KCl gradient between 100 and 130 mm KCl, a much lower salt concentration than expected for most specifically bound proteins. The concentration of MHC κB site bound to the CNBr-Sepharose matrix was 2.5 nmol/ml resin, about average for this methodology. To determine if the purification problem was due to limiting capacity of the conventional DNA affinity matrix, an 80-fold higher capacity matrix, having 200 nmol of the MHC κB site oligonucleotide/ml, was made by a novel method (Larson and Verdine, 1992) and tested. The increased capacity per volume necessitated the use of higher competitor DNA concentrations, in the range of 0.5–1.0 mg/ml. This condition allowed effective purification of H2TF1 from the relevant heparin-Sepharose column fractions. On the first passage on the high capacity matrix, H2TF1 activity eluted between 200 and 300 mm KCl, with a recovery of 30–60%. The purification was greater than 30-fold (Table I).
The eluate containing H2TF1 from the first affinity purification was chromatographed a second time over the same affinity column, under identical conditions. The H2TF1 activity again eluted between 200 and 300 mm KCl, with a recovery of 30–60% (Fig. 2A). The purification in the second affinity step was about 25-fold (Table I). On a third affinity step, a protein of Mr = 110,000 was the only major species detected by silver staining following electrophoresis on a denaturing gel (Fig. 2B). This protein co-eluted with H2TF1 activity. The DNA binding specificity of this highly purified H2TF1 was as expected (Fig. 2C). The DNA-binding specificity of the purified H2TF1 protein was compared with that of purified recombinant NF-κB1 p105. NF-κB1 p105 (baculovirus-expressed murine p105 purified to near homogeneity from SF9 insect cells; a gift of Takashi Fujita and David Baltimore) bound to the MHC κB site, but unlike H2TF1 this binding was effectively competed with the κB site,2 suggesting that the two proteins were different.
H2TF1 Is a 110,000 M, Polypeptide
A UV cross-linking experiment was done to determine if the 110,000 Mr polypeptide co-purifying with H2TF1 activity was the polypeptide binding the MHC κB site with H2TF1 specificity. H2TF1 was bound to a BrdUrd-substituted MHC κB site oligonucleotide (Fig. 3A) in the presence and absence of competitor DNA species and then UV irradiated. The competitor DNA species included the MHC κB site (MHC WT), a double point mutant of the MHC κB site known not to specifically bind H2TF1 (MHC MT; Baldwin and Sharp (1987)), a canonical immunoglobulin kappa κB site (NF-κB), and an unrelated Blue-script SK II polylinker fragment generated by BamHI-XhoI digestion (BSCRIPT; see “Materials and Methods” for sequences). This experiment demonstrated cross-linking of a Mr = 110,000 polypeptide which was specifically competed with the MHC κB site (Fig. 3B). Substitution and labeling of either DNA strand gave identical results (data not shown). This result suggested that the H2TF1 factor is composed of a single 110,000 Mr polypeptide.
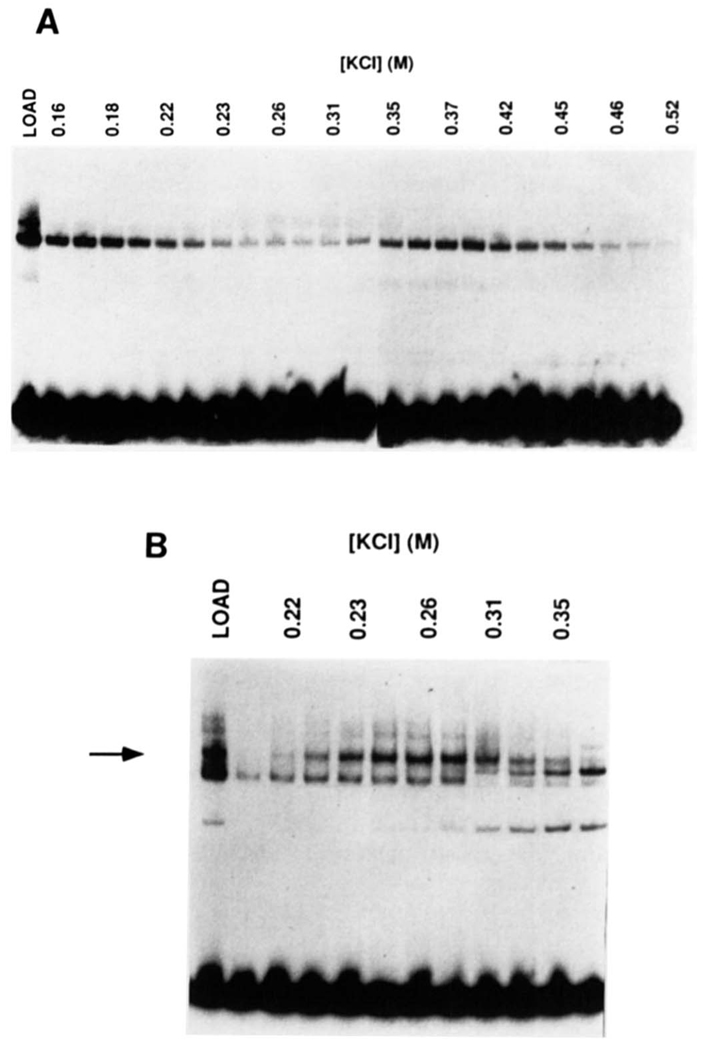
To determine whether H2TF1 DNA-binding activity could be attributed to a single polypeptide, proteins were eluted and renatured from horizontal sections of an SDS denaturing polyacrylamide gel lane representing discrete ranges of molecular weight. About 1.5 pmol of H2TF1 activity from a second affinity step was subjected to SDS-PAGE. Proteins eluted from gel sections were denatured by guanidine HCl treatment and renatured by dialysis to remove the guanidine HCl. The dialyzed fractions were assayed for MHC κB site binding activity and H2TF1 specificity (Fig. 4). The H2TF1 activity was found in a section spanning from 100 to 120 kDa, but not in surrounding sections (data not shown). These experiments confirmed that H2TF1 was approximately 110,000 molecular mass and, more importantly, that polypeptides smaller than 100 kDa were not part of the complex. Thus, the smaller members of the Rel family, including NF-κB1 p50, Rel, RelA, RelB, and NF-κB2 p49 are not necessary for the 110,000 Mr polypeptide to bind DNA and generate a complex with the expected mobility and specificity of H2TF1. To further confirm that H2TF1 did not consist of an NF-κB p50 homodimer or an NF-κB1 p105/p50 heterodimer, gel shift mobilities were determined for purified recombinant p50 and p105 (gifts of Takashi Fujita and David Baltimore) and for both proteins denatured and renatured together as a p105/p50 heterodimer. The p105 complex exactly comigrated with the H2TF1 complex, while the p50 complex migrated considerably faster, and the p105/p50 complex migrated with an intermediate mobility (data not shown). These results indicated that H2TF1 was not a heterodimer of full-length and truncated Rel proteins.
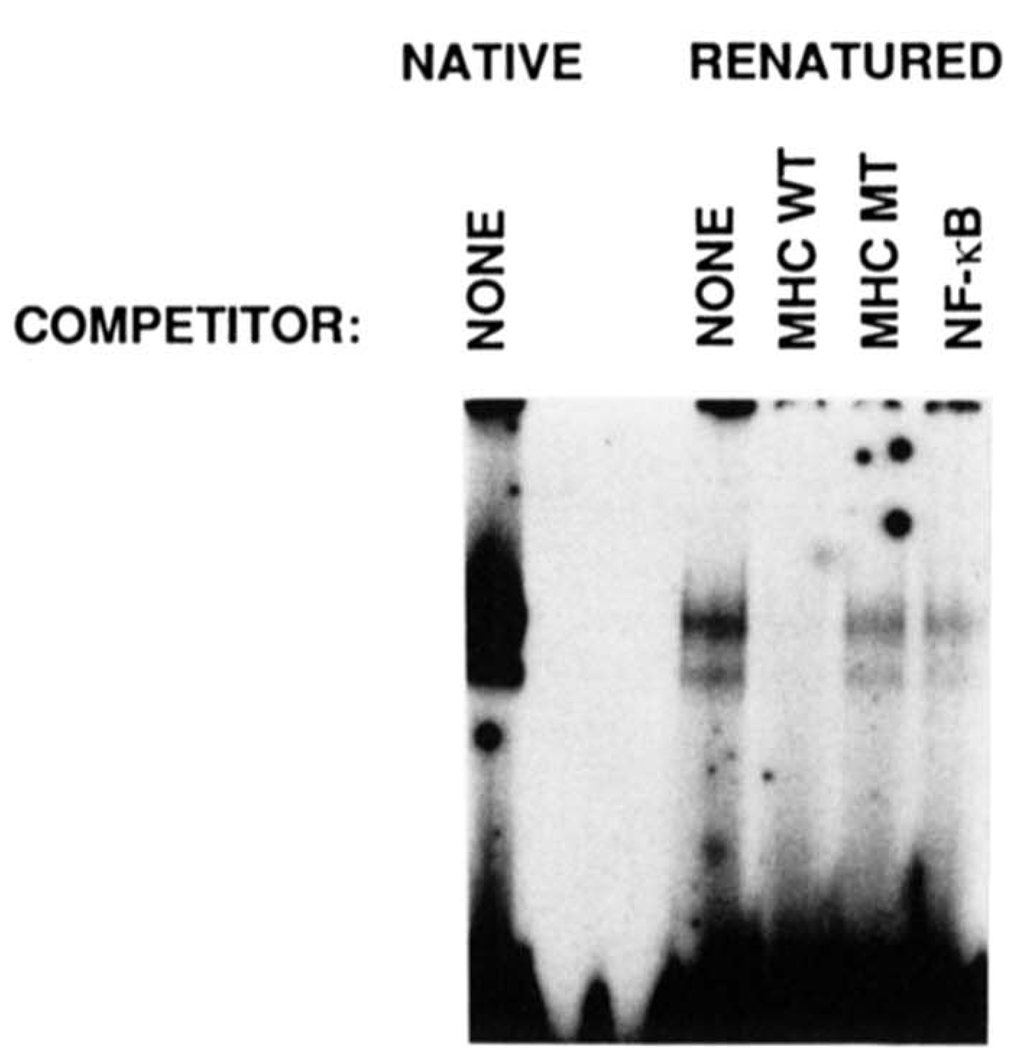
FIG. 4. Renaturation of H2TF1 sized by SDS-PAGE and demonstration of the DNA-binding specificity of the renatured protein.
H2TF1 was renatured from horizontal sections of an SDS-polyacrylamide gel according to a protocol modified from those of Baeuerle and Baltimore (1988) and Hager and Burgess (1980), described under “Materials and Methods.” Renatured H2TF1 was incubated with wild type MHC κB probe in the presence or absence of competitor DNA and assayed for DNA-binding by gel shift analysis. The mobility of the renatured protein. DNA complex (RENATURED) was identical to the starting material (NATIVE). Either wild type MHC κB site (MHC WT), double point mutant MHC κB site (MHC MT), NF-κB site(NF-κB), or no (None) competitor was added to the gel shift assays, and the result demonstrated that the renatured H2TF1 had the same DNA-binding specificity as the native material. The competitor DNA species were added at a 50-fold excess over the probe.
Determination of the Equilibrium Binding Affinity of H2TF1 for the MHC κB Site
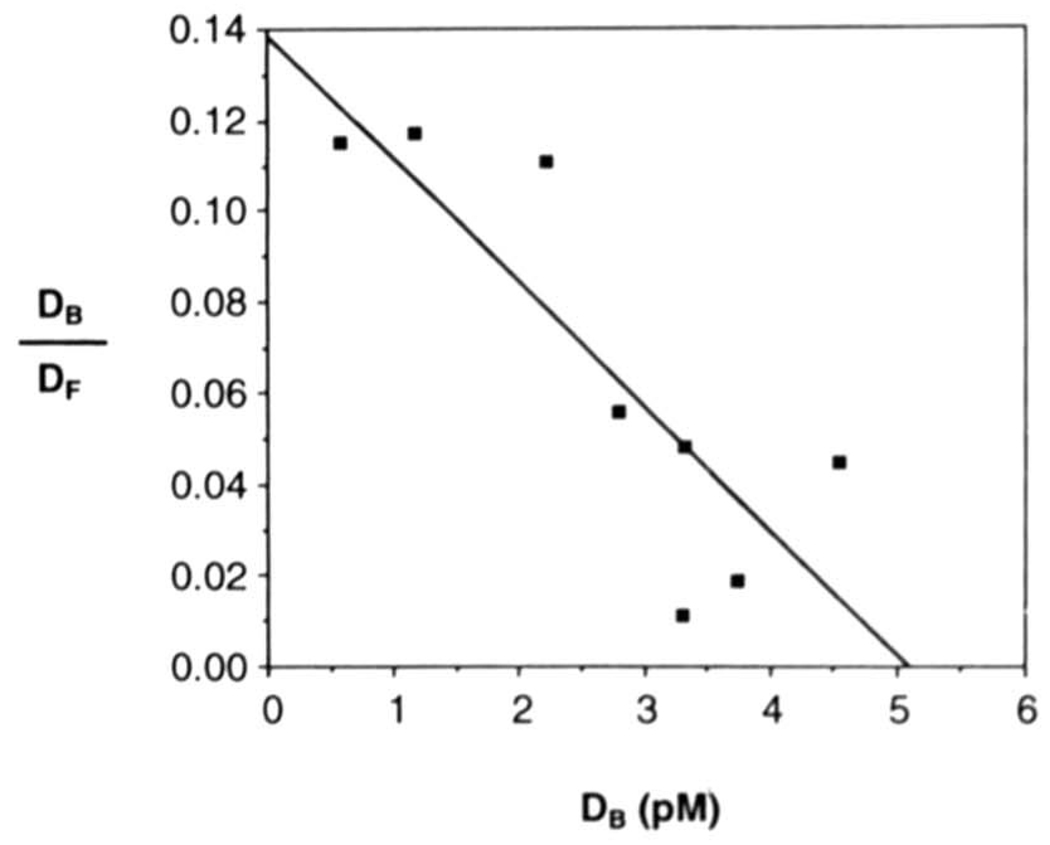
The half-life of the H2TF1-MHC κB site complex was measured to be about 130 min at 30 °C (data not shown) and the measurement of equilibrium binding affinity was therefore made at three half-lives or about 6 h. The H2TF1 concentration, determined by saturation binding with an excess of probe (data not shown), was kept constant for these measurements, at 80 pm, about 5-fold below the KD. Radiolabeled MHC κB binding sites were bound at concentrations ranging from 5 × 10−11 to 2 × 10−10 m, which generated multiple data points below the KD. By Scatchard analysis (Fig. 5), the KD was measured to be 3 × 10−11 m.
FIG. 5. Scatchard analysis of the binding of H2TF1 and the MHC κB site.
The equilibrium affinity of H2TF1 for the MHC κB site was determined by gel shift analysis. H2TF1 purified by three affinity steps (SDS-PAGE shown in Fig. 2B) was incubated for ≈3 half-lives of the complex (6 h) with varying concentrations of the MHC κB site probe in the presence of 50 µg/ml poly(dI·dC):poly(dI·dC) competitor, with initial conditions otherwise as described under “Materials and Methods.” The MHC κB site probe MHC WT was titrated at concentrations of 5, 10, 20, 50, 70. 100, and 200 pm, while the H2TF1 concentration was kept constant at 8 pm. The concentrations of protein. DNA complexes and free probe were determined by quantitative EMSA assay, as described under “Materials and Methods.” Each point represents the average of two experiments. Assuming a simple bimolecular interaction between H2TF1 and the probe, the following expression is true at equilibrium: DB/DF = KA(PT − DB). where DB is the concentration of bound DNA, DF is the concentration of free DNA, KA is the bimolecular association constant, and PT is the total protein concentration. Therefore, from the plot of DB/DF on the ordinate and DB on the abscissa, it is possible to determine the bimolecular dissociation constant, KD, from the slope, −KA, which is equal to −(l/KD). The KD of H2TF1 and the MHC κB site was thereby measured to be 3 × 10−11m.
Peptide Sequencing of H2TF1 Identifies It as the Product of the nfkb2 Gene
H2TF1 was purified from cytoplasmic extracts of 180 g of HeLa cells (gift of Ajinomoto Corp.), as outlined in Table I. After the second affinity step, approximately 40 pmol (by EMSA, or 80 pmol by SDS-PAGE and silver stain comparison with standards) of H2TF1 was loaded on an SDS-PAGE denaturing gel. The H2TF1 migrated as a doublet on this gel, with bands at 110,000 and 105,000 Mr. Both bands were eluted onto nitrocellulose, digested with trypsin, and separated by HPLC on a C18 silica column. The bands gave similar profiles of trypsin-cleavage products (data not shown) allowing the pooling of corresponding peaks for peptide sequencing.
Sequences of three H2TF1 peptides were obtained (Fig. 6). These peptide sequences matched inferred protein sequences derived from three nfkb2 gene cDNA isolates (Schmid et al., 1991; Neri et al., 1991; Bours et al., 1992). All three peptides were located in the COOH-terminal half of the protein. Peptide 1 was located between the glycine hinge and the first ankyrin repeat, peptide 2 between the third and fourth ankyrin repeats, and peptide 3 within the fifth ankyrin repeat. The peptides consisted of amino acids 460–471,579–597, and 635–647, respectively. Of note, the sequence of peptide 1 is not contained in the translation of the isolate of Schmid et al. (1991). It appears that the cDNA isolated by Schmid et al. (1991) contains two frame shifts, one before and one after peptide 1. The sequence of peptide 1 also resolves a conflict between the cDNA sequences of Bours et al. (1992) and Neri et al. (1991), placing an alanine rather than an arginine at amino acid position 471.
FIG. 6. Sequencee of three peptides derived from affinity purified H2TF1 match sequences contained in NF-κB2 p100.
H2TF1, twice purified by DNA affinity chromatography on an MHC κB column, was electrophoresed by SDS-PAGE, transferred to a nitrocellulose filter, and digested with trypsin as described under “Materials and Methods.” The resulting tryptic peptides were purified by HPLC on a C18 silica column. Three nontrypsin peptides were sequenced by Edman stepwise degradation on an Applied Biosystems 477A pulse liquid protein sequenator. The sequences of the peptides are given above, following the arginine/lysine residue at the site of trypsin cleavage. Of note, peptides 2 and 3 were present as a mixture in a single HPLC absorbance peak, and were distinguished from each other on the basis of relative molar yields. The beginning of each tryptic peptide sequence is indicated by K/R indicating by inference the lysine or arginine amino acid residue preceeding trypsin cleavage. Indeterminant amino acid residues are indicated by X. Where there was sequence ambiguity between 2 amino acids, both are listed. The amino acid sequences determined unambiguously matched the predicted amino acid sequence inferred from cDNA sequence of the nfkb2 gene (Schmidt et al., 1991; Neri et al., 1991; Bours et al., 1992), with two exceptions noted under “Results.” The amino acid residue numbers of the H2TF1 peptides are amino acids 460–471 of NF-κB2 pl00 for peptide 1, 579–597 for peptide 2, and 635–647 for peptide 3 (the numbered residues beginning with the residue following arginine).
Confirming the identification of H2TF1 as the NF-κB2 p100 polypeptide, H2TF1 and recombinant p100 formed complexes with the MHC κB site having 1) identical mobilities and 2) equivalent specific supershifts with an antiserum raised against the NH2-terminal peptide of NF-κB2 p100 (Fig. 7A). Of note, the gel shift of the recombinant p100 protein preparation revealed a minor complex of the faster mobility expected for NF-κBP p52, consistent with the presence of some p52 in the extract (Fig. 7, B and C). This minor complex was also specifically supershifted by the antiserum, as expected. No complex of the intermediate mobility expected for a p100/p52 heterodimer was observed. To determine the relative molecular weight of the H2TF1 polypeptide recognized by the antiserum, H2TF1 purified by two affinity steps (Fig. 2A) was resolved by SDS-PAGE and analyzed by immunoblot with the above antiserum (Fig. 7, B and C). Only a single polypeptide was detected by the antiserum, of Mr = 110,000 (Fig. 7B), and this reactivity could be blocked by preincubating the antiserum with the peptide to which it was raised (Fig. 7C). The H2TF1 polypeptide recognized by the antiserum exactly comigrated with the recombinant NF-κB2 p100 control. This relative molecular weight also agreed with that estimated for H2TF1 by UV cross-linking (Fig. 3B). Of note, the antiserum did not react with the human NF-κB2 p50 control, demonstrating its specificity for NF-κB2 p100. We concluded from the immunological data regarding the NH2-terminal domain, peptide sequence data from the COOH-terminal domain, UV cross-link sizing, renaturation sizing, and gel shift complex mobility that H2TF1 is composed of NF-κB2 p100, the nfkb2-encoded polypeptide.
DISCUSSION
We report the purification of the MHC class I transcription factor H2TF1 from human cells and its identification as the NF-κB2 p100 product of the nfkb2 (lyt-10) candidate proto-oncogene. This report also demonstrates that the full-length form of the nfkb2-encoded protein is a high affinity DNA-binding protein, which had not previously been recognized, thereby raising the possibility that the p100 product may directly regulate transcription.
Purified H2TF1 and recombinant NF-κB2 p100 expressed in human cells shared properties including: 1) identical mobilities upon denaturing SDS-PAGE, with immunoblot recognition of both protein bands by an NF-κB2-specific antiserum, 2) identical mobilities as protein. DNA complexes upon native electrophoresis, with supershift recognition of both complexes by an NF-κB2-specific antiserum, and 3) identical DNA-binding preferences for the MHC κB site as compared to the canonical immunoglobulin κB site (data not shown). Additionally, the amino acid sequences of three internal tryptic peptides of H2TF1 matched sequences predicted by the cDNA sequence of the nfkb2 gene (Schmid et al., 1991; Neri et al., 1991; Bours et al., 1992). Taken together with the observation that binding of H2TF1 to the MHC κB site was independent of either truncated forms of itself or smaller Rel family polypeptides, these data establish H2TF1 as NF-κB2 p100, a polypeptide which had been thought to have little or no DNA-binding activity in its full-length form.
High affinity DNA binding of purified H2TF1 and recombinant expressed NF-κB2 was unexpected, as previous reports describing in vitro translated p100 suggested otherwise (Neri et al., 1991; Bours et al., 1992). Purified recombinant NF-κB1 p105 also bound with high affinity and specificity to the MHC κB site,2 again in contrast with previous reports which showed that full-length in vitro translated NF-κB1 p105 was incapable of binding to cognate sequences as a full-length protein, but could when translated with truncation of the COOH-terminal domain (Kieran et al., 1990; Ghosh et al., 1990). A possible inhibitory effect of the COOH-terminal half of p100 on DNA binding was also suggested by Neri et al. (1991), based on their finding that the DNA binding of NF-κB2 p100 also improved with truncation of the COOH-terminal domain. Of note, the KD for NF-κB2 p49 binding to the MHC κB site has recently been determined to be 1.57 × 10−11m (Duckett et al., 1993). This dissociation constant is comparable to that of H2TF1 for the same site (KD = 3 × 10−11m), suggesting that the carboxyl-terminal half of NF-κB2 p100 has little effect on DNA-binding affinity, under the conditions tested. Differences between the DNA-binding properties of in vitro translated NF-κB1 p105 and NF-κB2 p100 and their cellularly expressed counterparts might be due to differences in protein folding or post-translational modification.
H2TF1 binding activity has been repeatedly detected by gel shift in nuclear extracts (data not shown), but the majority of the activity is found in the cytoplasm of cells, raising the question of whether H2TF1 is an active nuclear factor. The finding that H2TF1 is composed of NF-κB2 p100 polypeptide(s) makes this question more interesting as it suggests an additional role for the well studied NF-κB1 p105 polypeptide, which was also thought not to bind DNA, and has been thought to be exclusively cytoplasmic on the basis of immunofluorescence studies (Blank et al., 1991; Henkel et al., 1992). A recent finding which casts some doubt on the assignment of p105 to an exclusively cytoplasmic role is that the COOH-terminal half of p105 is a strong transcription activator (Morin and Gilmore, 1992). Co-transfection of a construct containing a fusion of the Gal4 DNA-binding domain and the COOH-terminal half of p105 with a chloramphenicol acetyl-transferase reporter construct driven by the Gal4 site showed strong activation of the reporter. Of note, the fusion protein was transcriptionally active despite cytoplasmic rather than nuclear localization of the protein, determined by immunofluorescence, consistent with the results of others indicating that the COOH-terminal half of p105 has a cytoplasmic retention activity (Blank et al., 1991; Henkel et al., 1992). The results of Morin and Gilmore (1992) combined with our finding that p100 and p105 bind the MHC κB site with high affinity (KD = 3 × 10−11 m for H2TF1) suggest that these proteins may have a transcriptional role, rather than the inactive precursor role which they had been assigned based upon the assumptions that: 1) they have an exclusively cytoplasmic location and 2) do not bind DNA. Indeed, given the high affinity of H2TF1-MHC κB site binding, nuclear DNA binding sites for H2TF1 will saturate at concentrations of this factor well below those detectable in nuclei by immunofluorescence. Therefore, although the COOH-terminal domain may confer a predominantly cytoplasmic localization to p100 and p105, it may not preclude a transcriptional role for these factors. Of interest, it has recently been reported that translocation of Rel proteins to the nucleus may be stimulated in lymphoid cells by Zn2+ and that a 115,000 Mr protein specifically immunoprecipitated by a polyclonal Rel-specific serum is trans-located to the nucleus under these conditions (Storms and Bose, 1992).
The strongest indication that H2TF1 regulates the MHC promoter is the biological correlation between the level of an H2TF1-like factor and the expression of MHC mRNA in different cell lines. Neuroblastoma cells with amplified N-myc expression and AKR leukemia cells with the greatest malignant potential display reduced levels of both MHC class I protein and an H2TF1-like factor binding to the MHC κB site but not the immunoglobulin κB site (Lenardo et al., 1989; Bernards, 1991). An alternative explanation for this correlation is that reduction in the level of the NH2-terminal half of NF-κB2 p100, which has the same DNA-binding specificity,2 accounts for the reduction in transcription. In vivo cleavage of NF-κB2 p100 may produce an NH2-terminal p52 protein (Fig. 7B) which binds DNA with the specificity of H2TF1, but forms a faster mobility protein. DNA complex (data not shown). A complex with these properties was found to be reduced in neuroblastoma cells with down-regulated MHC class I expression (Bernards, 1991). Thus, it remains to be determined whether the full-length NF-κB2 p100, NF-κB2 p52, or both are regulators of MHC class I gene expression.
Conventional methodology utilizing a DNA affinity matrix with a binding site concentration of 1–2 nmol/ml failed to purify significant amounts of H2TF1 because it eluted it at low salt concentration, preventing adequate purification. A new method of DNA affinity purification was devised using a novel matrix with a binding site concentration of 200 nmol/ml (Larson and Verdine, 1992). This matrix provided the greatest fold-purification with a competitor DNA concentration of 0.5–1.0 mg/ml, much higher than the 50 µg/ml concentration usually used with conventional DNA affinity columns. The higher concentration of competitor DNA resulted in elution of H2TF1 from the column at significantly higher salt concentrations, with a greater than 30-fold purification of this factor on the first pass. The overall purification of H2TF1 in two steps of affinity chromatography was about 870-fold, comparing favorably with the 500–1000-fold reported for conventional column matrices (reviewed by J. T. Kadonaga (1991)). With an overall yield of 30–60% per pass, with fractions which had previously been purified only 4-fold from an S100 extract, this column may also allow higher overall recovery than purifications requiring more purification steps before the affinity steps. This high capacity matrix thus allows direct application of affinity chromatography to crude fractions early in the purification process, once extraneous specific DNA binding activities have been removed.
Acknowledgments
We thank Ajinomoto Corp. for the kind gift of HeLa cells. We thank Takashi Fujita and David Baltimore for purified recombinant NF-κB1 p105 and p50 proteins, Sankar Ghosh and David Baltimore for antiserum to NF-κB1 p50, Albert S. Baldwin for plasmids containing the MHC WT, MHC MT, and κB sequences, and Kenneth LeClair for purified NF-κB1 p50. We express our appreciation to Albert Baldwin, Kenneth LeClair, Takashi Fujita, Sankar Ghosh, and David Baltimore for helpful communications and comments. We thank Melissa Moore and Joel Pomerantz for reviewing the manuscript, Tom Kristie for advice on protein purification and the other members of the Sharp laboratory for their continued interest and support. Tryptic digestion of H2TF1, peptide separation, and protein sequencing were performed by Richard F. Cook, Heather LeBlanc, and Sandra Schultz at the Biopolymers Laboratory, Howard Hughes Medical Institute, Massachusetts Institute of Technology. We thank Margarita Sciafaca for expert assistance with preparation of the manuscript.
Footnotes
This work was supported by fellowship support of the Division of Hematology/Oncology, Brigham and Women’s Hospital, a grant from the Medical Research Council of Canada, National Cancer Institute Grant K08-CA-01562-01 (to D. A. P.), United States Public Health Service Grant P01-CA42063, a cooperative agreement (CDR-8803014) from the National Science Foundation (to P. A. S.), by Cancer Center Support (core) Grant P30-CA14051 from the National Cancer Institute, a Liebig scholarship from the Fond Der Chemischen Industrie West Germany (to P. E.), a grant from the Harvard Medical School/Hoffman-La Roche Institute of Chemistry and Medicine, a Searle Scholar Award, a National Science Foundation Presidential Young Investigator Award (to G. L. V.), and National Institutes of Health Grant R01-AI29179 (to G. J. N.).
The abbreviations used are: MHC, major histocompatibility complex; bp, base pair; BrdUrd, bromodeoxybromouridine; BSA, bovine serum albumin; DTT, dl-dithiothreitol; EMSA, electrophoretic mobility shift assay; HPLC, high pressure liquid chromatography; KD, equilibrium dissociation constant; PAGE, polyacrylamide gel electrophoresis; S100, supernatant of 100,000 × g (approximate) centrifugation.
D. A. Potter, C. J. Larson, P. Eckes, R. M. Schmid, G. J. Nabel, G. L. Verdine, and P. A. Sharp, unpublished data.
REFERENCES
- Baldwin AS, Jr, Sharp PA. Mol. Cell. Biol. 1987;7:305–313. doi: 10.1128/mcb.7.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS, Jr, Sharp PA. Proc. Natl. Acad. Sci. U. S. A. 1988;85:723–727. doi: 10.1073/pnas.85.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- Bernards R. Trends. Genet. 1987;3:298–301. [Google Scholar]
- Bernards R. EMBO J. 1991;10:1119–1125. doi: 10.1002/j.1460-2075.1991.tb08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet O, Bourge J-F, Zinszner H, Israёl A, Kourilsky P, Dausset J, Degos L, Paul P. Proc. Natl Acad. Sci. U. S. A. 1992;89:3488–3492. doi: 10.1073/pnas.89.8.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Kourilsky P, Israёl A. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Kourilsky P, Israёl A. Trends Biochem. Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Bours V, Villalobos J, Burd PR, Kelly K, Siebenlist U. Nature. 1990;348:76–80. doi: 10.1038/348076a0. [DOI] [PubMed] [Google Scholar]
- Bours V, Burd P, Brown K, Villalobos J, Park S, Ryseck R-P, Bravo R, Kelly K, Siebenlist U. Mol. Cell. Biol. 1992;12:685–695. doi: 10.1128/mcb.12.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. New York: Greene Publishing Associates and Wiley-Interscience; 1991. pp. 12.2.1–12.2.10. [Google Scholar]
- David-Watine B, Israёl A, Kourilsky P. Immunol. Today. 1990;11:286–292. doi: 10.1016/0167-5699(90)90114-o. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Leibovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CS, Perkins ND, Kowalik TF, Schmid RM, Huang E-S, Baldwin AS, Jr, Nabel GJ. Mol. Cell. Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Nolan GP, Ghosh S, Baltimore D. Genes & Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Gifford AM, Riviere LR, Tempst P, Nolan GP, Baltimore D. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. Cell. 1990;62:841–843. doi: 10.1016/0092-8674(90)90257-f. [DOI] [PubMed] [Google Scholar]
- Hager DA, Burgess RR. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Henkel T, Zabel U, van Zee K, Müller J, Fanning E, Baeuerle P. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- Henseling U, Schmidt W, Schöler HR, Gruss P, Hatzopoulos AK. Mol. Cell. Biol. 1990;10:4100–4109. doi: 10.1128/mcb.10.8.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israёl A, Kimura A, Kieran M, Yano O, Kanellopoulos J, Le Bail O, Kourilsky P. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2653–2657. doi: 10.1073/pnas.84.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israёl A, Le Bail O, Hatat D, Piette J, Kieran M, Logeat F, Wallach D, Fellous M, Kourilsky P. EMBO J. 1989;8:3793–3800. doi: 10.1002/j.1460-2075.1989.tb08556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT, Tjian R. Proc. Natl. Acad. Sci. U. S. A. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Methods Enzymol. 1991;208:10–23. doi: 10.1016/0076-6879(91)08004-2. [DOI] [PubMed] [Google Scholar]
- Kimura A, Israёl A, Le Bail O, Kourilsky P. Cell. 1986;44:261–272. doi: 10.1016/0092-8674(86)90760-9. [DOI] [PubMed] [Google Scholar]
- Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban MB, Kourilsk P, Baeuerle PA, Israёl A. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson CJ, Verdine GL. Nucleic Acids Res. 1992;20:3525. doi: 10.1093/nar/20.13.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M, Rustgi AK, Schievella AR, Bernards R. EMBO J. 1989;8:3351–3355. doi: 10.1002/j.1460-2075.1989.tb08497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F, Israёl N, Ten R, Blank V, Le Bail O, Kourilsky P, Israёl A. EMBO J. 1991;10:1827–1832. doi: 10.1002/j.1460-2075.1991.tb07708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R, Hatada EN, Hohmann H-P, Haiker M, Bartach C, Rothlisberger U, Lahm H-W, Schlaeger EJ, van Loon APGM, Scheidereit C. Proc. Natl. Acad. Sci. U. S. A. 1991;88:966–970. doi: 10.1073/pnas.88.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Gilmore TD. Nucleic Acids Res. 1992;20:2453–2458. doi: 10.1093/nar/20.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri A, Chang C-C, Lombardi L, Salina M, Corradini P, Maiolo AT, Chaganti RSK, Dalla-Favera R. Cell. 1991;67:1075–1087. doi: 10.1016/0092-8674(91)90285-7. [DOI] [PubMed] [Google Scholar]
- Nolan GP, Ghosh S, Liou H-C, Tempst P, Baltimore D. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- Ruben SM, Dillon PJ, Schreck R, Henkel T, Chen C-H, Maher M, Baeuerle PA, Rosen CA. Science. 1991;251:1490–1493. doi: 10.1126/science.2006423. [DOI] [PubMed] [Google Scholar]
- Ryseck R-P, Bull P, Takamiya M, Bours V, Siebenlist U, Dobrzanski P, Bravo R. Mol. Cell. Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheppler JA, Nicholson JKA, Swan DC, Ahmed-Ansari A, McDougal JS. J. Immunol. 1989;143:2858–2866. [PubMed] [Google Scholar]
- Schmid RM, Perkins ND, Duckett CS, Andrews PC, Nabel GJ. Nature. 1991;352:733–736. doi: 10.1038/352733a0. [DOI] [PubMed] [Google Scholar]
- Schrier PI, Bernards R, Vaessen RTMJ, Houweling A, van der Eb AJ. Nature. 1983;305:771–775. doi: 10.1038/305771a0. [DOI] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- Singer DS, Maguire JE. Crit. Rev. Immunol. 1990;10:235–257. [PubMed] [Google Scholar]
- Storms RW, Bose HR., Jr Virology. 1992;188:765–777. doi: 10.1016/0042-6822(92)90531-s. [DOI] [PubMed] [Google Scholar]
- Tempst P, Link AJ, Riviere LR, Fleming M, Elicone C. Electrophoresis. 1990;11:537–553. doi: 10.1002/elps.1150110704. [DOI] [PubMed] [Google Scholar]
- Tenaka K, Yoshioka T, Bieberich C, Jay G. Ann. Rev. Immunol. 1988;6:359–380. doi: 10.1146/annurev.iy.06.040188.002043. [DOI] [PubMed] [Google Scholar]