SUMMARY
Peptidoglycan recognition proteins (PGRPs) are mediators of innate immunity and recently have been implicated in developmental regulation. To explore the interplay between these two roles, we characterized a PGRP in the host squid Euprymna scolopes (EsPGRP1) during colonization by the mutualistic bacterium Vibrio fischeri. Previous research on the squid-vibrio symbiosis had shown that, upon colonization of deep epithelium-lined crypts of the host light organ, symbiont-derived peptidoglycan monomers induce apoptosis-mediated regression of remote epithelial fields involved in the inoculation process. In this study, immunofluorescence microscopy revealed that EsPGRP1 localizes to the nuclei of epithelial cells, and symbiont colonization induces the loss of EsPGRP1 from apoptotic nuclei. The loss of nuclear EsPGRP1 occurred prior to DNA cleavage and breakdown of the nuclear membrane, but followed chromatin condensation, suggesting that it occurs during late stage apoptosis. Experiments with purified peptidoglycan monomers and with V. fischeri mutants defective in peptidoglycan-monomer release provided evidence that these molecules trigger nuclear loss of EsPGRP1 and apoptosis. The demonstration of a nuclear PGRP is unprecedented, and the dynamics of EsPGRP1 during apoptosis provide a striking example of a connection between microbial recognition and developmental responses in the establishment of symbiosis.
INTRODUCTION
Peptidoglycan-recognition proteins, or PGRPs, are a protein family of innate immune system effectors and receptors conserved across the animal kingdom (Zhang et al., 2007, Coteur et al., 2007, Dziarski et al., 2006b, Werner et al., 2000, Goodson et al., 2005). PGRPs mediate host responses to the bacterial cell-envelope component, peptidoglycan (PGN). In-depth research on the PGRPs in the last 10 years has principally focused on biochemical characterizations of the activity and binding specificity of the purified proteins, as well as the genetics of their in vivo function in Drosophila melanogaster (Filipe et al., 2005, Park et al., 2007, Kaneko et al., 2004, Kaneko et al., 2006, Lim et al., 2006, Dziarski et al., 2006a, Lu et al., 2006). For example, in vitro studies have demonstrated that the characteristic PGRP domain binds specifically to PGN ligands (Swaminathan et al., 2006, Chang et al., 2006) and in some isoforms also cleaves the substrate (Steiner, 2004). Genetic analyses have implicated these behaviors in the regulation of downstream response pathways (Michel et al., 2001, Kaneko et al., 2004, Bischoff et al., 2006).
These studies have defined similarities and differences among the vertebrates and invertebrates. In both animal groups, some PGRPs can act to attenuate the inflammatory response by degrading the PGN through N-acetyl-muramyl-L-alanine amidase (NAMLAA) activity (Gelius et al., 2003, Zaidman-Remy et al., 2006, Bischoff et al., 2006), which requires a cysteine residue in the PGN-binding pocket. In this enzymatic behavior, the stem peptide is cleaved from the sugar backbone of PGN, resulting in products that are less immunogenic (Bischoff et al., 2006). Although all mammalian PGRPs described thus far are reported to be secreted proteins, the invertebrate homologs can act as receptors for PGN, a feature reflected in their localization. The invertebrate PGRPs are predicted to be either intracellular, membrane associated, or secreted (Royet et al., 2007), although few studies have defined precise cellular localization (Gupta, 2008, Kaneko et al., 2006, Dziarski et al., 2003). The more varied predicted localization of invertebrate PGRPs correlates with broader function. In Drosophila, PGRPs have been implicated in the regulation of apoptosis and development (Maillet et al., 2008, Bischoff et al., 2006), activation or dampening of the immune response (Yoshida et al., 1996, Takehana et al., 2002, Kaneko et al., 2006, Kaneko et al., 2004, Maillet et al., 2008), control of phagocytosis (Ramet et al., 2002), and as direct immune effectors (Mellroth et al., 2006).
PGRPs have been implicated not only in mediating responses to microbial pathogens, but also in the dynamics of mutualistic symbioses in invertebrates (Anselme et al., 2006, Bischoff et al., 2006, Anselme et al., 2008). The binary association between the Hawaiian bobtail squid Euprymna scolopes and the marine bacterium Vibrio fischeri has been used for the last 20 years as a model for the study of mutualistic symbioses [for reviews see: (Nyholm et al., 2004, Visick et al., 2006)]. The analysis of an EST database constructed from the symbiotic tissues of juvenile E. scolopes revealed the expression of four host PGRPs, EsPGRP1-4, in these bacteria-containing tissues (Goodson et al., 2005). This finding suggested the squid-vibrio system could be a model for the study of these proteins in mutualistic associations. In this naturally occurring, binary symbiosis, the partners can be experimentally manipulated. This feature facilitates the discovery of the molecular underpinnings of the cellular interactions between host and symbiont. In addition, molecular genetics have been developed in V fischeri, providing the opportunity to study changes in the host in response to genetic alterations in key symbiont characters (Visick et al., 2006).
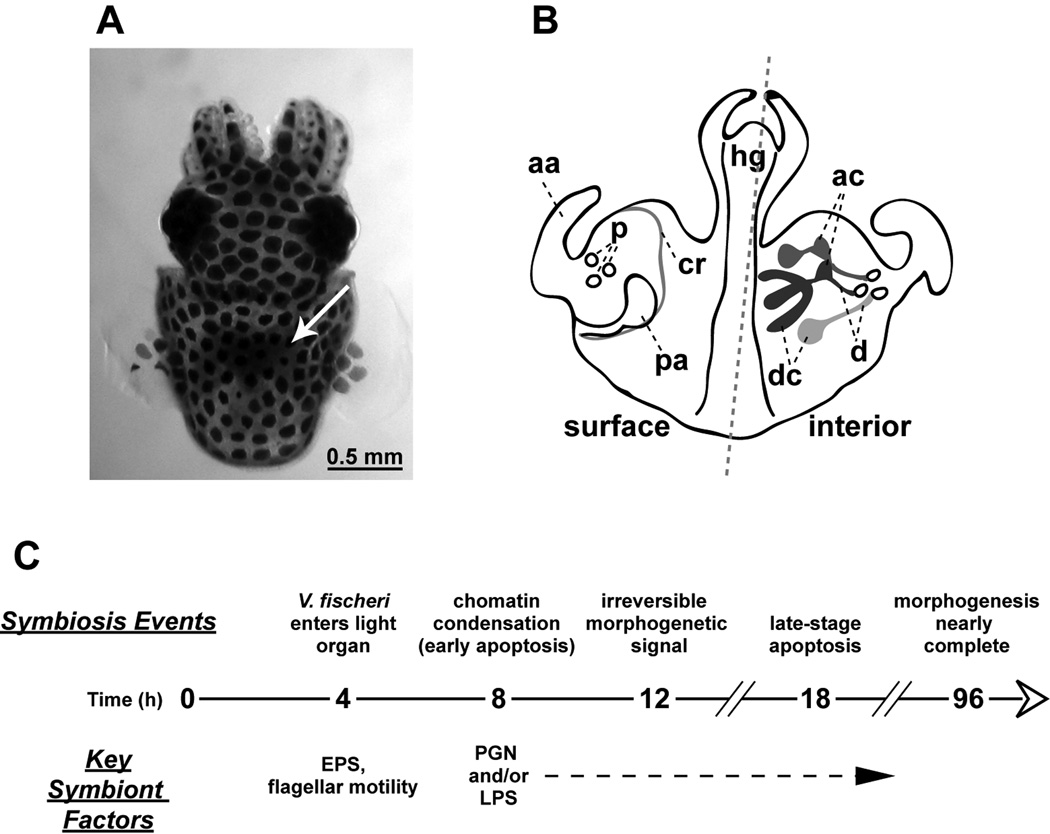
A key feature of this symbiosis is its exclusivity: only V. fischeri is capable of colonizing host tissues and inducing morphogenesis (McFall-Ngai et al., 1991). The exclusivity is achieved through the multiple steps required for colonization, including competitive dominance during aggregation in host mucus, locomotion to the light-organ pores, and resistance to the oxidatively stressful environment of the ducts (Nyholm et al., 2004). V. fischeri cells appear to signal morphogenesis from the light-organ interior, which is several cell layers away from the tissue layer that regresses (Fig. 1) (Montgomery et al., 1994, Doino et al., 1995). Thus, either the morphogenetic signals are sensed by the internal epithelia and responses are transduced through the tissues to the superficial epithelium, or the bacterial signal molecules are transported through host tissues to the sites of morphogenesis where they would act directly on host cells. In either case, the development is irreversibly triggered at 12 h and completed by 96 h (Doino et al., 1995).
Figure 1. Events in the early symbiosis of the squid/vibrio association.
A) A dorsal view of a hatchling E. scolopes. E. scolopes harvests V. fischeri cells from ambient seawater within hours of hatching and maintains a population of the symbiont in a specialized ‘light organ’. The light organ appears as a dark region in the center of the body cavity (white arrow). B) A diagram depicting the ventral view of the hatchling light organ; left of the dashed line surface features are shown, right of the dashed line interior structures are displayed. Ciliated epithelial fields specific to the surface of the juvenile light organ promote colonization by the symbiont. These ciliated epithelial fields that undergo morphogenesis include the anterior appendage (aa), the posterior appendage (pa), and the ciliated ridge (cr). C) A timeline illustrating relevant characteristics of the early symbiosis. V. fischeri cells aggregate near the ciliated epithelial fields and enter the light organ around 4 to 6 hours post hatching. Within hours of successful colonization of the deep crypts of the light organ, the symbionts deliver an irreversible signal that triggers an extensive developmentally-programmed tissue destruction (morphogenesis) of the light organ into a mature form lacking the ciliated epithelial fields. ac, antechamber; d, ducts; dc, deep crypts; EPS, exopolysaccharide; hg, hindgut; LPS, lipopolysaccharide; p, pores; and PGN, peptidoglycan.
Studies of these early developmental events implicated the cell-envelope constituents of V. fischeri, specifically derivatives of lipopolysaccharide (LPS) and PGN, in the triggering of host tissue morphogenesis. V. fischeri is one of the few bacterial species known to release the tetrapeptide peptidoglycan monomer, 'tracheal cytotoxin' (TCT) (Cloud-Hansen et al., 2006, Rosenthal et al., 1987), which mediates the destruction of epithelial tissues in certain pathogenic associations (Cookson et al., 1989, Cloud et al., 2002). Further, as stated above, only V. fischeri can enter the crypts of E. scolopes to present these morphogens. Light organ development can be triggered in the absence of V. fischeri by exposing juvenile squid to the synergistic activity of TCT and the lipid A component of LPS (Koropatnick et al., 2004). LPS alone fails to drive light organ development but will induce chromatin condensation without progression into the later stages of apoptosis, such as DNA fragmentation (Foster et al., 1998). When added as pharmacological agents, which are readily taken up into the crypt spaces, TCT synergizes with LPS to mimic symbiont-induced chromatin condensation and epithelial regression (Koropatnick et al., 2004). While the effects of TCT and LPS on late-stage apoptosis were not investigated, this study led to the model that these biomolecules are sufficient to induce light organ morphogenesis.
The finding that V. fischeri PGN is critical for morphogenesis, coupled with the discovery of expression of PGRPs in the juvenile light organ, suggested that the EsPGRPs are good candidates to mediate the host response to bacterial PGN products. EsPGRP1 is of particular interest because its 107 expression is up-regulated during early morphogenesis (Chun et al., 2008). The derived amino acid sequence and lack of a putative signal peptide predicted that EsPGRP1 is a 23.5-kDa intracellular protein (Goodson et al., 2005). The presence of conserved amino acid residues required for NAMLAA activity suggested EsPGRP1 has PGN amidase activity.
In this study, we sought to describe the role of EsPGRP1 during early post-embryonic development of the light organ. We observed this protein in host tissues of uncolonized animals and during colonization by wild-type V. fischeri or by TCT-production mutants, as well as during the host response to pharmacological exposure to PGN and LPS derivatives. The data presented here provide evidence that PGRPs are components of the host response to mutualistic microbial associations and are integral components of the developmental response to bacterial cues in this symbiosis.
RESULTS
Characterization of the EsPGRP1 protein and an anti-EsPGRP1 antibody
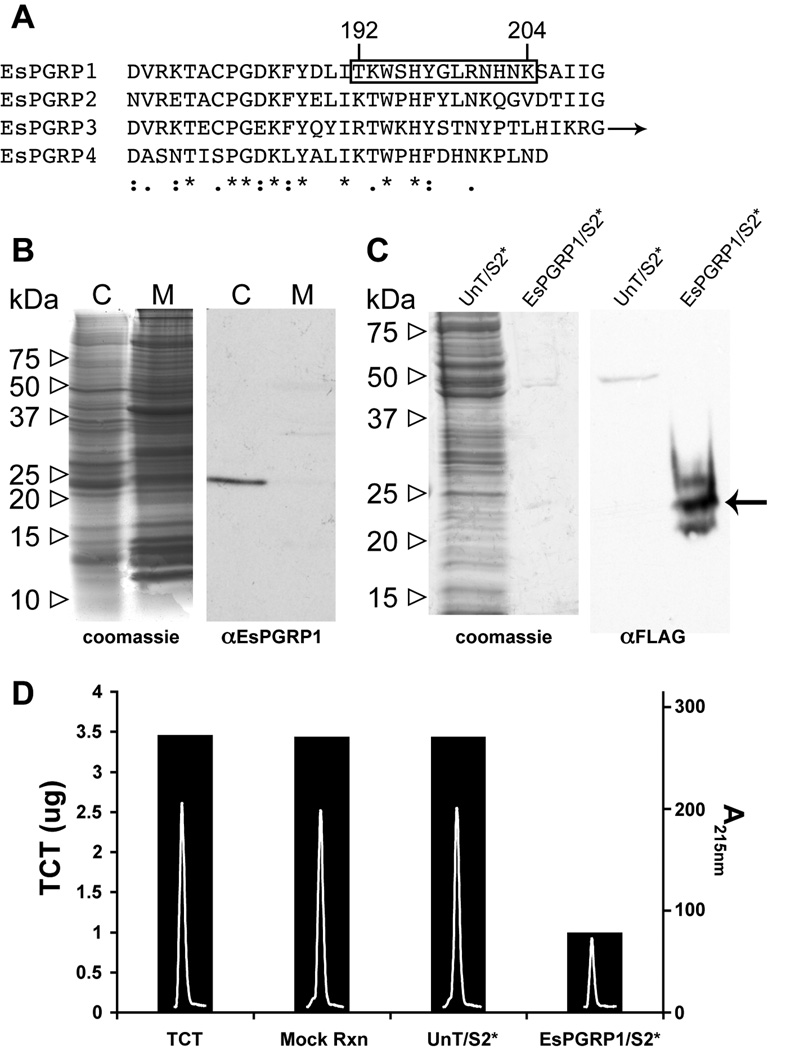
Western blot analyses determined that EsPGRP1 is present in the soluble fraction of lysed cells of whole newly-hatched animals. The EsPGRP1 antibody (Fig. 2A) reacted with a peptide at ~24 kDa, consistent with the molecular mass predicted by the derived amino acid sequence (Fig. 2B). To test the prediction that EsPGRP1 has amidase activity (Goodson et al., 2005), we transfected a FLAG-tagged EsPGRP1 expression plasmid into Drosophila S2* cells. A protein fraction enriched for EsPGRP1-FLAG was generated from these transfectants using anti-FLAG affinity chromatography (Fig. 2C). This fraction was co-incubated with TCT. Within 1 h, 64 ng of the EsPGRP1-enriched protein fraction degraded over 2.5 µg of TCT (Fig. 2D), in a manner consistent with PGRP amidase activity (Gelius et al., 2003). In contrast, 24 µg of crude protein extract of S2* cells was found to have no catalytic activity against TCT. The potential for direct interaction between EsPGRP1 and PGN was further supported by the appearance of zones of clearing on a PGN-gel zymogram at positions corresponding to EsPGRP1-bands on a companion immunoblot (Fig. S1).
Figure 2. Biochemical characteristics of the EsPGRP1 protein.
A) An alignment of the EsPGRP C-termini (arrow indicates additional C-terminal sequence for EsPGRP3). Boxed area highlights antigen sequence used for anti-EsPGRP1 antibody generation. *, identical residues; :, conserved substitutions; ., semi-conserved substitutions. B) A representative immunoblot of E. scolopes tissue. The anti-EsPGRP1 antibody reacts with a major band in the aqueous soluble fraction at ~24 kDa. C, cytoplasmic (aqueous) fraction; M, membrane (SDS soluble) fraction. C) Anti-FLAG immunoblot and companion gel of 24 µg crude protein extract from untransfected Drosophila S2* cells and 256 ng of an EsPGRP1-enriched fraction of S2* cells transfected with an EsPGRP1-FLAG expression construct. The arrow indicates an anti-FLAG reactive band at the expected molecular weight for EsPGRP1. D) An EsPGRP1 enriched EsPGRP1/S2* fraction degraded TCT in vitro, but a protein extract of untransfected S2* cells had no effect on TCT levels. Bars indicate mass of TCT (left axis) in each reaction, white peaks inside bars are the TCT peaks following treatment as the absorbance at 215nm on a reverse-phase HPLC chromatograph (right axis). UnT/S2*, untransfected S2* cell extract +; EsPGRP1/S2*, EsPGRP1-enriched fraction of S2* cells transfected with pJT28; mock rxn, TCT incubated in reaction buffer for 3 h; TCT, 7 µL 1mM TCT loaded directly onto HPLC column.
EsPGRP1 localized to epithelial nuclei
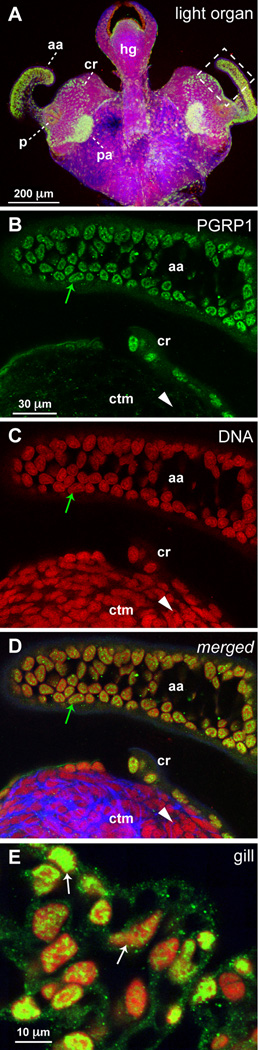
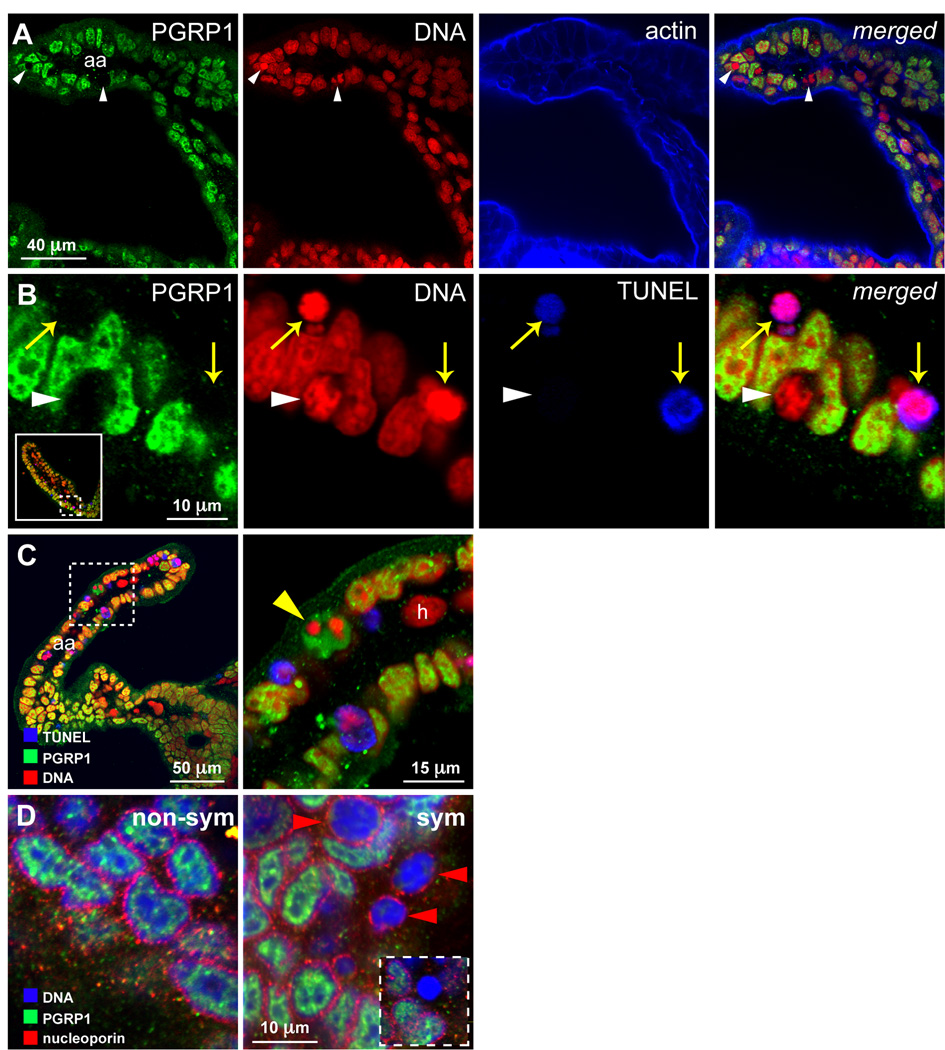
To gain insight into the possible functions of EsPGRP1 in the symbiosis, we performed confocal immunocytochemistry to localize the protein in tissues and cells. Reactivity to the EsPGRP1 antibody occurred in the nuclei of epithelia throughout the body (Fig. 3, Fig S2), including those of the light organ, gills, gut, and mantle. Unlike other epithelia, where variation in staining intensity occurs among cells, nuclear localization of EsPGRP1 in the superficial ciliated epithelium of the light organ was uniform from cell-to-cell in hatchling and in non-symbiotic animals (n > 100 individuals, from the eggs of 10 different females) (Fig. 3A–D). EsPGRP1 was detected at low levels in the connective tissue matrix of the light organ, and when present, appeared to be entirely non-nuclear (Fig. 3B, D). The nuclear distribution of EsPGRP1 was heterogeneous; anti-EsPGRP1 antibodies most intensely stained sub-nuclear regions that were weakly labeled for nucleic acids (Fig. 3–Fig 6), suggesting that EsPGRP1 does not directly bind to DNA. Pre-immune serum did not react detectably with host tissues at comparable gain settings on the confocal microscope (Fig. S3), and antibodies generated to the other three EsPGRPs did not show significant reactivity in the nuclei (data not shown).
Figure 3. Localization of EsPGRP1 in host tissue.
Confocal immunocytochemistry of hatchling and 24-h non-symbiotic E. scolopes localized EsPGRP1 to epithelial nuclei.
A) A low magnification image of the light organ. Anti-EsPGRP1 antibodies intensely labeled the ciliated epithelial fields on the lateral surfaces of the light organ. B–D) Representative higher magnification images displaying the anterior appendage and a portion of the more medial ciliated field (dashed box in A). The nuclei of these epithelial cells (aa and cr) labeled strongly with the anti-EsPGRP1 antibody (green arrows), whereas non-epithelial nuclei of the underlying tissues (ctm) appeared devoid of EsPGRP1 (white arrowheads). E) Epithelia across the body contain nuclear EsPGRP1. A representative image taken from gill tissue is shown, note variations of staining intensity in the nuclei (white arrows). aa, anterior appendage; cr, ciliated ridge; ctm, connective tissue matrix; hg, hindgut; p, pores; pa, posterior appendage. EsPGRP1, green; DNA, red; and actin cytoskeleton, blue.
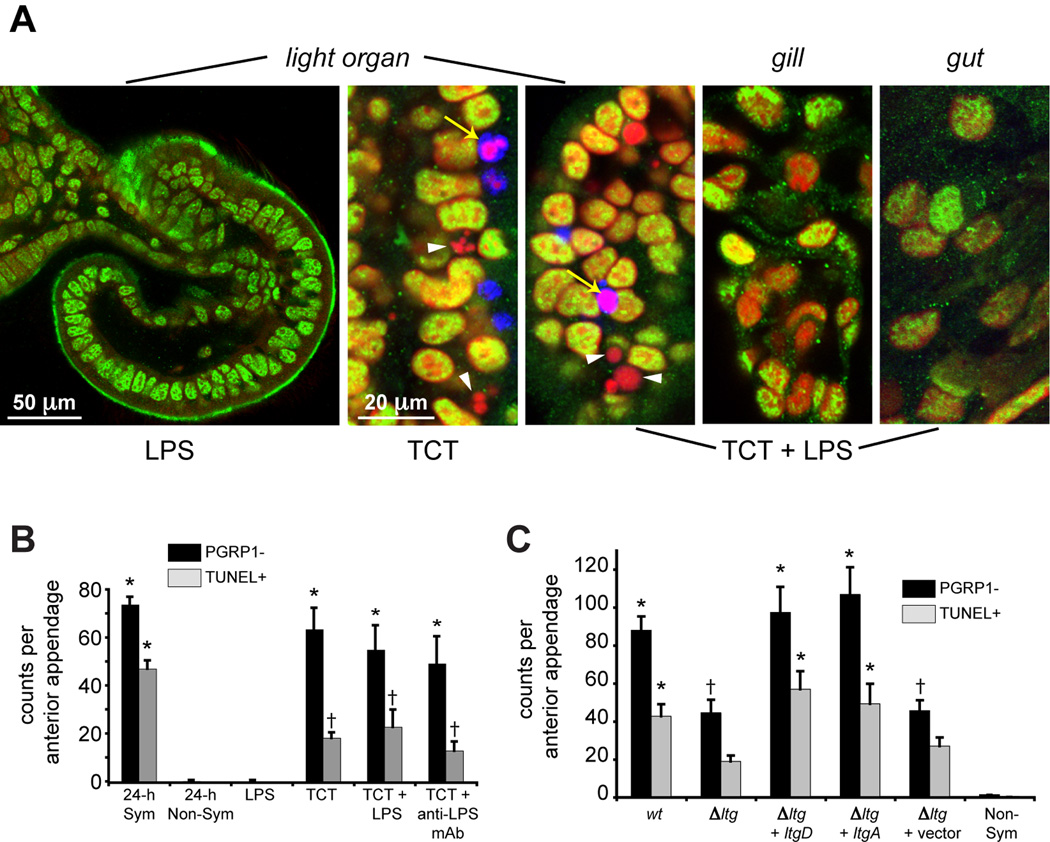
Figure 6. TCT induction of EsPGRP1 loss and late-stage apoptosis.
A) Representative confocal micrographs of E. scolopes epithelia. Hatchling E. scolopes were exposed to 10 ng/ml LPS and/or 10 µM TCT and assayed for EsPGRP1-negative and TUNEL-positive nuclei at 24 h. TCT induces EsPGRP1-negative (white arrowheads) and TUNEL-positive (yellow arrows) nuclei. EsPGRP1, green; DNA, red; TUNEL, blue. B) Direct-count estimates of EsPGRP-negative and TUNEL-positive nuclei in TCT/LPS treated light organs (n = 10). Symbols represent groupings that are statistically different from other groups. C) Direct-count estimates of EsPGRP1-negative and TUNEL-positive nuclei in light organs colonized by V. fischeri mutants defective in TCT secretion. Δltg represents mutant strain DMA388, which is defective for three lytic transglycosylases (ΔltgA, Δ ltgD, ltgY::erm). Complementation strains carry indicated gene on the plasmid pVSV107 (Dunn et al., 2006). Symbols represent groupings that are statistically different from non-symbiotic light organs († represents an intermediate level of EsPGRP1 loss that is different from both sym and non-sym). For both B and C, Statistical analysis included ANOVA with Tukey’s pairwise comparisons to identify statistically significant differences between time points (group error rate p ≤ 0.05). Error bars represent standard error.
Absence of nuclear EsPGRP1 was correlated with symbiosis-induced late-stage apoptosis in the light organ
To understand the function of EsPGRP1 during colonization, we first examined its localization at 24 h. All morphogenic processes of the superficial light organ epithelia are underway at this time. Whereas all epithelial cells of non-symbiotic animals stained similarly, a subset of the epithelial nuclei in 24-h symbiotic light organs did not stain for EsPGRP1 (Fig. 4A). The EsPGRP1-negative nuclei contained condensed chromatin, and cells with these phenotypes occurred at a frequency similar to that observed previously for apoptotic cells in the 24-h symbiotic light organ (Foster et al., 1998, Koropatnick et al., 2004). These data suggested that EsPGRP1-negative nuclei occur in the cells undergoing apoptosis. This hypothesis was supported by the finding that all nuclei undergoing DNA degradation, i.e., were TUNEL positive, a characteristic of the later stages of programmed cell death in this system (Foster et al., 1998), also lacked EsPGRP1 (Fig. 4B). The reverse did not occur, i.e., only a subset of EsPGRP1-negative nuclei was TUNEL positive. These observations provide evidence that nuclear loss of EsPGRP1 occurs prior to late-stage apoptosis.
Figure 4. Loss of EsPGRP1 during progression through apoptosis.
Confocal microscopy of symbiotic light organs revealed nuclear loss of EsPGRP1 and entry into late-stage apoptosis in anterior appendage epithelial nuclei.
A) Micrographs of an optical section of the anterior appendage; individual channels on the left, merged image on the right. Colonization of the light organ by V. fischeri for 24 h induces a subset of nuclei in the ciliated epithelial fields to lose EsPGRP1 staining (e.g., white arrowheads). B) High magnification micrographs of epithelial cells from the anterior appendage. TUNEL staining of cleaved DNA only occurs in nuclei that have lost EsPGRP1 staining (yellow arrows), but not all EsPGRP1-negative nuclei are TUNEL positive (white arrowhead). Dashed box in inset indicates area of the anterior appendage shown in high magnification. C) Micrograph of an anterior appendage. Dashed box indicates area of high magnification in right panel that displays a transient alteration of intranuclear (based on size and shape) localization of EsPGRP1 staining after chromatin condensation but prior to the loss of EsPGRP1-signal from the nucleus (yellow arrowhead) as seen in A. Note also that hemocytes (h), which have migrated to the light organ blood sinus, fail to stain with EsPGRP1. Color palette is the same as in B: EsPGRP1, green; DNA, Red; and TUNEL, Blue. D) High magnification micrographs of anterior appendage epithelial cells from non-symbiotic and symbiotic animals. Labeling of the nucleoporins (red) with the monoclonal Ab 414 indicates that loss of EsPGRP1 from the nucleus (blue) occurs prior to the dissolution of the nuclear membrane (red arrowheads). Inset in sym panel exhibits an EsPGRP1-negative nucleus with complete breakdown of the nuclear envelope.
To determine the relationship of EsPGRP1 loss to the integrity of the nucleus during apoptosis, we examined EsPGRP1 localization in the context of known nuclear apoptosis events. We observed an alternate intra-nuclear distribution of EsPGRP1, in which the DNA appeared condensed and EsPGRP1 appeared to fill the remainder of the nucleoplasm (Fig. 4C). The alternate EsPGRP1/DNA distribution was observed very rarely, suggesting that this distribution is highly transient. In addition, we investigated the temporal/spatial relationship between EsPGRP1 and nucleoporins, which during apoptosis undergo spatial redistribution in the nuclear membrane and are eventually degraded (Kihlmark et al., 2001). In EsPGRP1/nucleoporin co-staining experiments in symbiotic animals, EsPGRP1 was lost from the nucleus prior to the redistribution and breakdown of the nucleoporins (Fig. 4D). Taken together these data suggest that nuclear loss of EsPGRP1 rapidly follows chromatin condensation and precedes the breakdown of the nuclear envelope, linking nuclear loss of EsPGRP1 to the apoptotic pathway.
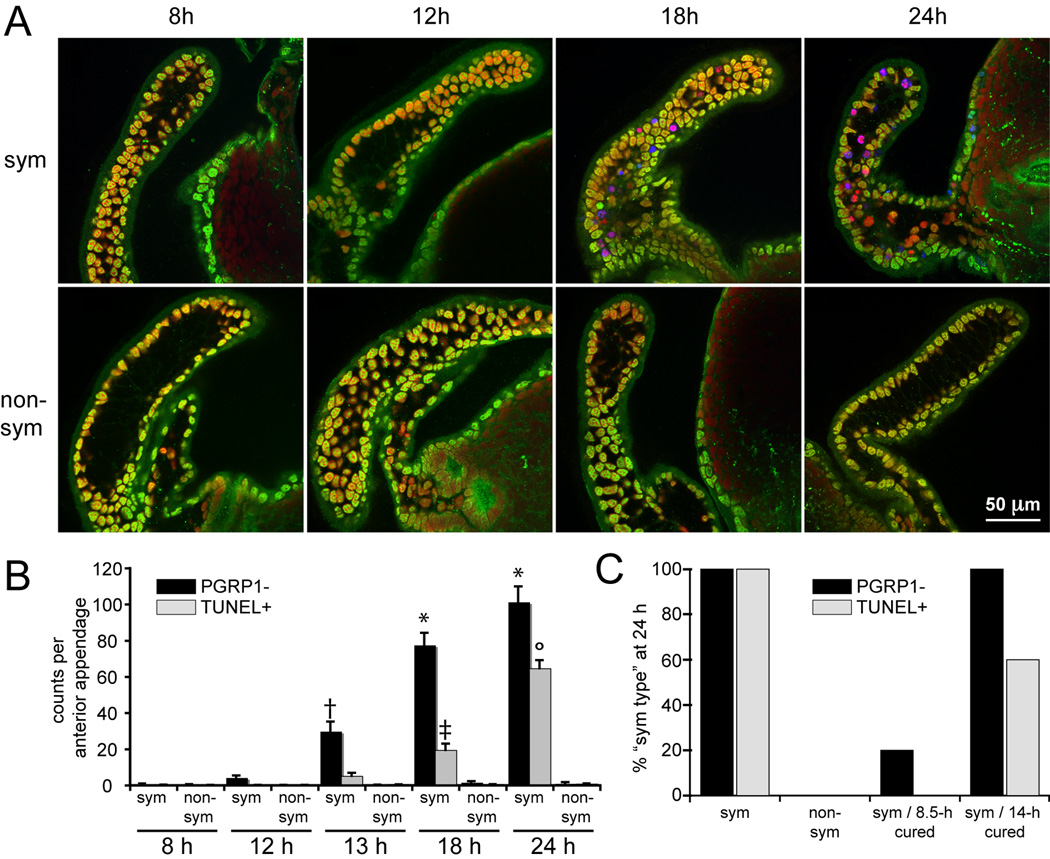
Timing of EsPGRP1 loss suggested a role in late-stage apoptosis and morphogenesis
As described above, all TUNEL-positive nuclei were EsPGRP1 negative while only a subset of EsPGRP1-negative nuclei was TUNEL positive (Fig. 4B), suggesting that nuclear loss of EsPGRP1 occurs prior to late-stage apoptosis. To characterize the timing of these processes, we enumerated EsPGRP1-negative and TUNEL-positive nuclei in anterior appendages from both symbiotic and non-symbiotic light organs at the following time points: 8 h, 12 h, 13 h, 18 h and 24 h post hatching (Fig. 5A,B). The loss of nuclear EsPGRP1 was first detected in symbiotic animals at 12 h, a time point coincident with the delivery of the irreversible signal for morphogenesis. In addition, we observed significantly greater numbers of EsPGRP-negative nuclei at 13 h, only 1 h after the morphogenesis signal. An increased level of EsPGRP1-negative nuclei was present in symbiotic animals at each time point up to 24 h. In contrast, TUNEL-positive nuclei were first detected in symbiotic light organs at 13 h, and greater numbers were not observed until 18 h, followed by another significant increase at 24 h. Greater numbers of both EsPGRP1-negative (13–24 h) and TUNEL-positive (18–24 h) nuclei were found in symbiotic compared to non-symbiotic light organs, consistent with these events being components of colonization-induced morphogenic tissue destruction. Beyond 24 h, cells were rapidly lost from the anterior appendage due to morphogenesis and counts from these later time points cannot be compared to the earlier time points. These data provide evidence that nuclear loss of EsPGRP1 precedes entry into late-stage apoptosis. As it is not possible to follow an individual cell through the entire process, it cannot be ruled out that a subset of EsPGRP1-negative nuclei never become TUNEL positive.
Figure 5. The timing of EsPGRP1 nuclear loss.
EsPGRP1 loss occurs before entry into late-stage apoptosis and is triggered by the irreversible morphogenesis signal.
A) Representative confocal micrographs from a timecourse of 8–24 h symbiotic and non-symbiotic hatchlings. EsPGRP1, green; DNA, red; and TUNEL, blue. B) Direct-count estimates of EsPGRP1-negative nuclei (black bars) and TUNEL-positive nuclei (grey bars) per anterior appendage (n=15). Statistical analysis included ANOVA with Tukey’s pairwise comparisons to identify statistically significant differences between time points (group error rate p ≤ 0.05). Symbols represent groupings that are statistically different from other groups. Error bars represent standard error. C) A qualitative comparison of light organs from a “curing experiment.” Symbiotic hatchlings were treated at 8.5 h or 14 h with 20 µg/ml chloramphenicol and scored for EsPGRP1-negative and TUNEL-positive nuclei in the anterior appendage at 24 h. The 95% confidence intervals (CI) for EsPGRP1-negative and TUNEL-positive nuclei were determined for 24-h symbiotic animals and the values for each animal from the two cured and the non-symbiotic groups were compared to the 24-h sym CI. If the number of EsPGRP1-negative or TUNEL-positive nuclei in a given anterior appendage fell within the 24-h sym CI, then that animal was considered “sym-type” for that category (protein localization or cell-death).
Because the onset of nuclear-EsPGRP1 loss coincided with the delivery of an irreversible morphogenic signal by V. fischeri at 12 h (Doino et al., 1995), we hypothesized that the EsPGRP1 phenotype was a component of the host response to this morphogenesis signal. To test this hypothesis, we performed “curing” experiments to determine whether EsPGRP1 loss is also a component of the irreversible morphogenic program. In these experiments, symbiotic juvenile E. scolopes were treated with the antibiotic chloramphenicol to clear the crypts of symbionts either before or after the 12-h time point and were subsequently assayed at 24 h (Fig. 5C). Previous experiments showed that only a small proportion of animals cured before 12 h progressed through morphogenesis, whereas 100% of animals cured at 12 h or later underwent development (Doino et al., 1995). Light organs cured of V. fischeri after 12 h were similar to fully symbiotic light organs with respect to nuclear loss of EsPGRP1 and entry into late-stage apoptosis. In contrast, 100% of light organs cured before 12 h were protected from cell death and 80% were protected from loss of EsPGRP1. These data implicate nuclear loss of EsPGRP1 in symbiotic light organs as a component of the developmental response to the irreversible morphogenic signal delivered by V. fischeri.
TCT induced nuclear loss of EsPGRP1 in light organ tissues
Because TCT and LPS together irreversibly signal morphogenesis at 12 h (Koropatnick et al., 2004), we performed experiments to determine whether TCT and LPS are sufficient to trigger EsPGRP1 loss and entry into late-stage apoptosis. We took two approaches: (i) pharmacological treatment with TCT and/or LPS (or its active component lipid A), and (ii) colonization with V. fischeri mutants defective in TCT release. While treatment with V. fischeri LPS had no effect on either the nuclear localization of EsPGRP1 or late-stage apoptosis, the presence of 10 µM TCT induced a level of nuclear loss of EsPGRP1 close to that characteristic of symbiotic tissues (Fig 6A, B). Unlike regression and hemocyte trafficking 212 (Koropatnick et al., 2004), EsPGRP1 loss was not increased by the addition LPS/lipid A with TCT (Fig. 6B). Because PGN monomers were not previously reported to induce apoptosis in the squid light organ, we postulated that a small amount of LPS contaminating the filter-sterilized Instant Ocean could have been synergizing with the TCT to induce the levels EsPGRP1 loss and TUNEL-positive nuclei observed in all TCT treatments. Alternatively, because EsPGRP1 directly interacted with PGN in vitro (Fig. 2), it remained possible that this phenotype was distinct and dependent only on the presence of PGN fragments, such as TCT. To address this matter, we included a control using a polyclonal anti-LPS antibody to inhibit any effects from contaminating LPS as previously described (Koropatnick et al., 2004). The anti-LPS antibody had no statistically significant effect on either nuclear loss of EsPGRP1 or the induction of TUNEL (Fig. 6B), suggesting that all of the effects could be attributed to the activity of TCT. A mutant of V. fischeri lacking three lytic-transglycosylase genes (Δltg), which is unable to release TCT in vitro (Adin et al., 2008), did not induce full EsPGRP1 and TUNEL phenotypes by 24 h when compared to wild-type colonized hatchlings (Fig. 6C). Some residual activity was observed in light organs colonized by the Δltg mutant, which is likely due to PGN release during normal symbiont cell turnover. Partial complementation of the triple mutant with either of two single lytic-transglycosylase genes in trans restored phenotypes comparable to wild-type colonized animals. The genetic data, coupled with pharmacological application of TCT, conclusively demonstrate that peptidoglycan monomers are necessary for both normal localization changes of EsPGRP1 and induction of late-stage apoptosis in the morphogenesis program.
The TCT-induced loss of nuclear EsPGRP1 and late-stage apoptosis was limited to the light organ epithelia. In addition to light organ tissues, we also examined the epithelia and connective tissues of the gut, gills and epidermis. In the epithelia of these three tissues, neither TCT treatment nor symbiosis was observed to have any effect on either the nuclear localization of EsPGRP1 or the induction of apoptosis (Fig. 6A and Fig S2). We observed an average of 4 TUNEL-positive epithelial cells per hindgut of non-symbiotic animals (n = 8). Comparable levels of TUNEL-positive epithelial nuclei were found in hindguts from TCT-treated animals (n = 7). Every TUNEL-positive epithelial nucleus lacked staining for EsPGRP1, a characteristic that contrasted with nuclei from neighboring non-apoptotic gut epithelial cells, which strongly stained for EsPGRP1. These data show that TCT is a specific morphogen of the light organ and that other tissues are insensitive to its effects. In addition, it appears that nuclear loss of EsPGRP1 is a common component of the apoptotic pathway in E. scolopes epithelial cells whether they are responding to TCT or some alternate apoptotic stimulus.
DISCUSSION
In this study, we characterized the cellular dynamics of EsPGRP1 in symbiont-induced tissue destruction in the light organ of E. scolopes, a process in which apoptosis is a hallmark character (Fig. 1C). We provide evidence that: 1) EsPGRP1 is a nuclear protein of the E. scolopes epithelia; 2) in the light organ, the nuclear localization of EsPGRP1 is lost during the course of symbiont-induced morphogenesis; 3) the alteration of EsPGRP1 localization is morphologically, temporally and spatially correlated with programmed cell death as a component of light organ development; and 4) nuclear loss of EsPGRP1 and subsequent entry into late-stage apoptosis are induced by the light organ morphogen, TCT, implicating EsPGRP1 dynamics in light-organ development.
The nuclear localization of EsPGRP1 is unprecedented and, to our knowledge, no other PGRPs have been reported to localize to the nucleus. While the EsPGRP1 protein contains no obvious nuclear localization signals, some proteins with molecular weights less than ~50 kDa can passively diffuse into the nucleus (Talcott et al., 1999). Passive diffusion alone would likely result in an even distribution of protein between the cytoplasm and the nucleus. However, EsPGRP1 concentrates in the nucleus relative to the cytoplasm. This uneven distribution suggests that some mechanism other than passive diffusion must also contribute to the cellular localization of EsPGRP1, e.g., binding to nuclear elements may be responsible for sequestering EsPGRP1, or EsPGRP1 may specifically bind to an unknown protein with a strong nuclear localization signal resulting in nuclear import.
Multiple lines of evidence tie the nuclear localization dynamics of EsPGRP1 to the apoptotic process. EsPGRP1 was lost from nuclei with morphologies consistent with apoptosis, and these EsPGPR1-negative nuclei occurred with a frequency similar to that observed for apoptotic cells during light organ morphogenesis (Foster et al., 1998, Koropatnick et al., 2004, Foster et al., 2000). All TUNEL-positive nuclei were EsPGRP1 negative and loss of nuclear EsPGRP1 occurred shortly after chromatin condensation, and prior to the re-distribution and breakdown of nucleoporins (Fig. 4). The timing of EsPGRP1 loss from the nucleus was consistent with that reported for apoptosis in the light organ. Chromatin condensation peaks between 12 h and 14 h (Foster et al., 1998) and the nuclear loss of EsPGRP1 was first observed to increase significantly in this time frame (Fig. 5). Further, we observed the onset of nuclear loss of EsPGRP1 consistently ahead of the onset of DNA cleavage (TUNEL) suggesting that removal of EsPGRP1 is permissive to apoptotic progression (Fig. 5). The timing of DNA cleavage observed in this study, using a fluorometric TUNEL assay, is consistent with the previously published timing of DNA cleavage and late-stage apoptosis in the light organ based on observations of cellular morphology and colorimetric TUNEL assays in histological sections (Foster et al., 1998, Montgomery et al., 1994). Thus, the timing described here is unlikely to be due to differences in sensitivity of the TUNEL and EsPGPR1 nuclear loss assays. We conclude that nuclear loss of EsPGRP1 occurs during the apoptotic process, following chromatin condensation and preceding reorganization of the nuclear envelope and DNA cleavage.
An association between PGRPs and apoptosis has begun to gain recognition. Genetic studies of PGRPs have revealed that they can both activate and be activated by the transcription factor NF-κB, a known regulator of apoptosis (Michel et al., 2001, Kaneko et al., 2004, Lang et al., 2008, Beg et al., 1996). Drosophila PGRP-LF mutants are developmentally defective in wing formation, including the aberrant appearance of apoptotic cells in the wing imaginal discs (Maillet et al., 2008). The correlation of EsPGRP1 activity and apoptosis within E. scolopes epithelial cells further supports the relationship between the PGRP family of innate immunity proteins and animal development through control and/or interaction with apoptosis pathways.
In experiments with bacterial cell-envelope components known to induce morphogenesis in the light organ, TCT was capable of inducing a full EsPGRP1 response with little or no contribution from lipid A or LPS. However, treatment with TCT or TCT + LPS failed to induce the full apoptotic response that is induced by V. fischeri, i.e., including late stage apoptosis as visualized by TUNEL (Fig. 6B). Earlier studies of apoptosis induction by TCT + LPS used an early-stage cell-death indicator, chromatin condensation visualized by acridine orange staining (Koropatnick et al., 2004). In that study, a dose response analysis was performed to determine the concentrations of TCT + LPS that most closely mimicked the onset of cell death by the symbiont, which provided the basis for concentrations used in the present study. Thus, it is likely that the failure of the TCT + LPS to induce the full program is not due to their concentration. However, subtle differences may exist in the way these molecules are presented by intact bacteria compared to pharmacological treatment, such that purified cell-envelope constituents are fully capable of inducing early-stage, but not late-stage, apoptosis. Alternatively, the symbionts may be providing a third morphogenic signal, which complements TCT + LPS under natural conditions.
Candidates for additional morphogenic signals include other cell surface molecules of the symbiont, such as the flagellins and the Syp exopolysaccharide, both of which are important early in colonization (Millikan et al., 2004, Yip et al., 2005). Symbiont luminescence, another possible signal, has been shown to have specific effects on morphogenesis. Bioluminescence is induced following colonization of the deep crypts. V. fischeri luxA mutants, which fail to produce light, are attenuated in their ability to induce host hemocyte trafficking (Koropatnick et al., 2007) and cell swelling (Visick et al., 2000), and are delayed in regression (unpublished data, Koropatnick and McFall-Ngai). It is intriguing to speculate that the slowed morphogenesis in animals colonized by luxA mutants could be due to a reduction in the speed of apoptosis due to the lack of symbiotic bioluminescence. Future studies will investigate whether dark mutants of V. fischeri are also deficient in their induction of the nuclear loss of EsPGRP1 and late-stage apoptosis.
Microarray studies of the onset of the symbiosis revealed that ESPGRP1 mRNA levels are increased at 18 h following inoculation in animals colonized by V. fischeri, i.e., concurrent with the loss of EsPGRP1 from the nuclei of the superficial epithelial field. In addition, luxA mutants fail to induce the full increase in EsPGRP1 transcript levels characteristic of colonization with wild-type V. fischeri (Chun et al., 2008). The findings that the luxA mutants are delayed in regression and show lower levels of EsPGRP1 mRNA suggest that these symbiont-triggered increases in EsPGRP1 gene transcription are important for the induction of full morphogenesis. How this up-regulation interfaces with the eventual loss of the protein from the nuclei of the cells of the superficial epithelial field remains to be determined.
It is unclear whether the activity of TCT is the result of a direct interaction with EsPGRP1 or works through an intermediary receptor/signal transduction pathway. Genetic evidence from D. melanogaster suggests TCT can be transported across host cell membranes (Kaneko et al., 2006), so it is conceivable that TCT could gain access to the cytoplasm and the nucleus, via passive diffusion, of light organ epithelial cells. We attempted to determine the location of TCT in host tissue through a suite of approaches including confocal microscopy of hatchling E. scolopes treated with FITC-labeled TCT (data not shown) (Flak et al., 1998) and NanoSIMS tracing of stable isotope labeled TCT (Fig. S4) (Lechene et al., 2006). To our knowledge, the localization of TCT in host cells has not been accomplished in any system. In the present study, we were unable to detect TCT associated with the host tissue. However, apoptotic cells in tissues that are unresponsive to TCT undergo nuclear loss of EsPGRP1, suggesting that this process is a component of apoptosis, independent of TCT induction. Thus, it seems unlikely that EsPGRP1 is responding directly to TCT. EsPGRP4 is an obvious candidate for a TCT-receptor because this protein is expressed in the light organ, contains a putative transmembrane domain, and topology prediction programs suggest that the PGRP domain would be localized on the extracellular membrane surface (Goodson et al., 2005). However, the cytoplasmic domain of this putative integral membrane protein is small, providing few clues as to possible downstream signaling pathways that could potentially result in activation of apoptosis and morphogenesis. Recently, an intranuclear bacterial pathogen of bivalve mollusks, Candidatus Endonucleobacter bathymodioli, was characterized (Zielinski et al., 2009), suggesting a selection pressure that could explain the nuclear localization of EsPGRP1. It is easy to envision that rapidly inducing apoptosis upon intranuclear invasion by a pathogen would limit the infectious potential of such a pathogen by inhibiting its access to the energy-rich molecules of the nucleus. In such a scenario, it would seem likely that this immune associated apoptosis was co-opted for developmental regulation during the establishment of the squid/vibrio mutualism.
Whereas this is the first report of a nuclear PGRP associated with apoptosis, we expect that this phenomenon is not unique. An interesting avenue for further research will be the determination of mechanistic link to apoptosis. We predict that future studies of intracellular localization of PGRPs among the marine invertebrates will reveal other nuclear members of the PGRP protein family. Further, while the study of PGRPs has primarily focused on their roles in ameliorating pathogenesis, we demonstrate a role for these proteins in mutualistic associations of animals with their bacterial partners.
Experimental Procedures
General methods
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Adult E. scolopes were caught in shallow coastal waters of Oahu and maintained and bred in the lab as previously described (Montgomery et al., 1993). Juvenile squid used in experiments were collected within 10 min of hatching, washed 3 times in filter sterilized Instant Ocean (FSIO), and placed in scintillation vials containing either FSIO (non-symbiotic) or FSIO + 5×103 cfu/ml of V. fischeri ES114 grown in LBS or SWT broth at 28°C (symbiotic). The Δltg mutant (ES114-derivative strain DMA388: ΔltgA, ΔltgD, ltgY::erm) and its derivatives were used at 2×103 CFU/ml (Adin et al., 2008). In these experiments, ES114 was used at identical concentrations to V. fischeri mutants. All animal experiments were carried out on a 12-h light/dark cycle and colonization by V. fischeri was monitored by following luminescence using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) (Ruby et al., 1993).
In experiments in which animals were cured of their symbionts, symbiotic juveniles were exposed 364 to 20 µg/ml of chloramphenicol in FSIO beginning at 8.5 h or 14 h post colonization (Doino et al., 1995) and were scored at 24 h post colonization. Non-symbiotic juveniles exposed to chloramphenicol were also analyzed as a control for any effects of the antibiotic on EsPGRP1. For experiments involving bacterial cell envelope components, juvenile E. scolopes were exposed to 10 µM TCT and/or 10 ng/ml of either V. fischeri LPS or lipid A in FSIO for 24 h. Cell envelope components were purified from V. fischeri as previously described (Cookson et al., 1989, Apicella et al., 1994).
Antibody generation and western blots
Rabbit polyclonal antibodies were raised against an EsPGRP1 synthetic peptide conjugated to ovalbumin (Harlan Biosciences, Indianapolis, IN). The synthetic peptide of 13 amino acids, WSHYGLRNHNKSA, was selected from the C-terminal region of EsPGRP1 because it is predicted to be highly antigenic (Lasergene Protean, DNA Star, Madison, WI) and because it contains little similarity to the other EsPGRPs (Fig. 2A, Fig S1) or other sequences in the light organ EST database. Upon western blot analysis to test the specificity of the antisera for the EsPGRP1 antigen, squid tissues were homogenized in 20 mM sodium phosphate pH 7.4, 100 mM sodium chloride. The aqueous soluble or cytosolic fraction was obtained as the supernatant of a centrifugation of the sample at 14,000 × g for 15 min. The pellet was then washed 3 times with the homogenization buffer and the membranes were extracted with homogenization buffer containing 1% SDS. Protein concentrations were determined using the Bradford assay. The proteins were then separated on 12.5% SDS-PAGE gel and blotted onto a PVDF membrane (Bio-Rad, Hercules, CA). The blot was blocked overnight in 20 mM Tris, pH 7.4, 300 mM sodium chloride, 5% bovine serum albumin (BSA), and 1% goat serum (Jackson ImmunoResearch Labs, West Grove, PA). Anti-EsPGRP1 antibody was diluted 1:100 in the block solution and incubated with the blot overnight at room temperature. The blot was then incubated with the secondary antibody, a goat anti- rabbit horseradish peroxidase-conjugate (Jackson ImmunoResearch Labs, West Grove, PA) at a 1:3000 dilution. Reactivity was visualized using Pierce ECL Chemiluminescent Western Blot kit (Pierce, Rockford, IL).
TCT degradation assays
The EsPGRP1 coding sequence was PCR amplified with a C-terminal FLAG-tag using the following DNA primers: JT73, ACAGTGGATCCATGCACCATCACCATCACCATATGCACGCGTTCGGAG; and JT74, ACTGTGCGGCCGCTTATTTATCGTCATCGTCTTTGTAGTCACCAATAATGGCACTTT. The PCR product was subsequently cloned into the pPAC-PL plasmid using the BamHI and NotI restriction enzymes. Plasmid pJT28 was constructed by subcloning the FLAG-tagged EsPGRP1 coding sequence into the pRmHa-3 expression plasmid downstream of a Cu2+-inducible metallothionien promoter by EcoRI and SalI restriction of the PCR product of the following primers: JT136, TACTTGAATTCATGCACCATCACCATCACCATATGCACGCGTTCG; and JT134, ACTCTGTCGACTTATTTATCGTCATCGTCTTTGTAGTC. Drosophila S2* cells were stably transfected with pJT28 as previously described (Silverman et al., 2000). EsPGRP1 protein was induced by addition of 500 µM CuSO4 into the media for 8–12 hours. Cells were then harvested, washed with PBS and frozen at −80°C.
The Cu2+-induced pJT28/S2* cells were resuspended in lysis buffer (20 mM Tris, pH 7.4, protease inhibitor cocktail III and 1mM PMSF) and incubated on ice for 10 min. Sodium chloride was then added to a final concentration of 100 mM and the insoluble material was separated by centrifugation at 20,000 × g for 10 min at 4°C. The supernatant was passed over an anti-FLAG antibody affinity column three times and the column was subsequently washed with 500 column volumes of TBS (20 mM Tris, pH 7.4, 150 mM Sodium chloride). Bound proteins were eluted from the column with 100 µM FLAG-peptide. Fractions containing high levels of the EsPGRP1 protein were determined by anti-FLAG immunoblots. These fractions were combined and dialyzed against storage buffer (20 mM Tris, pH 7.4, 300 mM Sodium chloride, 1mM DTT and 50% glycerol) and 411 stored at −20°C. Anti-FLAG immunoblots were performed as above except using using 4% non-fat dry milk/TBS as the blocking solution and a 1:1,000 dilution of rabbit polyclonal anti-FLAG antibodies for 1 h at room temperature.
To perform the TCT-degradation assay, ~370 ng of the FLAG-purified EsPGRP1 protein fraction was diluted into 500 µL of reaction buffer (50 mM ammonium acetate, pH 7, 550 mM Sodium chloride, 4 µM ZnSO4 and 1% BSA) and concentrated to 9.2 ng/µL on a 3-kDa molecular weight cutoff microcon column (Millipore, Billerica, MA). 64 ng of the EsPGRP1 protein fraction was mixed 1:1 with 1mM TCT. The reaction was incubated at room temperature for 1 h, diluted with 86 µL HPLC-grade water and loaded onto a Betasil C18 reverse phase column (Thermo Hypersil-Keystone, Bellefonte, PA). The TCT was mobilized using a 0–15% acetonitrile gradient and detected by absorbance at 215 nm. The crude S2* cell protein extract was generated as above, excluding the anti-FLAG affinity fractionation step. To control for S2* cell-derived amidase activity, which could be contaminating the EsPGRP1-enriched protein fraction, 24 µg of the crude S2* cell protein extract was incubated with TCT for 2 h.
Immunocytochemical detection of EsPGRP1 in E. scolopes tissues
Unless otherwise noted, all fluorophores were obtained from Molecular Probes (Invitrogen, Carlsbad, CA). To prepare juvenile squid for immunocytochemistry, they were anaesthetized in FSIO + 2% ethanol and fixed with 4% paraformaldehyde in mPBS (50 mM sodium phosphate, pH 7.4, 0.45M sodium chloride) for 18 h at 4°C. Post-fix samples were kept at 4°C throughout the remainder of the protocol. Fixed squid were washed 4 times for 30 min in mPBS. The light organs were removed from each juvenile, permeabolized with 1% Triton-X-100 in mPBS (mPBST) for 2 days and then blocked overnight (mPBST, 0.5% BSA, 1% goat serum). Light organs were then exposed to the EsPGRP1 antisera diluted 1:1000 in the block solution for 7 days. Light organs were washed 4 times for 1 h each wash with mPBST and incubated overnight again in blocking solution. Samples were then exposed overnight to fluorescent goat anti-rabbit antibody conjugates (FITC or TRITC, Jackson ImmunoResearch Labs; AlexaFluor 633) diluted 1:25 in blocking solution overnight. To counterstain the actin cytoskeleton, light organs were incubated with 165 nM Alexa Fluor 633-phalloidin in mPBST overnight. To counterstain nuclear DNA, light organs were incubated with 100 µg/ml RNaseA in 2×SSC (30 mM sodium citrate, pH 7.2, 300 mM sodium chloride) for 30 min at 37°C and then stained with either 1.5 µM propidium iodide for 5 min or 2 µM TOTO-3 for 30 min at room temperature. Apoptotic chromosomal cleavage was detected using the DeadEnd Fluorimetric TUNEL Assay kit (Promega, Madison, WI) according to manufacturer’s instructions. The nuclear pore complex was visualized by immunocytochemistry as above, except the anti-nucleoporin mAb 414 (Covance Research Products, Denver PA) was co-diluted 1:1000 with the anti-PGRP1 Ab. Light organs were mounted in Vectashield anti-bleaching medium (Vector Labs, Burlingame, CA) and viewed on an LSM510 laser scanning confocal microscope (Carl Zeiss Microimaging, Thornwood, NY).
Supplementary Material
Acknowledgements
We are grateful to Michael Apicella for kindly providing the V. fischeri LPS and lipid A used in this study. We thank N. Bekiares, M C. Brennan, J. Dillard, H. Goodrich-Blair, L. Knoll, Mandel, B. Rader and E. Ruby for helpful comments on the manuscript. This work was funded by NIH RO1-AI50661 to MMN, NSF IOS 0817232 to MMN and EG Ruby, NIH RR R01-12294 to EG Ruby, and by NIH NRSA AI55397 to JVT through the Microbial Pathogenesis and Host Responses Training Program.
Reference List
- Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2008 doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselme C, Perez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, Latorre A, et al. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol. 2006;72:6766–6772. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella MA, Griffiss JM, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994;235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-alpha -induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, Dillard JP. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect Immun. 2002;70:2752–2757. doi: 10.1128/IAI.70.6.2752-2757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- Cookson BT, Cho HL, Herwaldt LA, Goldman WE. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun. 1989;57:2223–2229. doi: 10.1128/iai.57.7.2223-2229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coteur G, Mellroth P, De Lefortery C, Gillan D, Dubois P, Communi D, Steiner H. Peptidoglycan recognition proteins with amidase activity in early deuterostomes (Echinodermata) Dev Comp Immunol. 2007;31:790–804. doi: 10.1016/j.dci.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Doinox JA, McFall-Ngai MJ. A Transient Exposure to Symbiosis-Competent Bacteria Induces Light Organ Morphogenesis in the Host Squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006a;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006b;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. doi: 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flak TA, Goldman WE. Muramyl peptide probes derived from tracheal cytotoxin of Bordetella pertussis. Anal Biochem. 1998;264:41–46. doi: 10.1006/abio.1998.2826. [DOI] [PubMed] [Google Scholar]
- Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- Foster JS, McFall-Ngai MJ. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev Genes Evol. 1998;208:295–303. doi: 10.1007/s004270050185. [DOI] [PubMed] [Google Scholar]
- Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem Biophys Res Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ. Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D. Peptidoglycan recognition proteins-maintaining immune homeostasis and normal development. Cell Host Microbe. 2008;3:273–274. doi: 10.1016/j.chom.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kihlmark M, Imreh G, Hallberg E. Sequential degradation of proteins from the nuclear envelope during apoptosis. J Cell Sci. 2001;114:3643–3653. doi: 10.1242/jcs.114.20.3643. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- Lang MF, Schneider A, Kruger C, Schmid R, Dziarski R, Schwaninger M. Peptidoglycan recognition protein-S (PGRP-S) is upregulated by NF-κB. Neurosci Lett. 2008;430:138–141. doi: 10.1016/j.neulet.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Mcfall-Ngai M. Embryonic-development of the light organ of the sepiolid squid Euprymna-scolopes berry. Biological Bulletin. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai M. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Park JW, Kim CH, Kim JH, Je BR, Roh KB, Kim SJ, et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci U S A. 2007;104:6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Rosenthal RS, Nogami W, Cookson BT, Goldman WE, Folkening WJ. Major fragment of soluble peptidoglycan released from growing Bordetella pertussis is tracheal cytotoxin. Infect Immun. 1987;55:2117–2120. doi: 10.1128/iai.55.9.2117-2120.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev. 2004;198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, et al. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc Natl Acad Sci U S A. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci U S A. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talcott B, Moore MS. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm. Bombyx mori. J Biol Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang SM, Zeng Y, Loker ES. Characterization of immune genes from the schistosome host snail Biomphalaria glabrata that encode peptidoglycan recognition proteins and gram-negative bacteria binding protein. Immunogenetics. 2007;59:883–898. doi: 10.1007/s00251-007-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski FU, Pernthaler A, Duperron S, Raggi L, Giere O, Borowski C, Dubilier N. Widespread occurrence of an intranuclear bacterial parasite in vent and seep bathymodiolin mussels. Environmental Microbiology. 2009 doi: 10.1111/j.1462-2920.2008.01847.x. In Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.