Abstract
To prevent duplicate DNA synthesis, metazoan replication origins are licensed during G1. Only licensed origins can initiate replication, and the cytoplasm interacts with the nucleus to inhibit new licensing during S phase. DNA replication in the mammalian one cell embryo is unique because it occurs in two separate pronuclei within the same cytoplasm. Here, we first tested how long after activation the oocyte can continue to support licensing. Because sperm chromatin is licensed de novo after fertilization, the timing of sperm injection can be used to assay licensing initiation. To experimentally skip some of the steps of sperm decondensation, we injected mouse sperm halos into parthenogenetically activated oocytes. We found that de novo licensing was possible for up to 3 hr after oocyte activation, and as early as 4 hr before DNA replication began. We also found that the oocyte cytoplasm could support asynchronous initiation of DNA synthesis in the two pronuclei with a difference of at least 2 h. We next tested how tightly the oocyte cytoplasm regulates DNA synthesis by transferring paternal pronuclei from zygotes generated by intracytoplasmic sperm injection (ICSI) into parthenogenetically activated oocytes. The pronuclei from G1 phase zygotes transferred into S phase ooplasm were not induced to prematurely replicate and paternal pronuclei from S phase zygotes transferred into G phase ooplasm continued replication. These data suggest that the one cell embryo can be an important model for understanding the regulation of DNA synthesis.
Keywords: Mouse pronuclear replication, DNA origin licensing, delayed sperm injection
Introduction
The 6 billion base pairs of the mouse genome is replicated in the cell with over 103 to 105 different start sites, called replication origins, dispersed throughout the genome [Pardoll et al., 1980; Vogelstein et al., 1980]. The entire genome is replicated completely, and each segment is only replicated once. This occurs in a highly coordinated manner during the 6 hour S phase of most mammalian somatic cells. Different parts of the genome are replicated at different times [Stubblefield, 1975], and in different structural foci in the nucleus [Berezney, 1991; Berezney, 2002]. In order to prevent any of these origins from firing more than once during S phase and “rereplicating” parts of the genome, each origin is licensed during G1 [Blow, 1993; Leno et al., 1992]. This is regulated by the sequential binding of several different proteins, beginning with the six protein origin recognition complex ORC1L-ORC6L [DePamphilis, 2005; Thomae et al., 2008], followed by CDC6 and CTD1, and ending with the helicases MCM2-7 [DePamphilis et al., 2006; Krude, 2006; Takeda and Dutta, 2005]. DNA synthesis is initiated by CDK2/CCNA2 and CDK2/CCNE1, which simultaneously removes CDC6, thereby constitutively inhibiting additional licensing during S phase [DePamphilis et al., 2006; Diffley, 2004]. Because only licensed origins can initiate replication, and new licensing is inhibited during S phase, this mechanism ensures that every segment of the entire genome is only replicated once.
Two unique features of fertilization raise questions about how licensing and DNA replication is regulated in the mammalian one-cell embryo. The first is that the maternal and paternal genomes are replicated in two distinct nuclei within the same cytoplasm [Sirlin and Edwards, 1959]. In somatic cells, the cytoplasm participates in the regulation of licensing. Cells at the G1 phase can be induced to prematurely initiate DNA synthesis (and thereby inhibit new licensing) by incubation in S phase cytoplasm [Krude, 2006; Rao and Johnson, 1970]. Because of this cytoplasmic regulation, the initiation of DNA synthesis in the two zygotic pronuclei might be expected to be synchronous. However, this is controversial. Several reports have suggested that in the mouse DNA replication begins in the male pronucleus up to 2 hr before it does in the female pronucleus in embryos generated by normal mating [Bouniol-Baly et al., 1997; Ferreira and Carmo-Fonseca, 1997; Howlett and Bolton, 1985; Luthardt and Donahue, 1973], while in in vitro fertilization (IVF) this asynchrony was less apparent [Aoki and Schultz, 1999; Howlett and Bolton, 1985]. This raises questions about the role of the cytoplasm in the regulation of DNA synthesis in the one-cell embryo.
The second unique feature of mammalian zygote is that the two genomes enter the cell cycle in very different forms. The maternal chromatin in the unfertilized oocyte remains in metaphase II of meiosis, which is not dissimilar from mitotic chromosomes. In mammalian somatic cells, licensing may begin very soon after the M/G1 transition [Stoeber et al., 2001], so the maternal chromatin is poised for licensing immediately after fertilization. The male genome, in contrast, must first go through a complicated remodeling process in which protamines are replaced by histones throughout the chromatin. Licensing would not be expected to be initiated until after the paternal pronucleus is fully formed, up to 4 hr after fertilization [Ajduk et al., 2006].
We have shown previously that when the male pronucleus is formed from sperm with damaged DNA, the complete chromosomal degradation ensues at the onset of DNA synthesis, while in the same zygote the female pronucleus replicates its DNA normally [Yamauchi et al., 2007]. These data indicate that it is possible for the two zygotic pronuclei to respond independently to replication signals from the same cytoplasm, but support a signaling role of the cytoplasm to initiate DNA synthesis in the zygote. In this work, we tested whether the oocyte maintains the ability to initiate DNA licensing after activation by injecting sperm and sperm halos into oocytes at various time points after parthenogenetic activation. We then explored the regulation of the DNA synthesis by the cytoplasm by transferring pronuclei from G1 zygotes to parthenogenetic S phase zygotes, and S phase pronuclei into G1 zygotes.
Materials and Methods
Chemicals
Mineral oil was purchased from Squibb and Sons (Princeton, NJ); eCG and hCG were from Calbiochem (San Diego, CA). All other chemicals were obtained from Sigma Chemical Co. (St. Louis, MO) unless otherwise stated.
Animals
B6D2F1 (C57BL/6J × DBA/2) mice were obtained at 6 wk of age from the National Cancer Institute (Raleigh, NC). The mice were fed ad libitum with a standard diet and maintained in a temperature and light controlled room (22°C, 14L:10D), in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and those prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Resources National Research Council. The protocol for animal handling and the treatment procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Hawaii.
Media
Medium T6 [Quinn et al., 1982] was used for IVF, and HEPES-buffered CZB medium (HCZB [Chatot et al., 1989; Kimura and Yanagimachi, 1995] was used for gamete handling and ICSI. Medium CZB [Chatot et al., 1989] was used for embryo culture. Both CZB and T6 were maintained in an atmosphere of 5% CO2 in air, and HCZB was maintained in air.
Preparation of swim-up spermatozoa
Caudae epididymides were removed from one male, and the epididymal fluid was forced out with a small pair of tweezers. Droplets of dense sperm mass fromthe caudae epididymides were allowed to sink to the bottom of a 1.5ml tube containing 400 µl of HCZB. Spermatozoa were allowed to swim up for 5 min at room temperature and were then taken for injections.
Preparation of sperm nuclear halos
Epididymal sperm were extracted into 1 ml of 50 mM Tris on ice, pH 7.4. Then, the sperm were homogenized with a pipette tip, then 0.5% SDS was added, and the suspension spun at 2,000 g × 5 min. The pellet was re-suspended in 200 µl of 2 M NaCl, 25 mM Tris, pH 7.4, and divided into two aliquots. One aliquot was diluted with another 100 µl of 2 M NaCl, and added dithiothreitol to 5 mM. Both aliquots were then kept on ice until injected into oocytes. Nuclei treated with dithiothreitol formed nuclear halos, while those without dithiothreitol (salt extracted nuclei) did not. Halo formation was confirmed by fluorescence microscopy, by adding ethidium bromide (100 µg/µl) [Nadel et al., 1995]. Although sperm nuclear halos were optically less dense than normal sperm nuclei, they could be easily seen with the Hoffmann interference-contrast microscope.
Collection and parthenogenetic activation of oocytes
Mature females, 8 to 12 wk old, were induced to superovulate with i.p. injections of 5 IU equine (eCG) and 5 IU hr human chorionic gonadotropin (hCG) given 48 hr apart. Oviducts were removed 14–15 hr after the injection of hCG and placed in HCZB [Kimura and Yanagimachi, 1995]. The cumulus-oocyte complexes were either used for IVFor released from the oviducts into 0.1% bovine testicular hyaluronidase (300 USP units/mg) in HCZB medium to disperse cumulus cells. The cumulus-free oocytes were washed with HCZB medium and either used immediately for ICSI or artificially activated by treatment with Ca2+-free CZB containing 5 mM SrCl2 for up to 4 hr at 37°C under 5% CO2 in air, washed, and incubated in CZB until sperm injection.
In vitro fertilization
The method for sperm capacitation and IVF using T6 medium has been reported by us before [Ajduk et al., 2006]. Briefly, 200 µl drops of T6 medium (fertilization drops) were overlaid with mineral oil in a plastic culture dish (60 mm diameter) and equilibrated overnight at 37°C in a humidified atmosphere of 5% CO2 in air. The volume of sperm suspension added to the fertilization drop was dependent on the concentration of spermatozoa after dispersion in capacitation drop. Generally, 10 µl of sperm suspension from capacitation drop was added to each fertilization drop to give final sperm concentrations of approximately 2–3 106/ml. The contents of four oviducts were released into each fertilization drop. Gametes were co-incubated for 60 min. Preliminary experiments demonstrated that this was a minimal time to maintain high fertilization rates. When gametes were co-incubated for shorter time (30 min), only ~20% of oocytes were fertilized. After gamete co-incubation, the oocytes were washed several times with HCZB medium, followed by at least one wash with CZB medium. Only morphologically normal oocytes were selected for culture and analysis. In contrast with ICSI, in IVF the exact fertilization time could not be clearly defined. Gametes were co-incubated for 60 min. The ‘‘0-min’’ timepoint reflecting approximate moment of sperm-oocyte fusion was defined as 30 min after initiation of gamete co-incubation.
Intracytoplasmic sperm injection
Injection of sperm was carried out as described recently by Szczygiel and Yanagimachi [Szczygiel and Yanagimachi, 2003]. Briefly, a small drop of sperm suspension was mixed thoroughly with an equal volume of HCZB containing 12% (w/v) polyvinyl pyrrolidone (Mr 360 kDa) immediately before ICSI. The injections were performed with Eppendorf Micromanipulators (Micromanipulator TransferMan, Eppendorf, Germany) with a Piezo-electric actuator (PMM Controller, model PMAS-CT150; Prime Tech, Tsukuba, Japan).
In the standard ICSI, a single spermatozoon was drawn, tail first, into the injection pipette and moved back and forth until the head-midpiece junction (the neck) was at the opening of the injection pipette. The head was separated from the midpiece by applying one or more piezo pulses. After discarding the midpiece and tail, the head was redrawn into the pipette and injected immediately into an oocyte. Sperm-injected oocytes were transferred into CZB medium and cultured at 37°C, 5% CO2 in air. The oocytes were taken for further processing at fixed at specific timepoints after injection. For evaluating the timing of DNA replication in maternal and paternal pronuclei, the oocytes were injected in groups of 5 to 10 eggs, and the time of injection per group was less than 10 min. The ‘‘0-min’’ timepoint reflecting the moment of fertilization was defined as the end of injection of one group of oocytes. In both nuclear halo and salt extracted sperm, sperm head and tail had separated during SDS treatment. The oocytes injected with halo and salt extracted sperm were activated immediately after injection by treatment with Ca2+-free CZB containing 5 mM SrCl2 for 4 hr and transferred into CZB at 37°C under 5% CO2 in air. In the delayed ICSI, swim-up sperm, halo, and salt extracted sperm were injected into oocytes 1 hr, 2 hr, 3 hr, and 4 hr after activation (i.e. after the oocytes were placed in medium containing SrCl2) and cultured in CZB at 37°C under 5% CO2 in air.
Pronuclear transfer (PN transfer)
Pronuclear transfer into artificially activated oocytes was performed with micromanipulator. Pronuclear stage oocytes providing the transferred pronuclei were produced by injecting swim up sperm. Oocytes either 4 hr or 7 hr after ICSI were placed into CZB with 5 µg/ml cytochalasin B for 10 min. Then, the oocytes were transferred into HCZB containing 5 µg/ml cytochalasin B. The male pronucleus (transferred pronucleus) was extracted from an oocyte into injection pipette (10 µm i.d.). The pronucleus was transferred into a drop of HCZB containing 12% PVP and freed from attached cytoplasm by drawing in and out of the injection pipette repeatedly. The pronucleus was then injected into an oocyte activated 3 hr or 7 hr earlier (Fig. 1). Maternal and paternal pronuclei were differentiated based on size and location within the oocyte. Paternal pronuclei are larger [Aoki and Schultz, 1999; Austin, 1961; Edwards and Sirlin, 1956; Sirlin and Edwards, 1959] and during the first hours after ICSI are located opposite to the opening in zona pellucida (easily distinguishable under Hoffman phase contrast). The maternal pronuclei are smaller and localize diagonally to paternal pronuclei.
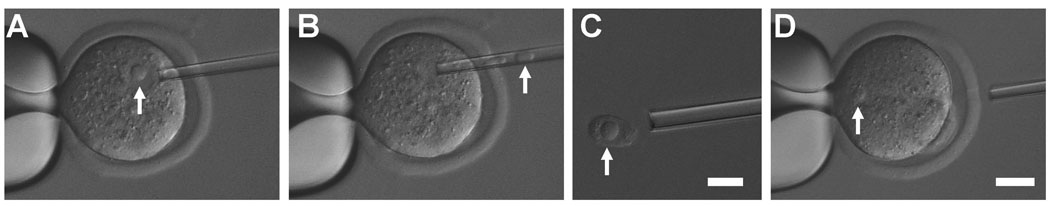
Figure 1. Transfer of Pronuclei from Transferred Oocyte to Acceptor Oocyte.
(A) A pipette is inserted next to a pronucleus in a donor zygote resulting from ICSI done 7 hr earlier. (B) The transferred pronucleus is taken into the pipette by suction and (C) extruded into the media to demonstrate its size (this was not routinely done). Note the large nucleolus (shown at higher magnification). (D) The transferred pronucleus was injected into the acceptor parthenogenetically activated oocyte. Arrow indicates the transferred pronucleus. Scales: for A, B, and D, Bar = 50 µm; for C, Bar = 20 µm.
Assessment of DNA Replication
DNA replication analysis was performed according to the procedure described previously [Adenot et al., 1997] with modifications [Ajduk et al., 2006]. Briefly, sperm injected oocytes or pronuclei transferred oocytes were transferred into CZB with 10 µM 5-bromo-2-deoxyuridine (BrdU) at different time points and incubated for 30 min or longer. BrdU was prepared as a 1 mM stock solution in DMSO, and 10 µl was added to 990 µl of media for BrdU incubation. Following incubation in BrdU, the oocytes were fixed with 2.5% paraformaldehyde in PBS (phosphate buffered saline, 10 mM Na1/2H2/1PO4) with 0.5 M NaOH, pH 7.3, for 15 min at room temperature. Fixed oocytes were washed in PBS containing 10% fetal bovine serum (FBS) and 0.2% Triton X-100 (TX-100) and blocked in the same solution for 30 min at 37°C. The oocytes were then washed in PBS containing 2% FBS and 0.1% TX-100 (PBS: 2% FBS/0.1% TX-100). Oocytes were then incubated in drops with anti-BrdU antibody conjugated with Alexa Fluor 488 (Invitrogen) and diluted 1:19 in PBS (2% FBS/0.1% TX-100) for 1 hr at 37°C. The oocytes were placed on poly-lysine (1 mg/ml)-coated microscope slides. The preparations were covered with VectaShield mounting media containing propidium iodide (Vector Laboratories, Burlingame, CA) and examined using a fluorescence microscope (Nikon Eclipse E600 at ×200 magnification) fitted with the appropriate filters.
Results
Licensing can occur up to three hours after parthenogenetic activation
We first tested whether the oocyte maintains the ability to initiate DNA replication origin licensing for several hours after activation. Spermatozoa contain unlicensed DNA. Therefore, if DNA replication occurs after delayed sperm injection, the licensing must be de novo, indicating that the oocyte still has the ability to license DNA for replication. We first verified that 1 hr oocyte incubation in SrCl2 is sufficient to result in oocyte activation; 92% (23/25) of the oocytes incubated in SrCl2 for this period of time extruded 2nd polar bodies and formed pronuclei. This agrees with a previous report that suggested that incubation for as little as 20 min in SrCl2 can activate mouse oocytes [Kishigami et al., 2004]. For all experiments described below, activation time was measured from the point when the oocytes were placed in medium containing SrCl2.
We injected spermatozoa and sperm halos into parthenogenetically activated oocytes up to 7 hr after activation, long after DNA synthesis is normally programmed to begin (Fig. 2, top diagram) and tested for the ability of the paternal pronuclei to replicate their DNA. The maternal pronucleus in each oocyte served as an internal control and in all experiments, only oocytes in which the maternal pronucleus incorporated BrdU were counted. After sperm injection, the oocytes were incubated in BrdU for 3 to 14 hr, and fixed. The percentage of oocytes with both pronuclei incorporating BrdU is shown in Fig. 2.
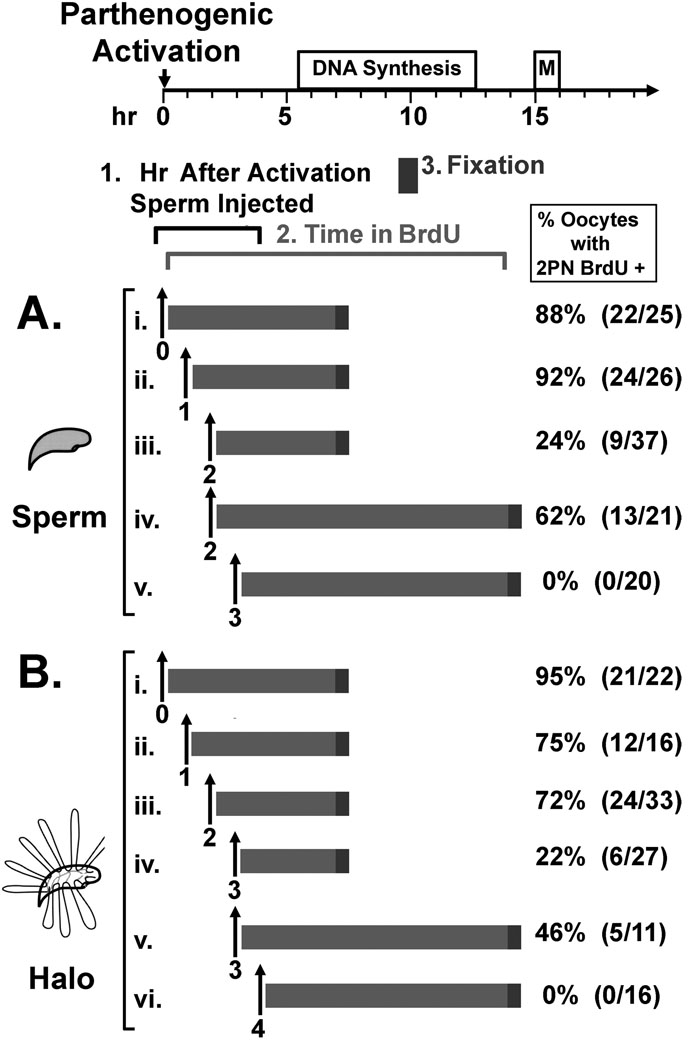
Figure 2. Delayed ICSI to Test Paternal DNA Replication.
Mouse sperm heads (A) or sperm nuclear halos (B) were injected into MII oocytes (i) or into oocytes 1 to 4 hr after (ii – vi) after parthenogenetic activation. After injection, the oocytes were incubated in BrdU for the indicated times (light grey bars) then fixed for 30 min (dark grey bars), and stained for BrdU using anti-BrdU antibodies. The percentages of oocytes that contained 2 pronuclei that were positive for BrdU are shown on the right. The number of oocytes that were positive/number injected are shown in parentheses. BrdU incubation bars and injection times are placed along the timeline for DNA synthesis and the first mitotic division of the mouse one cell embryo after ICSI, shown at the top of the diagram. In all cases, only oocytes in which the maternal parthenogenetic pronucleus was positive for BrdU (internal controls) incorporation were counted.
We have previously shown that when mouse sperm nuclei are injected into oocytes, DNA synthesis normally begins at about 5.5 hr after fertilization [Ajduk et al., 2006]. Previous work from two other laboratories demonstrated that when mouse sperm nuclei were injected into oocytes up to 2 hr [Maleszewski et al., 1999] or 3 hr [Kishigami et al., 2004] after parthenogenetic activation, DNA synthesis still occurred in the paternal pronucleus and a low percentage of these developed into pups when transferred to foster mothers. In our hands sperm nuclei injected 2 hr after oocyte activation developed into pronuclei that replicated DNA. A low percentage of these paternal pronuclei (24%) replicated their DNA at a similar time as the maternal pronuclei (Fig. 2A.iii.). We also repeated the previous report [Maleszewski et al., 1999] and showed that 2 hr post activation is the limit for which the mouse oocyte can support sperm decondensation, as none of the oocytes injected at 3 hr post activation developed male pronuclei that replicated DNA (Fig. 2A.v.). None of the oocytes injected with mouse sperm between 4 and 7 hr post activation replicated paternal DNA (data not shown).
We next tested whether we could increase the time after oocyte activation at which the paternal genome could be injected into oocyte and still form pronuclei that were able to replicate DNA. Discerning the timing of the licensing of the paternal genome after fertilization is complicated by the requirement for sperm chromatin to undergo remodeling before licensing can begin [Ajduk et al., 2006]. In an attempt to experimentally skip some of the steps of sperm this remodeling, we injected sperm halos into oocytes at various times after activation. Sperm halos are sperm nuclei devoid of all protamines and histones, with naked, protein free DNA attached to the nuclear matrix in loops at 50 kb average intervals [Nadel et al., 1995]. We have previously demonstrated that when halos were injected into MII oocytes paternal pronuclei formed, and the DNA was replicated and progressed to form normal paternal mitotic chromosomes, indicating that one complete round of DNA synthesis occurred [Mohar et al., 2002; Shaman et al., 2007]. Sperm halos were injected into MII oocytes up to 7 hr after oocyte activation, and the oocytes were incubated in BrdU for 7 to 12 hr to test for DNA replication in both pronuclei (Fig. 2B). We found that sperm halos could be injected up to 3 hr after activation and 22% of the paternal pronuclei that were formed were replicating DNA within the normal time period, as compared with maternal DNA replication (Fig. 2B.iv.). This confirmed that injecting decondensed sperm halos did increase the time at which the paternal genome could be added to the oocyte and still replicate. Mouse oocytes could no longer support the replication of paternal DNA when halos were injected at 4 hr (Fig. 2B.vi.) or 5 to 7 hr (data not shown) after activation.
These data demonstrate that de novo DNA licensing is possible for as long as 3 hr after oocyte activation. It is possible that oocytes could support de novo licensing after 3 hr, but that decondensation was no longer possible after this time (as described in the Discussion).
Both pronuclei initiate DNA synthesis synchronously in IVF and ICSI
Given the controversy discussed above regarding the synchrony of DNA synthesis initiation in the mouse one cell embryo [Aoki and Schultz, 1999; Bouniol-Baly et al., 1997; Ferreira and Carmo-Fonseca, 1997; Howlett and Bolton, 1985; Luthardt and Donahue, 1973], we tested this in zygotes generated in vitro. Embryos were produced by ICSI or IVF, then incubated with BrdU for 30 min at hourly intervals starting from “0-min” fertilization time point (Table 1). We found that the majority of both ICSI and IVF embryos initiated DNA synthesis between 5 and 6 hr after fertilization. More importantly, among of 196 embryos tested in all only 1 showed evidence for asynchronous DNA replication in that only one pronucleus incorporated BrdU. Our data agree with the synchrony of the initiation of DNA synthesis in the mouse pronuclei after IVF that was reported previously [Aoki and Schultz, 1999], and indicate that the same is true for ICSI.
Table 1. Mouse Pronuclei Initiate DNA Synthesis Synchronously.
Mouse one cell embryos were generated by ICSI or IVF, and incubated with BrdU at various times after fertilization. The numbers of oocytes with 0, 1 or 2 pronuclei (PN) that incorporated BrdU are shown.
| Exp. | Hr after Fertilization1 |
No. (%) Oocytes with BrdU Positive Pronuclei (PN) |
||
|---|---|---|---|---|
| 0 PN | 1 PN | 2 PN | ||
| ICSI | 5 | 17 (94) | 0 | 1 (6) |
| 6 | 9 (32) | 0 | 19 (68) | |
| 7 | 4 (19) | 0 | 17 (82) | |
| 8 | 0 | 0 | 34 (100) | |
| IVF | 5 | 29 (83) | 1 (3) | 5 (14) |
| 6 | 5 (19) | 0 | 21 (81) | |
| 7 | 1 (6) | 0 | 17 (94) | |
| 8 | 1 (6) | 0 | 15 (94) | |
Fertilization = approximate moment of sperm-oocyte fusion; in IVF this was 30 min after initiation of gamete co-incubation and in ICSI it was the end of ≤ 10 min long injection of one group of oocytes.
Mouse oocytes can support asynchronous initiation of DNA synthesis in the two pronuclei
We next tested whether the oocyte could support the asynchronous initiation of DNA synthesis by delayed sperm injection. When oocytes that were injected with sperm heads 2 hr after parthenogenetic activation were incubated in BrdU for 12 h, rather than for only 30 min, a much larger percentage (62% vs. 24%) replicated DNA in the paternal pronuclei (Fig. 2A.iv.). This demonstrated that most of the paternal pronuclei resulting from injecting oocytes 2 hr after activation initiated DNA replication at least 2 hr after the maternal pronucleus had initiated replication. When oocytes that were injected with sperm halos 3 hr after activation were incubated in BrdU for 12 hr, a larger percentage (46% vs. 22%), had two pronuclei that incorporated BrdU (Fig. 2B.v.), once again suggesting that asynchronous initiation of DNA synthesis is tolerated by the oocyte.
Replication in the paternal pronucleus is correlated with its size
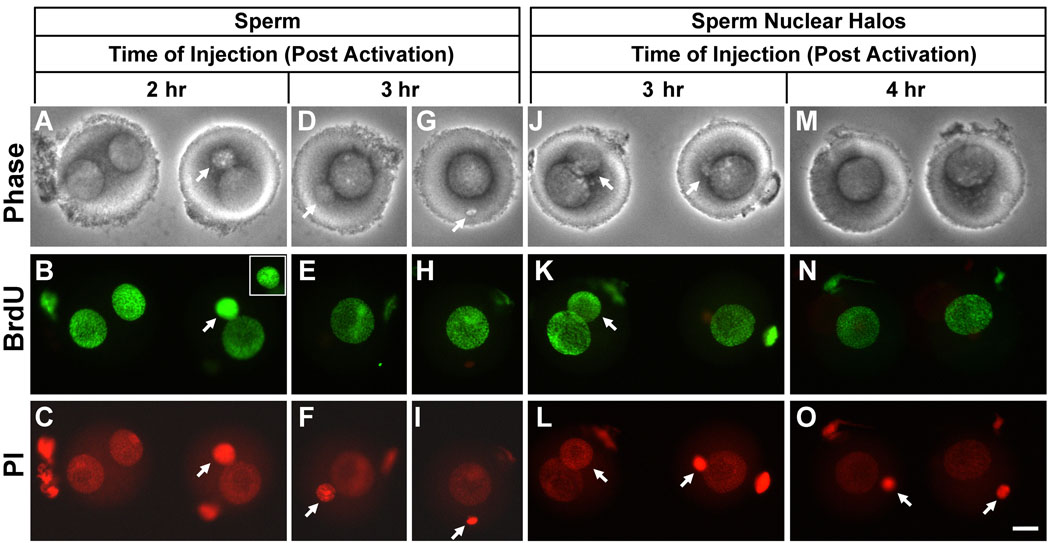
Fig. 3 shows some examples of BrdU incorporation into the pronuclei of embryos generated after delayed sperm and halo injection. All of the embryos shown in Fig. 3 were incubated in BrdU for 14 hr after injection, so the pronuclei are shown in the final stages of maturation and after the completion of DNA synthesis. When injections were performed 2 hr post activation, sperm heads formed various sized pronuclei, but all were smaller than the female pronucleus (Fig. 3A–C). We found that in some cases very small paternal pronuclei could still replicate their DNA (Fig. 3B, right embryo, arrow). However, this was about the smallest size pronucleus that was capable of DNA synthesis. Sperm injected 3 hr post activation remained condensed in 20/21 cases (as in Fig. 3I), and in one case changed its size to form a small round pronucleus (Fig. 3F). But none of these replicated. A similar correlation between size and replication was noted in halo injected oocytes. When injected 3 hr post activation, some halos developed into small pronuclei that incorporated DNA (Fig. 3K, left embryo) while others remained unchanged and did not replicate (Fig. 3K, right embryo). At 4 hr post activation, all the halos remained compact, and did not incorporate BrdU (Fig. 3M–O).
Figure 3. Examples of Paternal DNA Replication after Delayed Injection of Sperm Heads and Sperm Halos into Parthenogenetically Activated Oocytes.
All embryos were incubated in BrdU for 14 hr after injection. When it was possible to identify, the paternal pronucleus is shown by an arrow. (A–C) Sperm heads injected two hours after activation, corresponding to the experiment depicted Fig. 2A.iv. Inset in B shows the smaller paternal pronucleus on the right in a different focal plane to show the normal punctuate appearance of eukaryotic DNA replication. In both cases paternal pronuclei incorporated DNA. (D–I) Sperm heads injected 3 hr after activation, corresponding to Fig. 2A.v. None of paternal pronuclei incorporated BrdU. (J–L) Sperm halos injected 3 hr after activation, corresponding to Fig. 2B.v. Only the paternal pronuclei on the left incorporated BrdU. Note the difference in size of the two paternal pronuclei. (M–O) Sperm halos injected 4 hr after activation, corresponding to Fig. 2B.vi. Neither paternal pronucleus incorporated BrdU. All figures are shown at the same magnification, Bar = 20 µm.
Transfer of Licensed and Replicating Pronuclei into Activated Oocytes
Our delayed ICSI experiments indicated that the mouse oocyte can still initiate de novo DNA replication licensing as long as 3 hr after oocyte activation and that it tolerates asynchronous initiation of DNA replication in the same cytoplasm. Experiments in somatic cells indicate that the cytoplasm of S phase cells can prematurely initiate DNA synthesis in G1 phase nuclei, thereby inhibiting de novo DNA origin licensing [Krude et al., 1997]. We therefore explored the regulation of DNA synthesis in the oocyte cytoplasm by transferring pronuclei in G1 or S phase into S phase or G1 parthenogenic oocytes, respectively (Fig. 4).
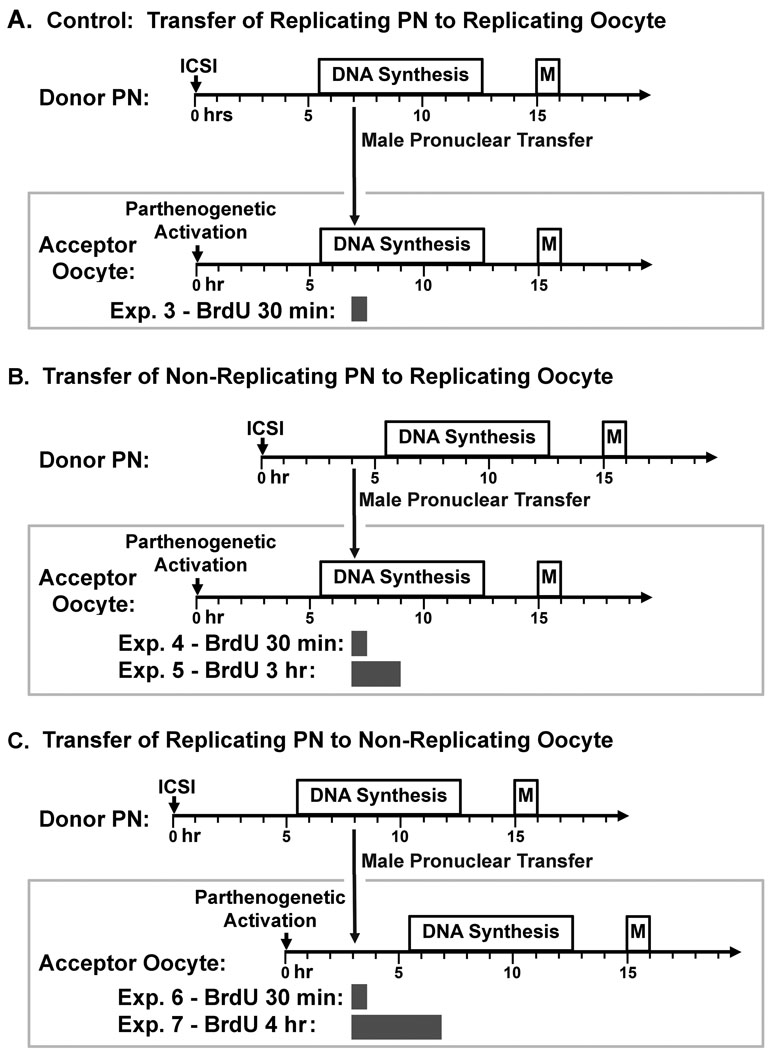
Figure 4. Experimental Design for Pronuclear Transfer Experiments.
(A) S phase pronuclei transferred to S phase parthenotes: As a control, S phase transferred pronuclei from zygotes 7 hr after ICSI were transferred by micromanipulation into acceptor oocytes that were parthenogenetically activated 7 hr earlier, and then incubated in BrdU for 30 min. The data from this experiment are shown in Fig. 5A–F, and Table 2, Exp. 3. (B) G1 pronuclei transferred to S phase parthenotes: G1 pronuclei from zygotes 4.5 hr after ICSI were transferred by micromanipulation into acceptor oocytes that were parthenogenetically activated 7 hr previously, and then incubated in BrdU for 30 min or 3 h. The data from this experiment are shown in Fig. 5G–L, and Table 2, Exps. 4 and 5. (C) S phase pronuclei transferred to G1 parthenotes: G1 pronuclei from zygotes 7 hr after ICSI were transferred by micromanipulation into acceptor oocytes that were parthenogenetically activated 3 hr previously, and then incubated in BrdU for 30 min or 3 h. The data from this experiment are shown in Fig. 5M–R, and Table 2, Exps. 6 and 7.
As a control for our transfer experiments, we confirmed that the maternal pronuclei in parthenogenetically activated mouse oocytes follow the same time course for DNA replication as normal ICSI. In all pronuclear transfer experiments the BrdU was added only after the transfer so any nuclear incorporation resulted from active DNA synthesis after transfer. The maternal pronuclei of parthenotes had no detectable BrdU incorporation at 4.5 hr after activation, but were replicating DNA by 7 hr after activation, as expected (Table 2, Exps. P1 and P2). We then tested our ability to transfer actively replicating male pronuclei from one oocyte to the other and whether they can maintain DNA synthesis after the micromanipulation (Fig. 4A). The majority of replicating pronuclei that were transferred into oocytes with replicating maternal pronuclei continued to incorporate BrdU after the transfer (Table 2, Exp. 3, and Fig. 5A–C). In rare cases, because of the large size of the male pronucleus, the pronucleus was fractured and only part of the paternal pronucleus was transferred, but the DNA continued to incorporate BrdU (Fig. 5, D–F, arrow). In these cases, the integrity of the nuclear membrane was likely to be disrupted, so there were no barriers to transfer of cytoplasmic factors to the transferred nucleus, but DNA was still replicated.
Table 2. Pronuclear Microinjection Experiments.
Donor zygotes were generated by ICSI and acceptor parthenotes were generated by activation of oocytes, as described in Methods. Male pronuclei were extracted from zygotes at indicated time points after fertilization and injected into acceptor oocytes at indicated times after activation. Age of the oocytes reflects time from the moment of sperm injection (Exp: ICSI) or moment when the oocytes were placed in SrCl2 containing medium to activate (Exp: P1, P2 and 3–7), and at which the oocytes were placed into media containing BrdU. Age of the transferred pronuclei indicates the time from the moment of sperm injection of ICSI to create the donor zygotes (Exp: 3–7). Oocytes were incubated in BrdU for 30 min (Exp. ICSI, P1, P2, 3, 4, and 6) for 3 hr (Exp. 5) or for 4 hr (Exp. 7), then fixed.
| Exp. ID | Age of Trans’d. PN (hr) |
Age of Oocyte (hr) |
No. Oocytes |
BrdU Positive | |||
|---|---|---|---|---|---|---|---|
| no PN | 1 PN (Trans’d., ♂) |
1 PN (Acceptor, ♀) |
2 PN (♂ + ♀) | ||||
| Controls | |||||||
| ICSI | NA | 7.0 | 22 | 3 (13.6%) | 0 | 1 (4.5%) | 18 (81.8%) |
| P1 | NA | 4.5 | 20 | 20 (100%) | NA | 0 | NA |
| P2 | NA | 7.0 | 28 | 1 (3.6%) | NA | 27 (96.4%) | NA |
| Tested groups | |||||||
| 3 | 7 | 7 | 47 | 0 | 0 | 10 (21%) | 37 (79%) |
| 4 | 4.5 | 7 | 23 | 0 | 0 | 23 (100%) | 0 |
| 5 | 4.5 | 7 | 27 | 0 | 0 | 2 (7.4%) | 25 (92.6%) |
| 6 | 7 | 3 | 37 | 13 (35%) | 24 (65%) | 0 | 0 |
| 7 | 7 | 3 | 21 | 1 (5%) | 1 (5%) | 0 | 19 (90%) |
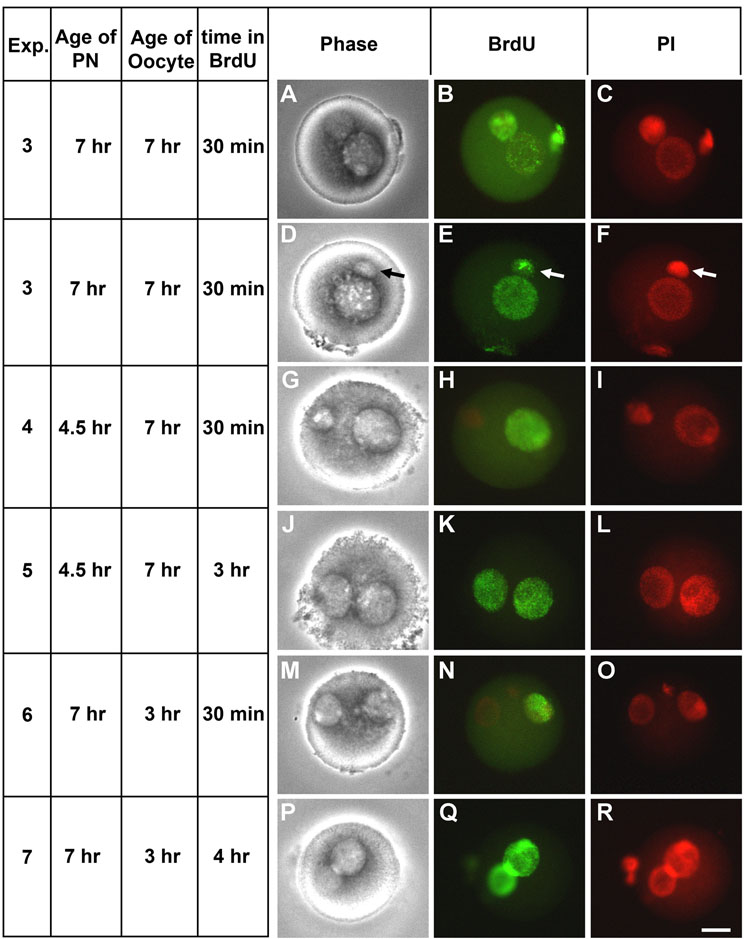
Figure 5. DNA Synthesis in Zygotes from Pronuclear Transfer.
(A–F) Zygotes were created by transferring S phase pronuclei into S phase parthenotes, as in Fig. 4A (arrow in D–F indicates pronuclear fragment that was transferred in this oocyte that continued to replicate); (G–L) G1 pronuclei into S phase ooplasm in parthenotes, as in Fig. 4B; (M–R) or by transferring S phase pronuclei into G1 ooplasm in parthenotes, as in Fig. 4C. The zygotes were then incubated in BrdU for 30, 3 hr or 4 h, as indicated on the left hand panel, then fixed and stained for BrdU incorporation and DNA with propidium iodide. Phase contrast, BrdU staining and propidium iodide staining are shown, as indicated. Bar = 20 µm.
We then tested the control of the oocyte cytoplasm on pronuclear DNA synthesis by transferring G1 male pronuclei into parthenogenetically activated S phase oocytes, and incubating the oocytes in BrdU for either 30 min or 3 hr (Fig. 4B). Transferred pronuclei were obtained from zygotes generated by normal ICSI which were then incubated for 4.5 hr in media. These were the earliest pronuclei that were identifiable by light microscopy for transfer. The paternal pronucleus was transferred by micromanipulation into an oocyte that had been parthenogenetically activated 7 hr earlier. By 30 min later, none of the transferred pronuclei had initiated DNA synthesis, while the maternal pronucleus of the activated acceptor oocyte incorporated BrdU in all oocytes tested (Table 2, Exp. 4, and Fig. 5G–I). When these oocytes were incubated for 3 h, the time at which the transferred pronuclei were normally scheduled to begin DNA synthesis, 93% of all oocytes incorporated BrdU into both pronuclei (Table 2, Exp. 5 and Fig. 5J–L).
We next performed the opposite experiment by transferring S phase pronuclei into G1 phase oocytes (Fig. 4C). In this case, we transferred male pronuclei 7 hr after ICSI into oocytes 3 hr after parthenogenetic activation. We found that replicating pronuclei continued to incorporate BrdU into DNA when placed into G1 phase oocytes (Table 2, Exp. 6, and Fig. 5M–O) but the maternal pronucleus did not begin to incorporate BrdU until later (Table 2, Exp. 7, and Fig. 5P–R).
Discussion
The data presented in this manuscript demonstrate that the mouse oocyte can support the asynchronous initiation of DNA replication in the two pronuclei, even though they normally initiate synchronously. Furthermore, de novo licensing of DNA origins for replication can occur for at least 3 hr after oocyte activation. As discussed below, the work suggests that the fertilized mouse oocyte may be an important model for studying mammalian DNA licensing.
The mouse oocyte can support asynchronous DNA Replication in the two pronuclei
In both sets of experiments, delayed ICSI and pronuclear transfer, it was clear that oocytes can support the asynchronous initiation of DNA synthesis in the two pronuclei. Previous studies have differed on whether this occurs in normal mating, IVF or ICSI [Aoki and Schultz, 1999; Bouniol-Baly et al., 1997; Ferreira and Carmo-Fonseca, 1997; Howlett and Bolton, 1985; Luthardt and Donahue, 1973], but our work clearly indicates it is possible when the oocyte is activated before fertilization. This suggests the pronuclei participate in the regulation of DNA synthesis. However, as described in the Introduction, it is also clear that the cytoplasm does play a role in its regulation.
De novo licensing of replication origins can occur late in zygote G1
A major difference between the regulation of DNA synthesis in somatic cells and that of the one cell zygote is the state of the chromatin at the start of the cell cycle. In the zygote, the sperm chromatin is packaged so tightly by protamines that most of the DNA is virtually inaccessible to binding by other proteins [Balhorn, 2007]. Although it has not been tested, we expect that the early licensing proteins ORCL1-6 are not bound to this compact chromatin (we are currently testing this prediction in our laboratory). Moreover, even if ORCL1-6 were bound to sperm DNA, they would be removed, together with protamines, during extraction with 2 M NaCl and 10 mM dithiothreitol needed for halo preparation. Thus, while the replication of the paternal chromatin in the sperm cell probably requires de novo licensing, that of sperm halos almost certainly does.
Our experiments show that de novo licensing is still possible as late at 3 hr after sperm halo injection. Because we assayed DNA replication at 7 hr post activation in these experiments, the possible window for de novo licensing is 4 hr (Fig. 2B.iv.), but may be even smaller. The data shown in Fig. 3J–K suggest that one probable reason that oocytes could not support paternal DNA synthesis when sperm were injected after 3 hr post activation was because the sperm were not fully decondensed. Therefore, it is possible that even after 3 hr, the oocyte remains capable of de novo licensing of DNA origins.
We did not test whether the paternal DNA synthesis that resulted from delayed ICSI replicated the entire genome completely, because we were interested in the timing of the initiation of DNA synthesis and not on other stages of development. To our knowledge, this is the first report that the delayed ICSI also resulted in delayed DNA synthesis initiation. Two related studies, however, focused on results which would require complete, paternal DNA synthesis. Maleszewski et al. [Maleszewski et al., 1999] showed that injection of normal sperm into oocytes as late as 2 hr after parthenogenetic activation resulted in the formation of normal paternal mitotic chromosomes and some live pups after transfer. Kishigami et al. [Kishigami et al., 2004] reported that normal ICSI could be delayed for as long as 3 hr after parthenogenetic activation and still lead to live pups. The differences between the two reports may be due to differences in experimental detail, but both laboratories agree that delayed ICSI can result in normal pups. Thus, the de novo licensing of DNA replication origins in delayed ICSI must result in complete paternal DNA replication.
Pronuclei may contain licensed origins by 4.5 hr after fertilization
Our experiments with transferring pre-replicative pronuclei into replicating oocytes (Fig. 4B and Fig 5G–L) suggest that either the licensing in pronuclei is complete enough by 4.5 hr after fertilization to initiate DNA synthesis or that continued licensing in that pronucleus is not inhibited by the S phase cytoplasm. In the normal cell cycle, licensing of new DNA replication origins is inhibited by the initiation of S phase through the CDK2/CCNA2 and CDK2/CCNE1 pathway [DePamphilis et al., 2006; Diffley, 2004]. Because these complexes can freely diffuse through the nuclear membrane [Jackman et al., 2002] it might be expected that the oocyte S phase cytoplasm would arrest any ongoing licensing in G1 pronuclei that were transferred into it. We have shown that the pronuclear transfer process itself can sometimes disrupt the nuclei (Fig. 5D–F) exposing it to the cytoplasmic factors rapidly, yet this did not change nuclei’s fate. The ability of the oocyte to support asynchronous initiation of DNA synthesis in the two pronuclei, discussed above, also suggests the possibility that licensing may not be complete in one pronucleus while DNA synthesis is ongoing in the other. Future experiments designed to understand how the oocyte cytoplasm participates in the regulation of two pronuclei with independent control of DNA synthesis should also provide insight into somatic cell DNA synthesis regulation.
Sperm nuclear halos as partially decondensed nuclei
The fact that replicating paternal pronuclei can be formed from sperm halos injected into oocytes one hour later after parthenogenetic activation than sperm nuclei suggests that sperm halos can mimic an early stage in sperm decondensation. Mammalian spermatozoa go through a complicated process of chromatin remodeling after fertilization to remove the sperm-specific DNA binding protamines to replace them with the normal histones [Dean, 1983; Mahi and Yanagimachi, 1975; Perreault et al., 1987]. Maleszewski et al. [Maleszewski et al., 1999] concluded from their delayed ICSI experiments that the oocyte only retains the ability to remodel sperm chromatin for up to 2 hr after activation. By injecting halos rather than condensed sperm heads into the oocyte we obviated the need for the oocyte to remove the protamines, thereby narrowing the time required for remodeling. These sperm halos still have condensed sperm nuclear matrices that retain the original shape of the nuclei [Nadel et al., 1995], so even sperm halos require some degree of nuclear reorganization. But the evidence that halos can be injected at later time points after activation than normal spermatozoa supports our conclusion that they can be considered functionally partially decondensed sperm nuclei.
Summary
These data suggest that the unique nature of the mouse zygote, in that it contains two pronuclei in the same cytoplasm, makes it an important model for understanding the regulation of somatic cell DNA replication. The molecular mechanisms for DNA regulation are probably very similar between the zygote and somatic cells, but the presence of two pronuclei in the former allows for greater experimental manipulation to test hypotheses related to DNA regulation.
Acknowledgments
Grant Support: This work was supported by NIH grants HD28501 to W.S.W. and HD048845 to M.A.W., and by the Office of the Vice Chancellor for Research and Graduate Education at the University of Hawaii at Manoa
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Ajduk A, Yamauchi Y, Ward MA. Sperm Chromatin Remodeling after Intracytoplasmic Sperm Injection Differs from That of In Vitro Fertilization. Biol Reprod. 2006 doi: 10.1095/biolreprod.106.053223. [DOI] [PubMed] [Google Scholar]
- Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote. 1999;7:165–172. doi: 10.1017/s0967199499000532. [DOI] [PubMed] [Google Scholar]
- Austin CR. The Mammalian Egg. Oxford: Blackwell Scientific Publications; 1961. [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin Cell Biol. 1991;2:103–115. [PubMed] [Google Scholar]
- Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- Blow JJ. Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol-Baly C, Nguyen E, Besombes D, Debey P. Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res. 1997;236:201–211. doi: 10.1006/excr.1997.3708. [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Dean J. Decondensation of mouse sperm chromatin and reassembly into nucleosomes mediated by polyglutamic acid in vitro. Dev Biol. 1983;99:210–216. doi: 10.1016/0012-1606(83)90269-5. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Sirlin JL. Labelled pronuclei in mouse eggs fertilized by labelled sperm. Nature. 1956;177:429. doi: 10.1038/177429a0. [DOI] [PubMed] [Google Scholar]
- Ferreira J, Carmo-Fonseca M. Genome replication in early mouse embryos follows a defined temporal and spatial order. J Cell Sci. 1997;110(Pt 7):889–897. doi: 10.1242/jcs.110.7.889. [DOI] [PubMed] [Google Scholar]
- Howlett SK, Bolton VN. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. J Embryol Exp Morphol. 1985;87:175–206. [PubMed] [Google Scholar]
- Jackman M, Kubota Y, den Elzen N, Hagting A, Pines J. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol Biol Cell. 2002;13:1030–1045. doi: 10.1091/mbc.01-07-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biology of Reproduction. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Wakayama S, Nguyen VT, Wakayama T. Similar time restriction for intracytoplasmic sperm injection and round spermatid injection into activated oocytes for efficient offspring production. Biol Reprod. 2004;70:1863–1869. doi: 10.1095/biolreprod.103.025171. [DOI] [PubMed] [Google Scholar]
- Krude T. Initiation of chromosomal DNA replication in mammalian cell-free systems. Cell Cycle. 2006;5:2115–2122. doi: 10.4161/cc.5.18.3248. [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Leno GH, Downes CS, Laskey RA. The nuclear membrane prevents replication of human G2 nuclei but not G1 nuclei in Xenopus egg extract. Cell. 1992;69:151–158. doi: 10.1016/0092-8674(92)90126-w. [DOI] [PubMed] [Google Scholar]
- Luthardt FW, Donahue RP. Pronuclear DNA synthesis in mouse eggs. An autoradiographic study. Exp Cell Res. 1973;82:143–151. doi: 10.1016/0014-4827(73)90256-5. [DOI] [PubMed] [Google Scholar]
- Mahi CA, Yanagimachi R. Induction of nuclear decondensation of mammalian spermatozoa in vitro. Journal of Reproduction & Fertility. 1975;44:293–296. doi: 10.1530/jrf.0.0440293. [DOI] [PubMed] [Google Scholar]
- Maleszewski M, Borsuk E, Koziak K, Maluchnik D, Tarkowski AK. Delayed sperm incorporation into parthenogenetic mouse eggs: sperm nucleus transformation and development of resulting embryos. Mol Reprod Dev. 1999;54:303–310. doi: 10.1002/(SICI)1098-2795(199911)54:3<303::AID-MRD11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Mohar I, Szczygiel MA, Yanagimachi R, Ward WS. Sperm Nuclear Halos Can Transform Into Normal Chromosomes After Injection Into Oocytes. Mol Reprod Dev. 2002;62:416–420. doi: 10.1002/mrd.10147. [DOI] [PubMed] [Google Scholar]
- Nadel B, de Lara J, Finkernagel SW, Ward WS. Cell-specific organization of the 5S ribosomal RNA gene cluster DNA loop domains in spermatozoa and somatic cells. Biology of Reproduction. 1995;53:1222–1228. doi: 10.1095/biolreprod53.5.1222. [DOI] [PubMed] [Google Scholar]
- Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19:527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Perreault SD, Naish SJ, Zirkin BR. The timing of hamster sperm nuclear decondensation and male pronucleus formation is related to sperm nuclear disulfide bond content. Biology of Reproduction. 1987;36:239–244. doi: 10.1095/biolreprod36.1.239. [DOI] [PubMed] [Google Scholar]
- Quinn P, Barros C, Whittingham DG. Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. J Reprod Fertil. 1982;66:161–168. doi: 10.1530/jrf.0.0660161. [DOI] [PubMed] [Google Scholar]
- Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The Sperm Nuclear Matrix is Required for Paternal DNA Replication. J Cell Biochem. 2007;102:680–688. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- Sirlin JL, Edwards RG. Timing of DNA synthesis in ovarian oocyte nuclei and pronuclei of the mouse. Exp Cell Res. 1959;18:190–194. doi: 10.1016/0014-4827(59)90308-8. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- Stubblefield E. Analysis of the replication pattern of Chinese hamster chromosomes using 5-bromodeoxyuridine suppression of 33258 Hoechst fluorescence. Chromosoma. 1975;53:209–221. doi: 10.1007/BF00329172. [DOI] [PubMed] [Google Scholar]
- Szczygiel M, Yanagimachi R. Intracytoplasmic sperm injection. In: Nagy A, Gertsenstein M, Vintersten K, Behringer R, editors. Manipulation of the Mouse Embryo - A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2003. pp. 585–597. [Google Scholar]
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci U S A. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Pardoll DM, Coffey DS. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980;22:79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal Pronuclear DNA Degradation Is Functionally Linked to DNA Replication in Mouse Oocytes. Biol Reprod. 2007;77:407–415. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]