Abstract
Nanotechnology is finding its use as a potential technology in consumer products, defense, electronics, and medical applications by exploiting the properties of nanomaterials. Single-walled carbon nanotubes are novel forms of these nanomaterials with potential for large applications. However, the toxicity studies on this material are not explored in detail and therefore limiting its use. It has been earlier reported that single-walled carbon nanotubes induces oxidative stress and also dictates activation of specific signaling pathway in keratinocytes. The present study explores the effect of single-walled carbon nanotubes on stress genes in human BJ Foreskin cells. The results show induction of oxidative stress in BJ Foreskin cells by single-walled carbon nanotubes and increase in stress responsive genes. The genes included inducible genes like HMOX1, HMOX2, and Cyp1B1. In addition we validated increase for four genes by SWCNT, namely ATM, CCNC, DNAJB4, and GADD45A by RT-PCR. Moreover results of the altered stress related genes have been discussed and that partially explains some of the toxic responses induced by single-walled carbon nanotubes.
Keywords: BJ Foreskin Cells, Single-Walled Carbon Nanotube, Oxidative Stress, Gene Array, Anioxidants
1. INTRODUCTION
Nanotechnology is a rapidly evolving and expanding discipline with potential for large applications.1 Carbon nanotubes are one of such nanomaterial, which is attracting intense research efforts because of their unique properties that makes them suitable for many industrial uses.2 The nanomaterials has potential applications as strong, lightweight materials in the aerospace, electronics, biomedical, defense industries, and are now manufactured in bulk.2–3 The use of nanomaterials will be immense in a variety of applications and thereby creating more horizons for exposure. While the use of nanotechnology in large number of applications is being discussed but sufficient toxicological information that would allow drawing conclusive draft for an effective standard of exposure is still at its genesis.
Single-walled carbon nanotube (SWCNT) is a nanomaterial and toxicological studies have been reported both in animal and cell culture models.4–6 In rat model investigating acute lung toxicity of SWCNT has reported formation of multiple granulomas, however this observation failed to co-relate with toxicity profiling of the lavage fluid, cell proliferation, dose response relationship, and nonuniform distribution of lesions.7 In a more recent study SWCNT elicited acute inflammation coupled with early onset of progressive fibrosis and granulomas in lung of C57BL/6 mice further emphasizes the toxic potential of this material.5 Studies in human keratinocyte cells have largely indicated formation of free radicals and induction of oxidative stress by SWCNT in these cells.6,8 Oxidative stresses induced by SWCNT in keratinocytes were reflected by accumulation of peroxidative products, followed by antioxidant depletion and resulting in a loss of cell viability.6 In addition oxidative stress induced activation of nuclear transcription factor κB (NF-κB) and related signaling has been reported HaCaT cells.8 In contrast to these findings reports have shown no skin tumor formation by topical doses of fullerenes on mouse skin.9 Even though results that contradict the toxicity of nanomaterials, it still suggests unique health hazards due to its property and as shown in cell culture models warrants further evaluation.10
The reports as of today do provide evidence that shows SWCNT can induce stress in cells and also in tissues.4–8 Therefore in the present study we have evaluated the signaling plethora of stress genes induced by SWCNT in human BJ Foreskin cells using gene array imprinted on a nylon membrane. Our study shows a profile of genes activated by the stress induced by SWCNT in BJ Foreskin cells and urges further study to dissect the molecular events associated with these changes.
2. EXPERIMENTAL DETAILS
SWCNT (Catalogue Number 652512-G) was purchased from Sigma, St. Louis. In all the studies, SWCNT particles were dissolved in Dimethylformamide (DMF) and therefore in all control experiments cells were treated with equivalent volume of DMF. Human BJ Foreskin cells (CRL-2522) were purchased from American Type Culture Collection (Manassas, VA) Cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin.
Oxidative stress measurement in BJ Fore skin cells
To study the induction of oxidative stress in BJ Foreskin cells 1 × 105 cells were seeded in 96 well plate and grown overnight under standard culture conditions. The cells were then treated with 10 μM of dichlorofluorescin (5-(and-6)-carboxy-2, 7′-dichloro-dihydroxyfluorescein diacetate, H2DCFDA, C-400, Molecular Probes, Eugene, OR) for three hours in Hank's balanced salt solution (HBSS). Following three hours of incubation cells were washed with HBSS and SWCNT was added in presence of 10% FBS in DMEM medium. For studying effect of antioxidants in BJ Foreskin cells: cells were co-incubated with antioxidants in presence or absence of SWCNT (6 μg/ml). Cells were incubated in incubator for additional hours as detailed in the figure legend and fluorescence was measured at excitation wavelength of 485 nm and emission at 520 nm (Thermo Lab Systems, Franklin, MA). The values were normalized to the protein content that was determined for each well using BioRad Reagent (Catalog Number 500-0006, BioRad, Hercules, CA).
Gene Array
In an effort to investigate the changes in the stress associated genes we screened for stress genes using Stress and Toxicity Array (Super Array, Frederick, MD 21704). BJ Foreskin cells were treated with 6 μg/ml of SWCNT for 24 h with parallel control cells treated with DMF only. After 24 h of treatment cells were washed with chilled phosphate buffered saline (PBS) twice and cells were scrapped and immediately frozen. The screening of the Stress Toxicity Array (HS-012.2) was done by Super array (Frederick, MD 21704) and results were obtained after thorough analysis. In brief total RNA was extracted using Trizol reagent (Catalog Number 15596-018, Invitrogen, CA) according to the described procedure of the manufacturer. 5 μg of total RNA was used to synthesize c-DNA and was later used for labeling using AmpoLabelling-LPR kit according to the manufacturer's instruction (Super Array, Frederick, MD 21704). The biotinylated c-DNA was probed to nylon membrane and the hybridized copies of c-DNA were detected by incubating the membranes with streptavidin coupled to alkaline phosphatase followed by incubating with the substrate solution to yield chemiluminescence product. The membranes were exposed to X-ray film and the image was captured.
RT-PCR Analysis
The gene expression changes were validated by RT-PCR and the procedure was adopted as described by Sarkar et al., 2002.31 Cells were treated essentially as described for stress array and at the end of 24 h cells were scrapped and Trizol reagent was used to extract total RNA. For c-DNA synthesis 5 μg of total RNA from both control and SWCNT treated cells were reverse transcribed by reverse transcriptase in presence of a oligo dT primer using a c-DNA synthesis kit from Super Array (Catalog number C-01; Fedrick, MD). The first strand c-DNA synthesis reaction was carried out at 37 °C for 60 min and the reaction was terminated by heating the mixture for 2 min at 95 °C. The PCR reaction was done using specific primers purchased from Super Array and the amplification was carried out essentially as per the conditions described by the manufacturer. In brief the PCR mixture was denatured at 95 °C for 3 min followed by 30 cycles of annealing at 55 °C for 30 sec and extended at 72 °C for 30 sec the reaction was given additional extension of 3 min at 72 °C before terminating the PCR at 4 °C. The PCR product was mixed with loading dye and resolved in 1% agarose gel and the bands were stained with ethidium bromide to be visualized under UV light equipped with a camera. The intensity of the bands was then analyzed by Quantity One software from BioRad (Hercules, CA).
Gene expressions for three genes were performed to observe the kinetics of induction in BJ Foreskin cells upon treatment with SWCNT particles. Cells were seeded at a density of 1 × 108 cells/100 mm dish and allowed to grow. Cells were then deprived of serum and grown overnight. Following these cells were treated with SWCNT (6 μg/ml) and then collected by scrapping at 2, 4, 8, 12, and 24 h after incubation. The collected cells were washed with chilled phosphate buffer saline twice and collected by centrifugation at 4 °C. Hereafter the cell pellet was stored at −80 °C for RNA isolation. RNA was isolated and RT-PCR was performed as described earlier.31
3. RESULTS AND DISCUSSION
SWCNT Induces Oxidative Stress in BJ-Foreskin cells
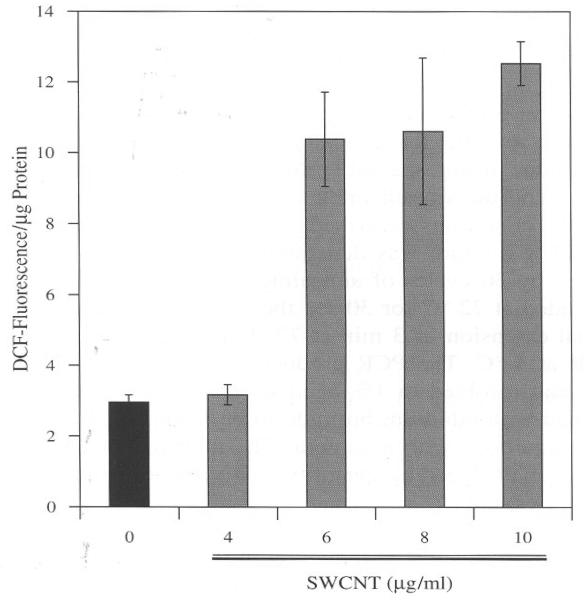
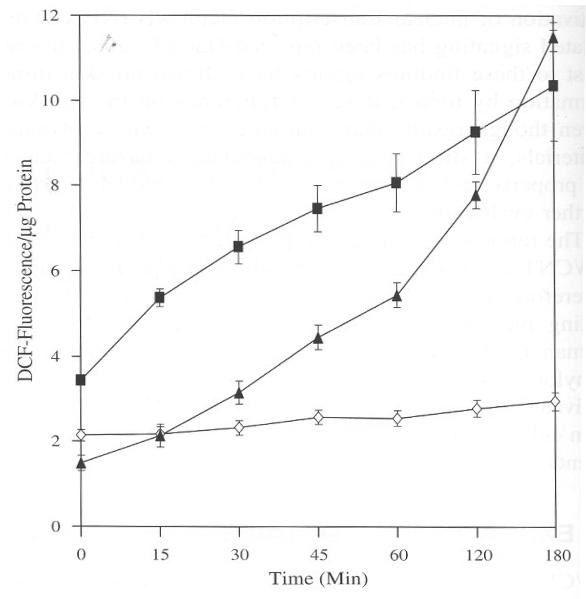
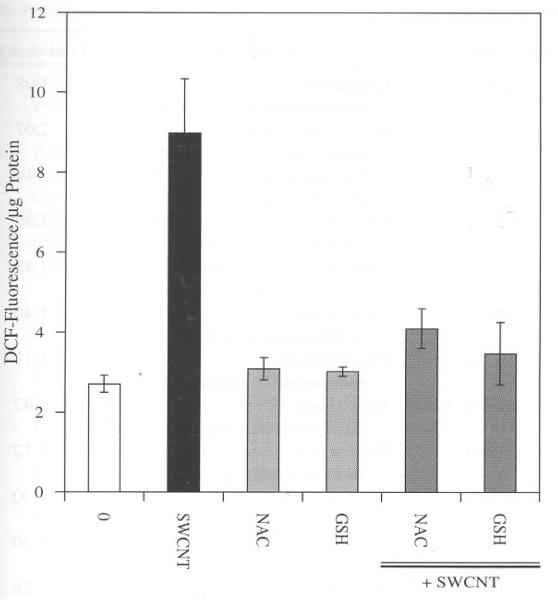
BJ Fore skin cells treated with SWCNT evoked oxidative stress as determined by increase in DCF fluorescence. The results from the assay clearly show the induction of reactive oxygen species (ROS) by SWCNT at concentrations 6, 8, and 10 μg/ml after three hours of incubation as compared to control (Fig. 1). This observation leads to the question whether the induction of ROS follows time kinetics and is required to unveil the response to a short period of time. Therefore for kinetics studies a dose of 6 μg/ml was selected, which was sufficient to observe changes in ROS in our experimental conditions (Fig. 1). The experimental set up was similar as mentioned for Figure 1 except changes in fluorescence was measured at different time points. We also included H2O2 (100 μM) as a positive control to compare the rate kinetics with a known oxidant. As shown in Figure 2 there was a steady rise in ROS across a time span of three hours induced by SWCNT particles. In the same assay H2O2 also showed a steady increase in ROS indicating the validity of the assay. Under similar conditions control cells incubated with the vehicle alone did not show any significant change in fluorescence. These results show that SWCNT induces ROS in a time dependent manner and these particles need to enter into the cells by crossing the membrane and that expends time. The sharp increase in ROS formation after 1 h, implies that the uptake into the cells was sufficient to elicit the observed response. It has been reported that protein and DNA conjugate of SWCNT particles take up to three hours to penetrate inside the cells through receptor mediated endocytosis.11 According to this finding it is conceivable to observe a significant increase in ROS after three hours of incubation considering the fact that this study used pure SWCNT particles. The oxidative stress reported in this study is a real time analysis of ROS generation in live cells. This in part therefore validates and supports similar observations obtained from different experimental setup and models regarding ROS induction by SWCNT.6,8 Further we treated with 1 mM of N-Acetyl Cysteine (NAC) and glutathione (GSH) to counteract the oxidative stress generated by SWCNT in BJ Foreskin cells. Both NAC and GSH decreased the oxidative stress induced by SWCNT after 3 h of incubation (Fig. 3). The decrease in ROS induced by SWCNT was approximately 2, 2.5 fold for NAC and GSH, respectively. These results suggest that the failure of the antioxidants may be the cause of SWCNT induced ROS in BJ Foreskin cells.
Fig. 1.
SWCNT induces ROS in BJ Foreskin Cells. 10,000 cells/well was seeded in a 96 well plate and incubated for 18 h. Cells were then washed with Hank's Balanced Salt Solution (HBSS) followed by incubation in CO2 incubator with 10 μM DCF for 3 h in HBSS. The cells were then washed with HBSS and incubated with different concentrations of SWCNT and 100 μM H2O2 in DMEN containing 10% FBS. Fluorescence was read at the end of 3 h. The fluorescence values were normalized with the protein content in each well. Values are mean±SD of 8 wells and is a representative from three experiments performed independently.
Fig. 2.
Time dependent induction of ROS by SWCNT (6 μg/ml) in BJ Foreskin cells. Cells were incubated in CO2 incubator and treated as described in Figure 1. Fluorescence was measured after each time interval and the values were normalized with the protein content in each well. Values are mean±SD of 8 wells and is a representative from three experiments performed independently. (◇) DMF; (▲) SWCNT(■); H2O2.
Fig. 3.
NAC and GSH counteracts SWCNT induced ROS in BJ Foreskin cells. Cells were incubated in CO2 incubator and in presence of SWCNT (6 μg/ml), NAC (1 mM), and GSH (1 mM) for three hours. NAC and GSH alone were also treated for same interval of time to observe its effect on the cells. All treatments were performed as detailed in Figure 1. Values are mean±SD of 8 wells and is a representative from three experiments performed independently.
SWCNT alters stress gene expression in BJ Foreskin Cells
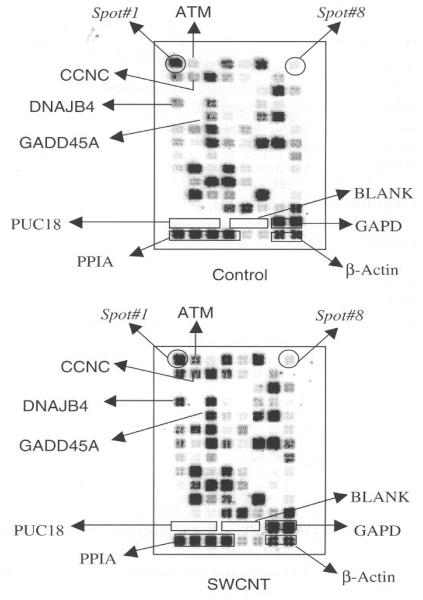
Oxidative stress can affect multiple signaling pathways, which can be transcriptional activation to inactivation of genes, phosphorylation, and dephosphorylation of proteins. 12–13 As indicated in Figure 1 and 2 SWCNT induced significant increase in ROS in BJ Foreskin cells, thereby allows to speculate, considerable changes in stress response genes. A representative array showing the alterations in gene expressions is shown in Figure 4. The complete profile of the changes in all the 96 genes is shown in Table I. The spot intensities were normalized to beta-actin and the ratio of the changes has been calculated against the respective genes. 28 genes showed significant change in SWCNT treated cells as compared to control. Surprisingly none of the genes showed a significant decrease in SWCNT treated cells as compared to control. Such observations indicate that SWCNT induced a battery of stress response genes involved in signaling. The spectrum of the differential gene expression is summarized in Table I, and shows increase in number of stress genes in response to SWCNT treatment. 28 genes showed significant increase with a ratio ranging from 1.5 to 3 and this was represented from the family of genes involved in apoptosis, xenobiotic metabolism, DNA repair genes, oxidative stress, and chemokines. NFκB increased by 1.5 fold and this represents a positive control for the array because we have earlier shown this transcription factor to be activated by SWCNT in HaCaT cells.8 55 genes showed a ratio ranging from 1 to 1.4 while only 13 genes were less than a ratio of 1 but not significantly decreased as compared to the control (Table I). Therefore the global trend in the gene expressions was an increased response to the stress induced by the SWCNT particles. The gene that responded with highest ratio was HMOX2 (3.14) followed by HMOX1 with a ratio of 2.07. HMOX-1 is an inducible gene that increased by 2.07 fold in cells treated with SWCNT and this indicates that inducible genes are possibly activated under the stress of this material.15–16 Heme oxygenases are microsomal enzymes that catalyze the oxidative cleavage of the porphyrin ring to generate biliverdin, free heme iron, and carbon monoxide (CO).16 Evidences from studies have shown that the catalytic end products contributes to the part of physiological functions of HMOX1 such as antioxidative, anti-inflammatory, anti-proliferative, and anti-apoptotic effects.15,17 HMOX1 is inducible isoform, is evolutionary conserved and ubiquitously distributed in tissues.14 HMOX1 expression is triggered by stress stimuli, ultraviolet radiation, hypoxia, and inflammation.18–20 SWCNT induced oxidative stress in BJ Foreskin cells and thus induction in HMOX1 may be a protective adaptation to the stress. This is because it has been shown that HMOX1 mediates protection of cells and tissues in several animal models of oxidant injury.21–23 HMOX-2 that showed the most significant increase in response to SWCNT treatment has been implicated in protection against oxidative stress, ischemia, and traumatic brain injury.24 Closely observing the gene expression profile it would suggest that HMOX-1, HMOX-2, and Catalase (ratio 1.5) increased to counteract the oxidative stress induced by the SWCNT in BJ Foreskin cells.
Fig. 4.
SWCNT alters stress response genes in BJ Foreskin cells. Representative Array showing the differential gene expression from control and SWCNT (6 μg/ml) treated BJ Foreskin cells. The spot 1 and 8 are indicated to aid alignment of the array with the spot numbers. The box marks indicates genes that served as internal and for non-specific hybridization controls. The position of the genes that were used for validation by RT-PCR are also shown in the array.
Table I.
Analysis of Stress Responsive Genes Induced by Single-Walled Carbon Nanotubes in BJ Foreskin Cells
| Spot # | Gene symbol | Gene name | Fold changea |
|---|---|---|---|
| 1 | ANXA5 | Annexin A5 | 1.24 |
| 2 | ATM | Ataxia telangiectasia mutated | 1.57 |
| 3 | BAX | BCL2-associated X protein | 1.41 |
| 4 | BCL2L1 | BCL2-like 1 | 1.43 |
| 5 | BCL2L2 | BCL2-like 2 | 0.91 |
| 6 | CASP1 | Caspase 1, apoptosis-related cysteine protease | 1.27 |
| 7 | CASP 10 | Caspase 10, apoptosis-related cysteine protease | 1.21 |
| 8 | CASP8 | Caspase 8, apoptosis-related cysteine protease | 1.54 |
| 9 | CAT | Catalase | 1.50 |
| 10 | CCNC | Cyclin C | 1.59 |
| 11 | CCND1 | Cyclin Dl (PRAD1: parathyroid adenomatosis 1) | 1.23 |
| 12 | CCNG1 | Cyclin G 1 | 1.78 |
| 13 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 0.96 |
| 14 | CHEK2 | CHK2 checkpoint homolog (S. pombe) | 1.06 |
| 15 | CRYAB | Crystallin, alpha B | 1.01 |
| 16 | CSF2 | Colony stimulating factor 2 | 1.80 |
| 17 | CYP1A1 | Cytochrome P450, family 1, subfamily A, | 1.34 |
| 18 | CYP1B1 | Cytochrome P450, family 1, subfamily B, | 1.84 |
| 19 | CYP2E1 | Cytochrome P450, family 2, subfamily E, | 1.43 |
| 20 | CYP7A1 | Cytochrome P450, family 7, subfamily A, | 1.57 |
| 217 | CYP7B1 | Cytochrome P450, family 7, subfamily B, | 1.27 |
| 22 | DDB1 | Damage-specific DNA binding protein 1, | 1.35 |
| 23 | DDIT3 | DNA-damage-inducible transcript 3 | 1.46 |
| 24 | DNAJA1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | 1.55 |
| 25 | DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | 1.55 |
| 26 | E2F1 | E2F transcription factor 1 | 1.17 |
| 27 | EGR1 | Early growth response 1 | 1.51 |
| 28 | EPHX2 | Epoxide hydrolase 2, cytoplasmic | 1.16 |
| 29 | ERCC1 | Excision repair cross-complementing group 1 | 1.38 |
| 30 | ERCC3 | Excision repair cross-complementing group 3 | 1.68 |
| 31 | ERCC4 | Excision repair cross-complementing group 4 | 2.10 |
| 32 | ERCC5 | Excision repair cross-complementing group 5 | 1.94 |
| 33 | FMO1 | Flavin containing monooxygenase 1 | 1.54 |
| 34 | FMO5 | Flavin containing monooxygenase 5 | 0.94 |
| 35 | GADD45A | Growth arrest and DNA-damage-inducible, alpha | 1.40 |
| 36 | GADD45B | Growth arrest and DNA-damage-inducible, beta | 1.27 |
| 37 | GPX1 | Glutathione peroxidase 1 | 1.03 |
| 38 | GSR | Glutathione reductase | 1.23 |
| 39 | GSTM3 | Glutathione S-transferase M3 (brain) | 1.08 |
| 40 | HMOX1 | Heme oxygenase (decycling) 1 | 2.07 |
| 41 | HMOX2 | Heme oxygenase (decycling) 2 | 3.14 |
| 42 | HSF1 | Heat shock transcription factor 1 | 1.36 |
| 43 | HSPH1 | Heat shock 105 kDa/110 kDa protein 1 | 1.13 |
| 44 | HSPA1A | Heat shock 70 kDa protein 1A | 1.40 |
| 45 | PTGS1 | Prostaglandin-endoperoxide synthase 1 | 1.11 |
| 46 | HSPA1L | Heat shock 70 kDa protein 1-like | 1.47 |
| 47 | HSPA2 | Heat shock 70 kDa protein 2 | 1.12 |
| 48 | HSPA4 | Heat shock 70 kDa protein 4 | 1.30 |
| 49 | HSPA5 | Heat shock 70 kDa protein 5 | 1.80 |
| 50 | HSPA6 | Heat shock 70 kDa protein 6 | 1.26 |
| 51 | HSPA8 | Heat shock 70 kDa protein 8 | 1.18 |
| 52 | HSPA9B | Heat shock 70 kDa protein 9B | 1.07 |
| 53 | HSPB1 | Heat shock 27 kDa protein 1 | 0.83 |
| 54 | HSPCA | Heat shock 90 kDa protein 1, alpha | 1.04 |
| 55 | HSPCB | Heat shock 90 kDa protein 1, beta | 0.92 |
| 56 | HSPD1 | Heat shock 60 kDa protein 1 | 1.43 |
| 57 | HSPE1 | Heat shock 10 kDa protein 1 | 2.35 |
| 58 | IGPBP6 | Insulin-like growth factor binding protein 6 | 1.19 |
| 59 | IL18 | Interleukin 18 | 1.77 |
| 60 | IL1A | Interleukin 1, alpha | 1.27 |
| 61 | IL1B | Interleukin 1, beta | 1.03 |
| 62 | IL6 | Interleukin 6 (interferon, beta 2) | 0.81 |
| 63 | LTA | Lymphotoxin alpha | 0.81 |
| 64 | MDM2 | Mdm2, transformed 3T3 cell double minute 2, | 1.31 |
| 65 | MIF | Macrophage migration inhibitory factor | 1.45 |
| 66 | PRDX1 | Peroxiredoxin 1 | 1.26 |
| 67 | PRDX2 | Petoxiredoxin 2 | 1.25 |
| 68 | MT2A | Metallothionein 2A | 1.21 |
| 69 | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 0.85 |
| 70 | NFKBIA | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 1.46 |
| 71 | NOS2A | Nitric oxide synthase 2A | 1.33 |
| 72 | PCNA | Proliferating cell nuclear antigen | 0.92 |
| 73 | GDF15 | Growth differentiation factor 15 | 1.76 |
| 74 | POR | P450 (cytochrome) oxidoreductase | 1.48 |
| 75 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1.16 |
| 76 | RAD23A | RAD23 homolog A (S. cerevisiae) | 1.10 |
| 77 | RAD50 | RAD50 homolog (S. cerevisiae) | 1.03 |
| 78 | CCL21 | Chemokine (C-C motif) ligand 21 | 1.40 |
| 79 | CCL3 | Chemokine (C-C motif) ligand 3 | 1.25 |
| 80 | CCL4 | Chemokine (C-C motif) ligand 4 | 1.34 |
| 81 | CXCL10 | Chemokine (C-X-C motif) ligand 10 | 1.23 |
| 82 | SERPINE1 | Serine (or cysteine) proteinase inhibitor | 1.19 |
| 83 | SOD1 | Superoxide dismutase 1 | 0.92 |
| 84 | SOD2 | Superoxide dismutase 2, | 0.88 |
| 85 | TNF | Tumor necrosis factor | 1.14 |
| 86 | TNFRSF1A | TNF receptor superfamily, member 1A | 0.91 |
| 87 | TNFSF10 | TNF (ligand) superfamily, member 10 | 1.38 |
| 88 | FASLG | Fas ligand (TNF superfamily, member 6) | 0.92 |
| 89 | TP53 | Tumor protein p53 (Li-Fraumeni syndrome) | 1.58 |
| 90 | TRADD | TNFRSF1A-associaled via death domain | 1.03 |
| 91 | UGT1A10 | UDP glycosyltransferase 1 family | 1.03 |
| 92 | UNG | Uracil-DNA glycosylase | 0.77 |
| 93 | XRCC1 | X-ray repair complementing defective repair in Chinese hamster cells 1 | 1.19 |
| 94 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | 1.21 |
| 95 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 | 1.19 |
| 96 | XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 | 1.14 |
Fold Changes represent the ratio of spot density of treated samples divided those by control. The density values were normalized to beta actin before obtaining the fold changes.
ERCC4 that was increased by a ratio of 2.1 and this is an essential human gene in the nucleotide excision repair (NER) pathway, which is responsible for removing UV-C photoproducts and bulky adducts from DNA. Among the NER genes, ERCC4 and ERCC1 are also uniquely involved in removing DNA inter strand cross-linking damage.26 Apoptosis that results upon SWCNT exposure to cells has been documented and this might result in the induction of ERCC4 as a counter measure to protect the cells from DNA damage.
From the family of genes involved in apoptosis we found increased ratio of TP53 (ratio 1.58), Caspase 8 (ratio 1.54) and both these genes are involved in cell death via apoptosis.27 Cell death via apoptosis by multi-walled carbon nanotubes has recently been demonstrated in skin fibroblasts.28 Therefore it may be possible that SWCNT induces apoptosis through TP53 and caspase 8 mediated signal transduction. The genes that are involved in inflammatory response also showed significant alterations in cells treated with SWCNT. Inflammation related gene, Macrophage migration inhibitory factor (MIF) was increased by SWCNT treatment to BJ Foreskin cells by a ratio of 1.45. The protein from this gene is an evolutionary conserved and mediates multiple functions in innate and acquired immunity. This also upon leaderless secretion acts as a typical inflammatory cytokine.29 In addition to this increase in IL18 (1.77) was observed in SWCNT treated cells. IL18 is involved in cellular inflammation and has been described to play a role in several inflammatory skin diseases such as eczema and psoriasis.30 It has been reported that SWCNT induces inflammation in mouse lung when treated through inhalation and both this inflammation related genes might be involved in the inflammatory response.5
In summary the genes that responded to SWCNT indicates that this material at a dose of 6 μg/ml can elicit stress and is evident by the spectrum of genes getting altered by the treatment.
RT-PCR on selected Genes Altered by SWCNT Treatments in BJ-Foreskin Cells
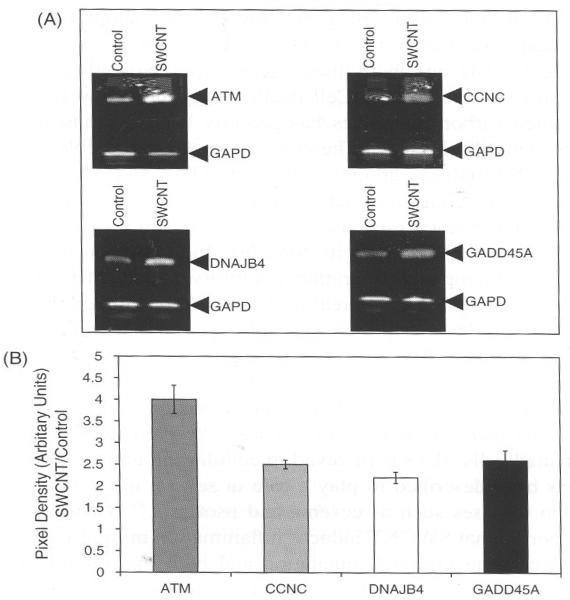
We were interested in validating few of the genes altered as SWCNT treatment in BJ Foreskin cells. The genes that were selected are shown in Figure 4 and also shown are there relative expression on the array. Figures 5(A) and (B) shows a representation of such analysis performed for genes Ataxia telangiectasia mutated (ATM), Cyclin C (CCNC), DnaJ (Hsp40) homolog (DNAJB4) and Growth arrest and DNA-damage-inducible, alpha (GADD45A). The results show a co-relation in the expression of the genes obtained previously in array and finally by RT-PCR analysis. The equal loading and normalization was done using the gene Glucose 6-phosphate dehydrogenase (GAPD), which was analyzed under similar experimental conditions as mentioned for the test genes.
Fig. 5.
Relative RT-PCR analysis of ATM, CCNC, DNAJB4, and GADD45A expression in BJ Foreskin cells treated with SWCNT. (A) BJ Foreskin cells were treated with SWCNT as detailed in text for 24 h. Total RNA was isolated and DNase treated before RT. Relative RT-PCR was performed as described in text and c-DNA was amplified using specific primers for indicated genes. PCR products were separated on 1% agarose gel and stained with ethidium bromide as shown in representative image. GAPD was used as internal control. (B) By densitometry analysis, ATM, CCNC, DNAJB4, and GADD45A gene expression was evident in 24 h. The band intensity of GAPD was used to normalize expression of genes.
ATM belongs a family of kinase that have sequence homology to phosphoinositide 3-kinase (PI3K).32 It is a protein kinase and is an important player in signaling that involves double strand break in DNA of higher eukaryotes.32 SWCNT increased ATM significantly and this possibly indicates that the cell cycle might have stalled and DNA damage also may be speculated due to the ROS generation.
The other gene validated was CCNC (Cyclin C) and this was of interest because it is involved in cell cycle (Fig. 4). Previous studies have shown loss of cell proliferation by SWCNT, which suggests alteration in cell cycle genes. CCNC interacts with CDK8 and this complex associates with carboxy-terminal domain of RNA polymerase II.33 Moreover its role in cell cycle has also recently demonstrated by Ren and Rollins.34 It was shown in this study that cyclin C combines with cdk3 to stimulate pRb phosphorylation at S807/811 during the G0/G1 transition, and that this phosphorylation is required for cells to exit G0 efficiently. At present we have no explanation how increase in this CCNC cell cycle kinase is affected by SWCNT in BJ-Foreskin cells but it is concluded that this material influences cell cycle associated genes.
DNAJA1 and DNAJB4 are represented from Hsp40 family. Both DNAJA1 (Ratio 1.55) and DNAJB4 (Ratio 1.55) were increased by SWCNT treatment in BJ Foreskin cells (Table I, Fig. 5). The Hsp70 chaperone system is formed by Hsp70 (DnaK-related) with its co-chaperones Hsp40 (DnaJ-related) and GrpE. Tins system assists many cellular processes involving proteins, including folding, transport through membranes, degradation, and escape from aggregation.35 Hsp70 gene was also up-regulated by SWCNT treatment implying that this family of chaperones responded to the stress generated by the treatment. This also cross validates the array analysis because both these family of genes are interrelated and dependent to each other to dispense their function.
GADD45A is induced under genotoxic stress and is up regulated in cells exposed to UV radiation.36–37 SWCNT induced GADD45A in BJ Foreskin cells as evident in both stress array hybridization and RT-PCR (Table I and Fig. 5). The increase in GADD45A would lead cells towards apoptosis and is shown in cells over expressing this gene.37 Therefore over expression of GADD45A by SWCNT may explain its role in apoptosis.
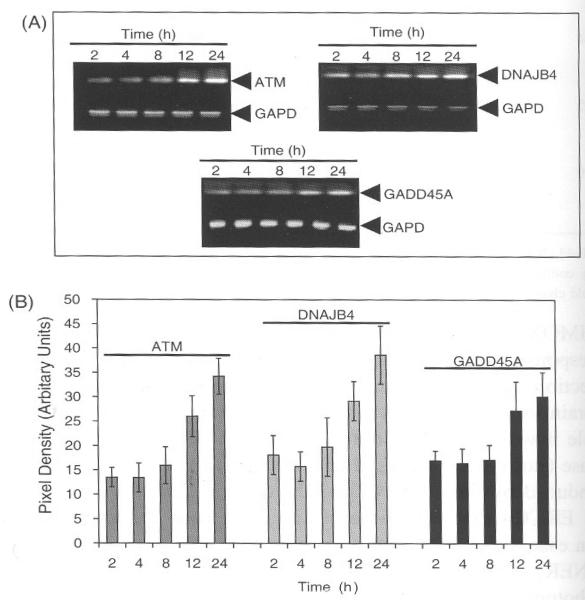
Time dependent changes in gene expressions by SWCNT in BJ-Foreskin cells
To observe the changes in gene expression as a function of time we exposed the cells to SWCNT and isolated RNA at different time intervals. Induction of ATM, DNAJB4, and GADD45A in BJ Foreskin cells treated with SWCNT showed significant inductions at 12 and 24 h (Figs. 6(A) and (B)). GAPD did not show any change in gene expression, which was analyzed for monitoring the efficiency of the PCR reaction and also accounts for equal amount of c-DNA used for PCR. The three genes (ATM, DNAJB4, and GADD45A) approximately induced at similar time intervals and none of these genes showed any change at early hours. The time dependent increase in all the three genes may have resulted due to accumulating stress over the time of exposure and allowing at these time points to detect the changes by RT-PCR. Both ATM and GADD45A have been shown to perform their function in concert with DNA damage.32–38 Therefore the increase in both these genes, showing similar profile of induction at least argues that a sequential signal transduction pathway is affected by treatment with SWCNT in BJ foreskin cells. In addition the uptake of SWCNT particle could adversely affect the cellular response and these mechanisms are still not completely understood. A study relating the uptake of DNA-conjugated SWCNT in cells was achieved after 1–3 h while silica, coated nanoparticles took almost 6 h to get inside the cells.11 In this study pure SWCNT was used and whether there exists a relation between the uptake of the SWCNT particles and cellular induction of genes remains unsolved. However, the induction of ROS by SWCNT in Figures 1 and 2 shows that oxidative stress was a time dependent phenomenon indicating thereby that as more of these particles move inside the cell the induction increases as a function of time. In conclusion more rigorous studies are still needed to understand the mechanism of induction of these stress response genes in BJ Foreskin cells by SWCNT.
Fig. 6.
SWCNT induced gene expression is time dependent. (A) Cells were treated with 6 μg/ml of SWCNT for different time periods and RNA was extracted as described in text. The c-DNA was amplified with specific primers and GAPD was used as internal control. (B) Densitometry analysis was done to detect the differences in band intensity of the PCR product. The band intensity of GAPD was used to normalize expression of genes.
4. CONCLUSIONS
The toxic effects induced by carbon nanoparticles on the biological systems and the importance of these effects in human and animal health is extensively reviewed and discussed.39–43 The toxicity of carbon nanomaterials is also dictated by its size and its geometrical structure. In a recent report Jia et al., 2005, have elucidated a comparative cytotoxicity study of various nanomaterials differing in size and geometrical shape.44 The results of the present study reflect that SWCNT treatment can induce significant ROS and influence stress response genes in BJ Foreskin cells. The induction of ROS in turn can induce several genes as a response to counteract the physiological changes in the cells. The ratio of such genes showing significant increase included genes involved in oxidative stress, apoptosis, DNA repairs genes, genes encoding for chaperon proteins and cytochrome p450 family. Approximately all these genes showed more than 1.5 fold increase in expression as compared to control and this indicates that SWCNT affects stress activated genes. The changes in gene expressions in BJ Foreskin cells by SWCNT treatment reveals that these particles are capable of manipulating signal transduction pathways through alterations in stress related gene expressions.
In addition the array results also show increase in inducible genes like HMOX1, CYP1A1, and CYP1B1 with ratios 2.07, 1.34, 1.84, respectively. HMOX1 is oxidative stress inducible gene and this substantiates the observation because SWCNT significantly increased ROS in BJ Foreskin cells (Figs. 1 and 2).8,23 The genes mainly ATM and GADD45A are more focused in this study because both of these genes are involved in DNA repair and apoptosis.32,38 Cell death by SWCNT and multi walled carbon nanotubes (MWCNT) has been reported.8,28 ATM and GADD45A might play role in cell death induced by SWCNT because both these genes were significantly up regulated in BJ Foreskin cells. In addition TP53 and Caspase 8 was also significantly up regulated by treatment with SWCNT and would suggest that induction of apoptosis is possible in these cells considering the change in expression of multiple genes involved in apoptosis. Evidences from various studies have shown loss of cell proliferation by nano-materials which further strengthens the observations made in this study by revealing the increase in some of the key genes involved in apoptosis.8,29,44
In summary, the results obtained from previous studies and together with the present results it is conceivable to argue that SWCNT particles at 6 μg/ml could elicit stress to cells in culture. The changes in the stress proteins are meaningful to understand the mechanism of toxicity induced by SWCNT particles and the results do explain some of the related toxic responses as reported elsewhere.4–8,29 However, how far the present study could be extended to animal model is the future research. This also warrants minimizing the unwanted effects of the fast growing nanotechnology towards its safe use both by the producer and its consumers.
Acknowledgment
This work was supported by NASA funding NCC 9-165: NCC-1-02038: NAG 9-1414: NIH/RCMI RR03045-18 (GR).
References and Notes
- 1. www.nano.gov.
- 2.Carbon Nanotubes-Worldwide Status and Outlook Applications, Applied Industries, Production, R&D, and Commercial Implications. 2002 [Google Scholar]
- 3.Martin CR, Kohli P. Nat. Rev. Drug Discov. 2003;2:29. doi: 10.1038/nrd988. [DOI] [PubMed] [Google Scholar]
- 4.Warheit D, Laurence B, Reed K, Roach D, Reynold G, Webb T. Toxicol. Sci. 2004;77:117. doi: 10.1093/toxsci/kfg228. [DOI] [PubMed] [Google Scholar]
- 5.Shvedova AA, Kisin ER, Mercer R, Murray AR, Johnson VJ, Potapovich AI, Tyurina YY, Gorelik O, Arepalli S, Schwegler-Berry D, Hubbs AF, Antonini J, Evans DE, Ku BK, Ramsey D, Maynard A, Kagan VE, Castranova V, Baron P. Am. J. Physiol. Lung Cell Mot. Physiol. 2005;289:698. doi: 10.1152/ajplung.00084.2005. [DOI] [PubMed] [Google Scholar]
- 6.Shvedova AA, Castranova V, Kisin ER, Schwegler-Berry D, Murray AR, Gandelsman VZ, Maynard A, Baron P. J. Toxicol. Environ. Health A. 2003;66:1909. doi: 10.1080/713853956. [DOI] [PubMed] [Google Scholar]
- 7.Lam CW, James JT, McCluskey R, Hunter RL. Toxicol. Sci. 2004;77:126. doi: 10.1093/toxsci/kfg243. [DOI] [PubMed] [Google Scholar]
- 8.Manna SK, Sarkar S, Barr J, Wise K, Barrera EV, Jejelowo O, Rice-Ficht AC, Ramesh GT. Nano Lett. 2005;5:1676. doi: 10.1021/nl0507966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson MA, Domann FE, Bowden GT, Hooser SB, Fernando Q, Carter DE. Toxicol Ind. Health. 1993;9:623. doi: 10.1177/074823379300900405. [DOI] [PubMed] [Google Scholar]
- 10.Magrez A, Kasas S, Salicio V, Pasquier N, Seo JW, Celio M, Catsicas S, Schwaller B, Forro L. Nano Lett. 2006;6:1121. doi: 10.1021/nl060162e. [DOI] [PubMed] [Google Scholar]
- 11.Kam NW, Dai H. J. Am. Chem. Soc. 2005;127:6021. doi: 10.1021/ja050062v. [DOI] [PubMed] [Google Scholar]
- 12.Juranek I, Bezek S. Gen. Physiol. Biophys. 2005;24:263. [PubMed] [Google Scholar]
- 13.Kimura H, Sawada T, Oshima S, Kozawa K, Ishioka T, Kato M. Curr. Drug Targets Inflamm. Allergy. 2005;4:489. doi: 10.2174/1568010054526287. [DOI] [PubMed] [Google Scholar]
- 14.Durante W. J Cell Physiol. 2003;195:373. doi: 10.1002/jcp.10274. [DOI] [PubMed] [Google Scholar]
- 15.Tenhunen R, Marver HS, Schmid R. Proc. Natl. Acad. Sci. USA. 1968;61:748. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morse D, Choi AM. Am. J. Respir. Cell Mol. Biol. 2002;27:8. doi: 10.1165/ajrcmb.27.1.4862. [DOI] [PubMed] [Google Scholar]
- 17.Applegale LA, Luseher P, Tyrrell RM. Cancer Res. 1991;51:974. [PubMed] [Google Scholar]
- 18.Janssen YM, Marsh JP, Absher MP, Gabrielson E, Borm PJ, Driscoll K, Mossman BT. Am. J. Respir. Crit. Care Med. 1994;149:795. doi: 10.1164/ajrccm.149.3.8118652. [DOI] [PubMed] [Google Scholar]
- 19.Keyse SM, Tyrrell RM. Proc. Natl. Acad. Sci. 1989;86:99. doi: 10.1073/pnas.86.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motterlini R, Foresli R, Bassi R, Calabrese V, Clark JE, Green CJ. J. Biol. Chem. 2000;275:13613. doi: 10.1074/jbc.275.18.13613. [DOI] [PubMed] [Google Scholar]
- 21.Rensing H, Jaeschke H, Bauer I, Patau C, Datene V, Pannen BH, Bauer M. Crit. Cure Med. 2001;10:1962. doi: 10.1097/00003246-200110000-00019. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto K, Ohta K, Yachie A, Yang Y, Shimizu M, Goto C, Toma T, Kasahara Y, Yokoyama H, Miyata T, Seki H, Koizumi S. Kidney Int. 2001;60:1858. doi: 10.1046/j.1523-1755.2001.01000.x. [DOI] [PubMed] [Google Scholar]
- 23.Ryter SW, Choi AM. Antioxid. Redox. Signal. 2005;7:80. doi: 10.1089/ars.2005.7.80. [DOI] [PubMed] [Google Scholar]
- 24.Dore S, Goto S, Sampei K, Blackshaw S, Hester LD, Ingi T, Sawa A, Traystman RJ, Koehler RC, Snyder SH. Neuroscience. 2000;99:587. doi: 10.1016/s0306-4522(00)00216-5. [DOI] [PubMed] [Google Scholar]
- 25.Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH. Mol. Cell Biol. 1996;16:6553. doi: 10.1128/mcb.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadenbach B, Arnold S, Lee I, Huttemann M. Biochim. Bio-phys. Acta. 2004;1655:400. doi: 10.1016/j.bbabio.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Ding HF, Fisher DE. J. Pediatr. Hematol. Oncol. 2001;23:185. doi: 10.1097/00043426-200103000-00014. [DOI] [PubMed] [Google Scholar]
- 28.Ding L, Stilwell J, Zhang T, Elboudwarej O, Jiang H, Selegue JP, Cooke PA, Gray JW, Chen FF. Nano Lett. 2005;12:2448. doi: 10.1021/nl051748o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiele M, Bernhagen J. Antioxid, Redox. Signal. 2005;9–10:1234. doi: 10.1089/ars.2005.7.1234. [DOI] [PubMed] [Google Scholar]
- 30.Wittmann M, Purwar R, Hartmann C, Gutzmer R, Werfel T. J. Invest Dermatol. 2005;124:1225. doi: 10.1111/j.0022-202X.2005.23715.x. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. J. Biol. Chem. 2002;277:36585. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 32.Shiloh Y. Nat. Rev. Cancer. 2003;3:155. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 33.Rickert P, Scghezzi W, Shanahan F, Cho II, Lees E. Oncogene. 1996;12:2631. [PubMed] [Google Scholar]
- 34.Ren S, Rollins BJ. Cell. 2004;117:239. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 35.Borges JC, Ramos CH. Protein Pept. Lett. 2005;12:257. doi: 10.2174/0929866053587165. [DOI] [PubMed] [Google Scholar]
- 36.Tong T, Fan W, Zhao H, Jin S, Fan F, Blanck P, Alomo I, Rajasekaran B, Liu Y, Holbrook NJ, Zhan Q. Exp. Cell Res. 2001;269:64. doi: 10.1006/excr.2001.5312. [DOI] [PubMed] [Google Scholar]
- 37.Zhan Q, Kontny U, Iglesias M, Alamo I, Jr., Yu K, Hollander MC, Woodworth CD, Fornace AJ., Jr. Oncogene. 1999;18:297. doi: 10.1038/sj.onc.1202310. [DOI] [PubMed] [Google Scholar]
- 38.Sheikh MS, Hollander MC, Fornance AJ. Biochem. Pharmacol. 2000;59:43. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- 39.Xing X, He X, Peng J, Wang K, Tan W. J. Nanosci. Nanotechmol. 2005;10:1688. doi: 10.1166/jnn.2005.199. [DOI] [PubMed] [Google Scholar]
- 40.Fiorito S, Serafino A, Andreola F, Togna A, Togna G. J. Nanosci. Nanotechnol. 2006;6:591. doi: 10.1166/jnn.2006.125. [DOI] [PubMed] [Google Scholar]
- 41.Paul JA, Borm, Wolfgang K. J. Nanosci. Nanotechnol. 2004;4:21. [Google Scholar]
- 42.Wei-Xian Z, Tina M. Environ. Sci. Technol. 2003;37:102A. [Google Scholar]
- 43.Mihail CR, Barbara K. Environ. Sci. Technol. 2005;39:106A. [Google Scholar]
- 44.Jia G, Wang H, Yan L, Wang X, Pei R, Yan T, Zhao Y, Guo X. Environ. Sci. Technol. 2005;39:1378. doi: 10.1021/es048729l. [DOI] [PubMed] [Google Scholar]