Abstract
The complete ectodomain of integrin αIIbβ3 reveals a bent, closed, low-affinity conformation, the β-knee, and a mechanism for linking cytoskeleton attachment to high affinity for ligand. Ca and Mg ions in the recognition site, including the synergistic metal ion binding site (SyMBS), are loaded prior to ligand binding. Electrophilicity of the ligand-binding Mg ion is increased in the open conformation. The β3 knee passes between the β3-PSI and αIIb-knob to bury the lower β-leg in a cleft, from which it is released for extension. Different integrin molecules in crystals and EM reveal breathing that appears on pathway to extension. Tensile force applied to the extended ligand-receptor complex stabilizes the closed, low-affinity conformation. By contrast, an additional lateral force applied to the β subunit to mimic attachment to moving actin filaments stabilizes the open, high-affinity conformation. This mechanism propagates allostery over long distances and couples cytoskeleton attachment of integrins to their high affinity state.
Introduction
Integrins are cell adhesion receptors that transmit bidirectional signals across the plasma membrane and link the extracellular environment to the actin cytoskeleton. The conformation of the integrin extracellular domain and its affinity for ligand are dynamically regulated by a process termed inside-out signaling. By coupling to the actin cytoskeleton, integrins promote firm adhesion and provide traction for lamellipodium protrusion and locomotion. In migrating cells the adhesiveness of integrins is spatially and temporally regulated so that integrins are activated near the leading edge to support lamellipod extension and deactivated near the trailing edge to facilitate uropod retraction and internalization (Alon and Dustin, 2007; Arnaout et al., 2005; Broussard et al., 2008; Evans and Calderwood, 2007; Luo et al., 2007).
Integrin αIIb β3, the most abundant receptor on platelets, binds to fibrinogen and von Willebrand factor, and mediates platelet aggregation and association with injured vessel walls. Inherited mutations in the αIIb or β3 subunits result in the bleeding disorder Glanzmann’s thrombasthenia. Antagonists to αIIb β3 are prescribed for the prevention of thrombosis (see (Springer et al., 2008; Xiao et al., 2004) and references therein).
The integrin α and β subunits have large N-terminal extracellular domains, single-pass transmembrane domains, and usually short C-terminal cytoplasmic domains. The entire ectodomain of αV β3 crystallized in a bent conformation revealed 10 of 12 domains (Xiong et al., 2001; Xiong et al., 2004; Xiong et al., 2002). A ligand-binding head formed by both subunits is followed in each subunit by legs that connect to transmembrane domains. The knees between the upper and lower legs are extremely bent. Integrin epidermal growth factor-like (I-EGF) domains 1 and 2 at the β-knee were disordered in the previous αV β3 structure. Crystals of β2 leg fragments containing I-EGF domains 1 and 2 have been solved in two different orientations (Shi et al., 2007), but the conformation of these domains in the bent integrin conformation remains unknown.
Subsequent to the αV β3 crystal structure, mutational studies on cell surface integrins and EM studies on αV β3, αL β2, and αX β2 integrins demonstrated that the bent conformation is the physiologically relevant, low affinity integrin conformation (Nishida et al., 2006; Takagi et al., 2002). Nonetheless, a cryo EM study on αIIb β3 revealed a different, less compact conformation with a different arrangement of leg domains (Adair and Yeager, 2002). Furthermore, two recent studies have revealed extended conformations of αIIb β3 but failed to find a bent conformation (Rocco et al., 2008; Ye et al., 2008). Crystal structure studies on αIIbβ3 are important to resolve these controversies. Revealing the structure within a complete ectodomain of the bent β-knee is important for understanding the mechanism of integrin extension. Moreover, no bent integrin crystal structure to date has been described in light of current knowledge that this conformation occurs on cell surfaces, and corresponds to the low-affinity state.
Most studies find that upon activation, integrins extend (Luo et al., 2007). Upon extension, the headpiece can remain in the closed conformation, as when bent, or transition to an open conformation with high affinity for ligand, as shown in crystals of the αIIb β3 headpiece bound to ligands (Xiao et al., 2004). In contrast, a “deadbolt model” posits that activation can occur in the absence of extension (Arnaout et al., 2005). Binding of cytoskeletal proteins such as talin and kindlins to the integrin β cytoplasmic domain appears to interfere with α/β cytoplasmic domain association, and induce integrin extension (Wegener and Campbell, 2008). However, there is currently no known feature of integrin structure that would enable cytoskeleton binding to couple to the extended, open conformation with high affinity for ligand. This would appear to be important to fulfill the key role of integrins in integrating the extracellular and intracellular environments.
Three closely linked metal ion binding sites in the β I domain are especially important for ligand binding. Mg2+ at the central, metal ion-dependent adhesion site (MIDAS) directly coordinates the acidic sidechain shared by all integrin ligands. In previous unliganded, bent αV β3 structures, the MIDAS and one adjacent site were unoccupied, and it was proposed that metal binding was either caused by integrin activation or induced by ligand binding (Xiong et al., 2002); however, crystals have not been reported with a combination of the two metal ions important for integrin ligand binding, Mg2+ and Ca2+. Therefore, in current comparisons between low and high affinity β I domain conformations, the changes associated with ligand binding and metal binding cannot be deconvoluted.
Here, we describe the structure of platelet integrin αIIb β3 in the bent conformation. Crystals with Ca2+ and Mg2+ show that physiologically in the low affinity state the metal binding sites in the β I domain are fully occupied. Furthermore, the conformation is revealed of I-EGF domains 1 and 2 at the β-knee, at the epicenter of conformational change. The arrangement of the legs within the bent structure and variation among structures in interdomain orientation have profound implications for the mechanism of integrin activation. Use of this information in models of extended integrins experiencing forces at sites of cell adhesion reveals how integrin affinity is regulated by force exerted parallel to the membrane by a motile actin cytoskeleton. Integrin structure and mechanochemistry provide a natural mechanism for increasing integrin affinity upon cytoskeleton attachment and decreasing it upon cytoskeleton disassembly.
Results and Discussion
αIIb β3 crystal structure and negative stain EM
A 2.55Å resolution crystal structure of the complete αIIb β3 ectodomain with Ca2+ and Mg2+ has been refined to an Rfree of 26.8% (Fig. 1A, Table 1). In comparisons to αV β3 below, differences in resolution and refinement should be kept in mind. The 3.1Å αV β3 structure is refined to an Rfree of 36.7% (Xiong et al., 2004). αIIb β3 has 95% and 0.4% residues in favored and outlier Ramachandran regions, respectively, and geometry in the 98th percentile (where 100 is the best); whereas αV β3 has 76% and 6.7% residues in favored and outlier regions, respectively, and geometry in the 21st percentile; as reported by MOLPROBITY (Davis et al., 2007). Waters, which are important in hydrogen bonding and metal coordination, are in the αIIb β3 but not the αV β3 structure, as appropriate for their respective resolutions. No cis-prolines are present in the αV β3 structure, whereas 6 are present in αIIb β3. Two cis-prolines, Pro-163 and Pro-169, are in the ligand-binding β3 I domain. The region around cis-Pro-169 has electron density typical for the αIIb β3 structure (Fig. S1). There is a shift in the sequence-to-structure register between αV β3 and αIIb β3 at β3 167–176, in the specificity-determining loop that forms the outer rim of the ligand-binding pocket in the β3 I domain. Thus, the αIIb β3 structure provides details about backbone conformation, hydrogen bonding, and sidechain packing that are important for understanding ligand and metal binding; and for accurate molecular dynamics simulations and structure-guided mutagenesis. Furthermore, for the first time, the structure factors for an integrin ectodomain have been deposited, opening access to the experimental electron density upon which the atomic models are based.
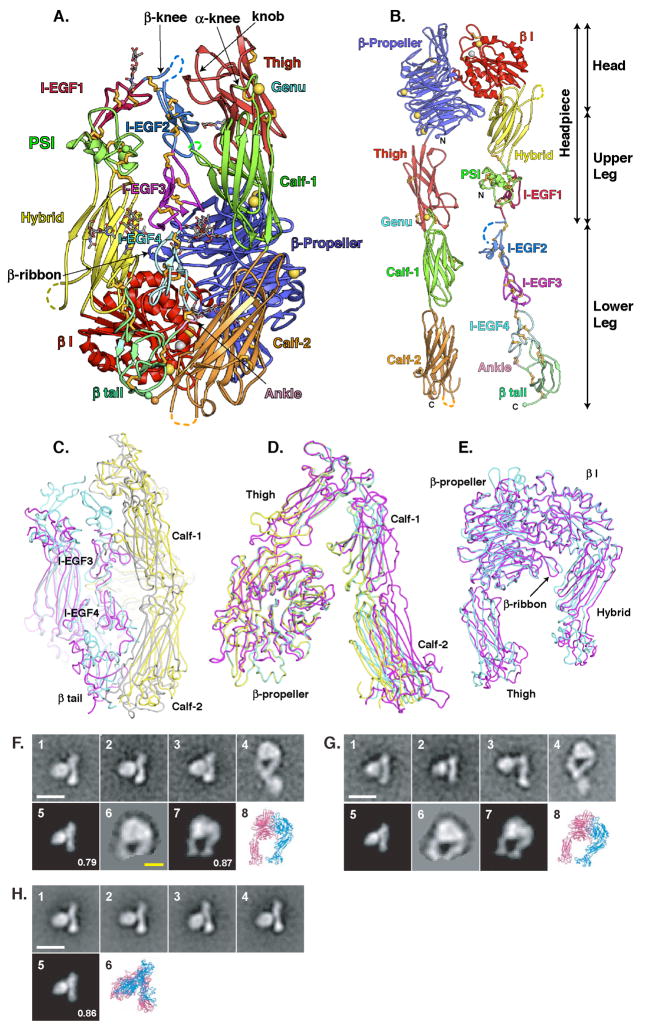
Figure 1.
The αIIb β3 crystal structure. A. Cartoon diagram of molecule 1 in αIIb β3 crystals. Ca and Mg ions are shown as gold and silver spheres, respectively. Disulfides are shown as gold sticks and glycans are displayed as thinner sticks with grey carbons. C and N-termini are shown as small spheres. Loops with missing density are shown as dashes. B. A model of αIIb β3 extended by torsion at the α and β-knees. C-E. Superpositions of molecules 1 and 2 of αIIb β3 and αV β3 (Xiong et al., 2004) showing breathing. C. A view showing variation in the distance of the lower α-leg from the lower β-leg, opening its cleft, and variation in the lower β-leg: αIIb β3 molecule 1 (αIIb grey, β3 cyan) and αV β3 (αV yellow, β3 magenta). D. A view of the α-subunit only, rotated about 90° from the view in C, showing variation in the distance of the lower α-leg from the upper α-headpiece: αIIb β3 molecule 1 (yellow) and molecule 2 (cyan); αV β3 (magenta). E. The headpieces of αIIb β3 molecule 1 (cyan) and αVβ3 (magenta), showing breathing at the β I/hybrid domain interface. All figures are made with PYMOL. F-H. Negatively stained αIIb β3 EM projection averages. F. αIIb β3 with a C-terminal coiled-coil clasp. G. αIIb β3 with the clasp removed. H. αIIb β3 disulfide-bonded near the C-termini of the β-tail and calf-2 domains. Panels 1–4 show representative class averages. Panel 5 shows the 20 Å resolution-filtered αIIb β3 crystal structure projection that best cross-correlates with panel 1. Panel 6 in F and G shows the masked headpiece region from panel 4, and panel 7 shows the corresponding best-correlated αIIb β3 headpiece crystal structure projection. Ribbon diagrams in panels 6 and 8 are in the same orientation (although enlarged) as the projections to their left. Numbers in panels 5 and 7 are normalized cross-correlation coefficients. White and yellow scale bars are 100 and 50 Å, respectively.
Table 1.
X-ray diffraction data and refinement
| Protein | αIIb β3 ectodomain | αIIb β3 headpiece |
|---|---|---|
| Spacegroup | P41 | P62 |
| Unit cell (a, b, c) (Å) | 81.3, 81.3, 654.6 | 332.1, 332.1, 88.3 |
| (α, β, γ) (°) | 90, 90, 90 | 90, 90, 120 |
| Wavelength (Å) | 0.97934 | 0.9760 |
| Resolution (Å) | 50-2.55 | 45-2.90 |
| Number of reflections (total/unique) | 614,293/135,066 | 1,251,268/122,126 |
| Completeness (%) | 98.6/93.9* | 98.3/93.9* |
| I/σ(I) | 12.2/2.1* | 17.4/3.0* |
| Rmerge (%)¶ | 7.1/56.6* | 9.7/60.2* |
| Rwork¶¶/Rfree‡‡ | 0.233/0.268 | 0.174/0.196 |
| RMSD: Bond (Å) | 0.003 | 0.006 |
| Angle (°) | 0.736 | 0.659 |
| Ramachandran plot** | 95.0%/4.6%/0.4% | 96.9%/2.9%/0.2% |
| PDB code | (prev. 1TYE) |
Asterisked numbers correspond to the last resolution shell.
Rmerge = Σh Σi |Ii(h) −<I(h)> |/Σh Σi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
Rwork = Σh||Fobs (h)|−|Fcalc (h)||/Σh|Fobs (h)|, where Fobs (h) and F calc (h) are the observed and calculated structure factors, respectively. No I/cutoff was applied.
Rfree is the R value obtained for a test set of reflection consisting of a randomly selected 1.3% subset of the data set excluded from refinement.
Residues in favorable, allowed, and outlier regions of the Ramachandran plot as reported by MOLPROBITY (Davis et al., 2007).
Overall bent structure
The overall arrangement of domains in the two independent αIIb β3 molecules in the asymmetric unit is similar to that in αV β3 crystals (Fig. 1C), except for differences in angles between domains (Table 2) that give insights into breathing. A similar bent conformation in solution with physiologic divalent cations is seen for three distinct αIIb β3 constructs in negative stain EM with class averaging (Fig. 1F–H and S2). The bent integrins from the three types of constructs are indistinguishable from one another (Fig. 1F panels 1–3, G panels 1–2, H panels 1–4) and show excellent cross-correlation with the αIIb β3 crystal structure (Fig. 1F panels 1 and 5, G panels 1 and 5, and H, panels 1, 5 and 6). One construct was clasped by appending to the α and β ectodomain C-termini 15-residue linkers containing TEV protease sites, followed by an α-helical coiled-coil (Nishida et al., 2006). Association near the C-termini of the α and β subunit ectodomains provided in vivo by association between the αIIb and β3 transmembrane domains (Luo et al., 2004) is mimicked by the clasp (Takagi et al., 2002). The clasped αIIb β3 particles were 64% bent and 32% extended (with 4% unclassified) (Fig. 1F). Unclasped particles, in which the clasp was removed with TEV protease, were 44% bent and 52% extended (Fig. 1G). A third construct, which was identical to that used in crystallization, contained cysteines introduced in C-terminal portions of the αIIb and β3 subunits in positions that resulted in efficient disulfide bond formation in cell surface integrins (Fig. S3). The disulfide-bonded construct was 100% bent (Fig. 1H).
Table 2.
Variation in inter-domain angles in integrinsa.
| Domain interface | bent αIIb β3b | bent αIIb β3c | bent αIIb β3d | open αIIb β3e | bent β3f | frag β2g |
|---|---|---|---|---|---|---|
| bent αIIb β3 | bent αV β3 | open αIIb β3 | open αIIb β3 | frag β2 | frag β2 | |
| α β-propeller -α thigh | 0.5° | 9.7–9.9° | - | - | - | - |
| α thigh - α calf1 | 1.3° | 19–20° | - | - | - | - |
| α calf1 - α calf2 | 3.4° | 14–17° | - | - | - | - |
| β I -β hybrid | 0.2° | 6.7–6.7° | 58–70° | 1.2–12° | - | - |
| β hybrid - β PSI | 0.4° | 7.2–7.8° | 2.0–11° | 1.8–9.2° | 18–27° | 5.0–8.3° |
| β PSI - β I-EGF1 | 0.8° | - | 3.4–13° | 5.7–18° | 5.5–40° | 5.5–41° |
| β hybrid - β I-EGF1 | 0.5° | - | 6.8–23° | 11–26° | 29–51° | 3.7–46° |
| β I-EGF1 - β I-EGF2 | 0.4° | - | - | - | 140–170° | 67° |
| β I-EGF2 - β I-EGF3 | 1.2° | - | - | - | 8.4–8.5° | - |
| β I-EGF3 - β I-EGF4 | 2.3° | 5.4–7.2° | - | - | - | - |
| β I-EGF4 - β ankle | 1.2° | 10–11° | - | - | - | - |
| β I-ankle - β TD | 46°h | 18°I | - | - | - | - |
| α β-propeller - β I | 0.2° | 3.0–3.3° | 1.7–2.8° | 0.6–1.1° | - | - |
Each pair of domains from two molecules were superposed using the first domain, and the change in angle upon superimposing the second domain was calculated. Dashes indicate where no comparison is possible, because only one or no domain pairs are available.
Two molecules in current structure (1 × 1).
Two molecules in current structure versus PDB code IU8C (2 × 1).
Two molecules in current structure versus PDB 2VDR and three molecules in PDB ITYE (2 × 4).
Comparisons among 2VDR and three molecules in ITYE (3 × 4/2).
Two molecules in current structure, and PDB 1U8C versus PDB 1YUK, PDB 2P26, and PDB 2P28 (3 × 3 to 3 × 1, depending on fragment length).
comparisons among PDB 1YUK, 2P26, and 2P28 (3 to 1 comparisons depending on fragment length).
Residues common to molecules 1 and 2 in TD are used, 606–612
αIIb β3 molecule 1 compared to αV β3
The differing proportion of bent particles in the three preparations shows that tighter association near the C-termini correlates with maintenance of the bent conformation, and also, with resistance to activation on the cell surface (Fig. S3 and Suppl. Mat). This is in agreement with work on other soluble integrin preparations, and a large body of work on cell surface integrins, which has shown that association of the α and β subunit transmembrane and cytoplasmic domains stabilizes integrins in the low-affinity state and in the bent conformation (reviewed in (Luo et al., 2007)).
Significance of the bent conformation
Similar bent conformations have previously been described in EM studies of the resting states of αV β3, αX β2, and αL β2 (Nishida et al., 2006; Takagi et al., 2002). Furthermore, extensive studies using mutations and antibodies to ligand-induced binding sites show that αIIb β3 is compact on the cell surface when resting, and extended when activated (Honda et al., 1995; Luo et al., 2007). The similarity in packing of two independent examples of αIIb β3 and of αVβ3 in crystal lattices and similar appearance of multiple soluble integrins in EM, together with the work cited above, strongly suggest that the bent crystal structure determined here is representative of the resting state of most, if not all, integrins. However, three cryo EM, EM, and hydrodynamic studies of detergent soluble αIIbβ3 from platelets have reached conclusions that are incompatible with one another, and with the domain arrangement seen here (Adair and Yeager, 2002; Rocco et al., 2008; Ye et al., 2008). The difficulty in obtaining a consensus view on αIIbβ3 structure may reflect the delicate equilibrium between bent and extended structures (Fig. 1G–H), averaging over ensembles of bent and extended conformations, the poor association of the αIIb and β3 transmembrane domains in detergent (Wegener and Campbell, 2008), and dissociation of the heterodimer into the αIIb and β3 subunits (Carrell et al., 1985).
Conceptual advances since the previously described αV β3 crystal structures allow us to describe the bent αIIb β3 crystal structure in light of its physiological relevance as the low affinity integrin state, and as the starting point for integrin extension. Furthermore, the αIIb β3 structure reveals I-EGF domains 1 and 2, and a highly acute bend between them in the bent conformation (Fig. 1A). In contrast, I-EGF domains 2, 3, and 4 extend in an almost straight orientation, with an approximate 90 left-handed twist between successive domains, to cover most of the length of the lower β-leg (Fig. 1A, 2A). The β-knee, at the junction between I-EGF1 and I-EGF2, is flanked on one side by the PSI domain and on the other by a knob-like projection in the thigh domain (Fig. 1A). The PSI and the knob are like goalpost uprights, which define the passage for I-EGF1 and I-EGF2. The importance of the knob is emphasized by its structural conservation between αIIb and αV, in contrast to the flexibility of loops at the opposite end of the thigh domain, adjoining the β-propeller domain (Fig. 2D).
Figure 2.
Integrin leg domains. A. The knee and lower β-leg of αIIb β3. Dashes mark gimbal flexion positions. B. Superposition using SSM (Krissinel and Henrick, 2004) of αIIb β3 I-EGF domains 1–4 in the same color scheme as in A. C. Superposition using I-EGF2 of I-EGF 1 and 2 module pairs. Domains from αIIb β3 are in red, and those from β2 fragments (Shi et al., 2007) are in cyan and grey. D-F. Superposition of αIIb (yellow) and αV (magenta) (Xiong et al., 2004) thigh (D), calf-1 (E), and calf-2 (F) domains.
The I-EGF domains of the lower β-leg are deeply buried in a narrow crevice, between the upper α-leg on one side and the upper and lower α-leg on the other, with the β I and β-propeller domains helping to form the back of the crevice (Fig. 1A). The flexible calf-1 DX loop extends into the cleft and partially shields the lower β-leg (Fig. 1A, 2E). Exit of the lower β-leg from the crevice appears to be the key step in integrin extension.
Overall extended structure
In the extended conformation, the α and β-legs straighten at the knees, and extend away from the headpiece (Fig. 1F and G, panel 4). The closed headpiece from the crystal structure cross-correlates excellently with the headpiece in EM (Fig. 1F and G, panels 6–8) showing that with Ca2+ and Mg2+, extended αIIb β3 predominantly assumes the closed headpiece, rather than the open headpiece conformation (Fig. 3A). Most extended class averages, whether with clasped or unclasped αIIb β3, show the α-leg crossing over or under the β-leg (Fig. 1F, G, panel 4). Leg crossing appears to be a consequence of upper leg configuration in the bent conformation with the long axis of I-EGF1 pointing toward the α-knee (Fig. 1A). When the bent crystal structure is extended at the α and β-knees, leg crossing results (Fig. S5A). However, the legs are highly flexible (see below), and for clarity are shown side-by-side in Fig. 1B. Extended integrins with crossed and uncrossed legs have also been seen for activated αV β3, αX β2, and detergent soluble αIIb β3 integrins (Iwasaki et al., 2005; Nishida et al., 2006; Takagi et al., 2002).
Figure 3.
Metal ion rearrangements in β I domain activation. A. Superposition of headpieces from our unliganded-closed structure and liganded-open αIIb β3 (Springer et al., 2008). The β I and hybrid domains are yellow (open) and magenta (closed) while PSI and I-EGF1 domains are red and green, respectively. The α-headpieces are cyan (open) and grey (closed). B. Enlarged view of β I domains with major differences in yellow (open) and magenta (closed). C and D. β I domain metal coordination sites in unliganded-closed αIIb β3 (C) and liganded-open αIIb β3 (D). Ca (gold) and Mg (green) ions are large spheres; waters (red or pink) are smaller spheres. N atoms are blue and O atoms are red or pink. Metal coordination and hydrogen bonds are dashed. The loop bearing M335 moves far away in (D). E. Superposition at the β I MIDAS. F. Superposition at the α I MIDAS of unliganded-closed (PDB code 1LFA) and liganded open αL (PDB code 1T0P), in the same orientation as the β I MIDAS in D. In C-F, carbons for unliganded-closed and liganded-open integrins and for ligands are wheat, grey, and cyan, respectively. G and H. Electrostatic potential surfaces at the unliganded (G) and liganded (H) binding sites.
After physiological activation of αIIb β3 on platelets or treatment with high concentrations of ligands, multiple ligand-induced binding site (LIBS) epitopes are exposed. These epitopes map to the lower β-leg, and to the PSI domain (Honda et al., 1995). The lower β-leg is buried in a cleft in the bent conformation (Fig. 1A), but will be exposed in the extended conformation (Fig. 1B and S5A). Similarly, the LIBS epitope in the PSI domain, mapped to residues 1–6 (Honda et al., 1995), is masked by I-EGF2 in the bent conformation (Fig. 1A). By contrast, this epitope is exposed after extension at the I-EGF1/I-EGF2 interface in the β-knee brings I-EGF2 away from the PSI domain (Fig. 1B). The previous functional studies, together with the location of these epitopes within the αIIb β3 structure, demonstrate that bent and extended αIIb β3 represent latent and activated integrins, respectively, contradict suggestions that αIIb β3 is extended in the resting state (Rocco et al., 2008; Ye et al., 2008), and agree with electron tomography of active, detergent soluble αIIbβ3 showing that it is extended (Iwasaki et al., 2005).
The ligand binding site is preloaded with metals
Integrins bind ligands at the interface between the α subunit β-propeller domain and β subunit I domain (Xiao et al., 2004; Xiong et al., 2002). These domains associate over an interface far larger than between other integrin domains (Table S1), to form the integrin head (Fig. 1B and 3A). Three metal binding sites formed by loops in the β I domain underpin the ligand binding site (Fig. 3A–C). Strong densities at all three sites reveal that they are occupied when physiologic divalent cations, Ca2+ and Mg2+, are present (Fig. 3C). Mg2+ at the central MIDAS site and Ca2+ at the two flanking sites are assigned by the coordination chemistry at these sites and the stronger electron densities at the two Ca2+ sites. In contrast, in previous αV β3 crystals in absence of ligand, only one divalent cation, either Mn2+ or Ca2+, was present, and neither the ligand-associated metal binding site (LIMBS) or MIDAS was occupied (Xiong et al., 2001; Xiong et al., 2002).
Since our results show that the LIMBS is not a ligand-associated or induced metal binding site, new nomenclature is required. We propose to rename this Ca2+-binding site the synergistic metal ion binding site (SyMBS). Low concentrations of Ca2+ synergize with low concentrations of Mg2+ for ligand binding to integrins (Marlin and Springer, 1987), and mutational studies show that the SyMBS has a positive regulatory effect on ligand binding, and that the SyMBS is the site responsible for Ca2+ synergy (Chen et al., 2003; Mould et al., 2003). The SyMBS designation also honors the finding with αVβ3 crystals that ligand binding at the MIDAS synergizes with Mn2+ binding at the SyMBS when Ca2+ is absent (Xiong et al., 2002).
Fig. 3C reveals the structural basis for synergy between the SyMBS and MIDAS. Although coordination at the MIDAS is octahedral, with six ligands, four of these are waters. An unusually low number of two Mg2+ oxygen ligands come from protein, donated by the sidechains of β3 Ser-121 and Glu-220. The sidechain of Glu-220 orients between the SyMBS and MIDAS, with one oxygen coordinating the SyMBS Ca2+ and the other coordinating the MIDAS Mg2+ (Fig. 3C). In the absence of either of these metals, the Glu-220 would likely reorient, and not form a proper coordination to the other site, explaining the basis for synergy.
It is conceptually attractive to find that the physiologically important metals Ca2+ and Mg2+ are pre-loaded prior to ligand binding. The Asp sidechain of integrin ligands such as Arg-Gly-Asp (RGD) directly coordinates the MIDAS Mg2+ (Fig. 3D). In the absence of Mg2+, approach of this Asp would be electrostatically repelled by Glu-220, Asp-119, and Asp-251 around the MIDAS. Furthermore, the SyMBS site is completely buried, and would be difficult to occupy after ligand binding.
The major differences between low and high affinity ligand binding site conformations are the movements of the β1- α1 loop and its bound Adjacent to MIDAS (AdMIDAS) Ca2+ toward the MIDAS Mg2+, as previously described (Xiao et al., 2004; Xiong et al., 2002) (Fig. 3C–E). In a similar movement of the β1- α1 loop in α I domains, the MIDAS metal ion moves 2 Å away from an Asp and towards a Thr in the high affinity state, and thus becomes more electrophilic for the acidic ligand residue (Arnaout et al., 2005; Luo et al., 2007) (Fig. 3F).
The current structure enables comparison for the first time of MIDAS metal ion and water positions between the low and high affinity states of β I domains. Our structure reveals that in contrast to α I domains, there is no lateral movement of the MIDAS metal ion across the ligand-binding pocket (Fig. 3E,F). In α I domains, the second Ser of the DXSXS motif (αL Ser-141 in Fig. 3F) moves to the position of the Mg2+ ion, and pushes the Mg2+ toward Thr-206 in the high affinity conformation (Fig. 3F). In β I domains, the Mg2+ is already in a ligand-binding position in the low affinity state, and the second Ser of the DXSXS motif, Ser-123, moves toward the Mg2+ from secondary coordination and displaces a water to occupy the primary coordination sphere (Fig. 3C–E).
What then is the basis for the increase in affinity of β I domains in the open conformation? In part, this must be due to the movement toward the MIDAS of the β1- α1 loop bearing the DXSXS motif, enabling two of its backbone N atoms to form stabilizing hydrogen bonds to the two O atoms of the ligand Asp sidechain (Fig. 3D). Additionally, movement in the open state of the AdMIDAS Ca2+ toward the MIDAS brings it into primary coordination with Asp-251, enabling Asp-251 to polarize toward the AdMIDAS Ca2+ (Fig. 3D) rather than toward the MIDAS Mg2+ (Fig. 3C). Furthermore, there is a significant increase of 1 Å in separation between the Mg2+ and Asp-119 in the open state (Fig. 3E and Supplement). The backbone, AdMIDAS, and Asp-119 movements all increase the positive potential in the environment of the MIDAS, and promote greater electrophilicity of the Mg2+ for ligand in the high-affinity, open state. The overall increase in positive potential near the MIDAS in the high affinity state is apparent in electrostatic potential surfaces (Fig. 2G, H).
The flexible β-knee and β-leg
The structure shows that all 56 cysteines in the integrin β3 subunit are disulfide-bonded and the disulfides exhibit no rearrangements with respect to integrin fragment structures (Beglova et al., 2002; Shi et al., 2007). As shown below, the disulfides are compatible with large inter-domain rearrangements, and there is no need for disulfide reduction for integrin activation as previously suggested (Yan and Smith, 2001).
Integrin EGF domains
Eight cysteines are disulfide bonded in a C1-C5, C2-C4, C3-C6, and C7-C8 pattern in the small, 37–50 residue I-EGF domains (Fig. 2B). The exception is I-EGF1, which lacks the C2-C4 disulfide. This enables the C1-C3 loop to occupy a position different than in other I-EGF domains (Fig. 2B) and prevents clashes and enables greater flexibility at interfaces with the PSI and I-EGF2 domains.
The connection between tandem I-EGF domains is gimbal-like, since two flexion points are revealed at the highly acute bend between I-EGF1 and I-EGF2 at the β-knee (dashed lines, Fig. 2A). Between tandem I-EGF domains, and also at the PSI/hybrid and hybrid/I-EGF1 junctions, only one residue intervenes between the last Cys of one domain and the first Cys of the next, limiting flexion at this C-X-C junction (dashed line i., Fig. 2A). However, the disulfide unique to I-EGF domains between C1 at the N-terminus, and C5 in the β-hairpin turn between the two β-strands (Fig. 2B,C), is surprisingly flexible. Most of the movement at the β-knee occurs at a second point of flexion within the tip of I-EGF2, in the C1-C5 disulfide and in the disordered loop connecting C1 and C2 (dashed line ii., Fig. 2A). This is evident from superpositions of β3 I-EGF domains 1–4 (Fig. 2B), and β2 leg fragments (Fig. 2C). The position of C1 is highly variable in these superpositions, demonstrating flexiblity of the N-terminal tip of the I-EGF domain, particularly in I-EGF2. Although previous examples of I-EGF1/I-EGF2 interfaces in β2 leg fragments had been termed bent and extended (Shi et al., 2007), the two fragments differ in angle by 140 to 170 from I-EGF1/I-EGF2 in αIIb β3, and both are extended compared to the highly bent conformation revealed here (Table 2 and Fig. 2C). Thus the gimbal-like connection between I-EGF domains is permissive of extreme rotations, and a wide range of inter-domain orientations.
A corollary to the gimbal-like junction at I-EGF domains is that flexibility should be related to the length and disorder of the polypeptide chain between C1 and C2 (or between C1 and C3 in I-EGF1 which lacks the C2-C4 disulfide). As fitting for its role in extension of the β-leg, the C1-C2 loop of I-EGF2 is the only disordered I-EGF loop in our structure, and is located at the apex of the β-knee (Fig. 1A and 2A). Furthermore, alignment of integrin β subunits β1 to β8 in diverse vertebrates shows that I-EGF2 has the longest C1-C2 loop, with 9 to 13 residues. By contrast, the C1-C2 loop has 4 residues in I-EGF3, and 6 to 7 residues in I-EGF4. In I-EGF1 the 8–10 residue length of the C1-C3 loop is consistent with variations of up to 41° at the PSI/I-EGF-1 junction in comparisons to αIIb β3 headpiece and β2 leg fragments (Table 2, Fig. S7). Furthermore, disorder of I-EGF1 and I-EGF2 in the αVβ3 crystal structure (Xiong et al., 2001) implies flexibility at the PSI/I-EGF1 and I-EGF2/3 interfaces.
The β-ankle
A disulfide bonded loop between I-EGF4 and the β-tail domain, previously defined as part of I-EGF4, has no equivalent in other EGF domains, and is termed here the β-ankle. It is not integrated by backbone hydrogen bonds into I-EGF4 or the β-tail, and is likely to be flexible in extended integrins.
The β-tail
Flexibility between the β-ankle and β-tail is substantial, with up to 46° variation (Table 2). Flexibility within the N-terminal α-helix of the β-tail is suggested by loss of density in molecule 2 midway through this helix, and by the weak density of the β-tail domain in molecule 1. The better order of the β-tail in αV β3 appears due to an unusually large lattice contact of 860 Å2, which is larger than any domain-domain junction within the β-knee or lower β-leg (400–550 Å2, Table S1).
A contact between the CD loop of the β-tail domain (termed the “deadbolt”) and the 7-helix of the β I domain has been proposed to inhibit integrin activation (Arnaout et al., 2005); although the size of this interface at 60 Å2 is too small to be significant (Janin, 1997). There is no such contact in αIIb β3, either in molecule 1 where the orientation of the β-tail differs, or in molecule 2 where this part of the β-tail is disordered. Since the β I domain in both molecules is in the inactive, closed conformation, the CD loop does not act as a deadbolt to restrain integrin activation. In agreement, mutation or deletion of the CD loop has no effect on activation of cell surface αV β3 or αIIb β3 integrins (Zhu et al., 2007a).
Integrin breathing and extension
Breathing in the bent conformation
In molecule 2 of αIIb β3 compared to molecule 1, the lower α-leg swings outward at the genu, away from both the upper α- and β-legs, thus widening the crevice in which the lower β-leg is buried (Fig. 1D). The I-EGF4 and β-tail domains also swing with the α-leg, away from the upper β-leg. In αVβ3 crystals, the lower α-leg swings out further, and the I-EGF domains 3 and 4 in the lower β-leg move away from the hybrid domain in the upper β-leg (Fig. 1C, D). About 10% of αV β3 and αIIb β3 particles exhibit substantially more opening between the lower legs and headpiece than other particles (Fig. 1G, panel 3) (Takagi et al., 2002). This opening is similar in directionality, but greater in amplitude than in the crystal structure comparisons.
A second component of breathing motion is swinging of the hybrid domain relative to the β I domain. The hybrid domain is more swung out in αV β3 than in αIIb β3 (Fig. 1E) and has a smaller interface with the β I domain. Swinging opens the crevice in which the lower β-leg is buried, and has the same directionality as transit to the open headpiece.
A flexible β-ribbon extension of the β2- β3 loop of β-propeller blade 5 (Fig. 1A) also evidences breathing. It differs in position between αV β3 and αIIb β3 (Fig. 1E), and between αIIb β3 structures (Fig. 3A). An introduced disulfide bond between the β-ribbon and I-EGF4 stabilizes αVβ3 and αIIb β3 in the bent conformation and prevents integrin activation on cell surfaces (Takagi et al., 2002). The residues mutated to cysteine are close in αV β3 and not in αIIb β3, demonstrating that the β-ribbon can differ significantly in position in bent αIIb β3 (Supplement).
The main components of motion, hinging of the legs and swinging of the hybrid domain, provide a plausible pathway for integrin extension. Each of these components is confirmed to be a low frequency and hence important normal mode (not shown). Both types of motions will allow release of the lower β-leg from its crevice between the upper β-leg and lower α-leg (Fig. 1A).
Extension
After release of the lower β-leg, the highly flexible I-EGF1/I-EGF2 interface could transit from its bent to extended conformation, while at the same time, extension occurs at the α-genu, resulting in a switchblade-like opening of the integrin. Flexibility described above at the PSI/I-EGF1, I-EGF1/I-EGF2, and β-ankle/β-tail interfaces is also important to enable the lower α and β-legs to extend without clashing near the knees, where the long axis of I-EGF1 points toward the α-knee (Fig. 1A). Reorientation of the ligand-binding head so that it points away from, rather than toward the cell surface, and extends further above it, will greatly facilitate ligand binding (Fig. 1B). Furthermore, extension frees the hybrid domain from extensive interfaces in the bent conformation (Table S1), making hybrid domain swing-out to the open conformation less energetically costly. However, it is sterically possible for hybrid domain swing-out to occur simultaneously with, or precede, extension.
Integrin ectodomain structure provides a mechanism for force-induced integrin activation and deactivation
Binding of the actin cytoskeleton-associated proteins talin and kindlins to specific residues in the integrin β subunit cytoplasmic domain is crucial for inside-out activation of integrins and bidirectional signal transmission (Moser et al., 2008; Wegener and Campbell, 2008). Talin and kindlins bind through their FERM domains to distinct NPX(Y/F) motifs in integrin β subunit cytoplasmic domains that are membrane proximal and distal, respectively. Talin binding to the β subunit cytoplasmic domain has been proposed to sterically interfere with α and β subunit cytoplasmic domain association. One model is that talin binding would alter the depth in the membrane of the β subunit transmembrane domain or its angle with respect to the α subunit transmembrane domain. Another model is that talin binding would cause dissociation of the α and β subunit transmembrane domains, i.e. separation in the membrane (Luo et al., 2004; Wegener and Campbell, 2008). However, steric interference with α and β subunit association is more difficult to envision for kindlins, which bind to a membrane-distal motif that is disordered in α β cytoplasmic complexes (Wegener and Campbell, 2008).
Crosslinking the integrin α and β subunit transmembrane domains, and fluorescence resonance between probes attached to the cytoplasmic domains, have each demonstrated that both inside-out and outside-in signaling require separation of the α and β subunit transmembrane domains, and are associated with separation of the cytoplasmic domains (Luo et al., 2007). Transmembrane and cytoplasmic domain separation would lead to separation of the lower α and β-legs, which in turn would destabilize their interfaces with the head and upper legs, and trigger integrin extension. However, extended integrins can have either the closed or open headpiece with low or high affinity for ligand, respectively. This begs the question of whether the open, high affinity headpiece conformation could be enforced by signals within the cell. The complete integrin ectodomain structure described here now allows an analysis of how force is transmitted between the ligand binding site and the transmembrane domains to regulate transition between low and high affinity states.
There is great interest in the concept that force is important in regulating the adhesiveness and conformation of integrins (Alon and Dustin, 2007; Astrof et al., 2006; Evans and Calderwood, 2007; Puklin-Faucher et al., 2006). However, a key factor that has not previously been considered is the lateral force exerted by actin cytoskeleton treadmilling or contraction. Talin and kindlins link integrins through a molecular clutch to the actin cytoskeleton. As a consequence, integrins move laterally on the cell surface at speeds up to 130 nm/s, in the same direction as actin filaments (Hu et al., 2007; Kaizuka et al., 2007). Thus, integrin binding to talin and kindlins is essentially synonymous with attachment to the cytoskeleton, and exertion of a lateral force on the β-subunit cytoplasmic domain.
Let us envision at the molecular level the consequences of dragging an integrin by its β-tail across the cell surface. Bent integrins have a cross-section of 80 × 100 Å near the plasma membrane that is unusually large for a cell surface glycoprotein. The dense packing of extracellular domains on the cell surface is one of the major barriers to diffusion (Sheetz, 1993). Thus, as an integrin is dragged across the cell surface, it would be buffeted by collisions with other cell surface glycoproteins. The thin, lower β-leg is shielded in its cleft by the robust α-subunit and upper β-leg and head (Fig. 1A), which will bear the brunt of buffeting. Thus the integrin will be forced through a gauntlet of other cell surface proteins, that will strip the α subunit and upper β-leg away from the lower β-leg that they shield. Furthermore, the calf-2 domain has an unusually broad base for a cell surface domain, and two long unstructured loops near the plasma membrane, one of which is cleaved during biosynthesis (Fig. 2F). Frictional forces due to interactions of these segments and the α subunit transmembrane domain with the plasma membrane will also pull the α-leg away from the β-leg.
These forces are on pathway with the breathing movements described above, and will shift the equilibrium toward integrin extension. Although the interfaces buried in the bent conformation are extensive (Table S1), they have low shape complementarity, are mainly hydrophilic, and are readily replaced by water, as shown by the shift in equilibrium toward extension upon C-terminal clasp cleavage (Fig. 1F–H) (Nishida et al., 2006; Takagi et al., 2002). Because of the central role of the lower β-leg in these interfaces (Table S1, Fig. 1A), its removal from the cleft will destabilize the bent conformation far more than clasp removal, and result in extension. Furthermore, extension places the bulky head of the integrin above the height of most cell surface glycoproteins, reducing frictional drag and favoring maintenance of extension.
Once the integrin is extended, lateral pulling on the β-tail will align the integrin so that β orients toward and α away from the pulling direction. Orientation will be enhanced by the frictional resistance of the three leg domains in α, which, at 140–170 residues and with extra β-strands compared to Ig domains, are unusually stout (Supplement) and will act as a sea anchor. In contrast, β-leg domains are at the lower size limit found on cell surfaces.
Extended and not bent integrins are competent for binding large biological ligands such as fibronectin and fibrinogen (Zhu et al., 2007a). Once a ligand in the extracellular matrix or on the surface of another cell is bound, resistance to lateral pulling by the β-subunit cytoplasmic domain will increase greatly, with a corresponding increase in the lateral force. Because the β-subunit hybrid domain extends laterally away from the ligand binding site in the open headpiece conformation (Fig. 3A), the lateral force should stabilize the open headpiece conformation.
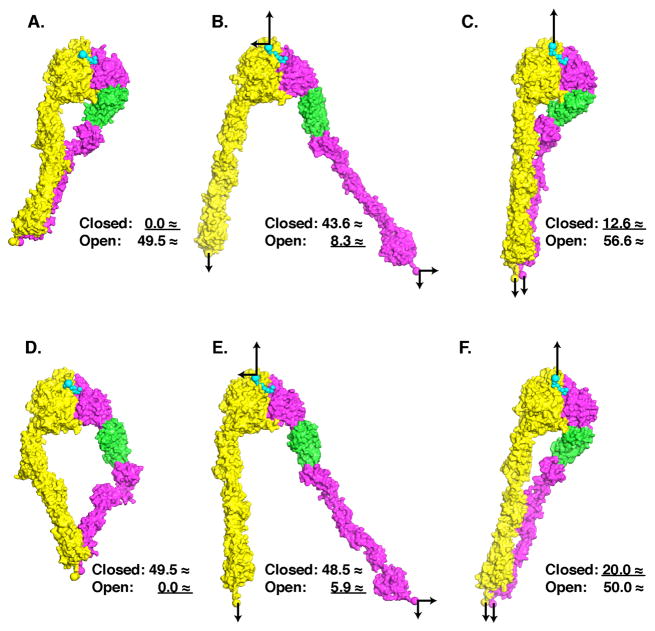
To test how forces regulate the conformation of the integrin headpiece, we used the complete ectodomain structure and the liganded, open headpiece structure (Springer et al., 2008; Xiao et al., 2004) to build extended integrin models with the headpiece closed or open and bound to the specific peptide recognition motif in fibrinogen (Fig. 4A and D). The C-termini of the α and β subunit ectodomains were tethered to a plane to mimic the plasma membrane. To mimic a cell pulling away from ligand in matrix or on the surface of another cell, a tensile (pulling) force normal to the membrane was applied to the α and β ectodomain C-termini, and an equal and opposite resisting force was applied to ligand (Fig. 4C and F). This mimics forces experienced by the integrin and ligand during cell adhesion in the absence of attachment of the integrin to the cytoskeleton. Alternatively, the same membrane normal forces were applied, and an additional lateral force parallel to the membrane was applied to the C-terminus of the β-subunit ectodomain that was resisted by an opposite force applied to ligand (Fig. 4B, E). This mimics forces experienced during cell adhesion in the presence of attachment of the integrin β subunit to talin or kindlins and the actin cytoskeleton.
Figure 4.
Regulation of integrin conformation by tensile force. Molecular surfaces show αIIb in yellow and β3 in magenta with hybrid domain in green. Fibrinogen peptide ligand is shown in cyan as Cα spheres. A and D are starting models. B and C are derived from A, and E and F from D, after applying tensile forces (arrows) in steered molecular dynamics simulations to αIIb and β3 C-terminal and ligand N-terminal atoms shown as large spheres. Models in A-F are aligned by superposition on the β-propeller and β I domains. Numbers show the distance after superposition of C-terminal residue 433 of the hybrid domain from the closed (A) and open (D) conformations. The underlined distance shows the conformation that models most closely resemble.
In the presence of both the membrane-normal force exerted by ligand and the lateral force exerted by the cytoskeleton, the extended-open integrin remained open (Fig. 4E). In the case of the extended-closed integrin (Fig. 4B), the lateral force was transmitted through the lower β-leg domains to the hybrid domain, which swung out and assumed an orientation similar to that in the open headpiece (Fig. 4B). Thus, binding to the actin cytoskeleton provides an active mechanism for separating the integrin α and β legs and inducing the extended-open integrin conformation with high affinity for ligand at sites of actin polymerization and contraction.
In the presence of only the force pulling the integrin away from ligand, the α and β legs remained together, and were extended (Fig. 4C, F). The closed-extended integrin remained closed (Fig. 4C). In the case of the open-extended integrin, the greater leg extension induced by tensile force caused the hybrid domain to swing inward, towards the closed headpiece conformation (Fig. 4F). Thus, a tensile force exerted on the ligand-integrin complex in the absence of cytoskeleton engagement stabilizes the closed, low-affinity state of the headpiece, opposite to previous expectation (Alon and Dustin, 2007). Thus, force-induced stabilization of the low-affinity state provides an active mechanism for downregulating integrin adhesion in migrating cells at sites of actin cytoskeleton disassembly, including the uropod, and enables integrins and plasma membrane to be internalized for transport in intracellular vesicles toward the front of the cell (Broussard et al., 2008).
Implications for integrin-mediated cell adhesion and migration
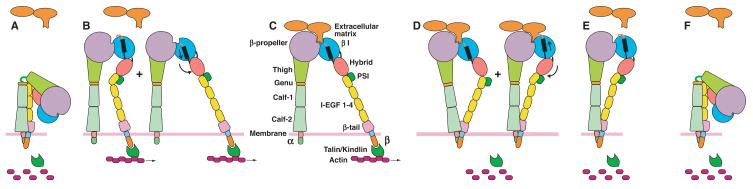
The above results show that inherent in integrin structure is a mechanism for activating the ectodomain by attachment of the actin cytoskeleton to the β subunit cytoplasmic domain, explaining the structural basis for inside-out signaling (Fig. 5). New sites of attachment of integrins to matrix are formed in the lamellipodium, where actin polymerization and branching is rapid (Broussard et al., 2008; Choi et al., 2008). Actin filaments in this region actively move backward toward the lamella. Integrins that associate with the actin cytoskeleton move rapidly along the cell surface, as observed with nanobeads (Giannone et al., 2003) and would be extended by the frictional buffeting forces described above (Fig. 5B). Once the integrins bind ligand, the ligand will resist the pulling force, increase the lateral tensile force, and thereby stabilize the high affinity state (Fig. 5C). Moreover, there is a positive feedback loop, because ligand binding reinforces integrin attachment to the cytoskeleton, and is important in the maturation of nascent adhesions (Broussard et al., 2008; Giannone et al., 2003). Thus, firm adhesion to the matrix is established at sites in the lamellipodium where actin polymerization occurs and contact with ligand is made, further reinforcing attachment between integrins and the cytoskeleton. This provides traction near the lamellipodium to support further protrusion of the leading edge of the cell. As new membrane is added and the cell moves forward, the nascent adhesion sites will be left behind, and find themselves in the lamella. Here, double-headed myosin II is present, which contracts actin filaments, and nascent adhesions can mature to focal adhesions, or disassemble (Broussard et al., 2008; Choi et al., 2008).
Figure 5.
The integrin cycle. A. In the bent conformation, integrins have low affinity for ligand. B. At sites where actin filaments are formed, the integrin β subunit cytoplasmic domain binds through talin or kindlins. Lateral translocation on the cell surface and buffetting cause integrin extension. Both open and closed headpiece conformations are putatively present. C. Binding to an immoblized extracellular ligand greatly increases the lateral force, and markedly favors the high-affinity, open headpiece conformation. D. Disassembly of the actin cytoskeleton removes the lateral force. Tensile force between the ligand and the integrin cytoplasmic domains favors the closed headpiece conformation, and ligand dissociation. E. Ligand dissociates, further favoring the closed headpiece conformation. F. In the absence of ligand and tensile force, the bent conformation is favored, completing the cycle, and the integrin returns to the same state as shown in (A).
For cell migration, deadhesion at the rear of the cell is just as important as adhesion at the leading edge. When integrins are artificially locked in the high affinity state, or actin disassembly in the uropod is blocked with rho kinase inhibitors, uropod retraction is blocked, and cells become highly elongated and stop migrating (Smith et al., 2007). As cells migrate over a substrate, the integrins that remain bound to ligand on the substrate will eventually find themselves in the uropod. Cytoskeleton disassembly in the uropod is coordinated with destabilization of adhesions (Broussard et al., 2008). In the absence of a lateral force on the β subunit, the normal force exerted on the integrin as the uropod pulls away from ligand stabilizes the closed headpiece with low affinity for ligand (Fig. 5D). This favors ligand dissocation (Fig. 5E). The integrin can thus return to the low affinity, bent conformation, completing the integrin adhesion/deadhesion cycle (Fig. 5F).
Activation of the high affinity state by the lateral pull exerted by the actin cytoskeleton provides an elegant solution to the problem of coordinating ligand binding by integrins to attachment to the cytoskeleton. The importance of the cytoskeleton for integrin function is supported by many studies. For example, actin cytoskeleton-disrupting agents inhibit integrin adhesion stimulated by inside-out signaling, i.e., affinity regulation (although not avidity regulation) (Kim et al., 2004). Actin filament movement is highly coordinated with movement of ligand-bound integrins into the ring-shaped immunological synapse, and actin poisons rapidly disperse integrins but not other adhesion molecules from the synapse (Kaizuka et al., 2007).
Our simulations used comparable forces normal and lateral to the cell surface as might be found in the lamellipodium; however, in focal contacts where actin stress fibers terminate, the lateral force exerted on the β tail by the cytoskeleton may be much greater than the normal force. This would only increase the tendency of lateral force to stabilize the open headpiece conformation with high affinity for ligand. In geometries in which no force is applied to the α tail, applying tensile force to the β tail alone will stabilize the high affinity state, since in the open headpiece the β subunit is much more extended than in the closed headpiece, with distances between the ligand-binding MIDAS in the β I domain and C-terminus of the hybrid domain of 83 and 64 Å, respectively (Astrof et al., 2006). In typical representations integrins extend approximately normal to the cell surface (Fig. 5). However, the direction of integrin extension will be determined by the force vectors. When lateral force is stronger than normal force, integrins will extend more in the direction parallel than normal to the membrane, i.e. they will lean over. Such a change in integrin orientation is consistent with measurements of cell-substrate distances by interference reflection microscopy and EM (Verschueren, 1985). The height to which integrins extend above the surface of 200 to 250 Å in our simulations is similar to the distance of 300 Å between the plasma membrane and extracellular matrix at close contacts in the lamellipodium of migrating cells. However, a membrane-matrix distance of 100 to 150 Å is found at focal contacts, consistent with an extended, leaned-over integrin conformation in the presence of strong lateral force.
We believe that application of lateral force is the most physiologically relevant mechanism for activating integins, but not the only one. Separation of the transmembrane and cytoplasmic domains, in the absence of an applied force, appears sufficient to induce integrin extension. This should enable at least a small fraction of integrins to transition to the extended-open conformation, and may be sufficient for integrin adhesiveness in many commonly employed assays. Disruption of association between membrane-proximal cytoplasmic regions may be sufficient to induce extension, and may explain why deletion of either the integrin α or β subunit cytoplasmic domains is activating (Lu et al., 2001; Lub et al., 1997). However, in contrast to other activation mechanisms, association with the actin cytoskeleton uniquely enforces selection of the extended-open conformation over the bent and extended-closed conformations, and selectively enhances the adhesiveness of those integrins that are experiencing tensile forces as a consequence of simultaneous binding to ligand and the cytoskeleton (Fig. 5C). Thus, this mechanism exquisitely supports the function of integrins in integrating cell adhesion and cell migration.
The mechanism described here is mechanochemical, since force alters the chemical equilibrium between conformational states and drives integrins from a bent, closed-headpiece, low affinity state to an extended, open-headpiece, high affinity state. The mechanism is also allosteric, with force as the allosteric effector. Previously, it has not been thought possible to transmit signals through flexible protein domains, because allostery involves relative changes in orientation and position, and flexibility decouples the relative positions of sending and receiving domains. However, tensile force extends and imparts stiffness to flexible proteins, and thus enables signal transduction through otherwise flexible protein regions. The outward swing of the hybrid domain, and with it the PSI domain in the upper β-leg, transmitted through stiffened β-knee and β-leg domains, elegantly couples rearrangements at the ligand-binding site in the β I domain to lateral force exerted by the actin cytoskeleton.
The integrin mechanochemical mechanism works well whether talin or kindlins link integrins to actin microfilaments that are moving as a consequence of myosin contraction, extension/polymerization at the leading edge, or treadmilling. This mechanism would not work for reception of signals from soluble ligands, but is uniquely well suited for adhesion receptors. This structural mechanism for linking cytoskeleton binding to ligand binding appears to be at the heart of the integrating function that gives integrins their name, and enables them to provide the traction for cell migration. Outside-in signaling by integrins requires both α and β subunit transmembrane domain separation, and clustering (Miyamoto et al., 1995; Zhu et al., 2007b). Cytoskeleton association may contribute to both of these components, first by inducing transmembrane domain separation, and second by cooperating with binding to multivalent ligands in inducing clustering.
No doubt further levels of complexity are added by the many members of the integrin family, including the distinctive β4 subunit; the large number of proteins that interact with integrin β cytoplasmic tails, including many such as talin and kindlins that contain FERM or protein tyrosine-phosphate binding domains, and other proteins that bind to the α tail (Wegener and Campbell, 2008).
Experimental Procedures
Crystallography
αIIb and β3 ectodomains were fused to C-terminal segments containing a protease site, coiled-coils, and tags, with or without αIIb-L959C and β3-P688C mutations to introduce a disulfide bond, and expressed in CHO Lec 3.2.8.1 cells. Purified αIIb-L959C/β3- P688C with the C-terminal tag removed by protease in buffer containing 1mM CaCl2 was crystallized in 10% PEG 3350, 50 mM magnesium acetate, and 0.1 M imidazole, pH 7.0. Diffraction data collected at 19-ID of APS was solved using molecular replacement in spacegroup P41. Final refinement with REFMAC5 utilized TLS and NCS. Crystals contain two molecules per asymmetric unit. Density is present for all ectodomain residues (αIIb 1–959 and β3 1–690) except for five loops, and in one molecule, the C-terminal portion of the β-tail domain. Thirteen or 18 N-linked carbohydrate residues are visualized in each molecule. I-EGF1 from the complete αIIb β3 ectodomain was used to model density for this domain in re-refined αIIb β3 headpiece structures with (Springer et al., 2008) or without Fab (Table 1).
Negative stain EM
The clasped and unclasped αIIb β3 was purified on a Superdex 200 HR column in Tris saline, 1 mM Ca2+, 1 mM Mg2+. The peak fraction was adsorbed to glow discharged carbon-coated copper grids, stained with uranyl formate, and inspected with an FEI Tecnai 12 electron microscope operated at 120 kV. Images were acquired at a nominal magnification of 67,000 x. Imaging plates were scanned and digitized with a Ditabis micron imaging plate scanner (DITABIS Digital Biomedical Imaging System, AG, Pforzheim, Germany) using a step size of 15 μm and 2 × 2 pixels were averaged to yield a final pixel size of 4.46 Å at the specimen level. 2,000–5,000 particles were interactively collected, windowed into 75 × 75-pixel individual images, and subjected to ten cycles of multi-reference alignment and classification. Images were processed and cross-correlated using SPIDER (Frank et al., 1996) as described (Nishida et al., 2006).
Steered molecular dynamics
To build an extended model with closed headpiece, bent αIIb β3 was extended at the junction between thigh and calf-1 domains in α, and between I-EGF1 and I-EGF2 in β. For extended αIIb β3 with open headpiece, we substituted the headpiece bound to the 10-residue peptide from the C-terminus of fibrinogen γ subunit. The same peptide ligand was added to extended-closed αIIb β3. Smaller adjustments were made at the hybrid/I-EGF1, I-EGF2/3, and β-ankle/β tail interfaces, guided by angles found in other structures and the gimbal-like nature of I-EGF domain interfaces, to retain the calf-2/β-tail interface. To accelerate simulations with lateral force, the α- and β-legs were pre-separated by changing the angles between thigh and calf-1 domains and between I-EGF1 and I-EGF2 domains.
During simulations, the N-terminus of the ligand (chain C, residue 402) was kept fixed, and forces were applied to the C-termini of the integrin αIIb β3 ectodomain (αIIb, residue 963 and β3, residue 690). Models were aligned in the same coordinate system using the β-propeller and β-I domains. The coordinate system was chosen such that the C-termini are confined to the xy plane mimicking the cell membrane, and the centers of masses of β propeller and β I domains are aligned parallel to the y axis. To mimic talin binding, a lateral force of 15 pN was applied at the C-terminus of β in the -y direction, away from α, as would occur on cells because of frictional resistance to pulling by α. To mimic tensile force extending the ligand-integrin complex, models were pulled along the -z direction at the C-termini of the αIIb and β3 subunits, each with force of 15 pN. The protein structures were solvated in a 4 Å thick water shell and neutralized with Na ions. Molecular dynamics simulations were with NAMD (Phillips et al., 2005) using the CHARMM22 force fields. The supplement contains further details and rationale
Supplementary Material
Acknowledgments
Supported by NIH grant HL-48675 and NSF Teragrid allocation TG-MCB080088T. We thank Junichi Takagi for a critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adair BD, Yeager M. Three-dimensional model of the human platelet integrin αIIb β3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R, Dustin ML. Force as a Facilitator of Integrin Conformational Changes during Leukocyte Arrest on Blood Vessels and Antigen-Presenting Cells. Immunity. 2007;26:17–27. doi: 10.1016/j.immuni.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- Astrof NS, Salas A, Shimaoka M, Chen JF, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglova N, Blacklow SC, Takagi J, Springer TA. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat Struct Biol. 2002;9:282–287. doi: 10.1038/nsb779. [DOI] [PubMed] [Google Scholar]
- Broussard JA, Webb DJ, Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr Opin Cell Biol. 2008;20:85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Carrell NA, Fitzgerald LA, Steiner B, Erickson HP, Phillips DR. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J Biol Chem. 1985;260:1743–1749. [PubMed] [Google Scholar]
- Chen JF, Salas A, Springer TA. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat Struct Biol. 2003;10:995–1001. doi: 10.1038/nsb1011. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA, Calderwood DA. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Giannone G, Jiang G, Sutton DH, Critchley DR, Sheetz MP. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J Cell Biol. 2003;163:409–419. doi: 10.1083/jcb.200302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Tomiyama Y, Pelletier AJ, Annis D, Honda Y, Orchekowski R, Ruggeri Z, Kunicki TJ. Topography of ligand-induced binding sites, including a novel cation-sensitive epitope (AP5) at the amino terminus, of the human integrin β3 subunit. J Biol Chem. 1995;270:11947–11954. doi: 10.1074/jbc.270.20.11947. [DOI] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Mitsuoka K, Fujiyoshi Y, Fujisawa Y, Kikuchi M, Sekiguchi K, Yamada T. Electron tomography reveals diverse conformations of integrin αIIb β3 in the active state. J Struct Biol. 2005;150:259–267. doi: 10.1016/j.jsb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Janin J. Specific versus non-specific contacts in protein crystals. Nature Struc Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci U S A. 2007;104:20296–20301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Carman CV, Yang W, Salas A, Springer TA. The primacy of affinity over clustering in regulation of adhesiveness of the integrin αLβ2. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Lu C, Takagi J, Springer TA. Association of the membrane-proximal regions of the α and β subunit cytoplasmic domains constrains an integrin in the inactive state. J Biol Chem. 2001;276:14642–14648. doi: 10.1074/jbc.M100600200. [DOI] [PubMed] [Google Scholar]
- Lub M, van Vliet SJ, Oomen SP, Pieters RA, Robinson M, Figdor CG, van Kooyk Y. Cytoplasmic tails of b1, b2, and b7 integrins differentially regulate LFA-1 function in K562 cells. Mol Biol Cell. 1997;8:719–728. doi: 10.1091/mbc.8.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Imm. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Akiyama SK, Yamada KM. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ. Role of ADMIDAS cation-binding site in ligand recognition by integrin α5β1. J Biol Chem. 2003;278:51622–51629. doi: 10.1074/jbc.M306655200. [DOI] [PubMed] [Google Scholar]
- Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J Cell Biol. 2006;175:349–360. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco M, Rosano C, Weisel JW, Horita DA, Hantgan RR. Integrin conformational regulation: uncoupling extension/tail separation from changes in the head region by a multiresolution approach. Structure. 2008;16:954–964. doi: 10.1016/j.str.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP. Glycoprotein motility and dynamic domains in fluid plasma membranes. Annu Rev Biophys Biomol Struct. 1993;22:417–431. doi: 10.1146/annurev.bb.22.060193.002221. [DOI] [PubMed] [Google Scholar]
- Shi M, Foo SY, Tan SM, Mitchell EP, Law SK, Lescar J. A structural hypothesis for the transition between bent and extended conformations of the leukocyte β2 integrins. J Biol Chem. 2007;282:30198–30206. doi: 10.1074/jbc.M701670200. [DOI] [PubMed] [Google Scholar]
- Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol Rev. 2007;218:135–146. doi: 10.1111/j.1600-065X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen by the platelet integrin aIIbb3. J Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Verschueren H. Interference reflection microscopy in cell biology: Methodology and applications. J Cell Sci. 1985;75:279–301. doi: 10.1242/jcs.75.1.279. [DOI] [PubMed] [Google Scholar]
- Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (review) Mol Membr Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Wang J-h, Coller BS, Springer TA. Structural basis for allostery in integrins and binding of fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Goodman SL, Arnaout MA. A novel adaptation of the integrin PSI domain revealed from its crystal structure. J Biol Chem. 2004;279:40252–40254. doi: 10.1074/jbc.C400362200. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Yan B, Smith JW. Mechanism of integrin activation by disulfide bond reduction. Biochemistry. 2001;40:8861–8867. doi: 10.1021/bi002902i. [DOI] [PubMed] [Google Scholar]
- Ye F, Liu J, Winkler H, Taylor KA. Integrin alpha IIb beta 3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J Mol Biol. 2008;378:976–986. doi: 10.1016/j.jmb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Boylan B, Luo BH, Newman PJ, Springer TA. Tests of the extension and deadbolt models of integrin activation. J Biol Chem. 2007a;16:11914–11920. doi: 10.1074/jbc.M700249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Carman CV, Kim M, Shimaoka M, Springer TA, Luo BH. Requirement of α and β subunit transmembrane helix separation for integrin outside-in signaling. Blood. 2007b;110:2475–2483. doi: 10.1182/blood-2007-03-080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.