Abstract
Shape Signatures is a new computational tool that is being evaluated for applications in computational toxicology and drug discovery. The method employs a customized ray-tracing algorithm to explore the volume enclosed by the surface of a molecule and then uses the output to construct compact histograms (i.e., signatures) that encode for molecular shape and polarity. In the present study, we extend the application of the Shape Signatures methodology to the domain of computational models for cardiotoxicity. The Shape Signatures method is used to generate molecular descriptors that are then utilized with widely used classification techniques such as k nearest neighbors (k-NN), support vector machines (SVM), and Kohonen self-organizing maps (SOM). The performances of these approaches were assessed by applying them to a data set of compounds with varying affinity toward the 5-HT2B receptor as well as a set of human ether-a-go-go-related gene (hERG) potassium channel inhibitors. Our classification models for 5-HT2B represented the first attempt at global computational models for this receptor and exhibited average accuracies in the range of 73−83%. This level of performance is comparable to using commercially available molecular descriptors. The overall accuracy of the hERG Shape Signatures–SVM models was 69−73%, in line with other computational models published to date. Our data indicate that Shape Signatures descriptors can be used with SVM and Kohonen SOM and perform better in classification problems related to the analysis of highly clustered and heterogeneous property spaces. Such models may have utility for predicting the potential for cardiotoxicity in drug discovery mediated by the 5-HT2B receptor and hERG.
Introduction
The heart is a highly complex structure that ensures the survival of the organism. Consequently, xenobiotic-mediated interference with its role in homeostasis can have catastrophic effects manifesting in cardiotoxicity. For example, interference with ion homeostasis by channel (1) or exchanger blockade (2), altered coronary blood flow, oxidative stress, organellar dys-function, and apoptosis are all potential mechanisms of cardiotoxicity (3). Two different proteins, namely, the 5-HT2B receptor and the human ether-a-go-go-related gene (hERG)1 potassium channel, have raised particular concern primarily due to their association with cardiac valve disease or potassium channel blockade, respectively. Unintended activity at these two proteins independently by several drugs resulting in toxicity has prompted their withdrawal from the market by the FDA. An area of considerable interest in drug discovery research is the computational modeling of toxicity-related proteins to identify such potential problems as early as possible (4), especially as currently there are no X-ray structures for these proteins.
Serotonin is found in many physiological systems from the central nervous system to the intestinal wall and, in concert with its many receptors, plays a major regulatory function in cardiovascular morphogenesis. The 5-HT2 receptor family of G-protein-coupled receptors including 5-HT2B is expressed in cardiovascular, gut, and brain tissues as well as human carcinoid tumors (5). In recent years, this receptor has been implicated in valvular heart disease defects, caused by the now FDA-banned “fen-phen” and pergolide (6-9). The primary fenfluramine metabolite, norfenfluramine, potently stimulates 5-HT2B (10, 11). Computational modeling of this receptor has been very limited to date but is urgently needed to proactively identify drugs that may bind this receptor.
Numerous classes of drugs have been shown to prolong the QT interval, which reflects a slowing of repolarization of the ventricular myocardium (12, 13), where excessive prolongation can lead to the potentially life-threatening ventricular tachyarrhythmia, torsade de pointes. In cardiac tissue, inhibition of potassium channels is associated with QT interval prolongation (14, 15). The most common potassium channel linked to drug-induced QT interval prolongation is also responsible for the rapid component of the delayed rectifier potassium current (IKr). The focus of considerable research is hERG, which is believed to encode the protein that underlies the delayed rectifier potassium current IKr (16, 17), and many drugs associated with QT interval prolongation have been found to block hERG (18-20). Several drugs have been withdrawn from the market in the past decade due to cardiovascular toxicity associated with undesirable blockade of this channel. Since 2002, there have been numerous studies that have described individual three-dimensional (3D) quantitative structure–activity relationship (QSAR) models, statistical models, or pharmacophores for hERG (21-34). These different studies and others have encompassed a wide set of data generation and modeling techniques as well as an array of molecules for model building and testing as recently reviewed (35). There are some gross similarities in the suggested requirements for hERG inhibitors, namely, the requirement for hydrophobic features surrounding a positive ionizable/basic nitrogen feature. However, depending on the molecules and techniques used for model building, the pharmacophore or descriptors suggested may vary widely.
The focus to date has been primarily on individual hERG models of a “global” nature consisting of structurally diverse molecules across therapeutic targets (antipsychotics, antihista-mines, antibiotics, etc.) although “local” models have also been generated around narrow structural series (24). These ligand-based computational models, along with a growing number of homology models (36, 37), have provided insights that complement experimental studies such as site-directed mutagenesis (38, 39). We recently illustrated for the first time the comparison of multiple modeling approaches including Kohonen maps, Sammon maps, and recursive partitioning with the same training set, to assess whether one or a combination of approaches is preferable (40). All hERG models were assessed with a sizable external data set of published molecules and exhibited good predictivity. In addition, it was found that a structural similarity measure provides a valuable means to limit extrapolations far beyond the training set of the quantitative recursive partitioning model. The descriptors selected for the qualitative mapping methods provide further insight into the structural features of hERG inhibitors when compared with those generated by other available methods, suggesting that molecular shape or topological characteristics are also important for hERG inhibitors. Drugs for noncardiovascular indications that interact with either 5-HT2B or hERG are undesirable. Although there has been considerable modeling of hERG within pharmaceutical companies and less so for 5-HT2B, there is an unmet need for a computational platform that focuses on identifying molecules that bind to either of these proteins with affinity that may be clinically significant.
A new approach has recently been proposed that utilizes molecular shape-dependent signatures as the basis for molecular recognition (41). The Shape Signatures method employs a customized ray-tracing algorithm to explore the volume enclosed by the surface of a molecule, then uses the output to construct compact histograms (“shape signatures”) that encode for molecular shape, polarity, and other biorelevant properties (Figures 1 and 2). The method has already proven successful for a number of drug discovery programs when used for database similarity searching (41-45) and has several advantages over other approaches (Table 1). The goals of the present study were to extend the Shape Signatures tool into the domain of toxicology modeling. More specifically, we demonstrate that Shape Signatures can be employed to generate ensembles of 3D molecular descriptors useful for classifying compounds with respect to their experimentally tested activity at the 5-HT2B receptor and the hERG channel. These models were also tested against more traditional classification models with two-dimensional (2D) molecular descriptors. Our aim is to develop practical and accessible models that reliably predict whether a molecule is likely to exhibit cardiotoxicity mediated via these two proteins.
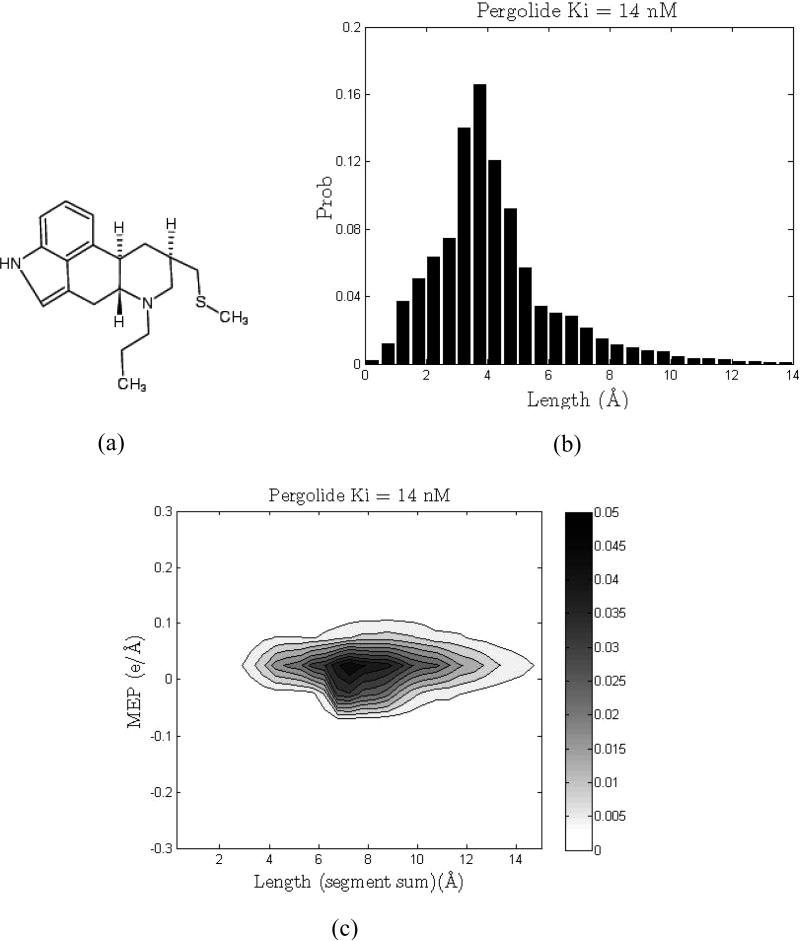
Figure 1.
One-dimensional (1D) and 2D shape signatures of pergolide, a 5-HT2B active (strong binder, Ki = 14 nM). (a) Chemical structure. (b) 1D (shape only) signature histogram. (c) 2D (shape and polarity) signature plot.
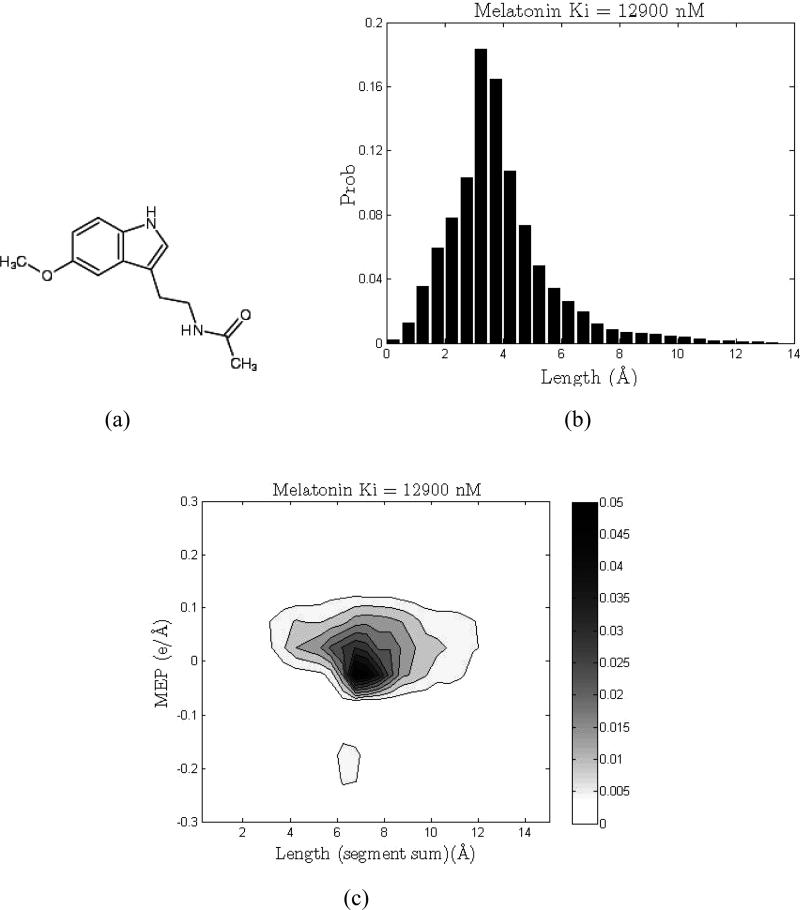
Figure 2.
1D and 2D shape signatures of melatonin, a 5-HT2B nonactive (weak binder, Ki = 12900 nM). (a) Chemical structure. (b) 1D (shape only) signature histogram. (c) 2D (shape and polarity) signature plot.
Table 1.
Comparison of Several Key Performance Measures for Traditional Descriptor-Based QSAR Approaches (1D, 2D, and 3D QSAR) vs Shape Signatures
| asset | traditional descriptor-based QSAR approaches | Shape Signatures |
|---|---|---|
| speed | ✓✓✓ | ✓✓✓ |
| accuracy | ✓✓ | ✓✓ |
| scalability | ✓ | ✓✓✓ |
| model requires reformulation as new data added | no reformulation needed as new data added | |
| coverage | ✓ | ✓✓✓ |
| descriptors must be available for chemical species | always works, i.e., organics, inorganics, organometallics, ions, etc. | |
| sensitivity | ✓ | ✓✓ |
| global model, lacks sensitivity (can also be used for local models) | local model, enhanced sensitivity | |
| domain applicability | ✓ | ✓✓✓ |
| model very sensitive to chemical (sub) structure of training set | much less sensitive to chemical (sub) structure of training set | |
| interoperability | ✓✓ | ✓✓✓ |
| integration with other QSAR models requires reformulation | fully compatible with other methods | |
| ease of use | ✓✓ | ✓✓✓ |
| preprocessing of queries requires time and know-how | no preprocessing, extremely simple to use |
Experimental Procedures
Data Acquisition
A database of >130 unique molecules was assembled for which patch clamp data for the hERG channel were available (40). Following the analysis of Ekins and co-workers, we selected 39 strong binders (IC50 < 1μM) and 44 weak binders (IC50 > 10μM) (40). In the case of 5-HT2B, the recent annotation of a database of binding information (Ki) for receptors (http://kidb.cwru.edu/) provided 182 molecules with documented binding properties (46). Among them, 116 compounds with Ki ≤ 100 nM were designated “active”, while 66 other compounds with low affinity (Ki ≥ 1μM) were “nonactive”. The full lists of compounds for each target class are available in the Supporting Information. The associated libraries of Shape Signatures required for classification were generated using the procedure outlined below.
Shape Signatures Method
In the Shape Signatures method, 3D molecular features, such as overall molecular shape and distribution of charges, are encoded in the form of the 1D and 2D dimensional histograms. The structure-related properties are regarded as key indicators of ligand–receptor molecular recognition and, thus, to the relative biological activity of the compound. These histogram-based fingerprints (“signatures”) have been used to compare the query molecule with other druglike compounds from precomputed databases. Shape similarity between the two molecules is then assessed by comparing their 1D signatures (Figures 1b and 2b). Matching the 2D signatures of the two compounds compares their overall molecular shapes and molecular electrostatic potentials (MEP) (Figures 1c and 2c). The closest matches are retrieved for further analysis. The process is fast and efficient, and the method benefits from its ability to capture the true 3D structure of the molecules without atom-based alignment of the molecules (43, 44).
A detailed description of the Shape Signatures technique can be found in the original publication (41), and we only briefly highlight the major steps of the algorithm. These descriptors are similar to the PEST shape/property descriptors described previously by Breneman et al. (47, 48). The procedure starts by generating a 3D structure of the molecule under investigation using CORINA (developed by J. Gasteiger et al., Molecular Networks GmbH, Nägelsbachstrasse 25, 91052 Erlangen, Germany; http://www.molnet.de), followed by computing partial charges for each atom using the Gasteiger–Marsili scheme (49). A solvent-accessible surface (SAS) is then constructed around the molecule, and the triangulated representation of this surface is subsequently generated by the SMART algorithm (50). Next, the ray-tracing process is initiated inside the cavity bound by the SAS, which encompasses the molecule. The ray is propagated from a randomly chosen point on the interior lining of the molecular compartment. As it strikes the opposite side, it is reflected and propagates in the direction determined by the law of optical reflection. As the ray bounces back and forth inside the enclosed molecular compartment, it generates a path composed of a number of straight line segments joined by the reflection points. For each reflection point, two quantities are calculated and stored in memory: the value of the truncated Coulomb potential at this geometric point created by the nearest atomic charges and the combined length of the incident and reflected ray segments. Given a sufficient number of reflections (100000 in this study), the trajectory of the ray will eventually explore the entire volume of the molecule. To prevent trapping of light inside some tight and unusually shaped parts of the molecular compartment, the ray-tracing procedure is periodically stopped and reinitiated from a different randomly selected point on the inner surface. At the end of the run, all recorded ray segments are binned by their length in a 1D histogram with the predefined bin width of 0.5 Å (Figures 1b and 2b). Simultaneously, a second histogram is also constructed; this one bins records by values of the MEP with a step of 0.05 e/Å and the associated total length of the two path segments joined by the reflection point, resulting in a 2D histogram (41) (Figures 1c and 2c). Both histograms are properly normalized. The first histogram encodes exclusively for the shape characteristics of the molecule (it represents the probability distribution of finding a ray segment of a particular length inside the SAS surrounding the molecule), whereas the second histogram reflects both the molecular shape and the 3D arrangement of atomic charges in this molecule (it expresses a joint probability distribution of finding a particular value of MEP with a certain length of the two ray segments connected by the reflection point). Once calculated, the resultant Shape Signatures fingerprints can be employed in a variety of problems in drug discovery and computational toxicology, which require matching chemical structures based on their shapes and polarities.
Shape Signatures Molecular Descriptors
For every molecule in this study, the heights of the corresponding normalized 1D and 2D shape signature bins comprise the sets of distinct molecular descriptors related to this particular structure. Consequently, each chemical has two sets of continuous descriptors: one based exclusively on molecular shape and the other reflecting both molecular shape and polarity. It is important to emphasize that although these features are pieces of 1D and 2D shape signatures, they are inherently 3D molecular descriptors since they encode for the 3D arrangements of atoms and atomic charges in a molecule. We also note that for all classification runs based solely on Shape Signatures, no additional descriptors were appended to the input Shape Signatures data vectors. As will be discussed later in the text, a mixed descriptor scheme with some combination of the traditional commercially available 2D descriptors [e.g., those in Molecular Operating Environment (MOE), Montreal, Canada: Chemical Computing Group Inc.] and the Shape Signatures-derived 3D descriptors seems an interesting continuation of the reported analysis in the future.
As a preliminary test, we used the suggested descriptor allocation scheme to cluster 22 bioactive compounds from our in-house libraries. The set included 10 nuclear receptor (five estrogen receptor and five androgen receptor) binders, five Pfmrk kinase inhibitors, and seven tubulin ligands. Clustering was based on molecular shape captured by the 1D Shape Signatures histograms, used simple Euclidean distances as similarity scores, and was performed by constructing and analyzing Kohonen self-organizing maps (SOM) (51). The results obtained (data not shown) indicated that structures from different target families occupied distinct regions of the SOM, and with only two clear misclassifications for the 22 molecules. These misclassifications were genistein [a frequent hitter (52-54)] and raloxifene, which was observed to lie closer to the androgen receptor molecules rather than the estrogen receptor compounds (55). Encouraged by these preliminary findings, we decided to build classification models for the 5-HT2B receptor and hERG potassium channel data sets.
Classification Procedures
We have investigated descriptors derived from the Shape Signatures representations using different classifier algorithms. For a simple classifier, we chose the k-nearest neighbors (k-NN) algorithm (56), which is very easy to implement. Despite its simplicity, this method has been shown to produce acceptable results for many applications (57-59). In this method, each query molecule from a given test set is compared in turn with all compounds in the training data set with known class affiliations, and similarity scores are calculated for every pair. The comparison is made between the corresponding 1D and 2D shape signatures of the two molecules, and we utilize the χ-square measure (56), widely employed for comparing discrete distributions, to compute similarity scores. At the end of each run, the entire training set is rank ordered with those more similar (to the query) structures being placed at the top of the list. The decision on which class a given query structure shall belong to is made based on a majority vote of its k nearest, that is, k most similar, neighbors. For highly unbalanced data sets, the weights of the neighbors are adjusted according to their class prior probabilities. For an unbalanced data set, k-NN with a straightforward majority vote will favor assignment to the larger of the two classes as the size of the NN list grows. To avoid such a situation, we need to adjust the majority vote rule accordingly (or equivalently assign different weights to molecules on the NN list belonging to different classes). It is therefore customary to vary k within some range, depending on the size of the training set, in search of the value with maximum prediction accuracy.
The support vector machine (SVM) method, based on the principle of structural risk minimization (60, 61), is a relatively recent addition to the family of supervised classification methods [discussed in detail in a recent book chapter (62)]. This technique has already gained recognition as one of the most robust and efficient classifiers (21, 56-58, 63). It can tackle nontrivial problems by projecting the original descriptor vectors to a higher dimensional feature space where a clearer division between the two classes of data becomes feasible. In such a high-dimensional feature space, a linear SVM routine is applied next to optimally position the separating hyperplane between the instances from the two classes (62). Minimization of the expected generalization error for the test data sets is achieved by finding a separating hyperplane with the maximal margin. Computationally, the transformation into a higher dimensional feature space is implicit as only the distances between the pairs of the transformed data are needed for training and these are computed using the predefined kernel functions K, the associated parameters for K, and the original input descriptor vectors. As such, this approach is less likely to suffer from “data overfitting” and can successfully handle situations involving hundreds or many thousands of descriptors. We used a well-tested and freely available program LIBSVM (C-SVM) (64). We worked with the radial basis function kernel, whose parameter γ and the penalty term C were determined in each case via a grid search procedure utilizing 5-fold cross-validations.
The data sets ultimately used in this study included 83 chemicals for the hERG potassium channel and 182 chemicals for 5-HT2B receptor. For each of these data sets, a pair of 1D and 2D shape signatures was constructed according to the procedure detailed above. There are on average about 20−60 nonzero bins/descriptors for the 1D (shape only) Shape Signatures histograms. For the 2D histograms (shape and polarity), this number is significantly higher, on the order of several hundred. Consequently, to avoid overfitting in the latter case, we applied the unsupervised forward selection (UFS) method of Livingstone and co-workers (65) to reduce the dimensionality of the problem. The UFS scheme, which was designed to eliminate redundancy and diminish multicollinearity of the input data, has been demonstrated to be fairly successful for a number of QSAR studies (65). The algorithm consists of two major steps. While processing the original descriptor data matrix (responses are not included), the routine first excludes descriptor columns with small standard deviations (ε < εmin) as contributing no significant information. It then analyzes the reduced data matrix, selects two least correlated descriptor columns, and rejects those with high pairwise correlation coefficients (R2 > Rmax2). The list of the selected descriptors is augmented by the column that has the smallest squared multiple correlation coefficient. This step is performed repeatedly, producing a growing list of nominated descriptor columns, which survive the rejection filter based on the squared multiple correlation coefficient with the columns picked in the previous step. The procedure stops when the list of columns is fully exhausted. For our experiments, we used the code available from Whitley et al. (65), with the default parameter settings for εmin and Rmax2 as 0.0005 and 0.99, respectively.
Neural Network Modeling Using Self-Organizing Kohonen Maps
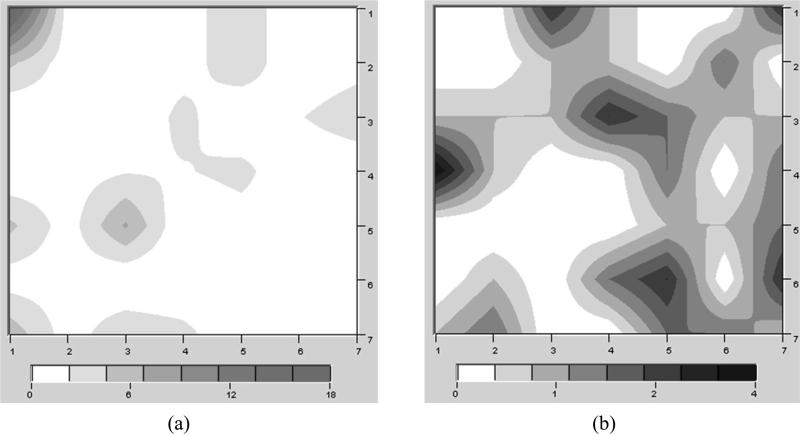
As we have described previously (40, 66), the general idea behind Kohonen maps (51) is to map a set of vectorial samples onto a 2D lattice in a manner that preserves the topology of the original space. Kohonen maps belong to a class of neural networks known as competitive learning or self-organizing networks. The Kohonen map consists of artificial neurons that are characterized by weight vectors with the same dimensionality as the descriptor set. The neurons are connected by a distance-dependent function. In an unsupervised training algorithm, the neurons self-organize until their pairwise neighborhoods represent the correct topology of the original data set. Kohonen maps have recently been applied to successfully model cytochrome P450-mediated drug metabolism (67, 68) and hERG inhibition (40). The generation of the Kohonen SOMs (51) was conducted using the Smart Mining software v1.01 (ChemDiv, Inc., San Diego, CA, www.chemosoft.com). A 7 × 7 node architecture was chosen to provide the studied molecules with the optimal distribution space. The 5-HT2B data set included 140 compounds (77% of the entire database) denoted as a “training” set (89 active and 51 nonactive) and 42 compounds (23% of the entire database) denoted as a “test” set (27 active and 15 nonactive). In total, 182 compounds (100%) were used for generation and validation of the SOMs. Similar to the SVM analysis, Kohonen networks were constructed using 102 2D Shape Signatures descriptors computed for each molecule in the set. The training parameters for the SOM were as follows: The classical algorithm based on the incremental learning method was applied for generation of the Kohonen maps, the neurons were studied using the normal distribution law encoded by the Gaussian probability function, the initial distribution of synaptic weights was randomly assigned, the number of interactions for the training runs was 3000, the starting adjustment radius for the training runs was 4, and the initial learning rate factor was 0.5. We have used 30 randomizations of the input training set for the Kohonen map generation. After the SOMs were generated, we studied the distribution of active and nonactive compounds within the best mapping. The resulting maps are shown in Figure 3.
Figure 3.
Separate distribution of 5-HT2B active (a) and nonactive (b) compounds within the Kohonen network (the best randomization). Darker intensities relate to more molecules in that area. The data have been smoothed for presentation purposes.
Model Testing
To carefully evaluate the performance of each Shape Signatures/classifier combination applied to the hERG or 5-HT2B data, three different types of statistical testing were undertaken. For the Shape Signatures paired with the SVM approach, we conducted straightforward 10-fold cross-validations on the entire data sets and subjected the systems to a series of leave-N-out runs. The leave-N-out tests were designed as follows. For either target, N compounds from the original data set were randomly picked to represent the hold-out test set, and the rest of the data constituted the training set for this particular data partition. The selection was carried out to approximately preserve the correct proportion of active and nonactive structures in both sets. In particular, for hERG, N = 20 (24% of the data set) with 10 active and 10 nonactive, and for 5-HT2B, N = 42 (23% of the data set) including 27 active and 15 nonactive molecules. Each classification algorithm was then trained on the training set and applied to predict class attributes of the compounds in the test set. Next, a set of statistical indicators of prediction accuracy were computed and stored. To obtain better statistical estimates, the described procedure was repeated 30 times, each time with a different composition of the test and training sets. For each target, the reported final statistical measures were averaged over the indicated number of repetitions.
Model Statistics
A broad spectrum of statistical indicators is available for assessing the performance of a given classification model (56). In this study, we report the most commonly encountered measures for estimating prediction accuracy of a classifier: sensitivity (SE), specificity (SP), and overall accuracy (Q). These quantities are defined in terms of the numbers of true positives (tp) and false positives (fp), indicating strong binders to either hERG or 5-HT2B in our case, and the numbers of true negatives (tn) and false negatives (fn), that is, presumably nonactive compounds. Sensitivity, SE = tp/(tp + fn), then expresses the prediction accuracy for molecules with high affinity to the considered targets, whereas specificity reflects the prediction accuracy for weak binders: SP = tn/(tn + fp). We also tabulate the overall prediction accuracy defined as Q = (tp + tn)/(tp + fp + tn + fn). In addition, following Ung et al. (58), we report the values of Matthew's correlation coefficient (69) C = [tp × tn - fp × fn]/[(tp + fn)(tp + fp)(tn + fp)(tn + fn)]1/2, which is another measure of the overall prediction performance. This indicator has interesting properties: For a perfect classifier (fp = fn = 0), C ) 1.0, while for random performance (resulting in tp ≈ fp and tn ≈ fn on average), C ≈ 0. A negative value C would imply worse than random performance.
Results
hERG Models
An initial evaluation of the Shape Signatures descriptors was performed with the hERG data set. The results of various classification schemes applied to discriminate between strong and weak blockers of hERG are summarized in Table 2. All of the reported models perform substantially better than random. The UFS-SVM model with shape and charge descriptors appears to perform slightly better than the k-NN models. The average prediction accuracy for the external test sets varies from 66 to 74%, which is comparable to the 70−85% established by summarizing the results of other predictive modeling studies of hERG reported in the literature (35).
Table 2.
Classification of hERG Active and Nonactive Compounds Using Shape Signatures Descriptors with Different Methods
| leave-20-out testingd |
||||||
|---|---|---|---|---|---|---|
| classification methoda | descriptorsb | 10-fold cross-validationc (%) | SE (%) | SP (%) | Q (%) | C |
| SVM | shape only | 77 | 70 | 68 | 69 | 0.390 |
| UFS-SVM | shape + charges | 78 | 73 | 74 | 74 | 0.488 |
| k-NN (k = 7) | shape only | 68 | 79 | 53 | 66 | 0.343 |
| k-NN (k = 3) | shape + charges | 69 | 79 | 56 | 67 | 0.367 |
The specified values of k for k-NN classifications are those that yield the maximum average overall prediction accuracies Q for the leave-20-out experiments.
“Shape only” label descriptor sets derived from the 1D Shape Signatures histograms, and “shape + charges” designates descriptors sets based on the 2D Shape Signatures histograms.
This column lists prediction accuracies estimated from 10-fold cross-validations performed on the entire data set.
The tabulated values of SE, SP, Q, and C are averaged over 30 different hold-out test sets.
5-HT2B Models
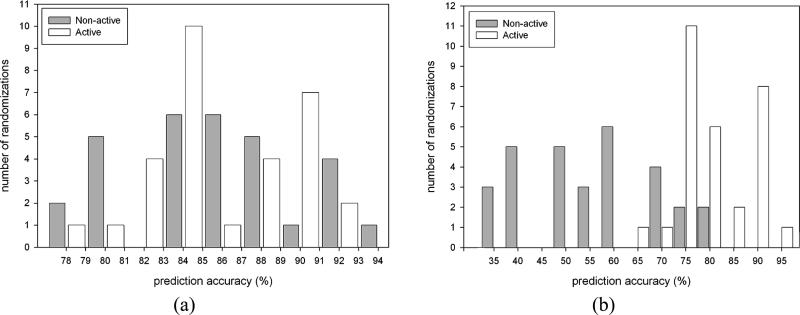
Our evaluation of the 5-HT2B data is shown in Table 3. As a direct comparison with Shape Signatures descriptors, we have used a set of 184 2D molecular descriptors available in MOE. The initial data matrix for these descriptors was also processed by the UFS algorithm (described above), and the resulting ensemble of 73 2D MOE descriptors was used for final calculations. Both of the UFS-SVM models with Shape Signatures or MOE descriptors perform similarly in terms of model statistics resulting in prediction accuracies of 87% after 10-fold cross-validation (Table 3). We have also used UFS with SOM and the Shape Signatures descriptors. Among 30 randomizations used in the SOM, the average percentage of correctly predicted compounds belonging to both classes included in the training set was 85% for active compounds and 86% for nonactive compounds. The average percentage of correctly predicted compounds from the test set was 78% for active compounds and 54% for nonactive compounds. On average, 5% of the tested compounds were assigned to the “unclassified” class. The ratio of correctly predicted compounds from both tested groups (active and nonactive) was in general well-balanced for the training sets (Figure 4a). At the same time, a clear bias toward better prediction accuracy for active structures was observed for the internal test sets (Figure 4b). One possible reason for such bias is an increased dissimilarity and smaller number of nonactive compounds in the training set as compared to active compounds. Therefore, the training results are statistically less significant for the nonactive subset.
Table 3.
Classification of 5-HT2B Active and Nonactive Compounds Using Shape Signatures or MOE Descriptors with Different Methods
| leave-42-out testingd |
||||||
|---|---|---|---|---|---|---|
| classification methoda | descriptorsb | 10-fold cross-validationc (%) | SE (%) | SP (%) | Q (%) | C |
| SVM | shape only | 80 | 81 | 59 | 73 | 0.424 |
| UFS-SVM | shape + charges | 87 | 91 | 69 | 83 | 0.638 |
| UFS-SVM | MOE (2D) | 87 | 91 | 70 | 84 | 0.640 |
| UFS-SOMe | shape + charges | 86 | 78 | 54 | 70 | 0.345 |
| k-NN (k = 3) | shape only | 69 | 86 | 46 | 72 | 0.352 |
| k-NN (k = 1) | shape + charges | 74 | 93 | 53 | 79 | 0.527 |
The specified values of k for k-NN classifications are those that yield the maximum average overall prediction accuracies Q for the leave-42-out experiments.
“Shape only” labels descriptor sets derived from the 1D Shape Signatures histograms, and “shape + charges” designates descriptors sets based on the 2D Shape Signatures histograms.
This column lists prediction accuracies estimated from 10-fold cross-validations performed on the entire data set.
The tabulated values of SE, SP, Q, and C are averages over 30 different hold-out test sets.
On average, one active and one nonactive molecule were unclassified.
Figure 4.
Prediction accuracy for 5-HT2B active and nonactive compounds, from the training (a) and test (b) sets used in the Kohonen network (SOM).
Discussion
Computational Methods
Several ligand-based and structure-based computational methods exist that implicitly or explicitly include some representation of molecular shape. The program UCSF DOCK (70, 71) packs spheres into a protein receptor site; candidate ligands can then be evaluated for shape compatibility with the site by checking for containment within the array of spheres. Comparative molecular field analysis (72) (CoMFA) represents the shape of molecules implicitly by mapping steric and electrostatic fields on a 3D grid that surrounds the molecule; biological activity is then correlated with variations of the fields at the grid points. Essentially all automated docking programs implicitly represent molecular shape via some form of energy calculations. Inasmuch as shape is directly related to molecular structure, tools that employ pharmacophore models (e.g., Catalyst, UNITY, etc.) represent molecular shape via interatomic distance constraints. The aforementioned shape-based methods, although invaluable, demand considerable computation involving either energy or distance–geometry calculations. Moreover, the matching of a compound to a receptor site or pharmacophore model typically involves some sort of simulation (genetic algorithm, Monte Carlo method, etc.) for generating orientations and configurations of the ligand. CoMFA requires manual alignment of the series of molecules, a highly subjective process that effectively limits the number of compounds to ≈150. When the goal is to screen large vendor or legacy databases of compounds, such methods may lead to prohibitive computational costs. What is needed is a method that can rapidly compare shapes of large databases of compounds to each other, or to a receptor site, with a minimum of computation, without requiring explicit 3D representation of shape and without actual ligand–receptor docking. This is just the sort of method embodied in Shape Signatures descriptors used for QSAR, which has several advantages over traditional molecular descriptor-based QSAR methods (Table 1).
hERG Models
While there have been many models for this potassium channel, our goal in this study was to use hERG as a test case to evaluate Shape Signatures descriptors with different classifier algorithms. A qualitative comparison of our hERG results with those extracted from other studies reported to date (35) suggests that the differences may be insignificant. Overall, in this study, the prediction accuracies of SVM based methods outperform the k-NN models. Although k-NN models yield better selectivity rates, this is achieved at the expense of much lower specificity values. In particular, k-NN classifiers suffer from a large number of false positive predictions and, thus, are less selective than the procedures that use SVM. We found that regardless of the classifier employed, the models based on the 2D Shape Signatures (shape and charge) are slightly more accurate than those derived from the 1D signatures (shape). This implies that for better selectivity, one may need to incorporate the polarity of the molecules into the model. This observation is consistent with the notion that the hERG channel can accommodate inhibitors of different size and shape. This may also relate to the position of the basic N atom found in many hERG inhibitors and suggested to be important in many of the published pharmacophores. Ekins et al. previously used a single external set of 21 compounds (seven active and 14 nonactive) to test their classification models based on Kohonen and Sammon mapping techniques (40). For Kohonen SOM, they obtained SE = 86%, SP = 79%, and Q = 81%, and for Sammon mapping, they reported SE = 86%, SP = 100%, and Q = 95%. These results are better than the average values obtained over the series of 30 leave-20-out experiments tabulated in Table 2. However, if we turn to some of the best-performing UFS-SVM models in our study, these models yield SE = 80−90%, SP = 80−100%, and Q = 85−95%, which is comparable to the Sammon mapping model previously described (40).
5-HT2B Models
Our results with the 5-HT2B classification models are the major focus of this study and are the first classification models for this receptor to be reported to date. Previous computational modeling of 5-HT2B has encompassed a traditional QSAR study, which used a small number of tetrahydro-β-carboline derivatives as antagonists with the rat 5-HT2B contractile receptor in the rat stomach fundus (73). A 3D QSAR with GRID-GOLPE using 38 (aminoalkyl)benzo and heterocycloalkanones as antagonists of the human receptor resulted in poor model statistics possibly due to the limited range of activity measured and the complexity of the functional response (74). Homology models based on the bacteriorhodopsin as well as rhodopsin X-ray structures have been used for the mouse and human 5-HT2B receptor and combined with site-directed mutagenesis. The models based on bacteriorhodopsin proved more reliable and confirmed an aromatic box hypothesis for ligand interaction along transmembrane domains 3, 6, and 7 with serotonin (75). A more recent 5-HT2B homology model with the rhodopsin-based model of the rat 5-HT2A together with molecular dynamics simulations was used to determine the sites of interaction for norfenfluramine. Site-directed mutagenesis showed that Val 2.53 was implicated in high affinity binding through van der Waals interactions and the ligand methyl groups (76).
We have found in this study that, similar to the hERG modeling described previously, for 5-HT2B, SVM generally outperforms k-NN methods (Table 3). Interestingly, the same observation has been documented in a number of classification studies across different classes of protein targets (57, 58). In comparison to the results with the hERG data set (Table 2), for 5-HT2B, we were able to achieve generally better overall prediction accuracies for the test sets within the range of 72−84%. Among the models based on Shape Signatures descriptors, the UFS-SVM procedure is again the best. On average, these models compare well with SVM classifications paired with the traditional 2D molecular descriptors computed with MOE. This observation further validates the applicability of the Shape Signatures-derived molecular descriptors. The 2D Shape Signatures classifiers appear superior to the models based on the 1D histograms, indicating that molecular polarity is likely necessary for generating more accurate predictions for 5-HT2B.
The results presented in Table 3 also demonstrate that the prediction accuracies achieved in the Kohonen modeling experiments (86% prediction accuracy estimated from 10-fold cross-validations), in general, were similar to those observed in the best UFS-SVM models (87%) and better than in k-NN models (74%), when the same 102 “shape + charges” descriptors were used. The iterative methods based on vector quantization algorithms, such as SVM and Kohonen SOM, perform better in classification tasks related to analysis of highly clustered and heterogeneous property spaces. Classification results certainly vary from one classifier to another, but if several different classification models using the same collection of molecular descriptors produce consistent results, it would certainly add to the credibility of the utilized descriptor set. We found that the average statistics produced for either hERG or 5-HT2B by k-NN, SVM, and SOM models are generally consistent overall, indicating that the Shape Signatures histograms constitute a useful set of new molecular descriptors for these types of classification problems. Their utility likely also rests in their ability to reliably capture the shape and charge requirements for molecules to fit to these proteins.
To further investigate the utility of the shape signatures-based molecular descriptors, we evaluated the SVM and SOM models with an additional external test set (77). This set included 20 compounds with documented activities toward 5-HT2B: six active and 14 nonactive molecules. We attempted to classify compounds in this data set using models built on the original data set of 182 5-HT2B molecules, which are described in Table 3. As before, all calculations were performed using the set of 102 2D Shape Signatures descriptors. The best UFS-SVM and SOM models yielded SE = 33%, SP = 71%, and Q = 60%. This prediction accuracy is lower than the corresponding values reported in Table 3 for the original data set, especially for the SVM models. The reason for this may be 2-fold. First, with the current settings, Shape Signatures may not perform well in separating close structural analogues in an external test set. However, the use of alternative descriptors, such as the electrotopological indices (calculated using the SmartMining program), led to similar prediction accuracy on the same test set. Second, and probably the most important point to consider, is that the compounds in the external test set may lie outside the chemical space occupied by the structures from the original 182 molecules data set. Indeed, similarity measures in the form of Euclidean distances, calculated using the ChemoSoft software between pairs of molecules, suggest significant structural differences between the original data set (182 structures) and the external test set (data not shown).
A major objective of the reported study was to thoroughly examine the quality of a novel set of molecular descriptors derived from the associated molecular Shape Signatures previously used as a virtual screening tool for drug discovery (41-45). These descriptors are inherently 3D and fundamentally different from other 2D/3D descriptor collections normally used in predictive QSAR modeling (1, 78). We have therefore extended the Shape Signatures methodology in the form of molecular classifiers for computational toxicology. Practical classification models for the 5-HT2B receptor and the hERG potassium channel have been constructed and validated. Our classification models for 5-HT2B offer the potential to predict cardiotoxicity earlier in drug discovery. In the case of 5-HT2B, we report the first Shape Signatures-SVM-based classification models, which exhibit average accuracies in the range of 73−83%. These findings are comparable with the results of the SVM classification using traditional 2D molecular descriptors available in the commercially available software MOE, which was also performed in this study. Further research is currently underway in our laboratories to examine the combination of Shape Signature histograms with traditional 2D descriptors (such as from MOE) to assess whether this improves the models. For hERG, the prediction accuracy is comparable with the results of alternative computational models published to date. Altogether, our study demonstrates that the reported classification models perform well in discriminating between hERG and 5-HT2B active and nonactive molecules and could be applicable to other protein targets. We also note that, as with any molecular descriptors and algorithms used for QSAR to date, it is important to understand the chemical space covered in both the training and the test sets for optimal predictions (i.e., the applicable prediction space). Our results certainly attest to the notion that molecular shape and polarity are indeed key characteristics that regulate molecular activity toward specific protein targets. Given the simplicity, physical transparency, and applicability of the Shape Signatures representation, this method encodes these main features in a compact and practical form. Because the procedure obviates direct 3D molecular alignment or grid generation [as in CoMFA and comparative molecular similarity analysis (CoMSIA) etc.], the algorithm is also relatively fast and efficient. Models based on Shape Signatures histograms can therefore accommodate structurally diverse compounds; once generated, they can be used for a variety of tasks that require molecular recognition, and no model refitting is necessary in going from one problem to another (Table 1). We are currently using Shape Signatures to aid in drug discovery projects while also evaluating the Shape Signatures descriptors for other physicochemical properties, as we believe this approach is generally applicable. Overall, we conclude that the Shape Signatures method offers a novel practical approach to classifying compounds with respect to their potential for cardiotoxicity. Further future studies will use these 5-HT2B models for mining databases to identify additional compounds for in vitro testing to prospectively validate them, a strategy that we have successfully undertaken for transporters (79, 80).
Supplementary Material
Acknowledgment
Support for this work has been provided by the U.S. EPA-funded Environmental Bioinformatics and Computational Toxicology Center (ebCTC), under STAR Grant GAD R 832721-010. W.J.W. gratefully acknowledges support for this work provided by the Defense Threat Reduction Agency, under contract HDTRA-BB07TAS020. This work was also funded in part by NIH R21-GM081394 from the National Institute of General Medical Sciences and by NIH Integrated Advanced Information Management Systems (IAIMS) Grant 2G08LM06230-03A1 from the National Library of Medicine. This work has not been reviewed by and does not represent the opinions of the funding agencies. We are sincerely grateful to Randy Zauhar, Ph.D., of the University of the Sciences in Philadelphia, for useful discussions pertaining to technical aspects of the Shape Signatures algorithm.
Footnotes
Abbreviations: CoMFA, comparative molecular field analysis; CoMSIA, comparative molecular similarity analysis; hERG, human ether-a-go-go-related gene; k-NN, k-nearest neighbors; QSAR, quantitative structure–activity relationship; SOM, self-organizing maps; SVM, support vector machines; UFS, unsupervised forward selection.
Supporting Information Available: SDF files and activity data for molecules used in this study for hERG and 5-HT2B models. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ekins S, Embrechts MJ, Breneman CM, Jim K, Wery J-P. Novel applications of Kernel-partial least squares to modeling a comprehensive array of properties for drug discovery. In: Ekins S, editor. Computational Toxicology: Risk Assessment for Pharmaceutical and Environmental Chemicals. Wiley-Interscience; Hoboken, NJ: 2007. pp. 403–432. [Google Scholar]
- 2.Keenan SM, DeLisle RK, Welsh WJ, Paula S, Ball WJ., Jr. Elucidation of the Na+, K+-ATPase digitalis binding site. J. Mol. Graphics Modell. 2005;23:465–475. doi: 10.1016/j.jmgm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Ramos KS, Melchert RB, Chacon E, Acosta JD. In: Toxicology the Basic Science of Poisons. Klaasen CD, editor. McGraw Hill; New York: 1985. pp. 597–652. [Google Scholar]
- 4.Ekins S, Swaan PW. Computational models for enzymes, transporters, channels and receptors relevant to ADME/TOX. Rev. Comp. Chem. 2004;20:333–415. [Google Scholar]
- 5.Nebigil CG, Choi DS, Dierich A, Hickel P, Le Meur M, Messaddeq N, Launay JM, Maroteaux L. Serotonin 2B receptor is required for heart development. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9508–9513. doi: 10.1073/pnas.97.17.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth BL. Drugs and valvular heart disease. N. Engl. J. Med. 2007;356:6–9. doi: 10.1056/NEJMp068265. [DOI] [PubMed] [Google Scholar]
- 7.Schade R, Andersohn F, Suissa S, Haverkamp W, Garbe E. Dopamine agonists and the risk of cardiac-valve regurgitation. N. Engl. J. Med. 2007;356:29–38. doi: 10.1056/NEJMoa062222. [DOI] [PubMed] [Google Scholar]
- 8.Zanettini R, Antonini A, Gatto G, Gentile R, Tesei S, Pezzoli G. Valvular heart disease and the use of dopamine agonists for Parkinson's disease. N. Engl. J. Med. 2007;356:39–46. doi: 10.1056/NEJMoa054830. [DOI] [PubMed] [Google Scholar]
- 9.Jahnichen S, Horowski R, Pertz HH. Agonism at 5-HT2B receptors is not a class effect of the ergolines. Eur. J. Pharmacol. 2005;513:225–228. doi: 10.1016/j.ejphar.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald LW, Burn TC, Brown BS, Patterson JP, Corjay MH, Valentine PA, Sun JH, Link JR, Abbaszade I, Hollis JM, Largent BL, Hartig PR, Hollis GF, Meunier PC, Robichaud AJ, Robertson DW. Possible role of valvular serotonin 5-HT(2B) receptors in the cardiopathy associated with fenfluramine. Mol. Pharmacol. 2000;57:75–81. [PubMed] [Google Scholar]
- 11.Rothman RB, Baumann MH, Savage JE, Rauser L, McBride A, Hufeisen SJ, Roth BL. Evidence for possible involvement of 5-HT(2B) receptors in the cardiac valvulopathy associated with fenfluramine and other serotonergic medications. Circulation. 2000;102:2836–2841. doi: 10.1161/01.cir.102.23.2836. [DOI] [PubMed] [Google Scholar]
- 12.Tan HL, Hou CJ, Lauer MR, Sung RJ. Electrophysiologic mechanisms of the long QT interval syndromes and torsade de pointes. Ann. Intern. Med. 1995;122:701–714. doi: 10.7326/0003-4819-122-9-199505010-00009. [DOI] [PubMed] [Google Scholar]
- 13.Thomas SH. Drugs, QT interval abnormalities and ventricular arrhythmias. Adverse Drug React. Toxicol. Rev. 1994;13:77–102. [PubMed] [Google Scholar]
- 14.Barry DM, Xu H, Schuessler RB, Nerbonne JM. Functional knockout of the transient outward current, long-QT syndrome, and cardiac remodeling in mice expressing a dominant-negative Kv4 alpha subunit. Circ. Res. 1998;83:560–567. doi: 10.1161/01.res.83.5.560. [DOI] [PubMed] [Google Scholar]
- 15.Jeron A, Mitchell GF, Zhou J, Murata M, London B, Buckett P, Wiviott SD, Koren G. Inducible polymorphic ventricular tachyarrhythmias in a transgenic mouse model with a long Q-T phenotype. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H1891–H1898. doi: 10.1152/ajpheart.2000.278.6.H1891. [DOI] [PubMed] [Google Scholar]
- 16.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 17.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for the cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 19.Rampe D, Murawsky MK, Grau J, Lewis EW. The antipsychotic agent sertindole is a high affinity antagonist of the human cardiac potassium channel HERG. J. Pharmacol. Exp. Ther. 1998;286:788–793. [PubMed] [Google Scholar]
- 20.Suessbrich H, Schonherr R, Heinemann SH, Attali B, Lang F, Busch AE. The inhibitory effect of the antipsychotic drug haloperidol on HERG potassium channels expressed in Xenopus oocytes. Br. J. Pharmacol. 1997;120:968–974. doi: 10.1038/sj.bjp.0700989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobita M, Nishikawa T, Nagashima R. A discriminant model constructed by the support vector machine method for HERG potassium channel inhibitors. Bioorg. Med. Chem. Lett. 2005;15:2886–2890. doi: 10.1016/j.bmcl.2005.03.080. [DOI] [PubMed] [Google Scholar]
- 22.Roche O, Trube G, Zuegge J, Pflimlin P, Alanine A, Schneider G. A virtual screening method for the prediction of the hERG potassium channel liability of compound libraries. ChemBioChem. 2002;3:455–459. doi: 10.1002/1439-7633(20020503)3:5<455::AID-CBIC455>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Pearlstein RA, Vaz RJ, Rampe D. Understanding the structure-activity relationship of the human ether-a-go-go-related gene cardiac K+ channel. A model for bad behavior. J. Med. Chem. 2003;46:2017–2022. doi: 10.1021/jm0205651. [DOI] [PubMed] [Google Scholar]
- 24.Pearlstein RA, Vaz RJ, Kang J, Chen X-L, Preobrazhenskaya M, Shchekotikhin AE, Korolev AM, Lysenkova LN, Miroshnikova OV, Hendrix J, Rampe D. Characterization of HERG Potassium channel inhibition using CoMSiA 3D QSAR and homology modeling approaches. Bioorg. Med. Chem. 2003;13:1829–1835. doi: 10.1016/s0960-894x(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien SE, de Groot MJ. Greater than the sum of its parts: combining models for useful ADMET prediction. J. Med. Chem. 2005;48:1287–1291. doi: 10.1021/jm049254b. [DOI] [PubMed] [Google Scholar]
- 26.Kesuru GM. Prediction of hERG potassium channel affinity by traditional and hologram QSAR methods. Bioorg. Med. Chem. Lett. 2003;13:2773–2775. doi: 10.1016/s0960-894x(03)00492-x. [DOI] [PubMed] [Google Scholar]
- 27.Ekins S, Crumb WJ, Sarazan RD, Wikel JH, Wrighton SA. Three dimensional quantitative structure activity relationship for the inhibition of the hERG (human ether-a-gogo related gene) potassium channel. J. Pharmacol. Exp. Ther. 2002;301:427–434. doi: 10.1124/jpet.301.2.427. [DOI] [PubMed] [Google Scholar]
- 28.Ekins S. Predicting undesirable drug interactions with promiscuous proteins in silico. Drug Discovery Today. 2004;9:276–285. doi: 10.1016/S1359-6446(03)03008-3. [DOI] [PubMed] [Google Scholar]
- 29.Ekins S. In silico approaches to predicting metabolism, toxicology and beyond. Biochem. Soc. Trans. 2003;31:611–614. doi: 10.1042/bst0310611. [DOI] [PubMed] [Google Scholar]
- 30.Cianchetta G, Li Y, Kang J, Rampe D, Fravolini A, Cruciani G, Vaz RJ. Predictive models for hERG potassium channel blockers. Bioorg. Med. Chem. Lett. 2005;15:3637–3642. doi: 10.1016/j.bmcl.2005.03.062. [DOI] [PubMed] [Google Scholar]
- 31.Cavalli A, Poluzzi E, De Ponti F, Recanatini M. Toward a pharmacophore for drugs inducing the long QT syndrome: Insights from a CoMFA study of HERG K+ channel blockers. J. Med. Chem. 2002;45:3844–3853. doi: 10.1021/jm0208875. [DOI] [PubMed] [Google Scholar]
- 32.Bains W, Basman A, White C. HERG binding specificity and binding site structure: evidence from a fragment-based evolutionary computing SAR study. Prog. Biophys. Mol. Biol. 2004;86:205–233. doi: 10.1016/j.pbiomolbio.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Aronov AM, Goldman BB. A model for identifying HERG K+ channel blockers. Bioorg. Med. Chem. 2004;12:2307–2315. doi: 10.1016/j.bmc.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Aptula AO, Cronin MT. Prediction of hERG K+ blocking potency: Application of structural knowledge. SAR QSAR Environ. Res. 2004;15:399–411. doi: 10.1080/10629360412331297353. [DOI] [PubMed] [Google Scholar]
- 35.Aronov AM, Balakin KV, Kiselyov A, Varma-O'Brien S, Ekins S. Applications of QSAR methods to ion channels. In: Ekins S, editor. Computational Toxicology: Risk Assessment for Pharmaceutical and Environmental Chemicals. John Wiley and Sons; Hoboken, NJ: 2007. pp. 353–389. [Google Scholar]
- 36.Osterberg F, Aqvist J. Exploring blocker binding to a homology model of the open hERG K+ channel using docking and molecular dynamics methods. FEBS Lett. 2005;579:2939–2944. doi: 10.1016/j.febslet.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 37.Rajamani R, Tounge BA, Li J, Reynolds CH. A two-state homology model of the hERG K+ channel: Application to ligand binding. Bioorg. Med. Chem. Lett. 2005;15:1737–1741. doi: 10.1016/j.bmcl.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez D, Ghanta A, Kauffman GW, Sanguinetti MC. Physicochemical features of the HERG channel drug binding site. J. Biol. Chem. 2004;279:10120–10127. doi: 10.1074/jbc.M310683200. [DOI] [PubMed] [Google Scholar]
- 39.Sanguinetti MC, Mitcheson JS. Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trends Pharmacol. Sci. 2005;26:119–124. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Ekins S, Balakin KV, Savchuk N, Ivanenkov Y. Insights for human ether-a-go-go-related gene potassium channel inhibition using recursive partitioning, Kohonen and Sammon mapping techniques. J. Med. Chem. 2006;49:5059–5071. doi: 10.1021/jm060076r. [DOI] [PubMed] [Google Scholar]
- 41.Zauhar RJ, Moyna G, Tian L, Li Z, Welsh WJ. Shape signatures: a new approach to computer-aided ligand- and receptor-based drug design. J. Med. Chem. 2003;46:5674–5690. doi: 10.1021/jm030242k. [DOI] [PubMed] [Google Scholar]
- 42.Nagarajan K, Zauhar R, Welsh WJ. Enrichment of ligands for the serotonin receptor using the Shape Signatures approach. J. Chem. Inf. Model. 2005;45:49–57. doi: 10.1021/ci049746x. [DOI] [PubMed] [Google Scholar]
- 43.Wang CY, Ai N, Arora S, Erenrich E, Nagarajan K, Zauhar R, Young D, Welsh WJ. Identification of previously unrecognized antiestrogenic chemicals using a novel virtual screening approach. Chem. Res. Toxicol. 2006;19:1595–1601. doi: 10.1021/tx060218k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kortagere S, Welsh WJ. Development and application of hybrid structure based method for efficient screening of ligands binding to G-protein coupled receptors. J. Comput.-Aided Mol. Des. 2006;20:789–802. doi: 10.1007/s10822-006-9077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meek PJ, Liu Z, Tian L, Wang CY, Welsh WJ, Zauhar RJ. Shape Signatures: Speeding up computer aided drug discovery. Drug Discovery Today. 2006;11:895–904. doi: 10.1016/j.drudis.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Roth BL, Kroeze WK, Patel S, Lopez E. The multiplicity of Serotonin receptors: Uselessly diverse molecules or an embarrassment of riches? Neuroscientist. 2000;6:252–262. [Google Scholar]
- 47.Lavine BK, Davidson CE, Breneman C, Katt W. Electronic van der Waals surface property descriptors and genetic algorithms for developing structure-activity correlations in olfactory databases. J. Chem. Inf. Comput. Sci. 2003;43:1890–1905. doi: 10.1021/ci030016j. [DOI] [PubMed] [Google Scholar]
- 48.Breneman C, Sundling C, Sukumar N, Shen LQ, Katt WP, Embrechts M. New developments in PEST shape/property hybrid descriptors. J. Comput.-Aided Mol. Des. 2003;17:231–240. doi: 10.1023/a:1025334310107. [DOI] [PubMed] [Google Scholar]
- 49.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 50.Zauhar RJ. SMART: A solvent-accessible triangulated surface generator for molecular graphics and boundary element applications. J. Comput.-Aided Mol. Des. 1995;9:149–159. doi: 10.1007/BF00124405. [DOI] [PubMed] [Google Scholar]
- 51.Kohonen T. Self-Organizing Maps. 3rd ed. Springer Verlag; New York: 2000. [Google Scholar]
- 52.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 53.McGovern SL, Shoichet BK. Kinase inhibitors: not just for kinases anymore. J. Med. Chem. 2003;46:1478–1483. doi: 10.1021/jm020427b. [DOI] [PubMed] [Google Scholar]
- 54.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 55.Welsh WJ, Kholodovych V, Chekmarev D, Georgopoulos P. International Science Forum on Computational Toxicology. EPA, Research Triangle Park; North Carolina: 2007. Shape Signatures: A powerful new technology for risk assessment. [Google Scholar]
- 56.Fielding AH. Cluster and Classification Techniques for the Biosciences. Cambridge University Press; New York: 2007. [Google Scholar]
- 57.Plewczynski D, Spieser SA, Koch U. Assessing different classification methods for virtual screening. J. Chem. Inf. Model. 2006;46:1098–1106. doi: 10.1021/ci050519k. [DOI] [PubMed] [Google Scholar]
- 58.Ung CY, Li H, Yap CW, Chen YZ. In silico prediction of pregnane X receptor activators by machine learning approaches. Mol. Pharmacol. 2007;71:158–168. doi: 10.1124/mol.106.027623. [DOI] [PubMed] [Google Scholar]
- 59.Shen M, Xiao Y, Golbraikh A, Gombar VK, Tropsha A. Development and validation of k-nearest neighbour QSPR models of metabolic stability of drug candidates. J. Med. Chem. 2003;46:3013–3020. doi: 10.1021/jm020491t. [DOI] [PubMed] [Google Scholar]
- 60.Cortes C, Vapnik V. Support vector networks. Machine Learn. 1995;20:273–293. [Google Scholar]
- 61.Vapnik V. Statistical Learning Theory. Wiley; New York: 1998. [Google Scholar]
- 62.Chen ZY, Yap CW, Li H. Current QSAR techniques for toxicology. In: Ekins S, editor. Computational Toxicology: Risk Assessment for Pharmaceutical and Environmental Chemicals. John Wiley and Sons; Hoboken, NJ: 2007. pp. 217–238. [Google Scholar]
- 63.Xue Y, Yap CW, Sun LZ, Cao ZW, Wang JF, Chen YZ. Prediction of P-glycoprotein substrates by a support vector machine approach. J. Chem. Inf. Comput. Sci. 2004;44:1497–1505. doi: 10.1021/ci049971e. [DOI] [PubMed] [Google Scholar]
- 64.Chang CC, Lin CJ. LIBSVM: A library for support vector machines. 2001.
- 65.Whitley DC, Ford MG, Livingstone DJ. Unsupervised forward selection: A method for eliminating redundant variables. J. Chem. Inf. Comput. Sci. 2000;40:1160–1168. doi: 10.1021/ci000384c. [DOI] [PubMed] [Google Scholar]
- 66.Balakin KV, Savchuk NP, Kiselyov A. Computer algorithms for selecting molecule libraries for synthesis. In: Ekins S, editor. Computer Applications in Pharmaceutical Research and Development. John Wiley and Sons; Hoboken, NJ: 2006. pp. 445–468. [Google Scholar]
- 67.Balakin KV, Ekins S, Bugrim A, Ivanenkov YA, Korolev D, Nikolsky Y, Skorenko SA, Ivashchenko AA, Savchuk NP, Nikolskaya T. Kohonen maps for prediction of binding to human cytochrome P450 3A4. Drug Metab. Dispos. 2004;32:1183–1189. doi: 10.1124/dmd.104.000356. [DOI] [PubMed] [Google Scholar]
- 68.Korolev D, Balakin KV, Nikolsky Y, Kirillov E, Ivanenkov YA, Savchuk NP, Ivashchenko AA, Nikolskaya T. Modeling of human cytochrome P450-mediated drug metabolism using unsupervised machine learning approach. J. Med. Chem. 2003;46:3631–3643. doi: 10.1021/jm030102a. [DOI] [PubMed] [Google Scholar]
- 69.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 70.Ewing TJA, Kuntz ID. Critical evaluation of search algorithms for automated molecular docking and database screening. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- 71.Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 72.Cramer RD, Patterson DE, Bunce JD. Comparative Molecular Field Analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 73.Singh P, Kumar R. Quantitative structure-activity relationship study on tetrahydro-beta-carboline antagonists of the serotonin 2B (5HT2B) contractile receptor in the rat stomach fundus. J. Enzyme Inhib. 2001;16:491–497. doi: 10.1080/14756360127570. [DOI] [PubMed] [Google Scholar]
- 74.Brea J, Rodrigo J, Carrieri A, Sanz F, Cadavid MI, Enguix MJ, Villazon M, Mengod G, Caro Y, Masaguer CF, Ravina E, Centeno NB, Carotti A, Loza MI. New serotonin 5-HT(2A), 5-HT(2B), and 5-HT(2C) receptor antagonists: Synthesis, pharmacology, 3D-QSAR, and molecular modeling of (aminoalkyl)benzo and heterocycloalkanones. J. Med. Chem. 2002;45:54–71. doi: 10.1021/jm011014y. [DOI] [PubMed] [Google Scholar]
- 75.Manivet P, Schneider B, Smith JC, Choi DS, Maroteaux L, Kellermann O, Launay JM. The serotonin binding site of human and murine 5-HT2B receptors: molecular modeling and site-directed mutagenesis. J. Biol. Chem. 2002;277:17170–17178. doi: 10.1074/jbc.M200195200. [DOI] [PubMed] [Google Scholar]
- 76.Setola V, Dukat M, Glennon RA, Roth BL. Molecular determinants for the interaction of the valvulopathic anorexigen norfenfluramine with the 5-HT2B receptor. Mol. Pharmacol. 2005;68:20–33. doi: 10.1124/mol.104.009266. [DOI] [PubMed] [Google Scholar]
- 77.Hamprecht D, Micheli F, Tedesco G, Donati D, Petrone M, Terreni S, Wood M. 5-HT2C antagonists based on fused heterotricyclic templates: design, synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 2007;17:424–427. doi: 10.1016/j.bmcl.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 78.Todeschini R, Consonni V. Handbook of Molecular Descriptors. Vol. 11. Wiley-VCH; Weinheim: 2000. [Google Scholar]
- 79.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab. Dispos. 2006;34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 80.Ekins S, Johnston JS, Bahadduri P, D'Souzza VM, Ray A, Chang C, Swaan PW. In vitro and pharmacophore based discovery of novel hPEPT1 inhibitors. Pharm. Res. 2005;22:512–517. doi: 10.1007/s11095-005-2505-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.