Abstract
In mammalian cells, endoplasmic reticulum (ER) stress has recently been shown to induce autophagy and the induction requires the unfolded protein response (UPR) signaling pathways. However, little is known whether autophagy regulates UPR pathways and how specific UPR targets might control autophagy. Here, we demonstrated that whereas ER stress-induced autophagy was suppressed by PI3KC3 inhibitor 3-methyladenine (3-MA), wortmannin and knockdown of Beclin1 using siRNA, only 3-MA suppressed UPR activation. We discovered that the UPR regulator and ER chaperone GRP78/BiP is required for stress-induced autophagy. In cells where GRP78 expression was knockdown by siRNA, despite spontaneous activation of UPR pathways and LC3 conversion, autophagosome formation induced by ER stress as well as by nutrition starvation was inhibited. GRP78 knockdown did not disrupt PI3KC3-Beclin1 association. However, electron microscopic analysis of the intracellular organelle structure reveals that the ER, a putative membrane source for generating autophagosomal double membrane, was massively expanded and disorganized in cells where GRP78 was knockdown. ER expansion is known to be dependent on the UPR transcription factor XBP-1. Simultaneous knockdown of GRP78 and XBP-1 recovered normal levels of stress-induced autophagosome formation. Thus, these studies uncover 3-MA as an inhibitor of UPR activation and establish GRP78 as a novel obligatory component of autophagy in mammalian cells.
Keywords: autophagy, GRP78, unfolded protein response
Introduction
The endoplasmic reticulum (ER) is an essential intracellular organelle for the synthesis and maturation of cell surface and secretory proteins, and maintenance of Ca2+ homeostasis. Disruption of these physiological functions leads to accumulation of unfolded proteins and induces the unfolded protein response (UPR).1,2 During this process, three ER transmembrane signaling molecules PERK, IRE1 and ATF6 are activated. Activated PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α), which transiently blocks most protein translation and reduces the influx of nascent proteins into the ER lumen.3 One exception is the transcriptional factor ATF4, whose translation under ER stress is selectively activated through internal ribosomal entry site. A known downstream target of ATF4 is C/EBP-homologous protein (CHOP), a transcription factor implicated in the control of translation and apoptosis.4 Activated IRE1 releases its endonuclease activity, resulting in splicing of Xbp-1 mRNA, which encodes an active transcriptional factor. Another downstream target of IRE1 is c-Jun N-terminal kinase (JNK), whose activation regulates cell death.5,6 Activated ATF6 translocates from the ER to the Golgi complex, where it is cleaved by S1P/S2P proteases and generates another active transcriptional factor. In concert or independently, ATF4, ATF6 and XBP-1 upregulate ER chaperone proteins, folding enzymes and protein degradation molecules, which in turn either prevent the aggregation of unfolded proteins, aid in their subsequent folding or in degradation of excessive misfolded proteins.
A major UPR upregulated target protein is the 78 kDa glucose regulated protein, GRP78, an ER molecular chaperone also referred to as BiP. GRP78 is involved in many cellular processes, including translocating newly synthesized polypeptides across the ER membrane, facilitating the folding and assembly of newly synthesized proteins, maintaining them in a state competent for subsequent folding and oligomerization and regulating Ca2+ homeostasis.7,8 In addition to its chaperoning function, GRP78 is a key regulator of ER stress transducers. GRP78 binds and inhibits PERK, IRE1 and ATF6 activation in non-stressed cells.9 Upon ER stress and malfolded protein accumulation in the ER, these molecules are released from GRP78 and become activated. Recent studies reveal that GRP78 is antiapoptotic and plays critical cytoprotective roles in early embryogenesis, oncogenesis, neurodegenerative diseases and atherosclerosis.10–16 Despite these advances, the mechanisms whereby GRP78 protects eukaryotic cells against cell death under a wide range of stress and pathological conditions remain to be explored.
Recently, it was discovered that autophagy is activated upon ER stress as a defensive mechanism for survival.17,18 Autophagy is an intracellular protein degradation system required for normal turnover of cellular components and for the starvation response. When autophagy is induced, a double membrane structure called autophagosome is formed de novo or from existing membrane to enclose the subcellular components. Upon fusion of the outer membrane of autophagosome with lysosomal membrane, the cytoplasm-derived contents are degraded together with the inner membrane of the autophagosome. While the contribution of the endomembrane organelles to autophagy is under active investigation, evidence is emerging that the ER provides membrane for autophagosome formation and that autophagy is critical for ER homoeostasis.19 Distinct classes of phosphatidylinositol 3′-kinases (PI3Ks) are involved in signaling pathways that control macroautophagy in mammalian cells.20 Initiation of the autophagy process requires class III PI3K (PI3KC3) and its complex formation with Beclin1 and the myristylation protein kinase p150. This initiation process could be suppressed by 3-methyladenine (3-MA), a specific inhibitor of endogenous lysosomal protein degradation that targets PI3KC3 but not the other PI3Ks,21 as well as wortmannin, another PI3K inhibitor. The further elongation of the autophagosome membrane is mediated by two ubiquitin-like conjugation systems. One of them mediates microtubule associated protein 1 light chain 3 (LC3) conversion from a free form (LC3-I) to a phosphatidylethanolamine conjugated form (LC3-II). The accumulation of LC3-II and its location to autophagosome (punctate dot formation) are commonly used as markers of autophagy.
In mammalian cells, autophagy has recently been linked to ER stress and the UPR pathways.19,22 However, little is known whether the process of autophagy regulates UPR pathways and how specific UPR targets might control autophagy. We report here that while 3-MA, wortmannin and knockdown of Beclin1 all suppress ER stress-induced autophagy, surprisingly only 3-MA suppresses UPR activation. When GRP78 expression was knockdown by siRNA, UPR pathways are activated, however, autophagosome formation by ER stress as well as nutrient starvation (NS) is inhibited. We further discovered that the ER, a putative membrane source for generating autophagic vacuole membranes,23,24 is massively expanded and disorganized in cells where GRP78 is knockdown. ER biogenesis and expansion are known to be dependent on the UPR transcription factor XBP-1.25 Simultaneous knockdown of GRP78 and XBP-1 recovers normal levels of stress-induced autophagosome formation. These studies uncover novel modulations of the UPR signaling and autophagy pathways by 3-MA and GRP78 in human cells.
Results
The autophagy inhibitor 3-MA is a novel suppressor of UPR activation
To compare autophagy formation and UPR activation induced by acute ER stress in human cell lines, HEK293 or HeLa cells were treated with ER stress inducers targeting different ER functions: thapsigargin (TG), a SERCA inhibitor which blocks Ca2+ reuptake to the ER; tunicamycin (Tun), which inhibits N-glycosylation or brefeldin A (BFA), which blocks protein transportation to Golgi. First, the endogenous LC3 conversion from LC3-I to LC3-II, indicative of the onset of autophagy, was analyzed by Western blot. As shown in Figure 1a, both TG and Tun increased LC3-II formation by 4 h of treatment. Induction of CHOP and GRP78, monitored in parallel, served as positive control for UPR activation. Second, formation of autophagosome upon ER stress and nutrient starvation was monitored and measured by formation of the punctuate dot structures (autophagosomes) in live cells transiently transfected with green fluorescence protein (GFP)-LC3 (Figure 1S). As summarized in Figure 1b, all three ER stress inducers (TG, Tun and BFA) resulted in autophagosome formation with similar or slightly higher magnitude as NS. The autophagosome formation by NS and ER stress was further confirmed by electron microscopy, where autophagic vacuoles containing different intracellular contents were clearly visible in the cytosol of the stressed cells (Figure 1c and d).
Figure 1.

ER stress induces endogenous LC3 conversion and autophagosome formation. (a) HEK293 cells were treated with thapsigargin (TG, 2 μM) or tunicamycin (Tun, 3 μM) for the indicated time. Cell lysates were analyzed by Western blot for LC3 conversion (LC3-I, 18 kDa; LC3-II, 16 kDa), CHOP and GRP78 induction and β-actin (as a loading control) using antibodies sequentially. (b) Quantitation of autophagosomes per cell. HeLa cells transiently transfected with GFP-LC3 were either non-treated (NT), nutrient starved (NS) for 2 h or treated with TG (2 μM), Tun (3 μM) or BFA (2.5 μg/ml) for 4 h. For each condition, 15 to 35 cells were examined. The experiments were repeated 3 times. The results were summarized and expressed as the mean with the indicated standard deviation. (c) Representative electron micrographs of HEK293 cells either non-treated (NT), NS for 2 h or treated with Tun (3 μM) for 5 h. Autophagosomes are indicated by black arrow heads, ER by white arrow heads and N=nucleus; Bar=1 μm. (d) Enlarged images of autophagic vacuoles shown in (c) from NS and Tun-treated cells; Bar=0.2 μm.
To test the dependence of UPR activation on autophagy formation in human cells, HEK293 or HeLa cells were pre-treated with 3-MA or wortmannin. As shown in Figure 2a, both 3-MA and wortmannin inhibited LC3-II conversion induced by NS, Tun and TG. Strikingly, 3-MA, but not wortmannin, inhibited CHOP induction by Tun and TG. GRP78 protein level was not affected by NS; 4 h treatment by TG or Tun did not substantially elevate GRP78 protein level and treatment with either 3-MA or wortmannin had no effect. This leads us to further investigate if other UPR pathways are affected by 3-MA or wortmannin. As shown in Figure 2b, in both human cell lines, in parallel with CHOP, 3-MA also inhibited Tun and TG induction of ATF4. In contrast, wortmannin was without effect (Figure 2c). In the two human cell lines, 4 h treatment of TG or Tun did not substantially elevate GRP78 protein level, and treatment with 3-MA or wortmannin was without effect (Figure 2b and 2c). Upon ER stress, the activation of eIF2α and JNK1 is demonstrated by their phosphorylation levels. As shown in Figure 2d, 3-MA blocked both eIF2α and JNK1 phosphorylation while having no effect on the steady state level of both proteins. A downstream target of IRE1 activation is the splicing of X-box binding protein 1 (Xbp-1) mRNA. In 3-MA treated cells, Xbp-1 splicing induced by Tun was suppressed (Figure 2d). This was not observed in wortmannin treated cells (Figure 2e). Collectively, these results reveal that 3-MA not only suppresses autophagy induced by ER stress but is also a novel inhibitor of UPR activation.
Figure 2.
Suppression of ER stress-induced autophagy by 3-MA inhibits the UPR. (a) HEK293 cells were either non-treated (NT) or pretreated with 3-methyladenine (3-MA, 10 mM) or wortmannin (WM, 100 nM) for 1 h. The cells were then either subjected to NS for 2 h, TG (2 μM) or Tun (3 μM) treatment for 4 h, either in the absence or continuous presence of 3-MA or WM as indicated. Cell lysates were analyzed for levels of LC3-I conversion to LC3-II, CHOP, GRP78 and β-actin by Western blot using antibodies sequentially. (b) HEK293 or HeLa cells were treated the same as in (a). Cell lysates were analyzed for CHOP, ATF4, GRP78 and β-actin expression by Western blot using antibodies sequentially. (c) Same as (b) except the cells were pretreated with wortmannin. (d) HEK293 cells were non-treated (0 h) or pretreated with 3-MA (10 mM) for 1 h, then treated with Tun (3 μM), either in the absence or continuous presence of 3-MA for the indicated time. Cell lysates were analyzed for levels of phospho-JNK1 (p-JNK1), JNK1, phospho-eIF2α (p-eIF2α) and eIF2α Further, total RNA were extracted and analyzed for expression of Xbp-1, spliced Xbp-1 (Xbp-1s) and β-actin (as a control) by RT-PCR. (e) HEK293 cells were non-treated (0 h) or pre-treated with wortmannin (100 nM) prior to Tun treatment and assayed for Xbp-1 splicing as described in (d). For all panels, the experiments were repeated 2 to 3 times.
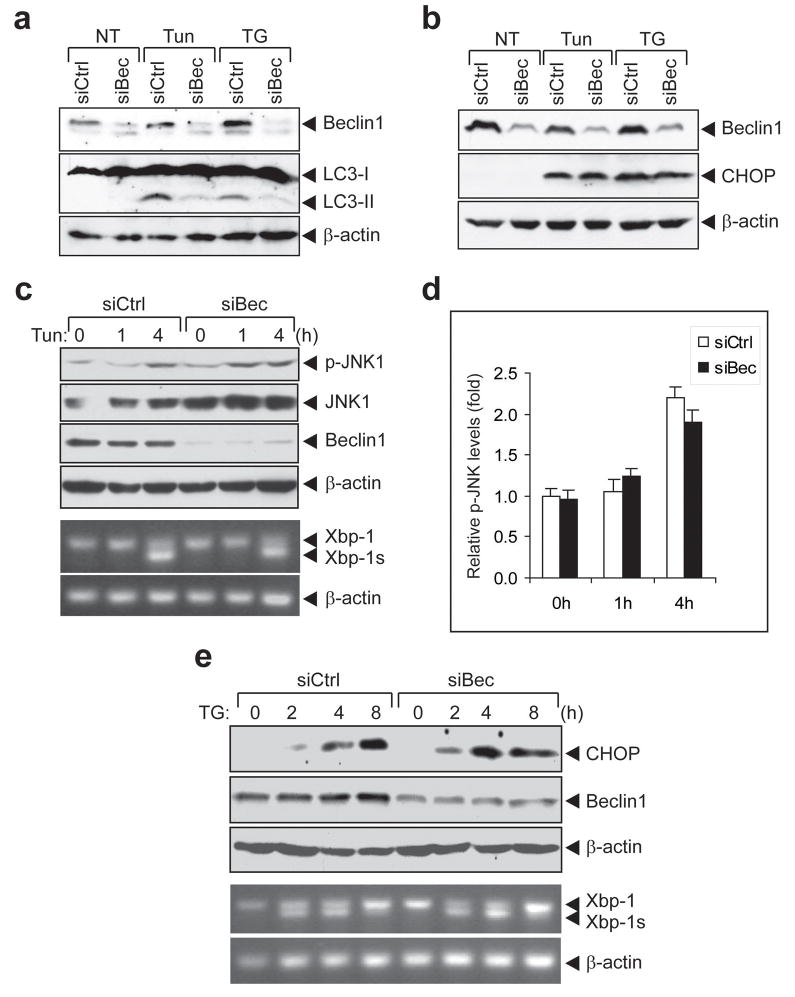
Figure 3.
Beclin1 is required for ER-stress induced autophagy but not for UPR activation. HEK293 cells were transfected with either control siRNA (siCtrl) or siRNA against Beclin1 (siBec). The cells were non-treated (NT) or treated with TG (2 μM) or Tun (3 μM) for 5 h. (a) Cell lysates were analyzed for levels of LC3-I conversion to LC3-II, Beclin1 and β-actin (as a loading control), or (b) CHOP, Beclin1 and β-actin by Western blot using antibodies sequentially. (c) SiRNA transfected HEK293 cells were treated with Tun (3 μM) for indicated time. Cell lysates were analyzed for levels p-JNK1 and JNK1 by Western blot using antibodies sequentially. Total RNA were extracted and analyzed for expression of Xbp-1, Xbp-1s and β-actin (as a control) by RT-PCR. (d) Quantitation of the p-JNK1 level shown in (c) after normalization with total JNK1 level during the time course of Tun treatment in cells transfected with either siCtrl or siBec. (e) Time course of TG induction of CHOP and Xbp-1 splicing in cells transfected with either siCtrl or siBec. A lower concentration of TG (0.3 μM) was used in this experiment. The levels of Beclin1 and β-actin were monitored in parallel.
Beclin1 is required for ER-stress induced autophagy but not for UPR activation
Beclin1 is required for NS-induced autophagy and is an interactive protein of PI3KC3, a putative target of 3-MA.26 Some endogenous Beclin1 has been localized to the ER.27 To test the role of Beclin1 in UPR activation, siRNA was used to knockdown Beclin1 in HEK293 cells. LC3-II conversion induced by Tun and TG was suppressed (Figure 3a). However, in contrast to 3-MA treatment, knockdown of Beclin1 did not affect TG or Tun-induced UPR activation, as evidenced by no change in CHOP induction (Figure 3b). While Beclin1 knockdown elevated JNK1 protein expression, it had no effect on Tun-induced JNK1 activation in form of phosphorylation (Figure 3c and d). Tun-induced Xbp-1 splicing was also not affected (Figure 3c). Time course analysis further confirmed that Beclin1 knockdown had no effect on TG induction of CHOP and Xbp-1 splicing (Figure 3e). These results show that Beclin1 is required for ER-stress induced autophagy but not for UPR activation. Further, the results imply that autophagy formation in the form of LC3-II conversion is not an absolute requirement for UPR activation by ER stress.
Knockdown of GRP78 spontaneously activates the UPR
GRP78 is a major chaperone protein in the ER. It also binds to UPR signal transducers in non-stressed cells and releases them upon ER stress.9 Prior to testing its involvement in stress-induced autophagy, we examined the consequence of GRP78 depletion in human cells. To achieve this, GRP78 was knockdown by transiently transfecting HEK293 or HeLa cells with siRNA specifically targeted against human Grp78 (siGrp78). Western blot analysis confirmed that GRP78 protein expression level was decreased by siGrp78 compared to control siRNA (Figure 4a). In the GRP78 knockdown cells, the expression of another ER chaperone GRP94 was highly induced. The transcription factor CHOP was also induced but not calreticulin (CRT) and protein disulphide isomerase (PDI) (Figure 4a). Similar results were observed in HeLa cells (data not shown). Consistent with its pro-survival role in cells, knockdown of GRP78 by siGrp78 significantly decreased cell viability as determined by clonogenic survival assay (Figure 4b). In non-stressed cells, knockdown of GRP78 led to spontaneous phosphorylation of JNK1, a downstream target of IRE1; and JNK1 phosphorylation was further increased after 4 h of Tun treatment (Figure 4c). Quantitation of phosphorylated JNK1 levels after normalization to JNK1 levels was summarized in Figure 4d. Moreover, GRP78 knockdown expedited Xbp-1 splicing after Tun treatment (Figure 4c). Thus, in the HEK293 cells, GRP78 knockdown led to spontaneous activation of some UPR pathways and targets or enhances their activation upon ER stress. This was also observed in HeLa cells (data not shown).
Figure 4.
GRP78 knockdown spontaneously activates the UPR and impairs cell viability. (a) HEK293 cells were either transfected with 40 nM siCtrl or siRNA against human Grp78 (siGrp78) for 48 h. Cell lysates were analyzed for GRP78, GRP94, calreticulin (CRT) and β-actin (as loading control) or CHOP, PDI and β-actin expression by Western blot using antibodies sequentially. The experiments were repeated 2 times. (b) After transfection with either 40 nM siCtrl or siGrp78 for 48 h, HEK293 cells were subjected to clonogenic survival assay. The survival fraction of siCtrl transfected cells was set as 100%. The standard deviation was shown, *, p<0.05. (c) HEK293 cells were transfected with 40 nM siCtrl or siGrp78 for 48 h. The cells were then treated with Tun (3 μM) for indicated time. Cell lysates were analyzed for levels of p-JNK1 and JNK1 or GRP78 and β-actin (as a loading control) by Western blot using antibodies respectively. Total RNA were extracted and analyzed for expression of Xbp-1, Xbp-1s and β-actin (as a control) by RT-PCR. (d) The levels of p-JNK1 as shown in (c) were quantitated, normalized against JNK1 and plotted with the relative level in non-treated (0 h), siCtrl transfected cells set as 1.
Knockdown of GRP78 blocks autophagosome formation induced by ER stress and NS
Next, the effect of GRP78 knockdown on ER-stress induced autophagosome formation was tested. HeLa cells were transiently transfected with GFP-LC3 alone or GFP-LC3 in combination with control siRNA or siGrp78. Forty-eight hours after transfection, the cells were treated with Tun for 4 h. The autophagosome formation in live cells was determined by fluorescence microscopy. Similar to cells transfected only with the GFP-LC3 plasmid, Tun treatment induced autophagosome formation in cells co-transfected with control siRNA (Figure 5a and b). However, in cells co-transfected with siGrp78, Tun-induced autophagosome formation was suppressed (Figure 5a and b). Downregulation of GRP78 protein levels by siGrp78 was confirmed by Western blots showing suppression of GRP78 and upregulation of GRP94 (Figure 5c).
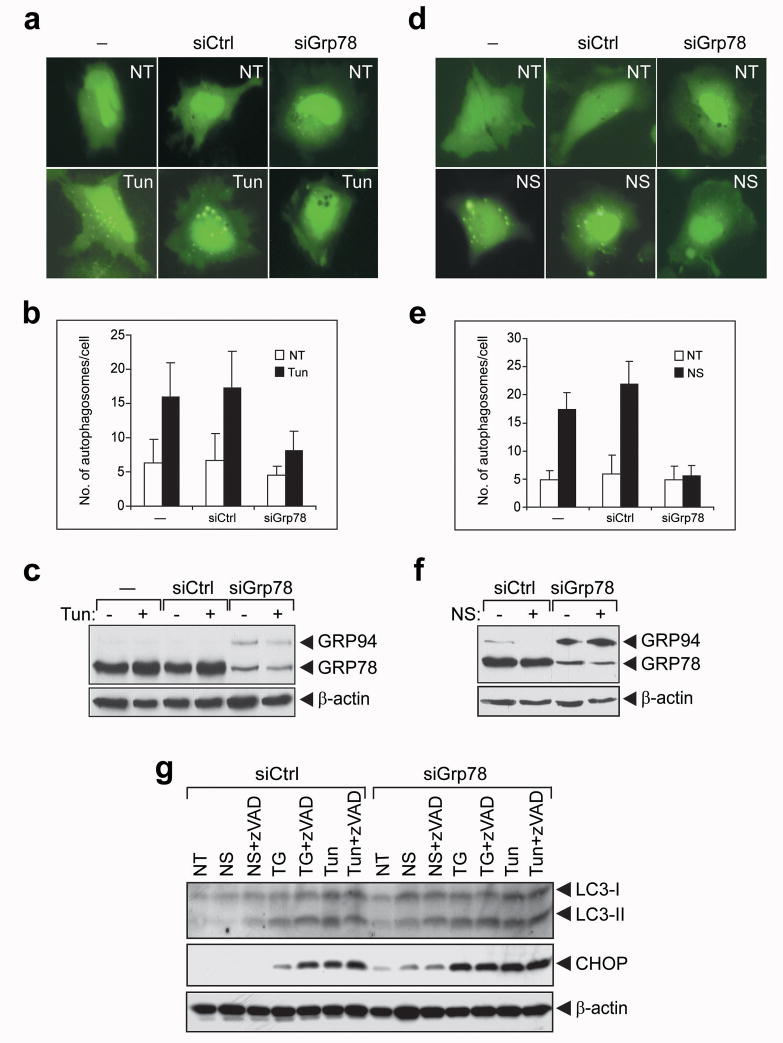
Figure 5.
GRP78 knockdown blocks autophagosome formation induced by ER stress or NS. (a) HeLa cells were transiently transfected with GFP-LC3 alone (−) or in combination with 40 nM of siCtrl or siGrp78. The cells were then either non-treated (NT) or treated with Tun (3 μM) for 4 h. Representative images of GFP-LC3 punctate dot formation in live cells captured by fluorescence microcopy are shown. (b) Quantitation of autophagosome formation in cells treated as in (a). The results from 3 independent experiments were summarized and expressed as the mean with the indicated SD. (c) Western blot analysis of GRP78, GRP94 and β-actin (as loading control) protein levels in cells treated as in (a). (d–f) Same as (a–c) except the cells were either non-treated (NT) or subjected to NS. (g) HeLa cells were transfected with 40 nM of either siCtrl or siGrp78 and subjected to NS treatment for 2 h, TG or Tun treatment for 4 h in the absence or presence of zVAD-fmk (40 μM) as indicated on top. The levels of LC3-I and II, CHOP and β-actin were detected by Western blot. The experiments were repeated 2 times.
To test if GRP78 also affects NS-induced autophagy formation, HeLa cells transfected with GFP-LC3 alone or in combination with control siRNA or siGrp78 were nutrient starved for 2 h. As shown in Figure 5d and e, NS increased autophagosome formation in cells transfected with GFP-LC3 alone or in combination with control siRNA. However, NS-induced autophagosome formation in cells co-transfected with siGRP78 was inhibited. Similarly, in cells where GRP78 was knockdown, GRP94 was upregulated (Figure 5f).
Next we investigated whether GRP78 knockdown inhibits LC3 conversion which in turn blocks autophagosome formation. HeLa cells were transfected with control siRNA or siGrp78. The transfected cells were either non-treated, treated with Tun or TG, or subjected to NS. To test the effect of suppression of apoptosis induced under these experimental conditions, the pan caspase inhibitor zVAD-fmk was added in parallel. As shown in Figure 5g, in cells transfected with control siRNA, LC3-I to LC3-II conversion was readily detected in cells treated with TG and Tun, and addition of zVAD-fmk had no major effect. For cells subjected to NS where the LC3-II conversion was barely detectable, zVAD-fmk addition enhanced the conversion. In cells transfected with siGrp78, LC3-II conversion was intact or even slightly enhanced under all experimental conditions, and similar results were observed in cells with or without zVAD-fmk treatment (Figure 5g). Thus, despite upregulation of GRP94 and intact LC3 conversion, depletion of GRP78 leads to major suppression of autophagosome formation initiated by nutrient deprivation or ER stress.
The blockage of autophagosome formation by GRP78 knockdown is independent of PI3KC3-Beclin1 complex formation
It was recently reported that BCL-2 prevents autophagy through interruption of the association of PI3KC3 and Beclin1 at the ER.27 To test if knockdown of GRP78 has similar effect, HEK293 cells were transfected with control siRNA or siGrp78. Forty-eight hours after transfection, Beclin1 was immunoprecipitated. PI3KC3 level in the immunocomplex was tested by Western blot. As shown in Figure 6a, knockdown of GRP78 did not inhibit, but rather increased modestly PI3KC3 association with Beclin1. Furthermore, the effect of overexpression of GRP78 on PI3KC3-Beclin1 association was examined. HEK293 cells were either transfected with vector alone or different amounts of GRP78 expression plasmid. As shown in Figure 6b, overexpression of GRP78 did not change the association of PI3KC3 with Beclin1. Neither knockdown nor overexpression of GRP78 affected endogenous PI3KC3 or Beclin1 level (Figure 6a and b). Therefore, the blockage of autophagosome formation by GRP78 knockdown is independent of PI3KC3-Beclin1 association.
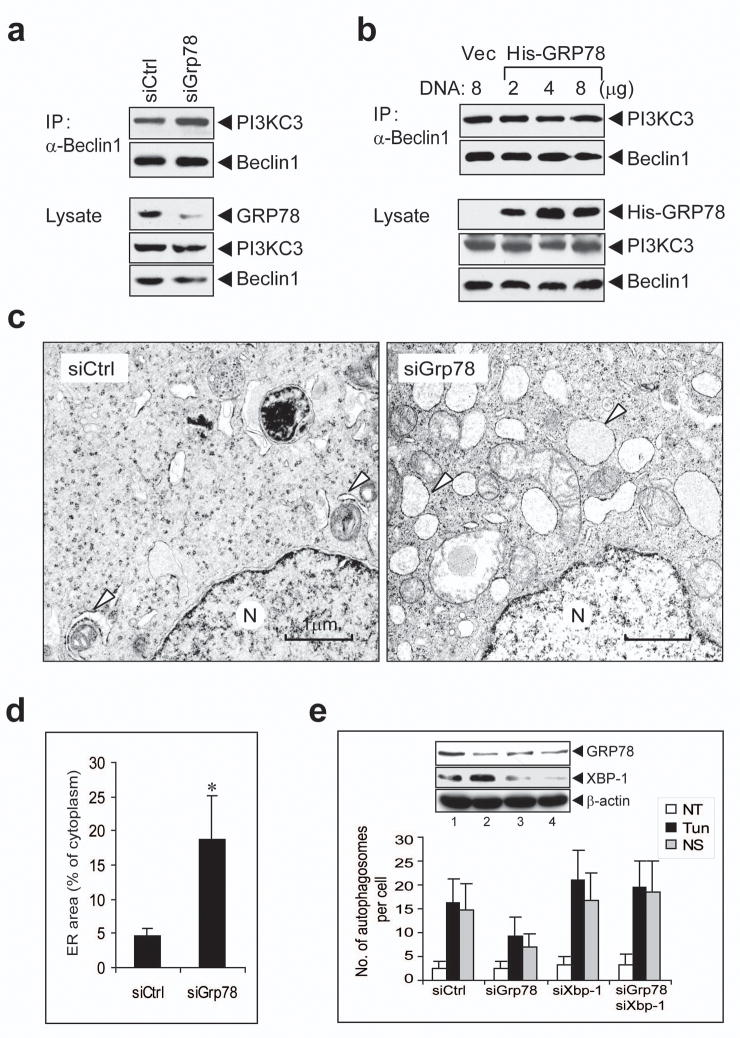
Figure 6.
GRP78 knockdown disrupts ER integrity but not PI3KC3-Beclin1 complex formation. (a) Cell lysates from siRNA transfected HEK293 cells were immunoprecipitated with anti-Beclin1 antibody. The immunocomplex was analyzed for PI3KC3 and Beclin1 levels by Western blot using antibodies sequentially. Part of the cell lysates were also analyzed for GRP78, PI3KC3 and Beclin1 expression by Western blot using antibodies sequentially. (b) HEK293 cells were transfected with pcDNA3 (vector control, vec) or indicated amount of pcDNA3-His-GRP78 (His-GRP78). Immunoprecipitation, analysis of immunocomplex and His-GRP78, PI3KC3 and Beclin1 expression were done the same as in (a). (c) Representative electron micrograph of cells transfected with siCtrl (left panel) showing sparse ER structures (white arrow heads) or siGrp78 (right panel) showing increased ER structures with expanded lumens (white arrow heads). N, nucleus; Bar=1 μm. (d) Quantitation of the ER area in the cytoplasm in cells transfected with either siCtrl or siGrp78. The standard deviation is shown, *, p<0.0001. (e) Knockdown of XBP-1 recovered normal levels of stress-induced autophagosome formation inhibited by knockdown of GRP78. The indicated siRNA(s) was transiently co-transfected with GFP-LC3 into HeLa cells. The cells were either non-treated (NT), treated with Tun (3 μM) for 4 h or NS for 2 h. The formation of autophagosome under each condition was assayed by fluorescence microscopy and quantitated. The experiments were repeated 2 times. The results were summarized and expressed as the mean with the indicated standard deviation. The insert shows Western blot analysis of GRP78, XBP-1 and β-actin levels in cells transfected with siCtrl (lane 1), siGrp78 (lane 2), siXbp-1 (lane 3) and siGrp78 plus siXbp-1 (lane 4) respectively.
Knockdown of GRP78 leads to massive ER expansion and disorganization
It has been reported previously that in yeast, prolonged ER stress could cause ER membrane expansion.28 Since GRP78 knockdown causes spontaneous UPR, GRP78 knockdown may also lead to ER membrane expansion. To test this, HEK293 cells were transfected with either control siRNA or siGrp78. Forty-eight hours after transfection, the cells were prepared for electron microscopic evaluation. As shown in Figure 6c, cells transfected with control siRNA showed sparsely distributed subcellular organelles such as ER and mitochondrion. However, in GRP78 knockdown cells, there was a dramatic increase of subcellular organelles. Not only was the number of ER structures increased, but the lumens of ER were expanded. Quantitation of the ER area in the cytoplasm of the two groups of transfected cells yielded a 3.5-fold increase of the ER in the GRP78 knockdown cells, p<0.0001 (Figure 6d). The same ER expansion was observed in HeLa cells where GRP78 knockdown was achieved using siRNA (data not shown). ER expansion is known to be dependent on XBP-1.25 To test whether knockdown of XBP-1 in GRP78 depleted cells recovers stress-induced autophagy, HeLa cells were transfected with GFP-LC3, along with siGrp78, siXbp-1 or control siRNA. Autophagosome formation was monitored by fluorescence microscopy. When added alone, siGrp78 inhibited and siXbp-1 had no effect on autophagosome formation; when siXbp-1 was added in combination with siGrp78, normal level of autophagosome was observed in both Tun-treated and NS cells (Figure 6e). The knockdown of XBP-1 and GRP78 protein level by siRNA was confirmed by Western blot.
Discussion
Evidence is emerging that the ER plays an important role in the process of autophagy. At the same time, autophagy is also critical for maintaining ER homeostasis. GRP78 is a major chaperone in the ER and is well established to be a central regulator of the UPR. In examining the role of GRP78/BiP in autophagy in human cells, we uncover several novel observations on the inter-relationship between the UPR and autophagy, which is summarized in Figure 7.
Figure 7.
Modulation of UPR signaling and autophagy pathways by 3-MA and GRP78 in human cells. Nutrient starvation (NS) as well as ER stress leads to LC3 conversion and autophagosome formation. This step requires Beclin1, which is an interactive partner of PI3KC3. Treatment of cells with 3-MA blocks both NS and ER stress-induced LC3 conversion, as well as UPR activation by ER stress. In contrast, treatment of cells with wortmannin or knockdown of Beclin1 blocks both nutrient and ER-stress induced LC3 conversion but has no effect on UPR activation. JNK activation and eIF2α phosphorylation have been reported to induce LC3 conversion, leading to autophagy. Knockdown of GRP78 results in ER stress, activation of the UPR pathways and massive ER expansion and disorganization. In GRP78 depleted cells, the complex formation between Beclin1 and PI3KC3 is intact and LC3 conversion is slightly enhanced. However, autophagosome formation induced by both NS and ER stress is inhibited.
The first unexpected result is 3-MA, a widely used inhibitor for autophagy formation, is an inhibitor of UPR pathways. Recent studies showed that ER stress-induced autophagy may act through activation of the UPR pathways. For example, expanded polyglutamine 72 (polyQ72) aggregates induced ER stress and autophagy. PolyQ72-induced LC3 conversion was inhibited in cells stably expressing dominant-negative PERK or in murine embryonic fibroblasts (MEFs) with an eIF2α phosphorylation mutant (eIF2αA/A) knockin.29,30 Similarly, autophagy induced by a misfolded dysferlin mutant that aggregates and accumulates in the ER was also inhibited in eIF2αA/A knockin MEFs.31 These studies imply that the PERK/eIF2α phosphorylation pathway is critical for ER-stress induced autophagy. On the other hand, in Ire1-deficient MEFs or wild-type MEFs treated with a JNK inhibitor, the ER-stress induced autophagy was inhibited.17 However, autophagy was induced after ER stress in PERK-deficient cells or in ATF6 knockdown cells to a similar level as in the wild-type cells. These results indicate that IRE1-JNK pathway rather than PERK or ATF6 pathway is required for autophagy activation after ER stress. Thus, while the precise requirement of specific UPR pathways to induce autophagy under different stress conditions await further resolution, our experiments showed that 3-MA inhibited eIF2α phosphorylation, ATF4 and CHOP induction which are downstream of PERK activation, as well as Xbp-1 splicing and JNK phosphorylation which are downstream of IRE1 activation (Figure 7). This implies that a new mechanism for the inhibition of ER stress-induced autophagy by 3-MA may act through inhibiting UPR pathways. This also raises the interesting question whether inhibition of PI3KC3, a known target for 3-MA, may block UPR activation. Interestingly, another PI3K inhibitor, wortmannin, was unable to suppress UPR activation. Knockdown of Beclin1, an obligatory partner of PI3KC3 in autophagy, also failed to block UPR activation. Thus, the 3-MA target(s) which mediates inhibition of the UPR may not involve PI3K and requires further investigation.
Towards examining how GRP78 regulates ER-stress induced autophagy in human cells, we employed siRNA to knockdown GRP78 expression which leads to spontaneous induction of GRP94, CHOP, JNK activation and enhancement of ER-stress induced Xbp-1 splicing, likely as a result of ER stress (Figure 7). This provides direct evidence that GRP78 suppresses their activation, in support of the generally accepted notion that when GRP78 is titrated away from the ER stress sensors, the UPR is triggered. Upregulation of GRP94 and CHOP has also been recently reported in HeLa cells using 2′-0-methyl-modified siRNA against GRP78.32 Our results further showed that UPR activation, including upregulation of GRP94, cannot compensate for the loss of GRP78 in the ER. Electron microscopy revealed that knockdown of GRP78 caused dramatic increase of ER number and ER lumen expansion. An earlier study demonstrated that yeast GRP78 was required for homotypic ER membrane fusion during karyogamy; depletion of GRP78 or expression of mutant GRP78 resulted in membranes that could not participate in fusion reactions.33 Similarly in mammalian COS cells, expressing ATPase-defective GRP78 mutant resulted in vesiculated ER.34 Thus, expansion of ER structures appears to be an evolutionarily conserved measure to compensate for loss of GRP78 function in the ER.
Our studies reveal that GRP78 depletion inhibited both ER stress and NS-induced autophagosome formation. Interestingly, while knockdown of GRP78 does not prevent LC3 conversion, formation of autophagosomes as measured by GFP-LC3 punctate dot formation is blocked. To address the mechanism of this inhibition, we investigated the effect of GRP78 knockdown on the formation of the PI3KC3 and Beclin1 complex. A previous report showed that antiapoptotic protein BCL-2 inhibited NS-induced autophagy by disrupting the association of Beclin1 and PI3KC3 at the ER.27 Our results showed that suppression of GRP78 mildly increased PI3KC3 association with Beclin1 and that overexpression of GRP78 had no effect. So the mechanism of GRP78 action is distinct from that of BCL-2. Here we demonstrate that GRP78 knockdown leads to ER expansion and increase in ER number. Evidence is emerging that proper function of the ER is required for autophagosome formation. First, the ER might be at least one of the donors that contribute to the mobilization of membrane lipid during autophagy.23,24 Second, proper ER trafficking may be required for the autophagic process. For example, disruption of ER organization in the yeast early secretion (sec) mutants blocked autophagy.35,36 In a recent study, it was reported that while about 80% of yeast cells expanded their ER under ER stress inducing conditions, only about 20% of the cells formed ER containing autophagosomes (ERAs). Significantly, none of the cells in the population containing ERAs had proliferated ER.28 In our study with human cells, Tun treatment for 5 h resulted in autophagosome formation but no ER expansion, whereas GRP78 knockdown resulted in ER expansion and blockage of autophagosome formation induced by ER stress and NS. Thus, in both yeast and human cells, ER integrity appears to be a prerequisite for autophagosome formation. Consistent with this notion, simultaneous knockdown of XBP-1 and GRP78 by siRNA in HeLa cells recovers normal levels of autophagosome formation in cells subjected to ER stress or NS.
Lastly, our discovery that GRP78 is required for autophagy provides a new mechanism for the protective function of GRP78 in stressed cells. Upon ER stress, unfolded proteins accumulate in the ER and are cleared by the proteasome-involved ER-associated degradation (ERAD) system. Recent evidence indicates that in yeast, when the ERAD system is saturated, autophagy removes both soluble and aggregated forms of unfolded proteins.37 The second role of autophagy is to help cells maintain a new steady-state level of ER abundance and cell survival even in the face of continuously accumulating unfolded proteins.28 Interestingly, yeast cell survival upon ER stress and ER-phagy does not require autophagosomal degradation, but rather ER encapsulation. The activation of autophagy for cell survival upon ER stress also applies to mammalian cells.17,18 GRP78, through maintenance of ER structure and homeostasis, facilitates autophagy. This, coupled with its well established anti-apoptosis property, explains why GRP78 is a potent pro-survival protein in cells under stress or/and in pathological conditions.
Materials and Methods
Cell cultures
The HEK293 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, L-glutamine and antibiotics. All cells were plated 1 day prior to drug treatment. For detection of autophagy, the cells were treated with ER stress inducers thapsigargin (TG, 2 μM), tunicamycin (Tun, 3 μM) or brefeldin A (BFA, 2.5μg/ml) for 4 h unless specified. For nutrient starvation, the cells were incubated with Earle’s balanced salt solution (EBSS) for 2 h.
Plasmids, antibodies and reagents
The pEGFP-LC3 expression vector was a gift from Dr. N. Mizushima (The Tokyo Metropolitan Institute of Medical Sciences, Tokyo, Japan). The plasmid pcDNA3/His-Grp78 has been previously described.38 Antibody against LC3 was either a gift from Dr. R. Kopito (Stanford University, Stanford, CA) or purchased from MBL International. Antibodies used for Western blot analysis include Beclin1 (goat polyclonal IgG for immunoprecipitation, Santa Cruz Biotechnology; mouse monoclonal IgG for blotting, BD Pharmingen), phospho-JNK1 (mouse monoclonal IgG, Promega), PI3KC3 (rabbit polyclonal IgG, Invitrogen), β-actin (AC15, mouse IgG, Sigma); CHOP (B3, mouse IgG1), ATF4 (rabbit polyclonal), JNK1 (rabbit polyclonal IgG), and XBP-1 (rabbit polyclonal) from Santa Cruz Biotechnology; phospho-eIF2α (rabbit polyclonal IgG) and eIF2α (rabbit polyclonal IgG) from Cell Signaling Biotechnology; GRP78 (mouse monoclonal IgG) from BD Pharmingen; GRP78 (rabbit polyclonal antiserum), GRP94 (rat monoclonal IgG) and KDEL (mouse monoclonal IgG) from Stressgen. 3-methyladenine (3-MA), TG, Tun, and BFA were purchased from Sigma. Wortmannin and zVAD-fmk were purchased from Calbiochem.
Western blot analysis
For blot in general, the cell lysates from treated or untreated cells were resuspended in NP-40 lysis buffer (0.5% Nonidet P-40, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, and proteinase inhibitor mixture (1 tablet/10 ml of lysis buffer, Roche Applied Science). For LC3 blot using the Kopito lab antibody, the cells were lysed in RIPA buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1% deoxycholic acid, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 10% glycerol and proteinase inhibitor mixture). After incubation on ice for 20 min, the homogenate was centrifugedat 14,000 rpm for 15 min at 4°C. Thirty microgramsof total protein of the clarified supernatants was separated by 10–13% SDS-PAGE and transferred to nitrocellulose membrane. For LC3 blot using the MBL antibody, the cells were lysed in 1× phosphate buffered saline (PBS) with 1% Triton X-100, 10% glycerol and proteinase inhibitor mixture and sonicated. Thirty micrograms of whole cell lysates was used. Western blot analysis was performed as described previously.39 All immunoblots were visualized by ECL (Amersham Biosciences). The protein levels were quantitated by densitometry (Bio-Rad Laboratories, Hercules, CA).
Co-immunoprecipitation assay
To immunoprecipitate endogenous Beclin1 in HEK293 cells, cells were lysed in NP-40 lysis as described above. Immunoprecipitation was performed by incubating 300 μg of cleared cell lysates with 1 μg anti-Beclin1 goat polyclonal antibody at 4°C overnight. Protein A sepharose beads (30 μl, Millipore) were added, incubated for 2 h at 4°C, precipitated and washed for 3 times with the NP-40 lysis buffer. Anti-Beclin1 immunoprecipitates were subjected to SDS-PAGE gel electrophoresis and PI3KC3 was detected by Western blot analysis as described above.
Transfection and RNA interference
Small interfering RNAs (siRNAs) were synthesized by our Microchemical Core Facility. The siRNA sequences for human Beclin1, sense 5′-AAG AUU GAA GAC ACA GGA GGC-3′ and antisense 5′-GCC UCC UGU GUC UUC AAU CUU-3′; for human Grp78, sense 5′-GGA GCG CAU UGA UAC UAG ATT-3′ and antisense 5′-UCU AGU AUC AAU GCG CUC CTT-3′; for human Xbp-1, sense 5′-ACA GCA AGU GGU AGA UUU ATT-3′ and antisense 5′-UAA AUC UAC CAC UUG CUG UTT-3′; for siRNA control, oligos with no matching Genebank sequence were used: sense 5′-AAG GAG ACG UAU AGC AAC GGU-3′ and antisense 5′-ACC GUU GCU AUA CGU CUC CUU-3′. The pair of siRNA oligos were annealed and prepared as a 20 μM stock. For transient transfection, cells were culturedin 12-well plates and transfected at 80% confluence with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection, the cells were left for another48 to 72 h before they were used for experiments.
Quantification of the GFP-LC3 punctate dots
GFP-LC3 alone or in combination with siRNA was transiently transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. Forty-eight hours after transfection, the cells were induced with ER stress or treated with NS, in which cell medium was replaced with EBSS after 3 times of 1× PBS washing. The GFP-LC3 punctate dot structures in individual live cells were imaged using a fluorescence microscope (Olympus IX70 inverted fluorescent light microscope system) equipped with a digital camera (Olympus DP70 digital camera system). The number of GFP-LC3 punctate dots per cell in GFP-LC3-positive cells was counted. A minimum of 15 cells per sample was counted per condition per experiment. Result was expressed as mean ± standard deviation (SD) for combined data from representative of three independent experiments.
Electron microscopy imaging
Cells were spun down into a beam capsule with a clinical centrifuge. Cell pellets were fixed in 2% paraformaldehyde and 0.1% gluteraldehyde in 0.1 M phosphate buffer (pH 7.4) at room temperature for 1 h. The cell pellets were then postfixed with 1% Osmium tetroxide for 2 h on ice and then rinsed with distilled water three times. The fixed cell pellets were dehydrated through an ethanol (ETOH) dilution series up to 100% ETOH and then immersed in propylene oxide (PO) for 2 min, three times. Pellets were then infiltrated in a 3:1 PO/Eponate resin mixture overnight and were subsequently embedded in 100% Eponate resin (Ted Pell Inc., Redding, CA) in beam capsules and allowed to harden in a 65°C oven overnight. After hardening, tissue blocks were ultra thin sectioned at a 70 nm thickness and placed on 300 mesh copper grids. Grids were next counterstained with saturated uranyl acetate and lead citrate and then viewed on a Zeiss EM 10 electron microscope (Zeiss, Thornwood, NY). For quantitation of the ER area in the cytoplasm, image analysis was performed on 16 representative cells from each group imaged at 5000× magnification using a Zeiss EM10 Transmission Electron Microscope. For each cell, the % area of the cytoplasm occupied by endoplasmic reticulum was determined on digitized images using Scion Image Analysis software (Frederick, MD).
RT-PCR analysis of Xbp-1 mRNA splicing
This was performed as described.40 Briefly, after cells were treated as indicated, total RNA was extracted using TRIZOL (Invitrogen) following the manufacturer’s instructions. First-strand cDNA was synthesized with the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen). To detect human unspliced and spliced Xbp-1 mRNA, PCR was performed using primers 5′-CTG GAA CAG CAA GTG GTA GA-3′ and 5′-CTG GGT CCT TCT GGG TAG AC-3′ as described.40 As a control, RT-PCR of β-actin was performed at the same condition using primers 5′-TCG TGC GTG ACA TTA AGG GG-3′ and 5′-GTA CTT GCG CTC AGG AGG AG-3′. Unspliced (Xbp-1, 398 bp) and spliced (Xbp-1s, 424 bp) Xbp-1 fragments were separated by 2% agarose gels, stained with ethidium bromide and photographed (Bio-Rad Fluor-S Multimager).
Clonogenic survival assay
After transfection with siCtrl or siGrp78 for 48 h, the cells were seeded into 6-cm-diameter dishes at the density of 300 cells per dish. Plating was performed in triplicate. After growing in fresh medium for 10–14 days, the colonies were washed with ice-cold PBS, fixed with methanol and stained with 10% Giemsa staining solution. The survival fraction was determined by dividing the number of surviving colonies of siGrp78 transfected cells by that of the siCtrl transfected cells.
Supplementary Material
Acknowledgments
We thank Drs. N. Mizushima for the gift of the pEGFP-LC3 expression vector, R. Kopito for antibody against LC3 and members of the Lee lab, in particular, Dezheng Dong, for assistance and helpful discussions. This work is supported in part by NIH grants CA027607 and CA111700 (to A.S.L) and a grant from the Arnold and Mabel Beckman Foundation (to D.H.) Electron microscopy was performed in the Cell and Tissue Imaging Core Facility supported by Cancer Center Support Grant 5P30 CA14089.
Abbreviations
- ATF
activating transcription factor
- BFA
brefeldin A
- CHOP
C/EBP-homologous protein
- eIF2α
eukaryotic translation initiation factor 2 α subunit
- ER
endoplasmic reticulum
- GFP
green fluorescence protein
- JNK
c-Jun N-terminal kinase
- LC3
microtubule associated protein light 1 chain 3
- NS
nutrient starvation
- PI3KC3
class III phosphatidylinositol-3′-kinase
- TG
thapsigargin
- 3-MA
3-methyladenine
- Tun
tunicamycin
- UPR
unfolded protein response
- Xbp-1
X box-binding protein 1
References
- 1.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 4.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei K, Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA. 2003;100:2432–2437. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 7.Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- 8.Hendershot LM. The ER function BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- 9.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z, Luo H, Fu W, Mattson MP. The endoplasmic reticulum stress-responsive protein GRP78 protects neurons against excitotoxicity and apoptosis: suppression of oxidative stress and stabilization of calcium homeostasis. Exp Neurol. 1999;155:302–314. doi: 10.1006/exnr.1998.7002. [DOI] [PubMed] [Google Scholar]
- 11.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6:45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 14.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 16.Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 17.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 19.Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- 20.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 21.Araki N, Hamasaki M, Egami Y, Hatae T. Effect of 3-methyladenine on the fusion process of macropinosomes in EGF-stimulated A431 cells. Cell Struct Funct. 2006;31:145–157. doi: 10.1247/csf.06029. [DOI] [PubMed] [Google Scholar]
- 22.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 23.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mijaljica D, Prescott M, Devenish RJ. Endoplasmic reticulum and Golgi complex: Contributions to, and turnover by, autophagy. Traffic. 2006;7:1590–1595. doi: 10.1111/j.1600-0854.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 27.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momoi T. Conformational diseases and ER stress-mediated cell death: apoptotic cell death and autophagic cell death. Curr Mol Med. 2006;6:111–118. doi: 10.2174/156652406775574596. [DOI] [PubMed] [Google Scholar]
- 30.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 31.Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, et al. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Lu J, Zahed M, Kita K, Suzuki N. Reduction of GRP78 expression with siRNA activates unfolded protein response leading to apoptosis in HeLa cells. Arch Biochem Biophys. 2007;468:1–14. doi: 10.1016/j.abb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Latterich M, Schekman R. The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- 34.Hendershot LM, Wei JY, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen-starvation induced apoptosis. Cancer Res. 2007;67:3734–3740. doi: 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 40.Shang J, Lehrman MA. Discordance of UPR signaling by ATF6 and Ire1p-XBP1 with levels of target transcripts. Biochem Biophys Res Commun. 2004;317:390–396. doi: 10.1016/j.bbrc.2004.03.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.