Abstract
Polyomaviruses such as BK virus and JC virus have been linked to several diseases, but treatments that thwart their propagation are limited in part because of slow growth and cumbersome culturing conditions. In contrast, the replication of one member of this family, Simian Virus 40 (SV40), is robust and has been well-characterized. SV40 replication requires two domains within the viral-encoded large tumor antigen (TAg): The ATPase domain and the N-terminal J domain, which stimulates the ATPase activity of the Hsp70 chaperone. To assess whether inhibitors of polyomavirus replication could be identified, we examined a recently described library of small molecules, some of which inhibit chaperone function. One compound, MAL2-11B, inhibited both TAg’s endogenous ATPase activity and the TAg-mediated activation of Hsp70. MAL2-11B also reduced SV40 propagation in plaque assays and compromised DNA replication in cell culture and in vitro. Furthermore, the compound significantly reduced the growth of BK virus in a human kidney cell line. These data indicate that pharmacological inhibition of TAg’s chaperone and ATPase activities may provide a route to combat polyomavirus-mediated disease.
Keywords: SV40, Hsp70, BK virus, Heat shock protein, J domain, Dihydropyrimidine
1. Introduction
Polyomaviruses are non-enveloped viruses with doubled-stranded DNA genomes that encode only a few proteins (Eash et al., 2006). Five human polyomaviruses have been identified, and a sixth, Simian Virus 40 (SV40), was inadvertently introduced into the population as a contaminant of the original polio vaccine. Two of the human polyomaviruses, BK virus (BKV) and JC virus (JCV), have been definitively linked to kidney disease (BKV associated nephropathy) and a disease of the central nervous system (progressive multifocal leukoencephalopathy), respectively (Barbanti-Brodano et al., 1998; Safak and Khalili, 2003; Vats et al., 2006). The three most recently identified polyomaviruses, KI, WU, and MCV (Allander et al., 2007; Gaynor et al., 2007; Feng et al., 2008) may also contribute to disease. Although SV40 induces tumors in rodents, it remains unclear whether this virus is also tumorigenic in humans (Poulin and DeCaprio, 2006).
The early region of the polyomavirus genome is transcribed immediately after infection and includes the gene encoding the large tumor antigen (TAg), which is conserved in each of the polyomaviruses. In order to promote cell cycle progression and prevent apoptosis, TAg interferes with the function of cellular regulators, including p53 and Rb (Ahuja et al., 2005; Ali and DeCaprio, 2001; Pipas and Levine, 2001). In addition, viral DNA replication is initiated when TAg interacts with the SV40 origin of replication (ori). ATP binding to the TAg ATPase domain allows for high affinity association with ori and catalyzes TAg hexamerization and dodecamer assembly (Mastrangelo et al., 1989). The TAg hexamer then partially melts the DNA (Borowiec and Hurwitz, 1988; Li et al., 2003) and associates with proteins required for replication, including DNA topoisomerase I (Top1), replication protein A (RPA), and DNA polymerase α primase (pol-prim) (Arunkumar et al., 2005; Jiang et al., 2006). After replication is initiated, a “polymerase switch” occurs and pol-prim is replaced by replication factor C (RF-C), proliferating cell nuclear antigen (PCNA), and DNA polymerase δ (Maga et al., 2000; Mossi et al., 2000; Yuzhakov et al, 1999). Finally, the TAg dodecamer acts as a bi-directional helicase, hydrolyzing ATP and unwinding the DNA during replication (Smelkova and Borowiec, 1997).
Another region in TAg that is important for viral DNA replication in infected cells is the J domain, which encompasses ∼100 residues at TAg’s N-terminus (Brodsky and Pipas, 1998; Kim et al., 2001). J domains are conserved, four-helical bundles that are most commonly found in Hsp40 molecular chaperones. The J domain mediates the interaction between Hsp40s and Hsp70 chaperones, which results in a stimulation of Hsp70’s ATPase activity and in the hand-off of polypeptide substrates from the Hsp40 to Hsp70 (Craig et al., 2006; Hennessy et al., 2005). Mutations in the TAg J domain reduce viral DNA synthesis and there is consequently little virus production (Collins and Pipas, 1995; Gluzman and Ahrens, 1982; Peden and Pipas, 1992). It remains unclear why the J domain is required for DNA replication in cells.
Based on the specific interaction between the J domain and Hsp70 chaperones (Gassler et al., 1998; Suh et al., 1998), it seemed likely that Hsp70 might also be important for TAg function and SV40 replication. Indeed, Hsp70 has been known for some time to associate with TAg (Sawai and Butel, 1989; Sawai et al., 1994). It was subsequently found that the TAg J domain-Hsp70 interaction aids in the dissociation of the Rb-E2F transcription factor complex (Fewell et al., 2002; Sullivan et al., 2000), which initiates the cell cycle. As a result, efficient formation of a TAg-Hsp70 complex is also important for TAg-mediated cellular transformation (Campbell et al., 1997; Srinivasan et al., 1997). Moreover, Hsp70 appears to be involved in viral uncoating after infection (Chromy et al., 2006) and in viral coat assembly late in infection (Cripe et al., 1995).
The efficacy of many anti-viral agents derives from their ability to inhibit DNA replication (De Clercq and Holy, 2005). Unfortunately, few anti-viral agents have been identified as potential treatments for polyomavirus infections, at least in part because BKV and JCV are difficult to grow in the laboratory. Nevertheless, the cytosine analog, cidofovir, inhibits the growth of mouse polyomavirus and SV40 (Andrei et al., 1997) and is currently being examined in clinical trials for its ability to treat BKVAN (Gorczynska et al., 2005; Hymes and Warshaw, 2006). In addition, the fluoroquinolones, which inhibit Type II topoisomerases by stabilizing the linkage between the enzyme and DNA, have been shown to inhibit the replication of BKV and SV40 (Ali et al., 2007; Portolani et al., 1988). Both classes of compounds lack specificity for TAg/SV40. Another attempt to identify polyomavirus inhibitors employed an in vitro assay in which the interaction between TAg and p53 could be assessed. Identified “hits” would be expected to trigger apoptosis in SV40-infected cells, but because apoptosis was not evident, the authors suggested that the most potent inhibitor identified from this in vitro screen might also block p53 function (Carbone et al., 2003). To our knowledge, there have been no subsequent studies based on this screen or on the in vivo effects of other hits from this analysis.
We now report on the characterization of pyrimidinone-peptoid hybrid compounds that were previously found to compromise the functional interaction between the SV40 TAg J domain and Hsp70 (Fewell et al., 2004; Wright et al., 2008). One of these compounds, MAL3-101, triggers apoptosis in breast cancer cells (Rodina et al., 2007), which is consistent with the known effect of Hsp70 inhibition in breast cancer cell lines (Nylandsted et al., 2000). More generally, these data indicated that the compound – and most likely several of its structural congeners – are membrane permeable and remain active in cell culture. Surprisingly, however, we found that MAL3-101 exhibited little effect on SV40 replication or viral DNA synthesis. In contrast, a MAL3-101 precursor, MAL2-11B, which inhibits TAg stimulation of Hsp70 with greater efficacy than MAL3-101, significantly reduces viral replication and DNA synthesis. MAL2-11B also inhibited the activity of the TAg ATPase domain. We suggest that the improved anti-viral potency of MAL2-11B derives not only from its effect on the TAg-enhanced stimulation of Hsp70 ATPase activity but also from the fact that the compound inhibits TAg’s endogenous ATPase activity. These results are consistent with data indicating that the TAg ATPase domain and J domain are required for SV40 replication, and suggest that MAL2-11B and related compounds may serve as a novel class of polyomavirus inhibitors.
2. Materials and methods
2.1. Reagents
The yeast Hsp70, Ssa1p, and TAg were purified as described previously (Cantalupo et al., 1999; McClellan and Brodsky, 2000). Purified human Hsp70, Ydj1p, Sse1p, and the wild type and the mutant form of TAg, were kind gifts from Doug Placais, Shruthi Vembar, Jennifer Goeckeler, and Paul Cantalupo, respectively, and were purified as published (Cyr et al., 1992; Cantalupo et al., 1999; Goeckeler et al., 2002; Youker et al., 2004). Anti-Hsp70 and anti-actin antibodies were obtained from Cell Signaling Technology and Sigma, respectively, and the anti-TAg antibody PAB419 has been described (Harlow et al., 1981). Anti-PARP antibody was purchased from Cell Signaling Technologies. The syntheses of MAL3-101, MAL2-11B, and MAL3-51 have been reported (Fewell et al., 2004; Wright et al., 2008).
2.2. ATPase assays
Steady-state ATPase assays using the indicated chaperones or TAg were modified from a previously published protocol (Cyr et al., 1992). In brief, purified protein and equal volumes of a test compound or dimethyl sulfoxide (DMSO) were preincubated on ice in 50 mM HEPES, pH 7.4,50 mM NaCl, 2 mM MgCl2, and 10 mM dithiothreitol(DTT)for15 min. The reaction was started by adding 1 nmol ATP and 0.2 µCi [α32P]ATP and immediately placing the reaction in a 30°C water bath. The activity (in pmol ATP hydrolyzed) was determined after analysis on a Fujifilm BAS-2500 phosphorimager and Image Gauge software (Fuji Film Science Lab). The amount of spontaneous ATP hydrolysis at t = 0 was subtracted from each time point, and the results were averaged and fit using a linear regression that originated at the origin. Data are reported as the percentage of activity relative to the control (DMSO).
The ability of single-stranded DNA to activate the ATPase activity of TAg was measured as described (Taneja et al., 2002) using M13 DNA. Reactions were performed in the presence of the indicated concentrations of MAL2-11B or the identical volume of DMSO.
Single turnover ATPase assays were performed as described (Fewell et al., 2004). Specifically, 25 µg of Hsp70 was combined with 25 µM ATP and 100 µCi [α32P]ATP in 25 mM HEPES, pH 7.5, 100 mM KCl, and 11 mM MgOAc to allow the formation of an Hsp70-ATP complex, and unbound ATP was removed by purification over a NICK column (GE Healthcare). For reactions in which the effects of compounds on the endogenous ATPase activity of Hsp70 were measured, the Hsp70-ATP complex was thawed at 30°C and immediately combined with DMSO or an equal volume of the test compound. For reactions in which the effects of compounds on the co-chaperone (J domain-protein)-stimulated ATPase activity of Hsp70 were measured, TAg or Ydj1p were added to the thawed Hsp70-ATP complex and after 60 s the compound or DMSO was added. Data analysis was performed as previously published (Fewell et al., 2004; Wright et al., 2008).
2.3. Viral plaque assay
Plaque assays were performed after a modification of a previously published protocol (Tremblay et al., 2001). A total of ∼5 × 104 BSC40 cells were plated in each well of a 96-well plate in 200 µL Eagle minimal essential medium supplemented with 10% fetal bovine serum (MEM-10% FBS) and grown overnight. Then, ∼1 × 105 pfu of wild type SV40 was added such that the multiplicity of infection (MOI) was ∼2. After 2–3 h, the medium was changed and equal volumes of DMSO or a test compound at the indicated concentration was added to each well. All treatments were performed in duplicate. After 2 d, the medium was changed again and the compounds or DMSO were re-added. After another 2 d, the 96-well plates were frozen at −20 °C and the cells were lysed by three successive freeze/thaw cycles to obtain a viral stock. The viral stocks were diluted 1000- and 10,000-fold in MEM-10% FBS. Then, 500 µL of the each dilution was added, in duplicate, to freshly confluent BSC40 cells in 6 cm dishes. After 2–3 h, the medium was removed and 4 mL of a 50/50 mixture of melted 1.8% Bacto-agar and 2 × MEM-10% FBS lacking phenol red was added to the dishes. The agar/media overlay was repeated on days 4 and 7, but neutral red was included at a final concentration of 50 µg/mL on day 7. Plaques were counted on days 8 and 9.
2.4. Viral replication assays in cell culture
Measurements of SV40 DNA replication were performed based on previously published protocols (Cantalupo et al., 2005; Ziegler et al., 2004). BSC40 cells were plated in a 24-well dish in MEM-10% FBS and allowed to grow to confluence. Next, SV40 was added at an MOI = 2 and after 2–3 h the medium was changed and DMSO or a test compound was added immediately and then again after 48 h (when the medium was changed, as above). At the indicated time points, SV40 DNA was isolated using Qiaprep Spin Columns (Qiagen), digested with BamHI, and visualized on an ethidium bromide-stained agarose gel. Data were quantified using a Kodak 440CF Image Station and the associated Kodak 1D (v3.6) software.
To determine the amount of loaded DNA (see Fig. 2B), DNA samples were prepared from 24-well dishes (∼2 × 105 cells/well) and the DNA was isolated as described above. Primers against the actin gene from the African green monkey (the source of BSC40 cells) were: GATGAACACTGACCACAAGG (forward) and GCACATTTTCCCCACCT (reverse), and the probe for the real-time polymerase chain reaction (PCR) was 5′/56-FAM/ATCAGGAACCCAGCACTCCACT/3IowaBlack Quencher (synthesized by IDT). The reaction was performed on an ABI 7300, with Quantas Biosciences PerfeCta qPCR Supermix, containing the ROX fluorophore. The reaction conditions were 95 °C for 10 min and then 55,15 s cycles at 95 °C interspersed with 1 min cycles at 60 °C. Primer efficiency was calculated as described (Ramakers et al., 2003) in order to determine the normalized amount of actin DNA.
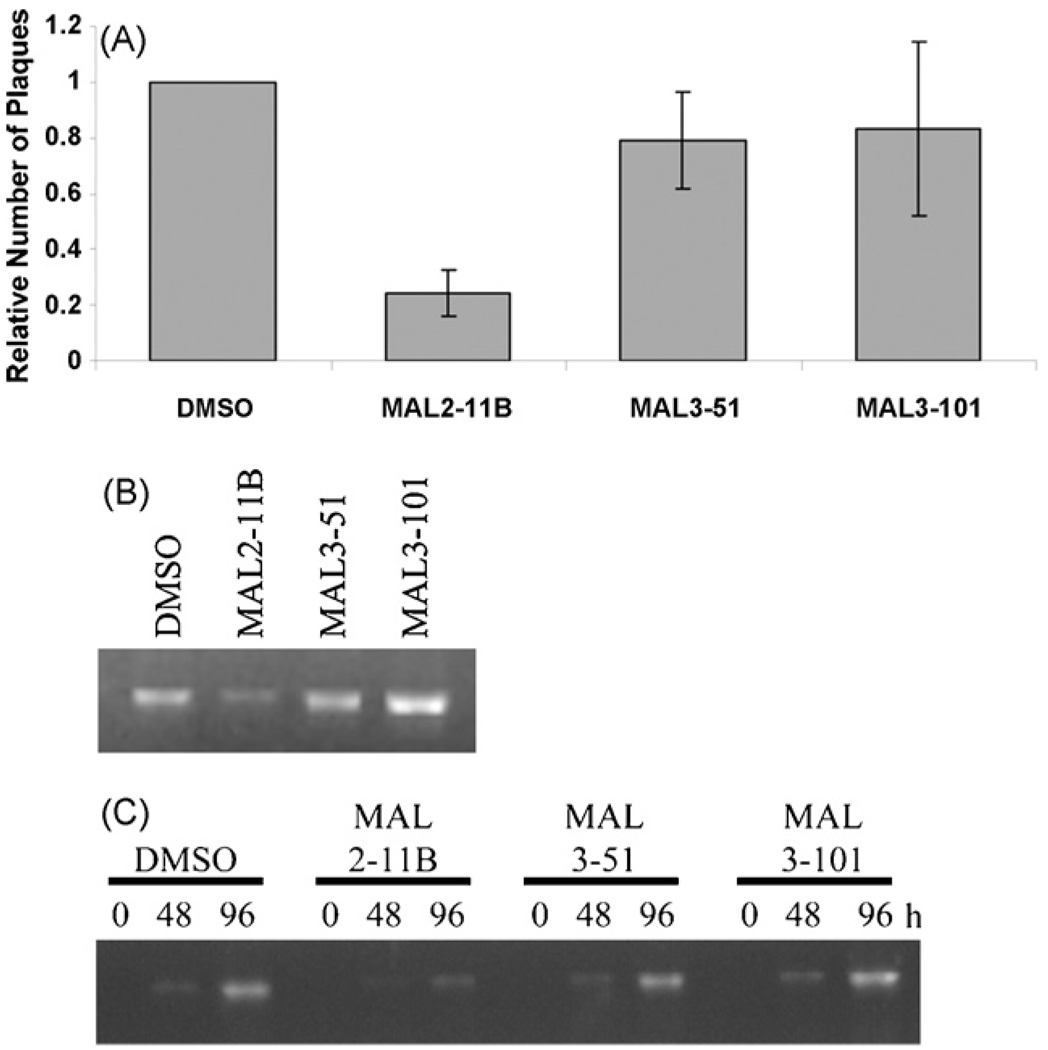
Fig. 2.
MAL2-11B inhibits SV40 viral replication and SV40 DNA synthesis. (A) MAL2-11B reduces viral propagation in plaque assays. The number of plaques visualized in duplicate reactions was averaged and the results were normalized to the DMSO control. The results of three independent experiments were then averaged and the means of the reactions ±the standard deviations of the data are shown. MAL2-11B compared to the DMSO control, p < 0.0002. (B) MAL2-11B inhibits SV40 DNA replication 48 h after treatment. Viral DNA was isolated from SV40-infected cells 48 h after treatment with 100 µM of the indicated compounds and digested with BamHI, and the products were visualized after resolution on an ethidium bromide-stained agarose gel. The fact that the product corresponded to viral DNA was confirmed based on its size (∼5.2 kb after BamH1 digestion) and by the fact that internal primers specific to SV40 DNA yielded the expected product after PCR (data not shown). (C) Time course of the effects of MAL2-11B, MAL3-101, and MAL3-51 on SV40 DNA synthesis. DNA was isolated at the indicated times and analyzed as described in part (B). Note: Less material was loaded than in part (B) in order to better visualize the time-dependent increase in SV40 DNA. To control for the DNA loaded in these gels, we used a real-time PCR analysis to detect the amount of the actin gene in the same samples used in part (B)(see Section 2). The amount of actin DNA in the DMSO control was set to “1.0”, and based on this analysis the relative amounts of total DNA analyzed were: DMSO, 1.0; MAL2-11B, 3.4; MAL3-51, 4.0; MAL3-101, 4.0.
The replication of BK virus (BKV) in the human kidney HK2 cell line was quantified by a quantitative, real-time polymerase chain reaction (PCR) assay using primers that bound to the gene encoding BKV TAg, as previously described (Farasati et al., 2005). The primers and probe used for the real-time PCR and the sequences were as follows: forward, GGACCCACCATTGCAGAGTTT; reverse, AGAGCCCTTGGTTTGGATAGATT; probe, 6-FAM (6-carboxyfluorescein)-5′-AAGCCAAACCACTGTGTGAAGCAGTCAAT3′-TAMRA (6-carboxytetramethylrhodamine). A cloned plasmid (pBKVT3) was used to generate the dilution curves for the quantitative real-time PCR assays, as described elsewhere (Bista et al., 2007). Non-confluent cells were treated with MAL2-11B at a final concentration of 15 µM or the DMSO control for 5 d at an MOI of 25.
2.5. Preparation of cells lysates and immunoblots
BSC40 cells were plated in 10 cm dishes, grown to confluence in MEM-10% FBS, and infected with SV40 at an MOI = 2 for 2 h. The medium was changed and DMSO or a test compound was added. All assays were conducted in duplicate. After 48 h, the cells were washed 3 times with PBS-EDTA (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.2, 137 mM NaCl, 2.7 mM KCl, 0.5 mM EDTA) supplemented with 200 µg/mL cycloheximide, removed with a scraper, and lysed in Branton’s Lysis Buffer (50 mM HEPES, pH 7.9,400 mM KCl, 0.5 mM EDTA, 0.1% NP40, 10% glycerol, 1 mM DTT, 1 µg/mL, 0.5 mM NaF, and a Complete EDTA-free Mini Protease Inhibitor Tablet [Roche]). The BioRad protein assay, with BSA as the standard, was utilized to determine the total protein concentration in the lysates. Equal amounts of total protein were resolved by SDS-PAGE and transferred to nitrocellulose and TAg and actin were decorated with 419 (Harlow et al., 1981) and anti-actin antibody (Sigma), respectively. The bound antibodies were visualized using anti-mouse antisera coupled to horseradish peroxidase and the Supersignal West Pico Chemiluminescent Substrate Kit (Pierce), and chemiluminescence was quantified as described above.
2.6. In vitro DNA replication assay
In vitro DNA replication assays were performed as previously described (Castellino et al., 1997). In brief, 25 µL reactions included 100 ng pUC.HSO, which contains the SV40 origin of replication, 90 µg of HeLa cell extract, 500 ng TAg, 50 mM creatine phosphate, 2.5 µg creatine phosphokinase, and 2.5 µCi [α32P] dCTP in 30 mM HEPES, pH 7.8, 7 mM MgCl2, 100 mM EDTA, 1 mM DTT, 4 mM ATP, 0.2 mM CTP, 0.2 mM UTP, 0.2 mM GTP, 25 µM dCTP, 100 µM dATP, 100 µM dTTP, 100 µM dGTP. Reactions were incubated for 2 h at 37 °C and 5 µL aliquots were removed every 30 min and mixed with 24 µL of Stop Solution (10 mM Tris, pH 8.0, 5 mM EDTA, 0.3 mg/mL tRNA, and 1 mg/mL proteinase K and 0.5% Sodium Dodecyl Sulfate [SDS]) for 1 h at 37 °C. Samples were placed on ice until the experiment was completed. The synthesized DNA was precipitated with ethanol, resolved on a 1% agarose gel, and stained with ethidium bromide to verify equal DNA loading. Next, the gel was dried and the amount of incorporated nucleotide was determined by phosphorimager analysis, as described above. When the effects of specific compounds were examined, half reactions were performed in triplicate at 37° C for1 h, at which time 50 µL of Stop Solution was added.
2.7. SV40 monopolymerase assay
Reaction mixtures (20 µL) with 600 ng of supercoiled pUC-HS plasmid DNA (2.8 kb) containing the complete SV40 origin, 500 ng of RPA, 750 ng of topoisomerase I, 600 ng of pol-prim, and 1400 ng of TAg in initiation buffer (30 mM HEPES-KOH, pH 7.9, 7 mM magnesium acetate, 10µM ZnCl2, 1 mM DTT, 4 mM ATP, 0.2 mM each of GTP, UTP, and CTP, 0.1 mM each of dGTP, dATP, and dCTP, 0.02 mM dTTP, and 40 mM creatine phosphate and 40 µg of creatine kinase/mL) supplemented with 3 µCi of [α32P] dTTP (3000Ci/mmol; Dupont NEN, Boston, MA). Four sets of reactions were carried out and the results were evaluated and statistically verified as described previously (Arunkumar et al., 2005).
3. Results
3.1. MAL2-11B inhibits the TAg-mediated stimulation of Hsp70 ATPase activity
Since polyomavirus replication requires the viral-encoded TAg, we reasoned that specific TAg inhibitors could represent a novel class of anti-viral agents. MAL3-101 (Fig. 1A) was previously shown to inhibit the TAg-mediated stimulation of Hsp70’s ATPase activity in single turnover assays (Fewell et al., 2004). In these assays, ATP is first pre-bound to Hsp70 and ligand-free ATP is removed by chromatography. The ATP-Hsp70 complex is then flash-frozen, and upon thawing in the presence/absence of TAg (or another J domain-containing protein) and/or a test agent, the ATP hydrolytic step (KCAT) can be monitored independent of ATP binding and release. For these assays, we routinely use yeast Hsp70 (Ssa1p) since it can be isolated in high quantities and is of greater purity than commercial preparations of Hsp70. MAL3-51, a structural analog of MAL3-101, has minimal activity in this assay (Fewell et al., 2004) and serves as a negative control. Another compound with some structural similarity is MAL2-11B, which represents the focus of this study (Fig. 1A).
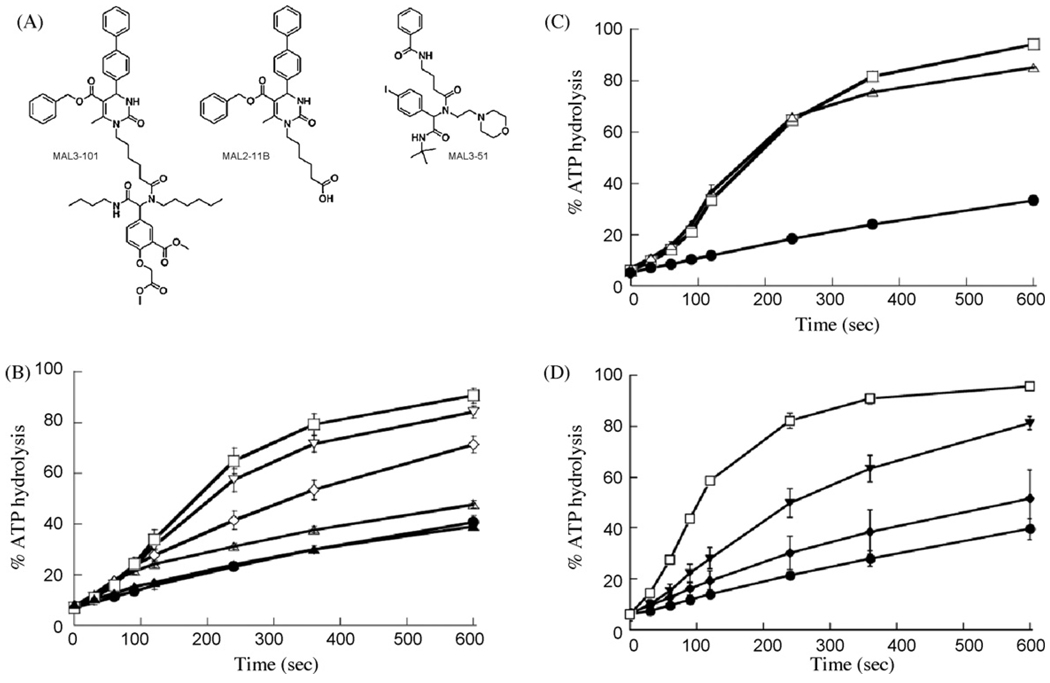
Fig. 1.
MAL2-11B inhibits the TAg-dependent stimulation of Hsp70 ATPase activity. (A) The chemical structures of MAL3-101, MAL2-11B, and MAL3-51. (B) MAL2-11B inhibits TAg stimulation of the yeast Hsp70, Ssa1p, in a concentration-dependent manner. Single turnover reactions were performed in the absence of TAg but supplemented with 300 µM MAL2-11B (filled triangles) or an equal volume of DMSO (filled circles), or in the presence of ∼0.2µM TAg and with the following concentrations of compound: DMSO control (open squares); 30 µM MAL2-11B (open inverted triangles); 100 µM MAL2-11B (open diamonds); 300 µM MAL2-11B (open triangles). (C) MAL2-11B has a minimal effect on Ydj1p-mediated stimulation of yeast Hsp70 in single turnover ATPase assays. Filled circles, DMSO; Open squares, ∼0.2µM Ydj1p plus DMSO; Open triangles, ∼0.2 µM Ydj1p and 300 µM MAL2-11B. (D) A mutation in the TAg J domain inhibits activation of Hsp70 ATPase activity Filled circles, DMSO; Open squares, TAg; Inverted filled triangles, TAg mutant 5061; Filled diamonds, D44N TAg. Error bars represent standard deviations of the data.
Recent work established that 300 µM MAL3-101 inhibits the ability of TAg to activate yeast Hsp70 by ∼40% (Wright et al, 2008). To compare the activity of MAL2-11B to MAL3-101, MAL2-11B was titrated into the single turnover assay 60s after the addition of TAg. As shown in Fig. 1B, 300 µM MAL2-11B virtually abolished the TAg-mediated stimulation of Hsp70 ATPase activity (∼75% inhibition) and also decreased the overall amplitude by almost 50%. A concentration-dependent inhibition of TAg-dependent stimulation of Hsp70 was apparent when MAL2-11B was examined at final concentrations of 30–300 µM and yielded an apparent IC50 of ∼100 µM. In contrast, the endogenous ATPase activity of Hsp70 was unaffected by MAL2-11B, even at a final concentration of 300 µM (compare the overlapping profiles of the closed symbols in Fig. 1B).
To determine if MAL2-11B blocked the ability of all J domain-containing proteins to activate the ATPase activity of an Hsp70, we tested the effect of MAL2-11B on the Ydj1p-dependent stimulation of Ssa1p in a single turnover assay. Ydj1p is a yeast Hsp40 and partners with Ssa1p in the yeast cytoplasm (Becker et al., 1996; Cyr et al., 1992; McClellan and Brodsky, 2000). Even though equimolar Ydj1p stimulated Ssa1p to a similar extent as TAg, 300 µM MAL2-11B had little effect on this activity (Fig. 1C). As a control for these assays, we found that a mutant TAg possessing a mutation in the J domain (D44N; Sullivan et al., 2000) exhibited significantly reduced stimulation of Hsp70 ATP hydrolysis, but an ATPase domain TAg mutant (5061, see below) was only partially defective for ATPase stimulation (filled diamonds and inverted triangles, respectively, Fig. 1D). These data indicate that MAL2-11B exhibits some degree of specificity for TAg over another, divergent J domain-containing protein.
We next examined the effects of MAL3-101, MAL2-11B, and MAL3-51 on the ATPase activities of other ATP-hydrolyzing chaperones at a final concentration of 300 µM. In contrast to the single turnover assays, these reactions were assembled so that we could assess whether the compounds affected ATP binding, ATP hydrolysis, and/or phosphate/ADP release (see Section 2). Furthermore, reaction mixtures were preincubated for 15 min on ice to allow the compound and chaperone to interact. To initiate hydrolysis, radiolabeled ATP was added and the reactions were shifted to 30 °C. The chaperones examined under these steady-state conditions included yeast and mammalian Hsp70 (Ssa1p and mHsp70), and the yeast Hsp110, Sse1p. As predicted, MAL3-51 had little effect on the activities of any of the ATPases tested (Table 1). Interestingly, despite similarities in the single turnover assays, very different effects were seen when MAL3-101 and MAL2-11B were compared. MAL3-101, which had a minor effect on endogenous Hsp70 ATP hydrolysis in the single turnover ATPase assay (Fewell et al., 2004), had a small effect on yeast and mammalian Hsp70 ATP hydrolysis. Similar data were obtained when the yeast Hsp110 was examined. MAL2-11B inhibited each chaperone to 25–64% of the control when tested at a final concentration of 300 µM. At this point, the step(s) at which MAL2-11B acts in the chaperones’ catalytic cycles is unknown.
Table 1.
Relative ATPase activities.
| Protein | DMSO | 300 µM MAL2-11B | 300 µM MAL3-51 | 300 µM MAL3-101 |
|---|---|---|---|---|
| 3.6 µM Ssa1p | 100 | 25 | 107 | 106 |
| 7µM mHsp70 | 100 | 57 | 107 | 114 |
| 3.2µM Sse1p | 100 | 64 | 100 | 99 |
| 0.6 µ M Tag | 10 0 | 9 | 103 | 100 |
The ATPase activities of unique chaperones are differentially affected by MAL2-11B, MAL3-51, and MAL3-101. Each protein was preincubated with 300 µM of the indicated compound and the formation of ADP was measured under steady-state conditions. The rate of ATP hydrolysis was calculated and is reported as percent hydrolysis relative to the DMSO control. Values represent the means of at least 2 independent reactions. Independent replicates of these reactions are shown in Table 2, last column.
3.2. MAL2-11B inhibits SV40 replication in kidney cells
MAL2-11B had potent effects on the TAg-dependent stimulation of Hsp70 ATPase activity under single turnover conditions when used at final concentrations of 30–100 µM (Fig. 1). We therefore investigated whether the chaperones’ ATPase activities were also affected at lower MAL2-11B concentrations. Significantly less pronounced effects on the enzymes were observed at lower MAL2-11B levels (Table 2). For example, at a final concentration of 100 µM, MAL2-11B minimally altered the activity of each chaperone tested except that the activity of human Hsp70 (mHsp70) dropped to 58% of the control.
Table 2.
Relative ATPase activities.
| Protein | DMSO | 30µM MAL2-11B | 100 µM MAL2-11B | 300 µM MAL2-11B |
|---|---|---|---|---|
| Ssa1p | 100 | 98 | 85 | 12 |
| mHsp70 | 100 | 84 | 58 | 49 |
| Sse1p | 100 | 105 | 100 | 57 |
| TAg | 100 | 81 | 51 | 2.5 |
The concentration-dependent effect of MAL2-11B on different ATPases. The relative steady-state ATPase activities for each enzyme in the presence of the indicated amount of MAL2-11B, or the equivalent volume of DMSO, are shown. All values are given relative to the DMSO control, which was set at 100%. The last column represents independent replicates of the experiments presented in Table 1, third column.
Based on these data, and based on the importance of TAg-Hsp70 interaction and Hsp70 activity during SV40 replication (see Section 1), we tested if MAL2-11B could inhibit viral replication using a well-established plaque assay (Fendrick and Hallick, 1983). BSC40 cells, an African green monkey kidney cell line permissive for SV40 replication, were plated and infected at a multiplicity of infection (MOI) of 2, and the infection was allowed to proceed for 2–3 h before the compounds were added at a final concentration of 100 µM. This allowed viral entry to occur independent of the compounds, and ensured that any inhibition would result from decreased viral replication and not from reduced viral uptake. To first establish the efficacy of these assays to screen for anti-viral compounds, the cytosine analog, cidofovir, was titrated onto SV40-infected BSC40 cells. Cidofovir showed a concentration-dependent effect on SV40 replication, and completely inhibited viral replication at ∼5 µg/mL (data not shown). The effect observed is similar to the previously noted impact of this compound on SV40 when used at a final concentration of 3.8 µg/mL (Andrei et al., 1997). Next, MAL2-11B, MAL3-51, or MAL3-101 were added to viral-infected BSC40 cells. At 100 µM, MAL2-11B inhibited viral replication by ∼4.5-fold, whereas MAL3-51 and MAL3-101 had little effect (Fig. 2A).
We then investigated whether MAL2-11B might also affect the endogenous ATPase activity of TAg, which is required for SV40 replication (Clark et al., 1983; Cole et al., 1986). Interestingly, MAL2-11B inhibited TAg ATPase activity by ∼50% at a final concentration of 100 µM and nearly abolished activity at 300 µM(Table 1 and Table 2, last row). As a control for these assays, we found that the endogenous ATPase activity of a mutant TAg, 5061, a full length protein in which a glycine at position 431 in the ATPase domain is replaced with three amino acids, was abolished in our assays (data not shown) (Castellino et al., 1997; Farber et al., 1987). These data suggest that the ability of MAL2-11B to inhibit SV40 replication might derive from its combined effects on TAg activation of Hsp70 and on the innate ATPase activity of TAg. Another possibility is that MAL2-11B affects the activity of mammalian Hsp70, thus compromising SV40 infection (see Section 4).
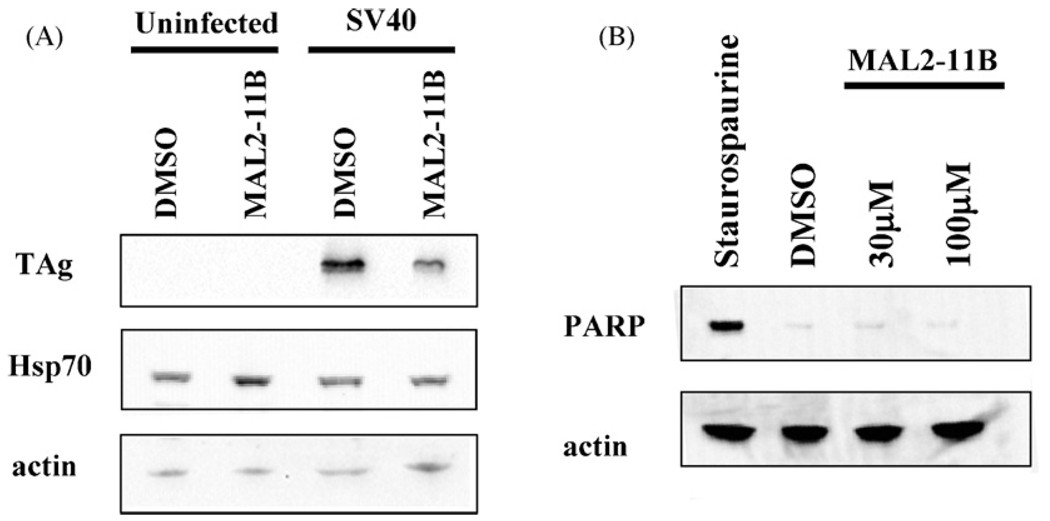
To substantiate the anti-viral effect of MAL2-11B, we next measured the amount of viral DNA present in cells treated with 100 µM MAL2-11B and found that there was a ∼50% reduction compared to the DMSO control; in contrast, there was no apparent reduction of SV40 DNA in lysates prepared from infected cells in the presence of MAL3-51 or MAL3-101 (Fig. 2B, C). A similar result was seen when the cells were infected at an MOI of ∼10 (data not shown). Also consistent with the data presented above, we discovered that MAL2-11B reduced the amount of TAg in infected cell lysates by 4.2-fold when present at a final concentration of 100 µM (Fig. 3A). As controls, we determined that MAL2-11B at an effective anti-viral concentration failed to elicit a non-specific, cellular stress (note the lack of significant Hsp70 activation, Fig. 3A) and did not induce PARP cleavage (Fig. 3B), which is an established marker for the apoptotic pathway.
Fig. 3.
MAL2-11B reduces TAg levels in SV40-infected BSC40 cells. (A) Freshly confluent BSC40 cells were infected with SV40 for 2 h and subsequently treated with 100 µM MAL2-11B for 48 h. Control cells that were uninfected, but treated with DMSO or MAL2-11B, were also examined. The cells were lysed and an equal amount of total cellular protein was resolved on a polyacrylamide gel. After transfer to nitrocellulose the blots were decorated with the indicated antisera. (B) Cells were treated as described in (A), except that 1 µM staurospaurine was added for 4 h, where indicated, or the indicated concentrations of MAL2-11B were included in the medium for 48 h, as above. Lysates were resolved, transferred to nitrocellulose, and probed with anti-PARP or anti-actin antibody.
3.3. MAL2-11B inhibits SV40 DNA replication in vitro
Since maximal DNA replication requires both the J domain and ATPase domain of TAg (see above), we postulated that inhibition of SV40 replication in the presence of MAL2-11B was due to decreased viral DNA synthesis. To test this hypothesis, we performed in vitro DNA replication assays in the presence of each of the compounds examined in this report. Purified TAg and HeLa cell extract were combined with radiolabeled nucleotide and a plasmid containing an SV40 origin of replication. At the indicated times the reactions were halted. The formation of TAg-derived, high molecular weight DNA species was monitored after gel electrophoresis and phosphorimage analysis. As shown in Fig. 4A, wild type TAg catalyzed a time-dependent rise in the formation of DNA species whose length increased as the reaction proceeded. As previously published (Castellino et al., 1997), an equal amount of the purified 5061 ATPase-defective mutant was unable to form these products (Fig. 4A). Next, MAL2-11B, MAL3-101, and MAL3-51 were titrated into the in vitro DNA replication assay and the reactions were stopped after 60 min. Consistent with the data shown in Fig. 2, MAL3-51 and MAL3-101 had no significant effect on DNA replication at any concentration tested, whereas MAL2-11B inhibited replication in a concentration-dependent manner (Fig. 4B). In fact, at 100 µM, MAL2-11B inhibited DNA replication by ∼5-fold, which was similar to the reduced level of viral replication seen in the plaque assays (Fig. 2A). We also determined that MAL2-11B at a final concentration of 100 µM had no effect on the growth or viability of BSC40 cells, as assessed using an MTS assay (Cory et al., 1991; Minguez et al., 2003) (G. Zhu and B. Day, personal communication), suggesting that decreased viral replication was not due to secondary effects on host cell homeostasis (also see Fig. 3). Together, these results suggest that MAL2-11B directly reduces viral DNA synthesis, which translates directly to decreased viral yield.
Fig. 4.
MAL2-11B abrogates DNA replication in vitro. (A) High molecular weight DNA products increase overtime in the presence of wild type TAg. A total of 0.5 µM of the indicated TAg constructs were used in the in vitro DNA replication assays. DNA synthesis was determined after 30,60,90, and 120 min, and the products were resolved on an agarose gel. The laddering present at the bottom of the gel represents DNA topoisomers of monomeric DNA (Li et al., 1986). (B) MAL2-11B, but neither MAL3-51 nor MAL3-101, inhibits SV40 DNA replication in vitro. A total of 0.5 µM TAg was incubated, in triplicate, with the indicated concentrations of MAL2-11B, MAL3-51, or MAL3-101, or with an equal volume of DMSO and the total DNA synthesized was determined after 60 min.
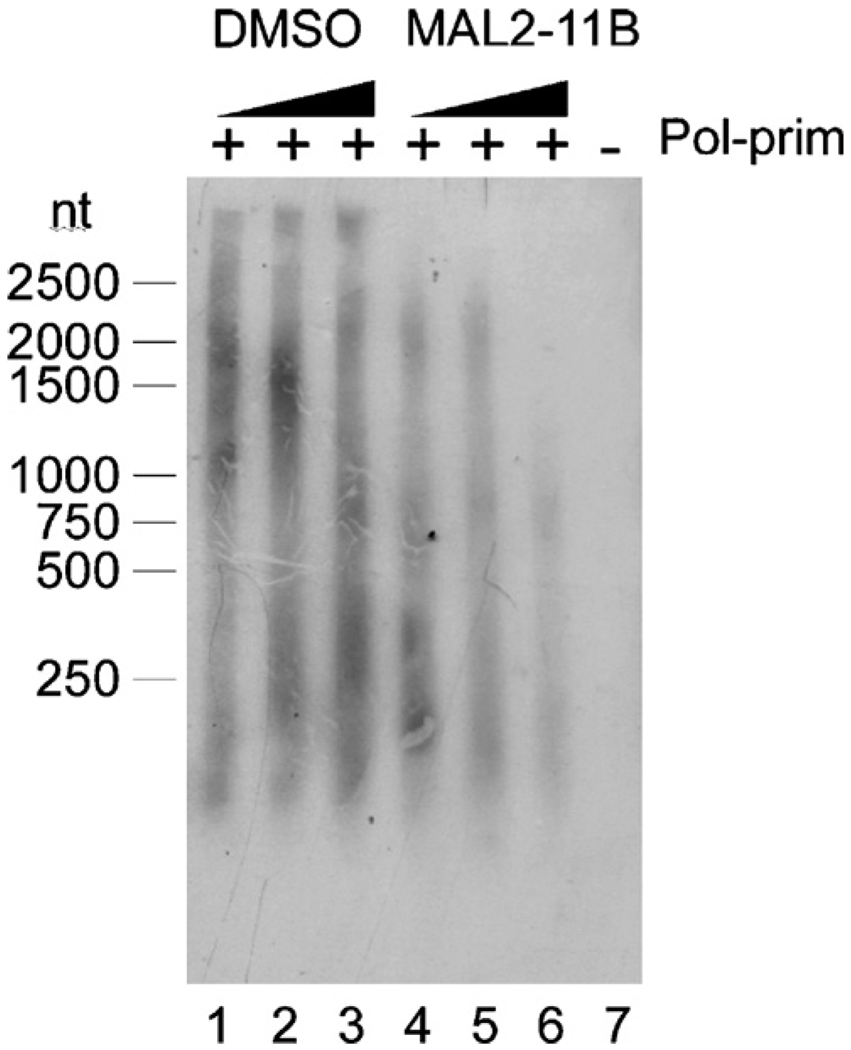
To better pin-point the step at which MAL2-11B inhibits SV40 replication, and to more definitively assess the specific action of this compound, we added MAL2-11B into a monopolymerase assay. This TAg-requiring reaction employs purified hRPA, Top1, and pol-prim in place of HeLa cell lysate. Thus, the impact of an agent on the initiation of DNA synthesis prior to the polymerase switch and elongation can be visualized (Matsumoto et al., 1990). As shown in Fig. 5, there was a concentration-dependent reduction in product upon MAL2-11B addition. In fact, by quantifying the amount of product formed we noted a 3.7-fold reduction in the presence of 100 µM MAL2-11B, which correlates well with the level of inhibition in the viral plaque assays and in the in vitro replication assays (Fig. 2A and Fig. 4B). Since neither hRPA, Top1, nor pol-prim is an ATPase, these results strongly suggest that MAL2-11B attenuates SV40 DNA synthesis by interacting specifically with TAg. Based on these collective data, we propose that MAL2-11B inhibits SV40 DNA synthesis and subsequent viral replication.
Fig. 5.
MAL2-11B inhibits the initiation of SV40 DNA synthesis. Increasing concentrations of MAL2-11B or an equal volume of DMSO were incubated with TAg, pol-prim, hRPA, and Top1, and the monopolymerase assay was performed as described in Section 2. The DNA products were resolved on an agarose gel. Lanes 1–3 contain increasing amounts of DMSO which correspond to the volume of MAL2-11B utilized in the reactions shown in lanes 4–6 (25 µM, 50 µM, and 100 µM, respectively). A control reaction performed in the absence of pol-prim is shown in lane 7, and size standards are indicated to the left of the figure.
The ATPase activity of TAg can be enhanced upon interacting with single-stranded DNA. Therefore, we assessed whether MAL2-11B blocks the ability of the oncoprotein to associate with DNA, which would explain the data presented in Fig. 4 and Fig. 5 and the anti-viral effect of the compound (Fig. 2). To this end, we first titrated MAL2-11B into steady-state ATPase assays in the presence or absence of M13 single-stranded DNA (Taneja et al., 2002) and found that DNA activated the hydrolysis of ATP regardless of whether MAL2-11B was present; however, the amount of ATP consumed decreased as higher concentrations of MAL2-11B were used, as expected (Fig. 6). Note that the profile of MAL2-11B-mediated inhibition is distinct from that in Fig. 2, in which single turnover ATPase assays were conducted. Nevertheless, these data suggest that the DNA-binding activity of TAg was unaffected by the compound. More generally, the data suggest that a DNA-dependent activation or conformational change of TAg was not significantly altered by MAL2-11B.
Fig. 6.
Effect of MAL2-11B on the DNA-dependent enhancement of TAg’s ATPase activity and binding to the SV40 origin of replication (ori). The ATPase activity of TAg in the absence (open bars) or presence (black bars) of single-strand DNA and in the presence of the indicated concentrations of MAL2-11B was assessed as described in Section 2. Note: In contrast to the experiments reported in Table 2, the compound was not preincubated in the presence of the TAg-DNA solution in these reactions. In this experiment and in parallel experiments, the SD was <10% of the means (data not shown).
3.4. Effect of MAL2-11B on BK virus replication
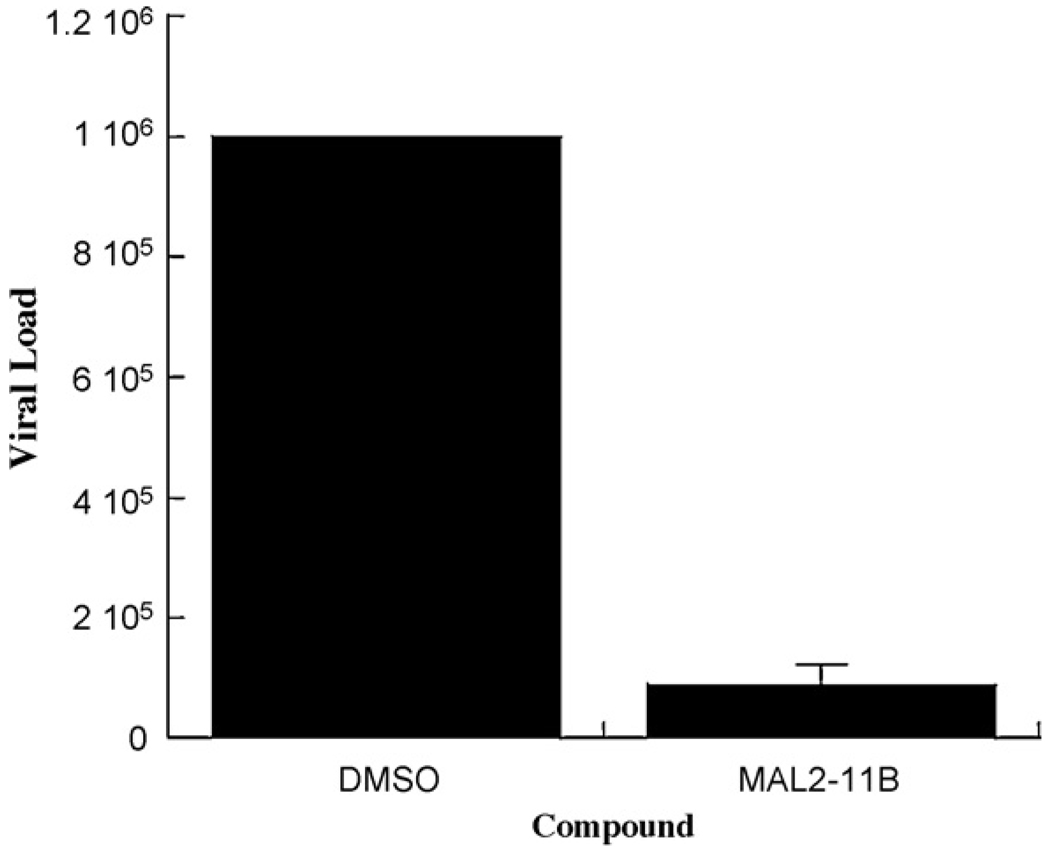
Finally, we wished to examine whether MAL2-11B inhibits the replication of a polyomavirus, BKV, which is known to cause disease. To this end, we employed an established protocol (Bista et al., 2007) in which the effects of anti-viral agents on BKV replication were previously examined (Farasati et al., 2005). We therefore incubated BKV-infected human kidney (HK2) cells with MAL2-11B at a final concentration of 15 µM, which had no apparent effect on cell viability. The cells were harvested, lysates were prepared, and a quantitative real-time PCR assay was performed to detect the viral load (see Section 2). As shown in Fig. 7, we found that the compound decreased the amount of viral DNA to ∼10% compared to the DMSO control.
Fig. 7.
MAL2-11B decreases the viral load in BKV-infected human kidney cells. Data shown represent the means of 3 independent determinations, ±SD. Results from a quantitative real-time PCR assay are shown (see Section 2). MAL2-11B was used at a final concentration of 15 µM and infected HK2 cells were treated with the compound for 5 d. Under these conditions, we noted that there was no effect on cell viability, as assessed by light microscopy (data not shown).
4. Discussion
We have identified a chemical probe, MAL2-11B, that targets the activities of the TAg J domain and ATPase domain and discovered that this agent inhibits the ability of TAg to initiate DNA synthesis and catalyze DNA replication in vitro. MAL2-11B also reduces the level of viral DNA and TAg in infected cells, and the anti-viral effect was confirmed by a comparison between MAL2-11B and an established inhibitor of SV40 replication, cidofovir (Andrei et al., 1997). MAL2-11B exerted similar effects on each of these diverse phenomena when an identical concentration of compound was employed (100 µM), suggesting specificity-of-action. Another compound, MAL3-101, which only inhibits the TAg-mediated stimulation of Hsp70 ATPase activity (Fewell et al., 2004) had no effect on TAg’s ability to hydrolyze ATP and correspondingly no impact on SV40 DNA replication. Similarly, MAL3-51 was without effect on SV40 replication in cultured cells or on viral DNA synthesis in vitro, which was expected based on its inability to significantly alter any biochemical activity examined. These data indicate that only select dihydropyrimidines exhibit anti-viral activity, and sets the stage for the development of compounds with greater potency. It is also important to re-emphasize that the effective concentration of MAL2-11B failed to elicit secondary effects (cell stress, apoptosis, mitochondrial defects) in treated cells. Accordingly, we conclude that the inhibition of SV40 replication by MAL2-11B arises from its multi-pronged modulation of TAg function.
Of additional interest, we noted that MAL2-11B inhibited the endogenous ATPase activity of mammalian Hsp70 by ∼40% when used at a final concentration of 100 µM (Table 2). Therefore, it is formally possible that the effect of the compound on SV40 replication (as assessed via the plaque assay; Fig. 2A) might have arisen in-part from modest inhibition of this chaperone. This concept is supported by the known requirement for Hsp70 on TAg-triggered processes, including DNA synthesis in vivo (Sullivan and Pipas, 2002). Hsp70 and Hsp40 chaperones have also been shown to play important roles in the replication of a number of other viruses that infect higher eukaryotes (Beck and Nassal, 2003; Glotzer et al., 2000; Lin et al, 2002; Mayer, 2005; Sock et al., 1999; Tanguy Le Gac and Boehmer, 2002). Interestingly, however, another Hsp70-modulator, SGC, has a potent effect on mammalian Hsp70 ATPase activity (Whetstone and Lingwood, 2003), yet failed to alter viral yield in the plaque assay utilized in Fig. 2 (data not shown).
To directly address whether the effect on SV40 DNA replication arose from Hsp70 inhibition, and to better define the specificity-of-action of MAL2-11B, we performed the monopolymerase assay, which utilizes purified hRPA, Top1, pol-prim and TAg and that lacks Hsp70. As noted above, the relative inhibition exhibited by the compound in this assay (Fig. 5) agreed well with the effect of MAL2-11B on each of the other diverse assays performed. Thus, while we cannot exclude the possibility that inhibition of Hsp70 contributes to the effect of MAL2-11B on SV40 propagation in cell culture, we are confident that Hsp70 inhibition is not the cause of MAL2-11B’s inhibition of SV40 DNA synthesis.
At present, we do not know how MAL2-11B exhibits its pleiotropic effects on diverse enzymes. MAL2-11B fails to alter the endogenous ATPase activities of either the yeast or mammalian Hsp70 in the single turnover ATPase assay (Fig. 1B, and our unpublished data) but can reduce the overall ATPase activity of an Hsp70, as was most apparent for the mammalian enzyme (Table 2). Therefore, MAL2-11B may impinge upon another aspect of the ATPase cycle, such as nucleotide release. To date, no small molecules that affect Hsp70 nucleotide release have been identified. At higher concentrations, MAL2-11B also affected other chaperones, suggesting that there might be unique affinities for MAL2-11B amongst various chaperones. For example, another Hsp70 modulator, 15-deoxyspergualin (DSG), was later found to interact with both Hsp70 and Hsp90 (Nadeau et al., 1994).
Several potential mechanisms of action of MAL2-11B on TAg can be proposed. First, MAL2-11B might completely unfold the TAg monomer, thus preventing the function of each TAg-embedded activity. This hypothesis is unsatisfying because the J domain and ATPase domain fold independently and are tethered to the origin-binding domain by a flexible linker. In addition, MAL2-11B did not inhibit the DNA-mediated activation of TAg’s ATPase activity (Fig. 6). Second, MAL2-11B might bind at two locations in TAg; thus, inhibition of J domain function and ATPase activity are distinct phenomena. However, the structures of these regions are distinct and it is unlikely that a single molecule could inhibit both activities. Third, MAL2-11B might inhibit a general TAg-intrinsic property that is required for both J domain and ATPase domain function. Some possibilities for this property include a MAL2-11B-mediated alteration of TAg’s ATP binding kinetics, oligomeric state, and/or proper domain orientation. In favor of this hypothesis, we note that the J domain and the ATPase domain are both implicated in the proper formation and activation of TAg oligomers (Valle et al., 2006; Castellino et al., 1997; Weisshart et al., 1996).
A comparison of the cellular and in vitro DNA replication assays indicates that the most likely mechanism underlying SV40 growth inhibition is reduced DNA replication. However, three other possibilities exist. First, inhibition of SV40 replication could be independent of TAg but dependent on another, unknown, protein. This explanation is in conflict with the fact that MAL2-11B inhibited initiation of DNA synthesis in the monopolymerase assay, which contains only four proteins (RPA, Top1, pol-prim, and TAg). Second, MAL2-11B might kill BSC40 cells, leading to reduced viral replication, but as noted in Section 3 BSC40 viability and physiology are unaffected by MAL2-11B. Third, SV40 replication could be inhibited at a stage in the SV40 life cycle that occurs before DNA replication, such as virion disassembly or cell cycle initiation. Future work is needed to address this hypothesis.
In summary, we have identified two classes of TAg modulators. The first class is represented by MAL3-101, which inhibits J-protein stimulation of Hsp70 in single turnover ATPase assays (Wright et al., 2008) but does not inhibit SV40 DNA replication or SV40 propagation. The second class is represented by MAL2-11B, which inhibits not only the J-protein stimulation of Hsp70 but also inhibits various ATPases, including TAg (a third class is defined by MAL3-51, which has no effect in these assays and underscores the biological selectivity of the dihydropyrimidine core). It is important to emphasize that MAL2-11B is not a “drug” but at this point represents a chemical probe. In fact, the relatively high concentration needed to see effects (100 µM) suggests that the compound alters protein–protein interfaces. By definition, these interfaces encompass large surface areas; thus, high concentrations of MAL2-11B, like other small molecule probes, are necessary in complex systems to act as an inhibitor. Nevertheless, given the effect on SV40 replication in cell culture, additional efforts are justified to better elucidate the anti-viral properties of dihydropyrimidines and improve the membrane permeability of these molecules.
Supplementary Material
Acknowledgments
The authors would like to thank Raj Chovatiya, Fengfeng Xu, and Guangyu Zhu for sharing unpublished data. This work was supported by grant MH84077 from the NIH and a Partnership for Cures and Goldman Philanthropic Partnerships grant to J.L.B. C.M.W. acknowledges the support of a Renal Epithelial Biology training grant from the National Institutes of Health (DK61296) and a Mellon Graduate Fellowship. E.F. acknowledges NIH grant GM52948, A.V. acknowledges NIH grant AI060602, and P.W. acknowledges grant P50-GM067082 from the NIH-NIGMS for support of the P50 Chemical Methodologies and Library Development Program at the University of Pittsburgh. This study was also supported by NIH grant DK79307, which funds the Pittsburgh Center for Kidney Research.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2008.12.018.
References
- Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24(52):7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Ali SH, Chandraker A, DeCaprio JA. Inhibition of Simian virus 40 large T antigen helicase activity by fluoroquinolones. Antivir. Ther. 2007;12(1):1–6. [PubMed] [Google Scholar]
- Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 2001;11(1):15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J. Virol. 2007;81(8):4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei G, Snoeck R, Vandeputte M, De Clercq E. Activities of various compounds against murine and primate polyomaviruses. Antimicrob. Agents Chemother. 1997;41(3):587–593. doi: 10.1128/aac.41.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E, Chazin WJ. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005;12(4):332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbanti-Brodano G, Martini F, De Mattei M, Lazzarin L, Corallini A, Tognon M. BK and JC human polyomaviruses and simian virus 40: natural history of infection in humans, experimental oncogenicity, and association with human tumors. Adv. Virus Res. 1998;50:69–99. doi: 10.1016/s0065-3527(08)60806-4. [DOI] [PubMed] [Google Scholar]
- Beck J, Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 2003;278(38):36128–36138. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996;16(8):4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bista BR, Ishwad C, Wadowsky RM, Manna P, Randhawa PS, Gupta G, Adhikari M, Tyagi R, Gasper G, Vats A. Development of a loop-mediated isothermal amplification assay for rapid detection of BK virus. J. Clin. Microbiol. 2007;45(5):1581–1587. doi: 10.1128/JCM.01024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec JA, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Pipas JM. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol. 1998;72(7):5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Mullane KP, Aksoy IA, Stubdal H, Zalvide J, Pipas JM, Silver PA, Roberts TM, Schaffhausen BS, DeCaprio JA. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11(9):1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 2005;79(20):13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P, Saenz-Robles MT, Pipas JM. Expression of SV40 large T antigen in baculovirus systems and purification by immunoaffinity chromatography. Methods Enzymol. 1999;306:297–307. doi: 10.1016/s0076-6879(99)06019-x. [DOI] [PubMed] [Google Scholar]
- Carbone M, Rudzinski J, Bocchetta M. High throughput testing of the SV40 Large T antigen binding to cellular p53 identifies putative drugs for the treatment of SV40-related cancers. Virology. 2003;315(2):409–414. doi: 10.1016/s0042-6822(03)00547-6. [DOI] [PubMed] [Google Scholar]
- Castellino AM, Cantalupo P, Marks IM, Vartikar JV, Peden KW, Pipas JM. trans-Dominant and non-trans-dominant mutant simian virus 40 large T antigens show distinct responses to ATP. J. Virol. 1997;71(10):7549–7559. doi: 10.1128/jvi.71.10.7549-7559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy LR, Oltman A, Estes PA, Garcea RL. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J. Virol. 2006;80(10):5086–5091. doi: 10.1128/JVI.80.10.5086-5091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R, Peden K, Pipas JM, Nathans D, Tjian R. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol. Cell. Biol. 1983;3(2):220–228. doi: 10.1128/mcb.3.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CN, Tornow J, Clark R, Tjian R. Properties of the simian virus 40 (SV40) large T antigens encoded by SV40 mutants with deletions in gene A. J. Virol. 1986;57(2):539–546. doi: 10.1128/jvi.57.2.539-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BS, Pipas JM. T antigens encoded by replication-defective simian virus 40 mutants dl1135 and 5080. J. Biol. Chem. 1995;270(25):15377–15384. doi: 10.1074/jbc.270.25.15377. [DOI] [PubMed] [Google Scholar]
- Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3(7):207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- Cripe TP, Delos SE, Estes PA, Garcea RL. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J. Virol. 1995;69(12):7807–7813. doi: 10.1128/jvi.69.12.7807-7813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 1992;267(29):20927–20931. [PubMed] [Google Scholar]
- De Clercq E, Holy A. Case History: Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat. Rev. Drug Discov. 2005;4(11):928–940. doi: 10.1038/nrd1877. [DOI] [PubMed] [Google Scholar]
- Eash S, Manley K, Gasparovic M, Querbes W, Atwood WJ. The human polyomaviruses. Cell. Mol. Life Sci. 2006;63(7–8):865–876. doi: 10.1007/s00018-005-5454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farasati NA, Shapiro R, Vats A, Randhawa P. Effect of leflunomide and cidofovir on replication of BK virus in an in vitro culture system. Transplantation. 2005;79(1):116–118. doi: 10.1097/01.tp.0000149338.97084.5f. [DOI] [PubMed] [Google Scholar]
- Farber JM, Peden KW, Nathans D. trans-Dominant defective mutants of simian virus 40 T antigen. J. Virol. 1987;61(2):436–445. doi: 10.1128/jvi.61.2.436-445.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrick JL, Hallick LM. Optimal conditions for titration of SV40 by the plaque assay method. J. Virol. Methods. 1983;7(2):93–102. doi: 10.1016/0166-0934(83)90095-2. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell SW, Pipas JM, Brodsky JL. Mutagenesis of a functional chimeric gene in yeast identifies mutations in the simian virus 40 large T antigen J domain. Proc. Natl. Acad. Sci. USA. 2002;99(4):2002–2007. doi: 10.1073/pnas.042670999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day BW, Brodsky JL. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 2004;279(49):51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA. 1998;95(26):15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3(5):595–604. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer JB, Saltik M, Chiocca S, Michou AI, Moseley P, Cotten M. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature. 2000;407(6801):207–211. doi: 10.1038/35025102. [DOI] [PubMed] [Google Scholar]
- Gluzman Y, Ahrens B. SV40 early mutants that are defective for viral DNA synthesis but competent for transformation of cultured rat and simian cells. Virology. 1982;123(1):78–92. doi: 10.1016/0042-6822(82)90296-3. [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1–151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell. 2002;13(8):2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczynska E, Turkiewicz D, Rybka K, Toporski J, Kalwak K, Dyla A, Szczyra Z, Chybicka A. Incidence, clinical outcome, and management of virus-induced hemorrhagic cystitis in children and adolescents after allogeneic hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 2005;11(10):797–804. doi: 10.1016/j.bbmt.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 1981;39(3):861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci. 2005;14(7):1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymes LC, Warshaw BL. Polyomavirus (BK) in pediatric renal transplants: evaluation of viremic patients with and without BK associated nephritis. Pediatr. Transplant. 2006;10(8):920–922. doi: 10.1111/j.1399-3046.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- Jiang X, Klimovich V, Arunkumar AI, Hysinger EB, Wang Y, Ott RD, Guler GD, Weiner B, Chazin WJ, Fanning E. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 2006;25(23):5516–5526. doi: 10.1038/sj.emboj.7601432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 2001;20(1–2):295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423(6939):512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- Li JJ, Peden KW, Dixon RA, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol. Cell. Biol. 1986;6(4):1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BY, Makhov AM, Griffith JD, Broker TR, Chow LT. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell. Biol. 2002;22(18):6592–6604. doi: 10.1128/MCB.22.18.6592-6604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Stucki M, Spadari S, Hubscher U. DNA polymerase switching: I. Replication factor C displaces DNA polymerase alpha prior to PCNA loading. J. Mol. Biol. 2000;295(4):791–801. doi: 10.1006/jmbi.1999.3394. [DOI] [PubMed] [Google Scholar]
- Mastrangelo IA, Hough PV, Wall JS, Dodson M, Dean FB, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338(6217):658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc. Natl. Acad. Sci. USA. 1990;87(24):9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Brodsky JL. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics. 2000;156(2):501–512. doi: 10.1093/genetics/156.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguez JM, Kim SY, Giuliano KA, Balachandran R, Madiraju C, Day BW, Curran DP. Synthesis and biological assessment of simplified analogues of the potent microtubule stabilizer (+)-discodermolide. Bioorg. Med. Chem. 2003;11(15):3335–3357. doi: 10.1016/s0968-0896(03)00186-x. [DOI] [PubMed] [Google Scholar]
- Mossi R, Keller RC, Ferrari E, Hubscher U. DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J. Mol. Biol. 2000;295(4):803–814. doi: 10.1006/jmbi.1999.3395. [DOI] [PubMed] [Google Scholar]
- Nadeau K, Nadler SG, Saulnier M, Tepper MA, Walsh CT. Quantitation of the interaction of the immunosuppressant deoxyspergualin and analogs with Hsc70 and Hsp90. Biochemistry. 1994;33(9):2561–2567. doi: 10.1021/bi00175a027. [DOI] [PubMed] [Google Scholar]
- Nylandsted J, Rohde M, Brand K, Bastholm L, Elling F, Jaattela M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. USA. 2000;97(14):7871–7876. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden KW, Pipas JM. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes. 1992;6(2):107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin. Cancer Biol. 2001;11(1):23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- Portolani M, Pietrosemoli P, Cermelli C, Mannini-Palenzona A, Grossi MP, Paolini L, Barbanti-Brodano G. Suppression of BK virus replication and cytopathic effect by inhibitors of prokaryotic DNA gyrase. Antivir. Res. 1988;9(3):205–218. doi: 10.1016/0166-3542(88)90004-6. [DOI] [PubMed] [Google Scholar]
- Poulin DL, DeCaprio JA. Is there a role for SV40 in human cancer? J. Clin. Oncol. 2006;24(26):4356–4365. doi: 10.1200/JCO.2005.03.7101. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339(1):62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, Wu N, Felts S, Wipf P, Massague J, Jiang X, Brodsky JL, Krystal GW, Chiosis G. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol. 2007;3(8):498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- Safak M, Khalili K. An overview: human polyomavirus JC virus and its associated disorders. J. Neurovirol. 2003;9:3–9. doi: 10.1080/13550280390195360. [DOI] [PubMed] [Google Scholar]
- Sawai ET, Butel JS. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J. Virol. 1989;63(9):3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai ET, Rasmussen G, Butel JS. Construction of SV40 deletion mutants and delimitation of the binding domain for heat shock protein to the amino terminus of large T-antigen. Virus Res. 1994;31(3):367–378. doi: 10.1016/0168-1702(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Smelkova NV, Borowiec JA. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 1997;71(11):8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Enderich J, Wegner M. The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol. Cell. Biol. 1999;19(4):2455–2464. doi: 10.1128/mcb.19.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 1997;17(8):4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA. 1998;95(26):15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Cantalupo P, Pipas JM. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 2000;20(17):6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Pipas JM. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 2002;66(2):179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja P, Gu J, Peng R, Carrick R, Uchiumi F, Ott RD, Gustafson E, Podust VN, Fanning E. A dominant-negative mutant of human DNA helicase B blocks the onset of chromosome DNA replication. J. Biol. Chem. 2002;277(43):40853–40861. doi: 10.1074/jbc.M208067200. [DOI] [PubMed] [Google Scholar]
- Tanguy Le Gac N, Boehmer PE. Activation of the herpes simplex virus type-1 origin-binding protein (UL9) by heat shock proteins. J. Biol. Chem. 2002;277(7):5660–5666. doi: 10.1074/jbc.M108316200. [DOI] [PubMed] [Google Scholar]
- Tremblay JD, Sachsenmeier KF, Pipas JM. Propagation of wild-type and mutant SV40. Methods Mol. Biol. 2001;165:1–7. doi: 10.1385/1-59259-117-5:1. [DOI] [PubMed] [Google Scholar]
- Valle M, Chen X, Donate LE, Fanning E, Carazo JM. Structural basis for the cooperative assembly of large T antigen on the origin of replication. J. Mol. Biol. 2006;357:1295–1305. doi: 10.1016/j.jmb.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Vats A, Randhawa PS, Shapiro R. Diagnosis and treatment of BK virus-associated transplant nephropathy. Adv. Exp. Med. Biol. 2006;577:213–227. doi: 10.1007/0-387-32957-9_16. [DOI] [PubMed] [Google Scholar]
- Weisshart K, Bradley MK, Weiner BM, Schneider C, Moarefi I, Fanning E, Arthur AK. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase alpha and replicate SV40 DNA in vitro. J. Virol. 1996;70(6):3509–3516. doi: 10.1128/jvi.70.6.3509-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstone H, Lingwood C. 3′Sulfogalactolipid binding specifically inhibits Hsp70 ATPase activity in vitro. Biochemistry. 2003;42(6):1611–1617. doi: 10.1021/bi026735t. [DOI] [PubMed] [Google Scholar]
- Wright C, Chovatiya R, Jameson N, Turner D, Zhu G, Werner S, Huryn D, Pipas J, Day B, Wipf P, Brodsky J. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg. Med. Chem. 2008;16:3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell. 2004;15(11):4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18(21):6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K, Bui T, Frisque RJ, Grandinetti A, Nerurkar VR. A rapid in vitro polyomavirus DNA replication assay. J. Virol. Methods. 2004;122(1):123–127. doi: 10.1016/j.jviromet.2004.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.