Abstract
A series of 30 RCO-HfR-NH2 derivatives show preference for the mouse MC1R vs. MC3-5Rs. trans-4-HOC6H4CH=CHCO-HfR-NH2 (13) [EC50 (nM): MC1R 83, MC3R 20500, MC4R 18130 and MC5R 935; ratio 1:246:217:11] is 11 times more potent than the lead compound LK-394 Ph(CH2)3CO-HfR-NH2 (2) and only 11 times less potent than the native tridecapeptide α-MSH at mMC1R. Differences in conformations of 2 and 13 are discussed.
Keywords: Melanocortin receptors, Melanocortin agonists, N-capping. Peptidomimetics, Conformational analysis, Molecular modeling
Melanocortin receptors (MCRs) are found in different tissues, have a plethora of physiological functions1 and are a target of intensive pharmacological research.2 Five MCRs (MC1R – MC5R) have been cloned. From the very onset of research in this area, the role of melanocortins in skin pigmentation has been firmly established and shown to be the result of activation of the MC1R. α-, β- and γ-Melanocyte stimulating hormones (MSH), the natural agonists of MC1, MC3, MC4 and MC5Rs, sharing the common melanocortin core sequence His6-Phe7-Arg8-Trp9 (HFRW), and their synthetic analogs (NDP-MSH, MT-II) with a similar His6-D-Phe7-Arg8-Trp9 (HfRW) core are very potent but non-selective. Although many peptide and non-peptide ligands of MCRs have been reported, pharmacological differentiation of properties of MC1, MC3, MC4 and MC5Rs is still a major challenge.1,2
| α-MSH | Ac-Ser-Tyr-Ser-Met-Glu-His6-Phe7-Arg8-Trp9-Gly-Lys-Pro-Val-NH2 |
| NDP-MSH | Ac-Ser-Tyr-Ser-Nle-Glu-His-D-Phe-Arg-Trp-Gly-Lys-Pro-Val-NH2 |
| LK-184 (1) | Ph(CH2)3CO-His-D-Phe-Arg-Trp-NH2 |
| LK-394 (2) | Ph(CH2)3CO-His-D-Phe-Arg-NH2 |
| Compounds 3–32 | RCO-His-D-Phe-Arg-NH2 |
Recently, we have described a superpotent hMC1R agonist LK-184 Ph(CH2)3CO-HfRW-NH2 (1) with an EC50 of 0.01 nM and ca. 500-fold selectivity for hMC1R compared to hMC3 and hMC4Rs.3a Truncation of 1 to Ph(CH2)3CO-HfR-NH2 gave a full hMC1 agonist LK-394 (2) with an EC50 of 5 nM and weak partial agonism at hMC3/4Rs.3b We have also shown that tetrapeptide 1 was more potent than α-MSH in repairing DNA damage resulting from exposure to solar UV radiation, in addition to stimulating pigmentation that confers further photoprotection and prevents mutagenesis.4a Such double-headed action can be used as an effective prevention strategy for the most fatal skin cancer - melanoma.4b The goal of this work was to explore modifications of the N-terminal end-capping group on the HfR-NH2 core sequence in order to increase potency and/or selectivity at MC1R compared to the lead compound 2 and provide insight into the interaction of short peptides with MCRs.
Our previous studies on the tetrapeptides RCO-HfRW-NH2 suggested that an aromatic π-binding site in the hMC1R conferred selectivity of the N-capped peptides for this receptor over other hMCRs.3 Thus, all tripeptides in this study contained an aromatic ring connected to the C=O group via C2 (3–10, 12–16 and 18–25), C3 (26–28) or C4 (29–31) spacer, a cyclic C2 mimic (11 and 17) or directly (32) (Table 1). For mapping the putative π-binding site, cinnamic derivatives 3–9, 12, 13, 15, 16, 18, 20 and 23–25 provided a shorter and more rigid C2 link compared to the freely rotating saturated C3 chain in 2, while the substituents in their phenyl rings provided an extended conjugated π system overlapping the conformational space with the π system of the phenyl (shown below) in lead compound 2. In addition, pyridine and imidazole derivatives 21 and 22 with saturated C2 chain served as flexible probes for potential basic or H-bonding sites at hMC1R near the N-terminal binding region. The N-capping groups with π-conjugated diene system (29 and 30) and the corresponding saturated C4 analog 31 were chosen to explore the space beyond Ph(CH2)3 in 2.
Table 1.
Agonist activity of N-capped tripeptides on mouse MC1, MC3, MC4, and MC5 receptors.
| RCO-His-D-Phe-Arg-NH2 | EC50(nM) ± EM | ||||
|---|---|---|---|---|---|
| # | R | mMC1R | mMC3R | mMC4R | mMC5R |
| 1 | Ph(CH2)3CO-His-D-Phe-Arg-Trp-NH2 (LK-184) | 1.60±0.29 | 26.0±4.0 | 0.54±0.19 | 0.33±0.14 |
| 2 | (LK-394) 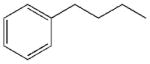
|
940±260 | 43700±1370 | 15140±8800 | 11800±5950 |
| 3 | 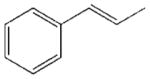 |
1200±390 | a | 52950±6580 | 30220±8200 |
| 4 | 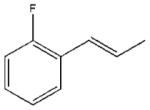 |
1160±570 | 28050±22600 | 14700±5800 | 6860±1200 |
| 5 | 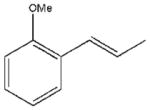 |
1690±465 | a | 13700±4700 | 12850±1710 |
| 6 | 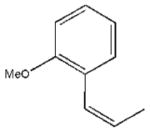 |
3660±1170 | a | 60400±19500 | 18400±7600 |
| 7 | 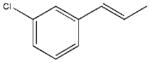 |
660±190 | 33800±13300 | 4010±240 | 6390±1100 |
| 8 | 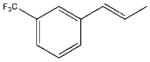 |
310±70 | 30300±17000 | 3700±500 | 6780±490 |
| 9 | 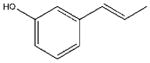 |
274±68 | a | 12740±4120 | 2770±330 |
| 10 | 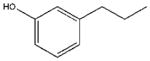 |
1120±216 | a | a | 9600±3940 |
| 11 | 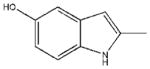 |
1175±330 | 7380±1240 | 17200±2700 | 2280±615 |
| 12 | 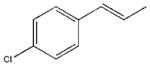 |
1080±300 | 58900±56000 | 5200±1700 | 6900±1400 |
| 13 | 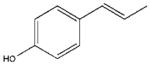 |
83.3±13.7 | 20500±12600 | 18130±3258 | 935±310 |
| 14 | 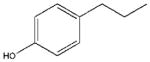 |
1160±88 | a | a | 10540±3200 |
| 15 | 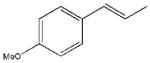 |
420±61 | a | 11500±4920 | 3410±360 |
| 16 |  |
780±230 | a | a | 6640±1830 |
| 17 | 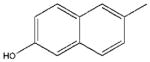 |
620±26 | a | a | 4760±1300 |
| 18 | 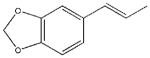 |
567±253 | a | 23600±1780 | 6187±1400 |
| 19 | 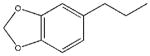 |
7280±4400 | a | 25500±3400 | 23000±4700 |
| 20 | 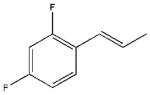 |
1250±130 | a | 23500±9900 | 17600±3090 |
| 21 | 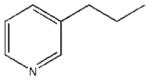 |
1670±437 | a | a | 18050±4510 |
| 22 | 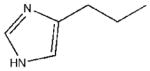 |
3200±670 | a | 38700±6370 | 70300±11350 |
| 23 | 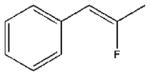 |
2790±650 | a | 19200±4100 | 19300±1340 |
| 24 | 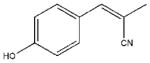 |
2060±330 | a | 42800±21600 | 8500±1720 |
| 25 | 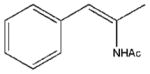 |
8200±2150 | a | 20300±6400 | a |
| 26 | 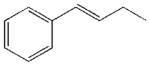 |
990±180 | 24100±4100 | 5020±1980 | 3080±210 |
| 27 | 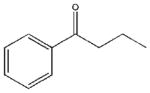 |
1745±685 | a | 22750±5870 | 16580±2307 |
| 28 | 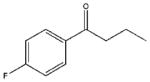 |
1230±320 | a | 12400±2240 | 10500±620 |
| 29 | 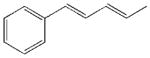 |
500±195 | 16800±4530 | 4710±1370 | 6830±1060 |
| 30 | 780±430 | 19500±2900 | 10050±1270 | 6230±790 | |
| 31 | 530±210 | 25900±15700 | a | a | |
| 32 | 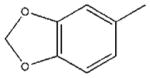 |
3030±1040 | a | 31100±21350 | 23600±1750 |
| α-MSH | 7.2±3.73 | 10.2±2.53 | 19.3±3.59 | 6.72±2.01 | |
| NDP-MSH | 0.014±0.002 | 0.09±0.02 | 0.14±0.03 | 0.17±0.08 | |
>100,000, indicates that no agonist response was observed at up to 100mM concentrations.
As seen in Table 1, the selectivity profile for reference compounds 1 and 2 differs from that previously reported for hMCRs.3 Tetrapeptide 1 has an EC50 in the nanomolar range at mMC1R, MC4R and MC5R (MC1/3/4/5Rs ratio 1:16:0.3:0.2), while its tripeptide analog 2 retains mMC1R selectivity (MC1/3/4/5Rs ratio 1:46:16:12) albeit with 590-fold loss in potency compared to 1. As we expected on the basis of our data for 1 and 23b and literature data on the melanocortin core sequence6, removal of Trp9 from HfRW-NH2 resulted in very low potency at mMC3R-MC5R. This desirable effect is most pronounced at mMC3R, where 2/3 of the synthesized tripeptides show no activity. Apparently, the binding site of mMC3R is larger than that of mMC1R, MC4R and MC5Rs and the Ph-C-C-group is too short to interact with the receptor. The elongation of the end-capping group (see the alignment above) due to aromatic substituents (4, 7, 8, 11–13 and 26) or spacer extensions (29–31 vs. 18–19) elicits a weak mMC3R response. This SAR is not straightforward and polar groups (5, 6, 9, 10, 14–19, 26 and 28) cancel the response.
In accordance with our initial reasoning that restrictions in spacer flexibility on the tripeptide core lacking tertiary structure might produce a conformation active at MC1R, we explored an unsaturated series consisting of 3–9, 12, 13, 15–18, 20 and 23–26. The direct unsaturated analog 26 of the parent tripeptide 2 was equipotent to 2 at mMC1R but showed some loss of selectivity (MC1/3/4/5Rs ratio 1:24:5:3). At the same time, the simple cinnamic derivative 3 was more promising - its potency at mMC1R was close to that of 2 and it showed much better selectivity (MC1/4/5Rs 1:44:25 and no activity at mMC3R). Substitution on the cinnamoyl aromatic ring revealed considerable tolerance of the mMC1R for structural changes. meta-Substitution (7–9) was favorable with a 2- to 3-fold increase in potency, while the para-OH derivative 13 was the most potent (11x) and selective (MC1/3/4/5Rs 1:246:217:11). The p-OH group was essential for activity since methylation (15) or substitution by the isosteric and isoelectronic Cl (12) resulted in a substantial drop in potency (Table 1).
In an attempt to pinpoint the location of the OH binding site in mMC1R, we synthesized free rotating analogs (14 and 10) of semi-rigid 13 and 9, as well as their rigid cyclic analogs (17 and 11). All of them were less potent than 13, i.e. the cinnamic scaffold provided an optimal positioning of the polar H-bonding group in the aromatic binding region of mMC1R.
To determine factors contributing to high potency of 13, we did a conformational search on 2, 13, 11 and 17 using the Monte Carlo Multiple Minimum (MCMM) mixed torsional/low-frequency sampling and the OPLS_2005 force field in water as implemented in Macromodel7. The results show that although the number of conformers for 2 (108 within 3 kcal/mol, 293 within 5 kcal/mol) and 13 (55 within 3 kcal/mol, 221 within 5 kcal/mol) does not vary drastically, their 3D distribution is totally different (Figure 1).
Figure 1.
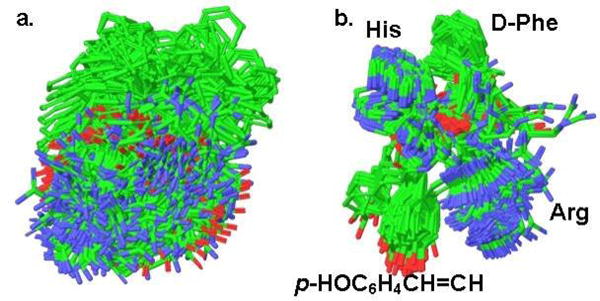
Conformations within 5 kcal/mol: a. Ph(CH2)3CO-His-D-Phe-Arg-NH2 (2), b. trans-4-HOC6H4CH=CHCO-His-D-Phe-Arg-NH2 (13).
While 2 has no preferred conformation (Figure 1a), all 221 conformers of 13 are very well aligned (Figure 1b). Even though the p-OH does not contribute to any intramolecular bonds, the semi-rigid cinnamic tail influences the rest of otherwise flexible tripeptide core placing the polar basic Arg and His on the same side of the molecule opposite to the non-polar aromatic D-Phe and cinnamoyl. Together with the existence of 13 in multiple but similar conformations that can morph into the biologically active conformation upon binding, such “pre-alignment” is beneficial for initial recognition of the ligand by the mMC1R that contains known compact acidic (Glu-94, Asp-117 and 121) and aromatic hydrophobic (Phe-175, 179, 196, and 257) binding pockets.8 The results of the conformational search for the cyclic analog 17 (not shown) are similar to 13, which demonstrates that its lower potency relative to 13 is related to misplacement of HO in the active site due to an increase in girth or rigidity of the end-capping group. It is interesting to note that the drop in potency of 17 is observed on all mMCRs subtypes with the total loss of activity at mMC3 and mMC4Rs.
Thus, the use of semi-rigid N-capping groups with the C2 spacer allows for a “soft” control of the peptide conformation without restricting dihedral angles of the backbone or side chains favorable for interaction with an active site. Application of this principle to a very flexible tripeptide 2 resulted in the compound 13 that was more selective than the tridecapeptide α-MSH and only 11 times less potent at mMC1R. Many of the N-capped HfR-NH2 derivatives show low potency or are inactive at mMC3 and/or MC4Rs (Table 1) and this could be relevant for future design of low molecular weight synthetic drugs acting only on targeted MCRs subtypes.10
Acknowledgments
This work was funded by grants RO1CA114095 (Z.A-M), RO1DK057080 (C.H-L) and Skin Cancer Foundation Henry W. Menn Memorial Award (JJK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.The Melanocortin Receptors; Cone RD, editor. Humana. Totowa: 2000. Abdel-Malek Z. Cell Molec Life Sci. 2001;58:1420. doi: 10.1007/PL00000868.Understanding G Protein-coupled Receptors and their Role in the CNS (The Molecular and Cellular Neurobiology Series); Pangalos M, Davies CH, editors. Oxford University Press; 2002. Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C. Pigment Cell Res. 2005;18:393. doi: 10.1111/j.1600-0749.2005.00278.x.Wikberg JES, Mutulis F. Nature Rev Drug Disc. 2008;7:307. doi: 10.1038/nrd2331.
- 2.Todorovic A, Haskell-Luevano C. Peptides. 2005;26:2026. doi: 10.1016/j.peptides.2004.11.024.Hadley ME, Hruby VJ, Jiang J, Sharma SD, Fink JL, Haskell-Luevano C, Bentley DL, Al-Obeidi F, Sawyer TK. Pigment Cell Res. 2006;9:213. doi: 10.1111/j.1600-0749.1996.tb00111.x.Insights into Receptor Function and New Drug Development Targets (Research and Perspectives in Endocrine Interactions); Conn M, Kordon C, Christen Y, editors. Springer; 2006. Ligand Design for G Protein-coupled Receptors (Methods and Principles in Medicinal Chemistry); Rognan D, Mannhold R, Kubinyi H, Folkers G, editors. Wiley-VCH; 2006. Current Top Med Chem. 2007;7No 11 – a special issue on Melanocortin Receptor Ligands and Their Potential Therapeutic Uses; Nozawa D, Chaki S, Nakazato A. Expert Opinion Ther Patents. 2008;18:403.
- 3.(a) Koikov LN, Knittel JJ, Solinsky MG, Cross-Doersen D, Ebetino FH. Bioorg Med Chem Lett. 2003;13:2647. doi: 10.1016/s0960-894x(03)00552-3. [DOI] [PubMed] [Google Scholar]; (b) 2004;14:3997. ibid. [Google Scholar]
- 4.(a) Abdel-Malek ZA, Kadekaro AL, Kavanagh RJ, Todorovic A, Koikov LN, McNulty JC, Pilgrim JJ, Jackson PJ, Millhauser GL, Schwenberger S, Babcock J, Haskell-Luevano C, Knittel JJ. The FASEB Journal. 2006;20:1561. doi: 10.1096/fj.05-5655fje. [DOI] [PubMed] [Google Scholar]; Abdel-Malek ZA, Kavanagh RJ, Koikov LN, Todorovic A, Haskell-Luevano C, McNulty JC, Jackson P, Millhauser G, Knittel JJ. Pigment Cell Res. 2004;17:449. [Google Scholar]; (b) Hearing VJ, Rouzaud F. Peptides. 2005;26:1858. doi: 10.1016/j.peptides.2004.11.041. [DOI] [PubMed] [Google Scholar]; Abdel-Malek ZA, Knittel JJ, Kadekaro AL, Starner R, Swope VB. Photochem Photobiol. 2008;84:501. doi: 10.1111/j.1751-1097.2008.00294.x. [DOI] [PubMed] [Google Scholar]
- 5.Haskell-Luevano C, Holder JR, Monck EK, Bauzo RM. J Med Chem. 2001;44:2247. doi: 10.1021/jm010061n. [DOI] [PubMed] [Google Scholar]
- 6.Hruby VJ, Wilkes BC, Hadley ME, Al-Obeidi F, Sawyer TK, Staples DJ, DeVaux A, Dym O, Castrucci AM, Hintz MF, Riehm JP, Rao KR. J Med Chem. 1987;30:2126. doi: 10.1021/jm00394a033. [DOI] [PubMed] [Google Scholar]; Haskell-Luevano C, Sawyer TK, Hendrata S, North C, Panahinia L, Stum M, Staples DJ, Castrucci AM, Hadley ME, Hruby VJ. Peptides. 1996;17:995. doi: 10.1016/0196-9781(96)00141-6. [DOI] [PubMed] [Google Scholar]; Sahm UG, Oliver GWJ, Branco SK, Moss SH, Pouton CW. Peptides. 1994;15:1297. doi: 10.1016/0196-9781(94)90157-0. [DOI] [PubMed] [Google Scholar]; Sahm UG, Quarawi MA, Oliver GWJ, Ahmed ARH, Moss SH, Pouton CW. FEBS Lett. 1994;350:29. doi: 10.1016/0014-5793(94)00725-x. [DOI] [PubMed] [Google Scholar]; Bednarek MA, Silva MV, Arison B, MacNeil T, Kalyani RN, Huang RRC, Weinberg DH. Peptides. 1999;20:401. doi: 10.1016/s0196-9781(99)00048-0. [DOI] [PubMed] [Google Scholar]; Bednarek MA, MacNeil T, Kalyani RN, Tang R, Van der Ploeg LHT, Weinberg DH. Biochem Biophys Res Communs. 1999;261:209. doi: 10.1006/bbrc.1999.0981. [DOI] [PubMed] [Google Scholar]; Holder JR, Xiang Z, Bauzo R, Haskell-Luevano C. J Med Chem. 2002;45:5736. doi: 10.1021/jm020296e. [DOI] [PubMed] [Google Scholar]; Bednarek MA, MacNeil T, Tang R, Tung MF, Cabello MA, Maroto M, Teran A. Biopolymers. 2007;89:401. doi: 10.1002/bip.20863. [DOI] [PubMed] [Google Scholar]
- 7.Schrodinger Suite 2008. Schrodinger LLC; New York, NY: 2008. [Google Scholar]
- 8.Haskell-Luevano C, Sawyer TK, Trumpp-Kallmayer S, Bikker JA, Humblet C, Gantz I, Hruby VJ. Drug Design Discovery. 1996;14:197. [PubMed] [Google Scholar]; Yang Y, Dickinson C, Haskell-Luevano C, Gantz I. J Biol Chem. 1997;272:23000. doi: 10.1074/jbc.272.37.23000. [DOI] [PubMed] [Google Scholar]; Haskell-Luevano C, Rosenquist A, Souers A, Khong KC, Ellman JA, Cone RC. J Med Chem. 1999;42:4380. doi: 10.1021/jm990190s. [DOI] [PubMed] [Google Scholar]; Holder JR, Haskell-Luevano C. Med Res Rev. 2004;24:325. doi: 10.1002/med.10064. [DOI] [PubMed] [Google Scholar]; Hogan K, Peluso S, Gould S, Parsons I, Ryab D, Wu L, Visiers I. J Med Chem. 2006;49:911. doi: 10.1021/jm050780s. [DOI] [PubMed] [Google Scholar]
- 9.Koikov L, Ruwe AR, Abdel-Malek Z, Haskell-Luevano C, Dirain ML, Portillo F, Xiang Z, Wortman M, Knittel JJ. Pept Sci; Abstract P263, American Peptide Symposium; 2009; Bloomington, IN. 2009. p. 362. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jois SDS, Koikov L, Abdel-Malek Z, Kavanagh R, Wortman M, Knittel JJ. ibid. :361. Abstract P259. [Google Scholar]
- 10.Nickolls SA, Cismowski MI, Wang X, Wolf M, Conlon PJ, Maki RA. J Pharmcol Exp Ther. 2003;304:1217. doi: 10.1124/jpet.102.044974. [DOI] [PubMed] [Google Scholar]; Sun H, Fry D. Curr Top Med Chem. 2007;7:1042. doi: 10.2174/156802607780906573. [DOI] [PubMed] [Google Scholar]
- 11.Compounds 1–32 (purity >95%) were obtained, analyzed and assayed as previously described.3,5 The EC50 values for compounds 1–32 (all of them are full agonists) in Table 1 are an average of at least three separate experiments with the associated standard error of the mean.


