Abstract
Traits related to energy balance and obesity are exceptionally complex, with varying contributions of genetic susceptibility and interacting environmental factors. The use of mouse models has been a powerful driving force in understanding the genetic architecture of polygenic traits such as obesity. However, the use of mouse models for analysis of complex traits is at an important crossroad. Genome-wide association studies in humans are now leading to direct identification of obesity genes. In this review, we focus on three areas representing the current and future roles of mouse models regarding genetics of complex obesity. First, we summarize increasingly powerful ways to harness the strength of mouse models for discovery of genes affecting polygenic obesity. Second, we examine the status of using a systems biology approach to dissect the genetic architecture of obesity. And third, we explore the effects of recent findings indicating increasing levels of complexity in the nature of variation underlying, and the heritability of, complex traits such as obesity.
Keywords: systems genetics, QTL, eQTL, collaborative cross, genetic variation
INTRODUCTION
Most phenotypes displaying continuous variation, including nearly all traits related to energy balance and obesity, are exceptionally complex, with varying contributions of genetic susceptibility and interacting environmental factors. The use of mouse models has been a powerful driving force in understanding the genetic architecture of polygenic traits such as obesity. In addition to the many mouse models of obesity caused by spontaneous mutations and targeted gene knockouts and insertions (not reviewed here), the commonly used inbred laboratory strains of mice constitute the primary mammalian model system and are an integral component of obesity research (40). Within these lines and their derivatives, such as recombinant inbred lines, genomewide congenic strains, chromosome substitution lines, advanced intercross lines, long-term selection lines, and heterogeneous stocks, there exists a vast array of obesity-relevant genetic and phenotypic variation (reviewed in 42). The study of such variation, in the form of complex trait analysis including candidate gene analysis and quantitative trait locus (QTL) mapping, has shed significant light on the genetic and genomic architecture of nearly all aspects of energy balance regulation and how body weight and body fat are controlled (42).
Despite these significant efforts, the use of mouse models for the analysis of complex traits such as obesity is at an important crossroad. The use of crosses between inbred strains and/or selection lines has been highly successful in thoroughly populating the mouse obesity predisposition map with QTLs for body weight, fatness, energy intake, and energy expenditure, but few if any of these QTLs have been robustly identified at the gene or nucleotide level, a situation that Flint et al. (16) refer to as “heading towards a crisis.” At the same time, genome-wide association studies experiments in humans have finally overcome many of the power obstacles that had rendered early studies unreliable, and direct identification of human genes for obesity-related traits (see 17) may call into question the continued use of mice as a model in such efforts.
In the current review, rather than dwell on a thorough summary of past findings, we focus on three areas representing the current and future roles of mouse models regarding genetics of complex obesity in mice. First, we summarize increasingly powerful ways to harness the strength of mouse models for the discovery of genes affecting polygenic obesity, with an emphasis on the Collaborative Cross (7). Second, we examine the status of using a systems biology (or more accurately in this case, systems genetics) approach to dissect the genetic architecture of obesity, with an emphasis on transcriptional analysis and expression QTL (eQTL) mapping. And third, we explore the effects of recent findings that indicate increasing levels of complexity in the nature of variation underlying, and the heritability of, complex traits such as obesity, including the importance of ribonucleic acid (RNA), other new sources of variation such as copy number variants, and transgenerational epigenetics.
NEW(ER) POLYGENIC MOUSE MODELS OF OBESITY
Although traditional two-strain backcrosses, F2 intercrosses, and recombinant inbred lines (RILs) may still be useful for poorly defined obesity phenotypes (e.g., voluntary exercise; 32), these tools have primarily run their course owing to the difficulty in moving from QTL to the underlying gene. Instead, new models and/or paradigms to exploit existing models have been (or are being) developed to enable much finer mapping of polygenes for complex traits. For example, a broadly powerful extension of the advanced intercross concept is the creation of heterogeneous stocks (HS). In this case, QTLs can be very finely mapped by exploiting historical recombinants that have accumulated in a genetically heterogeneous stock of mice descended from eight inbred progenitor strains (i.e., A/J, AKR/J, BALBc/J, CBA/J, C3H/HeJ, C57BL/6J, DBA/2J, and LP/J; 10). This HS resource has now been outbred for more than 50 generations. Although HS was initially intended for fine mapping of specific QTL regions, the current availability of very dense single-nucleotide polymorphisms (SNPs) and affordable high-throughput geno-typing facilitates use of these models for whole-genome discovery approaches. Using the HS, Valdar et al. (59) robustly mapped 843 QTLs with an average 95% confidence interval of 2.8 Mb. Many of these QTLs contribute to variation in obesity-relevant traits, including body weight gain, activity, type 2 diabetes, and blood chemistry (e.g., high-density lipoprotein, low-density lipoprotein, total cholesterol, and triglycerides).
The availability of extremely dense SNP genotyping tools has also enabled the “retrofitting” of standard inbred mouse strains as powerful gene discovery tools. Nearly 500 strains of mice have been documented in the Mouse Genome Informatics Database (27). Recently, a phenotypic survey of inbred lines has been formally championed by The Mouse Phenome Project, an international collaborative effort promoting the comprehensive characterization of a set of 40 commonly used and genetically diverse inbred strains and their derivatives. The Mouse Phenome Database (28) is being populated with data relevant to many complex human diseases, including most aspects of the metabolic syndrome. Data are available for metabolism, activity, food intake, body composition, effects of atherogenic diet, leptin and insulin levels, and other biological parameters that are being used to identify and characterize new mouse models for obesity research. By combining phenotyping of inbred strains with dense SNP genotyping, researchers are performing high-resolution in silico (i.e., biological experiments carried out entirely in a computer) QTL mapping with significant success. For example, Liu et al. (35) surveyed 173 mouse phenotypes across the 40 strains in the Mouse Phenotype Database. Using existing genotype data for ∼150K SNPs, they identified 937 QTLs for a variety of traits including many related to metabolism and obesity. About two-thirds of the QTLs were refined into genomic regions of 0.5 Mb with a 40-fold increase in mapping precision as compared with classical linkage analysis (35). Importantly, two QTLs were resolved to the causal gene level, both for atherogenic diet-induced obesity.
Although in silico QTL mapping has much promise, the most commonly used mouse resources harbor only a fraction of the genetic diversity of Mus musculus, which is not uniformly distributed, resulting in many blind spots. This was the conclusion reached by Roberts et al. (47) after analysis of resequencing data representing 8.3 million SNPs in 11 classical inbred strains and four wild-derived strains. Only resources that include wild-derived inbred strains from subspecies other than M. m. domesticus have no blind spots and a uniform distribution of the variation.
A new paradigm for complex trait analysis, the Collaborative Cross (CC; 7), is a large panel of RILs derived from a genetically diverse set of eight founder strains that has a distribution of allele frequencies resembling that seen in natural populations, such as humans, in which many variants are found at low frequencies and only a minority of variants are common (47). The ∼1000 RILs that will comprise the CC will be completed in 2009, and the eight parental inbred lines (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HiLtJ, CAST/Ei, PWK/PhJ, and WSB/EiJ) are estimated to capture more than 90% of the known variation present in mouse strains. Existing data on the founder strains and in the many F1 hybrid combinations demonstrate broad variability in obesity phenotypes (see 42), indicating that the CC will represent an excellent resource for identifying genes controlling predisposition to obesity and understanding the pathways, networks, and systems that control obesity.
By providing a large, common set of genetically defined mice, the CC may become a focal point for cumulative and integrated data collection for traits related to obesity in mice, giving rise to networks of functionally important relationships within and among diverse sets of biological and physiological phenotypes that can be altered by external factors such as diet and exercise. Furthermore, the CC has the potential to support studies by the larger scientific community incorporating multiple genetic, environmental, and developmental variables into comprehensive statistical models describing obesity susceptibility and progression. Equally important, the CC will be ideal as a test bed for predictive, or more accurately, probabilistic medicine, which will be essential for the deployment of personalized medicine.
Furthermore, the CC is highly extensible. It provides a community framework for data integration at all levels, from molecules to cells to physiological systems and across environments. Unlike many other resources that are primarily suited for gene discovery, the CC can support genomewide network analysis, which is the foundation of systems genetics.
APPLYING SYSTEMS BIOLOGY TO OBESITY GENETICS: EXPRESSION PROFILING
The advent of microarray technologies in the late 1990s (12, 53) revolutionized the way in which we approach complex genetic problems like obesity. Because of the complexity and scale of data generated by array analyses, new paradigms challenge biomedical research (65). This is largely because high-throughput interrogation of the thousands of transcripts contained on each microarray provides molecular evidence of genomewide interactions that underlie complex phenotypes in response to genetic and/or environmental perturbations. A well-designed array experiment not only identifies relevant candidate genes but also generates unique expression profiles from which multidimensional data are leveraged to provide biomarkers that lead to relevant therapies (8). The power of microarrays has been applied to understand the complex genetics of obesity in various rodent models.
The first study to use global analysis of transcriptional data to identify candidate genes underlying obesity QTLs involved microarray analysis of adipose from hypertensive rats (2). Here, linkage mapping of additional F2 rats was combined with data from a previous QTL study for hypertension to better define QTL boundaries. Microarray analysis of a congenic strain that localized the QTL interval from the hypertension-resistant strain onto the genetic background of the susceptible strain was used to discern relevant changes in gene expression by comparisons between the congenic and the parental strains. Using this strategy, CD36 was identified as a positional candidate for defective glucose and fatty acid metabolism, decreased high-density lipoprotein, and hypertension observed in the congenic strain. CD36 transgenic animals were then generated to validate the role of this candidate gene in metabolic syndrome. Additionally, CD36 deficiency was identified in several human populations with metabolic syndrome.
Although it is vital to identify individual candidate genes that underlie obesity phenotypes, so is understanding the genomic context in which multiple genes must interact. Perhaps this is why the greatest successes in microarray technologies are evidenced in the diagnosis and treatment of human cancers, where variations in patterns of differentially transcribed genes from primary lesions (as compared to healthy tissue) have been used to improve diagnosis (23), predict outcomes, and tailor therapies to improve survival (38). Similar methods have been applied to patterns of transcriptional variation for obesity-related phenotypes (Figure 1), which suggest useful predictors for diagnosis and subsequent therapeutic targeting. For example, transcription analysis was used to define the role of type 2 diabetes in obesity. Nadler et al. (36) profiled adipose from five obese strains of mice with varying degrees of hyperglycemia. Comparison of transcription profiles between the strains demonstrated a striking decrease in adipogenic genes, indicating that a reduction in adipocyte differentiation linked the dysregulation of lipid storage to aberrant glucose metabolism. Similarly, two independent groups (56, 63) profiled adipose from obese mice to explore the molecular origins of the chronic inflammatory response associated with obesity. Both studies observed an expression signature from proin-flammatory macrophages. Increased transcript abundance from inflammatory genes was positively correlated with increased adiposity and weight.
Figure 1.
Simple microarray analysis comparing at least two divergent but well-defined conditions in which the biological system is perturbed by genetic or environmental factors. (top) Two mice with very different obesity phenotypes are depicted. Although many platforms exist to assay global changes in gene expression, the methods behind microarray technology (center) rely on hybridization of labeled RNA (or RNA-derived products such as cDNA) to probes from thousands of expressed genomic sequences. To depict genomewide changes in gene expression, a heat map is often used (bottom). Here, genes that show a decrease in expression are assigned a color that changes in intensity as transcript abundance changes relative to other genes on the array. Statistical algorithms cluster together genes with similar patterns of expression within an experimental group. Each row represents expression of a single gene, and each column represents an experimental individual or group.
Timmons et al. (58) used expression profiling of preadipocytes in both white and brown mouse adipose to detect myogenic transcription factor signatures (Sirt1 in brown adipose and Tcf21 in white adipose) that suppress mito-chondrial energy expenditure to facilitate lipid storage. These authors postulate that therapeutic targeting of myogenic transcription factors in white adipose may provide a plausible approach to alter the uncontrolled lipid storage that occurs in obesity. Similarly, expression profiles from brown and white adipose demonstrated coordinate transcription of genes that exhibit circadian oscillations (69). Disruption of circadian patterns affected glucose and lipoprotein metabolism (45). Thus, circadian dysregulation should be accounted for when analyzing obesity phenotypes because changes in the expression of circadian genes could provide useful predictors of obesity predisposition.
APPLYING SYSTEMS BIOLOGY TO OBESITY GENETICS: SYSTEMS GENETICS AND EXPRESSION QUANTITATIVE TRAIT LOCUS ANALYSIS
The experimental basis for understanding heritable traits (such as susceptibility to obesity) has largely involved studying biological systems one gene (or transcript) at a time; however, the one-gene-at-a-time approach is insufficient for studies of mammalian genomes, which contain more than 20,000 genes and nearly limitless amounts of variation. This complexity is compounded by intricate interactions between genes (i.e., epistasis) and between genes and the environment. A far more efficient approach is factorial experimentation, a process by which many or all components of a complex system are altered simultaneously through randomization. This approach will allow researchers to perform analyses of all genes concurrently in order to illuminate mammalian biology well enough to synthetically reassemble biological knowledge.
Consequently, an entirely new paradigm is needed to understand the interactions between genes and the environment that lead to changes in disease susceptibility. This new experimental model must support mathematical models of highly complex mammalian biological systems that predict future disease susceptibility. This concept, called systems genetics, combines novel biological tools with innovative computational and statistical analyses.
One of the primary initial implementations of systems genetics is the mapping of eQTL. It is now possible to link expression data with genetic markers to create genetic maps for gene expression. In eQTL analysis— sometimes referred to as genetical genomics (26)—gene expression in a segregating population is treated like a classic phenotypic quantitative trait. eQTLs have several advantages over differentially expressed genes obtained through typical microarray experiments. These advantages stem from the fact that eQTL can be linked to the physical location of the expression transcript, thereby placing them at the interface between complex phenotypes and the sequence variants that underlie them. When further filtered through correlation analysis (e.g., transcript variation correlated with variation in obesity traits), the eQTLs become enriched candidate genes for obesity QTLs, containing significantly more information than just differential expression.
Several advantages of eQTL analysis were elegantly illustrated in a landmark paper by Schadt et al. (52), who were among the first to show how gene expression phenotypes can be treated like other classic traits (e.g., body weight, fatness) to identify gene expression patterns associated with obesity. They assayed livers from 111 C57BL/6 × DBA2/J F2 animals after four months of high-fat atherogenic diet feeding. Of the 23,574 genes on the array, 7861 were found to be differentially expressed. They detected 965 eQTL with LOD (logarithm of the odds, to the base 10) scores greater than 7.0, illustrating that viewing gene expression differences in the context of a segregating population can provide significantly stronger expression information than using differential gene expression analysis alone. In addition, they combined eQTL and obesity clinical traits in a manner that revealed a previously undetected level of heterogeneity in the F2 population. Profiles of differentially expressed genes for each fat accretion group presented two distinct patterns of expression not only between the high-and low-fat groups but also within the high-fat group. The ability to integrate clinical QTL and eQTL in a heterogeneous population is one of the stronger ramifications of eQTL analysis because complex diseases like human obesity are likely highly heterogeneous phenotypes.
One of the classic manifestations of genetical genomics is the presence of cis-acting loci (where the eQTL is mapped in close proximity to the physical location of the transcribed gene) and trans-acting loci (where the eQTL is not linked to the physical location of the transcribed gene). Cis eQTL are highly heritable and easier to identify because genetic control is generally highly robust. When a cis eQTL localizes to the confidence interval of a phenotypic QTL, it becomes a relevant positional candidate (43). Similarly, when the underlying gene expression phenotypes at a cis eQTL are highly correlated with obesity traits (Figure 2), this additional filter further strengthens the candidate gene hypothesis (41).
Figure 2.
Expression quantitative trail loci (eQTL) represent loci controlling variation in mRNA abundance that are genetically mapped by recombination within a segregating population. Each eQTL represents an expressed transcript whose underlying genomic location is known. When an eQTL’s position coincides with the transcript’s location (a), changes in expression are controlled by the regulatory regions of the transcript itself and are termed cis eQTL. Cis eQTL that underlie a confidence interval of a phenotypic QTL serve to better localize the QTL and identify positional candidates. Conversely, when genetic and genomic positions do not correspond, regulation of the transcript maps elsewhere in the genome and is termed trans eQTL. Trans eQTLs tend to cluster (b) at various locations within the genome, revealing transcription hot spots in which a master regulator controls transcription of multiple genes. LOD, logarithm of the odds (to the base 10).
Conversely, trans eQTLs are harder to detect because they demonstrate low heritability, generally have small allelic effect on expression variance, and are under polygenic regulation. However, many trans eQTL tend to cluster in expression hot spots (5, 67), which suggests coordinated control of many genes by a master regulator (41, 43). The biological roles of such master regulators are not yet clear but are the subject of active investigation.
Schadt et al. (50) utilized this eQTL interface between genetics, genomics, and transcription as an intermediate phenotype to infer a causal relationship between the sequence variants of eQTLs and obesity traits. They developed a likelihood-based causality model selection test to detect causal relationships between deoxyribonucleic acid (DNA) sequence variants underlying eQTL and correlated obesity-related traits. From this analysis, 10 eQTL candidate genes were identified for increased fat accretion. For three of the eQTL candidates (Hsd11b1, Lpl, and Mod1), previous studies implicated a regulatory role in modulating obesity phenotypes. To validate a causal role for the eQTL candidate Zfp90, transgenics were generated and subsequent phenotyping for fat-to-lean-mass ratios identified a significant increase in fat mass, implicating a causal role for this eQTL candidate in fat-mass accretion.
APPLYING SYSTEMS BIOLOGY TO OBESITY GENETICS: COEXPRESSION ANALYSIS
The molecular underpinnings of complex traits are composed of coordinated interactions between functionally related groups of genes. Although genetical genomics provides a relevant framework for the genetic influences involved in complex disorders, high-dimensional analyses of biological systems reveal the synergistic connections within complex living organisms that discern the causal mechanisms inherent to disease processes.
To predict how biological systems adapt to genetic or environmental perturbations requires information about the organizing principles that structure the interactions between the system’s components and emergent properties. Analysis of the mechanisms by which complex systems emerge from the numerous synergistic interactions is a current paradigm exploited in recent biomedical research to identify relevant therapeutic targets for common diseases such as obesity and is the basis of systems biology. For example, Lee et al. (31) used a systems biology approach to determine how clusters of coexpressed genes (Figure 3) organize themselves. Cluster analysis of the genes within the co-expression network showed consistent grouping of functionally related genes, implying that coexpression analysis is a feasible technique in studying the emergence of functional relationships for the transcriptome, metabolome, and proteome, especially in response to disease.
Figure 3.
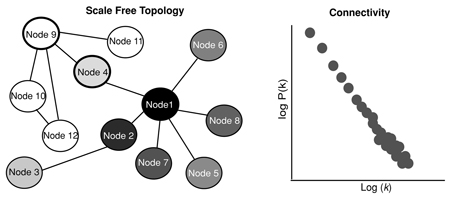
Pairwise correlations between mRNA abundance of genes demonstrate the coordinated expression of genes in response to genetic or environmental perturbations. Topological overlap matrix (TOM) plots (a) depict the clustering of correlated genes into distinct groups, or modules. Here, the expression of each gene is compared with all other genes in the assay, one by one. Each correlation between a pair of genes is assigned a color based upon the strength of the correlation measure (bottom bar). In this TOM, the stronger correlations are red; weaker ones are yellow. Coexpression modules represent the basic unit of regulation elicited from the coordinated expression of genes in response to environmental or genetic change. To better understand the relationships between transcripts within a module, expression networks are used to identify the overall organization of control within the module. Genes from the blue module from the TOM plot (b) are depicted in network to demonstrate several organizational principles common to all biological networks. Another organizational property of natural networks depicted by the blue module network is that of scale-free topology (pattern of connections between nodes). There are few connections between most nodes, although one or two nodes are highly connected.
Recent coexpression analyses of liver in two F2 crosses from diet-induced obese mice (18) used two different but related methods to find weight gain–associated biomarkers for therapeutic interventions. In the first analysis, functional relationships for a single trait defined clusters of coexpressed genes (modules) for body weight within both data sets. Additionally, QTL analyses were used to find expression hot spots that were highly correlated with the module’s coexpression profile (mQTL) and that localize the most relevant genes that drive expression within the module. A second analysis was used to compare modules between data sets from obese mice and lean mice in 135 females from one cross and previously published data from the other cross (18). Coexpression networks were constructed for obese and lean mouse data sets and evaluated to find differences in functional modules, transcript abundance, and variations in the number of connections between genes (see sidebar Co-expression Networks). Here, the aim was to identify the interactions and expression changes that trigger the phenotypic differences between the obese and lean mouse data sets. A highly significant mQTL was identified, and strong correlations to this mQTL, as well as increased connectivity to genes within and between modules and increased correlation to body weight, were used to prioritize eQTL candidates.
APPLYING SYSTEMS BIOLOGY TO OBESITY GENETICS: BRINGING IT ALL TOGETHER
The blending of genetic, genomic, transcriptome, proteome, and metabolome technologies (Figure 4) to identify the molecular basis for common diseases such as obesity, type 2 diabetes, and heart disease is accelerating rapidly (13, 51). New methodologies that merge analytical techniques from diverse disciplines such as computer science, mathematics, and engineering are currently being developed to keep pace with the quantity and multidimensional quality of the data from high-throughput technologies (65). An example was recently presented by Wentzell et al. (64), who used an integrated systems biology approach to demonstrate that variations in transcript abundance control metabolic variations in a segregating population of Arabidopsis. By delineating the effects of genotypic variation upon quantitative phenotypic alterations in both transcription and metabolite accumulation, they were able to better define the molecular and genetic basis of biosynthetic metabolism. These researchers showed that regulatory connections can feed back from metabolism to transcription, which suggests that natural variation in transcripts not only affects phenotypic variation, but also that natural variation in metabolites or their enzymatic loci can feed back to affect their transcripts (64).
Figure 4.
Complex trait analysis requires an approach that interrogates and integrates multiple levels of organization. Systems biology integrates multidimensional data from multiple experiments that examine global changes in an organism, tissue, or cell in response to alterations in genetic composition or environment. For example, (a) quantitative trail locus (QTL) mapping localizes the genetic influences that underlie a complex trait, and expression profiling identifies alterations in global patterns of expression. When integrated with transcriptome analysis (b) to identify regulatory sites for transcription [trans expression QTL (eQTL) mapping] and functional networks of transcript interaction, a detailed overview change in response to genetic or environmental perturbations emerges. But not all alterations in transcription translate into enzymatic or structural protein and metabolite modifications that are often the hallmark of complex genetic disorders. Therefore, including proteomic assays (c) such as 2-D gel analysis and metabolomic evaluation by mass spectrometry and/or nuclear magnetic resonance testing yields deeper insights into the synergistic interactions that give rise to disease phenotypes and also serve to identify biomarkers for the development of therapeutic agents.
A systems biology approach incorporates the synergistic connections between “omic” and environmental influences into a comprehensive framework. From this, key genetic, transcriptome, proteome, and/or metabolomic interactions can be modeled to provide an integrated approach for identifying critical sites in which to mediate therapeutic change. As the tools for assaying global changes in gene expression, protein expression, and metabolite content become more accessible to researchers, a paradigm shift in the way we approach and analyze complex traits is occurring (4a). Dissection of multi-faceted disorders such as obesity into extensive catalogs of proteins, genes, and metabolites is giving way to the identification of emergent traits. Because complex disorders involve the interactions of many genes, network analysis of coexpressed genes elucidates the context in which genetic and/or environmental perturbations synergize to define health and disease.When combined with high-throughput multidimensional proteomic and metabolomic analyses, the potential to unravel the genetic basis of disease to predict effects of change within a living system comes closer to reality.
Co-expression networks.
Co-expression networks consist of transcripts whose expression are correlated. Transcripts (nodes) are assigned to clusters (modules) within a network based upon similarity measured by correlation patterns patterns (e.g. tissue, genotype, treatments).
Structural (topological) properties of biological networks are used to quantitate the functional importance of interactions that contribute to multifactorial changes which underlie complex traits.
The degree to which one node is connected to another (connectivity - k) is a fundamental and quantifiable characteristic that measures the importance of each node in a network. Biological networks exhibit a scale free topology in which most nodes have few connections. This structure increases the robustness of the network because random elimination of multiple nodes will not cause the functional activity of the network to fail. However, elimination of a highly connected node, such as nodes 1 and 9 above (hubs), or nodes that connect one module to another (node 4) makes these networks susceptible to attack.
The probability that a given node will have k links, the degree distribution - P(k), is used to determine if a network is scale free with few highly connected nodes or randomly connected and therefore more susceptible to system failure if nodes are arbitrarily eliminated. As opposed to a random network that follows a Poisson distribution, scale free degree distributions approximate a power law where P(k) is proportional to k^ -γ. Here γ indicates how important the hubs are within the network, as it reflects how connected the hubs are to the rest of the network (3).
THE CHANGING FACE OF THE NATURE OF VARIATION IN OBESITY PHENOTYPES
Several important findings have recently emerged that indicate increasing levels of complexity in the nature of variation underlying, and heritability of, complex traits such as obesity. These include the burgeoning importance of RNAs, additional new sources of variation such as copy number variants, and transgenerational epigenetics.
“New” Sources of Genetic Variation: The Rise of RNA
Until recently, relevant genetic variation was thought to only exist within annotated genes and their closely linked regulatory regions (Figure 5). The annotated human genome contains 20,000 to 25,000 protein-coding genes, which is similar to reported estimates for the yeast, fly, and worm genomes. Since mammals presumably require more proteins to function than these lower animals, it has been thought that additional protein complexity is produced by alternatively splicing transcripts from the same locus. New genome-wide techniques are showing that several additional transcriptional mechanisms could account for protein diversity in mammals (Figure 5), which suggests that nucleotide sequence polymorphisms that affect development of complex phenotypes could exist in nearly any region of the genome.
Figure 5.
The transcriptome is large and complex. (a) Previously, it was thought that only genes were transcribed into functional transcripts. Genes were composed of a transcriptional start site, regulatory elements (not shown), exons (colored nodes), and introns, which were transcribed to produce primary transcripts that could be alternatively spliced to produce additional protein-coding transcripts. Thus, only polymorphisms within gene regions (and their closely linked regulatory regions) were thought to affect phenotypes. (b) The genome is pervasively transcribed. Annotated gene regions can produce many varieties of transcripts. Multiple transcriptional start sites appear to produce varieties of transcripts from overlapping DNA sequences, including short RNAs (colored rectangles). Interestingly, exons from different annotated gene regions and separated by long distances can be fused to form transcripts. Additional transcripts are produced from sense intronic (e.g., short RNAs), antisense DNA sequences and intergenic gene regions (not shown). Therefore, DNA sequence polymorphisms throughout the genome could affect phenotypes. MS, mass spectrometry; NMR, nuclear magnetic resonance.
The Functional Annotation of the Mouse (Fantom) Project and Encyclopedia of DNA Elements (ENCODE) Project consortia have recently published pilot data showing that the transcriptome is far larger and more biologically active than previously thought. Several lines of evidence from these studies support the idea that it is time to reconsider how we relate genetic variation to phenotypes (19). First, the Fantom3/Genome Network Project found that 66% of the mouse genome is transcribed (3), and the ENCODE Project Consortium estimated that as much as 93% of the human genome is capable of being transcribed, which is in stark contrast to previous estimates of 1% to 2% for messenger ribonucleic acid (mRNA) protein-coding regions (14). The ENCODE project found that a large population of these transcripts were unannotated and have no known function, they are referred to as transcripts of unknown function (TUFs). Second, >65% of the annotated genes have additional 5′ distal transcriptional start sites (TSSs) that appear to be tissue specific and relate to the production of TUFs (11). Third, annotated genes averaged 5.4 transcripts, with only 1.7 potentially coding for proteins (11). Many of these additional transcripts are antisense transcripts (from strand opposing the protein-coding strand) that overlap the same region of annotated genes from sense transcripts (from the protein-coding strand of DNA) (14, 29). Fourth, mature transcripts can be created by using exons from annotated introns and intergenic regions (48) as well as by fusing exons separated by long distances and containing intervening loci (48). Fifth, epigenetic chromatin structure and histone modifications can effectively predict the location and activity of TSSs (48).
Taken together, these results indicate that phenotypic variation is not necessarily limited to genetic variation within annotated gene regions, because transcripts can be made from DNA sequences outside classic gene regions. Therefore, SNPs outside of annotated gene regions could very well provide a large source of genetic variation contributing to the development of complex traits like obesity. Hence, the search for obesity genes may lead to genomic sequences that are not genes at all. And given the complexity of the genomic machinery that produces transcripts, it has been suggested that it may be more straightforward to treat functional RNA transcripts as the fundamental units of the genome (19) and potentially as the mechanism for translating genomic architecture to phenotype.
The identification of large numbers of TUFs raises questions concerning their function and the possibility that they are involved in biology of metabolism and obesity. We can get an idea of their biological role by considering what we already know about small RNAs such as microRNAs (miRNAs) and short interfering RNAs (siRNAs). miRNAs and siRNAs are known to be critically involved in regulating mRNA translation and degradation, and their regulatory pathways are collectively referred to as RNA interference (RNAi) (for review, see 60). The function of miRNA remains poorly understood, but generally it is thought that miRNA often reduces translation efficiency through imperfect base pairing to mRNA. Interestingly, miRNA appears to play an important role in metabolic disorders. For example, in Drosophila, miRNA Mir-14 regulates fat metabolism (66), and miRNA-278 regulates energy balance (57). In humans, miR-375 regulates insulin secretion (44). Based on these and similar results, an RNAi biotechnology industry has grown around efforts to target RNAs for pharmacological treatment of disorders such as obesity.
Given the emerging biological role of RNA transcripts and their potential relevance in metabolism and obesity, polymorphisms in the underlying DNA sequences that transcribe TUFs could be considered as a potentially rich source of functional genetic variation. For now, evidence is emerging that miRNA target sites may be an important polymorphic layer of functional control. Chen & Rajewsky (4) found that SNP polymorphisms in predicted miRNA binding sites are likely to be deleterious and are thus likely candidates for causal variants of human disease. Saunders et al. (49) analyzed publicly available SNP data in context with miRNAs and their target sites throughout the human genome and found a relatively low level of variation in functional regions of miRNAs, but an appreciable level of variation at target sites. And Yu et al. (68) showed that SNPs located in miRNA-binding sites affect miRNA target expression and function and are potentially associated with cancers.
Fortunately, emerging high-throughput and very dense genotyping platforms will, eventually, insure that all functional regions of the genome are included in scans for genetic variation associated with obesity-related complex traits. Although such scans will be inherently unbiased in terms of finding the general location of this underlying variation, strategies for subsequent identification of the causal variants may need to be broadened. And polymorphisms related to miRNAs potentially add complexity to the overall genetic architecture of complex traits such as obesity. A functional SNP in a region transcribing miRNA may have extensive pleiotropic effects while also interacting with SNPs at miRNA target sites, leading to increased epistasis.
“New” Sources of Genetic Variation: Copy Number Variants
Copy number variations (CNVs) are emerging as another important “new” source of genetic variation. As opposed to SNPs, which are polymorphisms in single nucleotides, CNVs are alterations involving insertions, deletions, duplications, and complex sequence rearrangements larger that 1 Kb. CNVs can alter phenotypes by altering gene dosage, interfering with gene expression, and disturbing gene regulation. CNVs were first identified in the 1980s but have been attracting more attention recently because they are more common than previously thought (54). They are also becoming highly associated with disease-related traits. For example, CNVs are associated with susceptibility to HIV-1 (24) and lupus (1), are likely associated with susceptibility to cancer (6), and probably play a role in modulating drug responses (37). An interesting example of CNVs related to nutrition was recently presented by Perry et al. (39), who found that copy number of the salivary amylase gene is positively correlated with salivary amylase protein level and that individuals from populations with high-starch diets have, on average, more salivary amylase gene copies than those with traditionally low-starch diets. This example of positive selection on a copy number–variable gene has implications for the role of CNVs in contributing to genetic variation for obesity and related traits.
Not surprisingly, CNVs are becoming recognized as important sources of genetic variability contributing to the expression of mouse phenotypes. The capability to produce mouse high-resolution maps of CNVs was recently demonstrated using inbred mice comprising the Mouse Phenome Database. Annotated genes within the CNVs associate with known mouse phenotypic traits (25). CNV maps have also been produced for 42 inbred mice comprising the Mouse Phenome Consortium (9). Food intake and CNV associations were tested to determine the utility of performing phenotype/CNV associations. Interestingly, mouse strains with either no amplification, duplications, or quadruplications across the candidate CNV locus had the highest, intermediate, and lowest food intake levels, respectively (9). These results show that different inbred mouse strains contain unique CNVs that can be associated with specific phenotypic traits and that CNVs may be a source of functional genetic variation contributing to complex obesity. Given emerging technologies to rapidly genotype CNVs on a genome-wide basis and the already dense map of genetic predisposition for obesity in mice (42), the importance of CNVs in genetic regulation of obesity should soon be elucidated.
“New” Sources of Phenotypic Variation: Epigenetics
It is widely accepted that most diseases and phenotypic traits develop from a complex interaction between the genome and environmental factors. Although the nature of this interaction remains poorly understood, epigenetic research has yielded remarkable insights into how the epigenome can modulate phenotypic plasticity. Epigenesis refers to chemical modifications to the genome above the DNA sequence level and falls into two main categories: DNA methylation and histone modification (for reviews, see 15, 46). The interaction between the genome and environment via epigenesis is a key factor determining phenotypic plasticity, which can be defined as the ability of cells to change their behavior in response to internal or external environmental cues (15). This ability is a key process of normal development that enables developing phenotypes to become tuned with expected environments.
Several lines of rodent research strongly suggest that early developmental epigenetic mechanisms are causally linked to metabolic disease susceptibility. Diet exerts particularly strong effects on metabolic phenotypes via complex epigenetic processes. For example, when inbred C57BL/6J mice are fed a high-fat diet and other environmental factors are tightly controlled, Koza et al. (30) found that adiposity and diabetes traits as well as gene expression underlying these traits are highly variable. As another example, rat dams given a low-protein diet during pregnancy produce offspring that develop abnormal metabolic function associated with gene methylation and histone acetylation of metabolically relevant receptors (33). Concurrently supplementing rat dam diets with folate, which promotes methyl group provision, prevents development of metabolic abnormalities of the offspring (34). Another interesting example comes from two studies showing that the phenotypic outcome induced by a transient cue at a late period of development is dependent upon earlier developmental environmental history. Using an in utero maternal undernutrition model, Vickers et al. (61) showed that the increased adult rat adiposity effects stemming from maternal undernutrition were offset by neonatal leptin treatment. They concluded that leptin treatment signaled adiposity to thin pups, which normalized their metabolic development to produce a phenotype adapted to a high-nutrition diet in adulthood. Using a similar experimental design, the same group showed that adult hepatic gene expression and epigenetic status were directionally dependent upon their in utero maternal diet (20). These results support a developmental origins hypothesis for elements of metabolic diseases (22, 62).
“New” Modes of Inheritance: Transgenerational Epigenetics
Although epigenetic mechanisms are critical for normal and disease-related somatic cell traits acquired over a single generation, a fascinating question—especially in regard to complex trait genetics—is whether epigenetic modifications within one generation can be transmitted to the next generation and beyond. A growing body of epidemiological and biological research shows that a range of parental traits can be inherited by offspring even in the absence of the cues that originally produced the parental trait. Environmental challenge during pregnancy has produced a number of interesting effects, several involving nutritional and endocrine manipulations, that could be explained by epigenetic inheritance (for review, see 21). One of the more interesting examples has been reported in humans. Pregnant women exposed to the Dutch famine of 1944/1945 gave birth to smaller babies, an effect that persisted in their grandchildren (55). This suggests that certain metabolic-disorder phenotypes may reflect a mismatch in predicted environments both within one generation, via epigenetic mechanisms, and between generations, presumably via transgenerational epigenetic inheritance.
According to the Neo-Darwinistic theory of evolution, new phenotypes are created only via random mutations in the DNA sequence; adaptive phenotypes are naturally selected and transmitted to offspring via the DNA sequence in germ cells. Evidence for transgenerational epigenetic inheritance calls into question the dogma that DNA variation is the only source of heritable variation. This adds an entirely new level of complexity into the science of understanding the genetics of obesity. Because of their rapid generation interval and their relative ease of experimental pliability, mouse models will play an instrumental role in elucidating the effect of transgenerational epigenetics on obesity-related traits.
ACKNOWLEDGMENT
The authors are grateful to Atila Van Ness for assistance with creation of several figures.
Glossary
- eQTL
expression quantitative trait loci
- RNA
ribonucleic acid
- SNPs
single-nucleotide polymorphisms
- CD36
thrombospondin receptor (also referred to as FAT, GP4, GP3B, GPIV, CHDS7, PASIV, SCARB3)
- DNA
deoxyribonucleic acid
- mQTL
module quantitative trait loci
- Fantom
Functional Annotation of the Mouse
- ENCODE
Encyclopedia of DNA Elements
- mRNA
messenger ribonucleic acid
- TUFs
transcripts of unknown function
- TSSs
transcriptional start sites
- miRNAs
micro ribonucleic acids
- siRNAs
short interfering ribonucleic acid
- RNAi
ribonucleic acid interference
- CNVs
copy number variations
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 2.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 3.Barabasi AL, Oltavai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 4.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 4a.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 6.Cho EK, Tchinda J, Freeman JL, Chung YJ, Cai WW, Lee C. Array-based comparative genomic hybridization and copy number variation in cancer research. Cytogenet. Genome Res. 2006;115:262–272. doi: 10.1159/000095923. [DOI] [PubMed] [Google Scholar]
- 7.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 8.Copland JA, Davies PJ, Shipley GL, Wood CG, Luxon BA, Urban RJ. The use of DNA microarrays to assess clinical samples: the transition from bedside to bench to bedside. Recent Prog. Horm. Res. 2003;58:25–53. doi: 10.1210/rp.58.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demarest K, Koyner J, McCaughran J, Jr, Cipp L, Hitzemann R. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav. Genet. 2001;31:79–91. doi: 10.1023/a:1010261909853. [DOI] [PubMed] [Google Scholar]
- 11.Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 13.Drake TA, Schadt EE, Lusis AJ. Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mamm. Genome. 2006;17:466–479. doi: 10.1007/s00335-005-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ENCODE Proj. Consort. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 16.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 17.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 20.Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 21.Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, et al. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc. Natl. Acad. Sci. USA. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 2007;61 doi: 10.1203/pdr.0b013e318045bedb. 5–10R. [DOI] [PubMed] [Google Scholar]
- 23.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 25.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 27.Jackson Laboratory. Mouse Genome Informatics Database. 2008 http://www.informatics.jax.org.
- 28.Jackson Laboratory. The Mouse Phenome Database. 2008 http://www.informatics.jax.org.
- 29.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 30.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lightfoot JT, Turner MJ, Pomp D, Kleeberger SR, Leamy LJ. Quantitative trait loci (QTL) for physical activity traits in mice. Physiol Genomics. 2008 doi: 10.1152/physiolgenomics.00241.2007. DOI: 10.1152/physiolgenomics.00241.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 34.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br. J. Nutr. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE. 2007;2:e651. doi: 10.1371/journal.pone.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc. Natl. Acad. Sci. USA. 2000;97:11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouahchi K, Lindeman N, Lee C. Copy number variants and pharmacogenomics. Pharmacogenomics. 2006;7:25–29. doi: 10.2217/14622416.7.1.25. [DOI] [PubMed] [Google Scholar]
- 38.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 39.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- 41.Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pomp D. In: Natural polygenic models. In Obesity: Genomics and Postgenomics. K Clement, T Sorensen., editors. New York: Informa Healthcare; 2007. pp. 125–142. [Google Scholar]
- 43.Pomp D, Allan MF, Wesolowski SR. Quantitative genomics: exploring the genetic architecture of complex trait predisposition. J. Anim. Sci. 2004;82 E-Suppl:E300–E312. doi: 10.2527/2004.8213_supplE300x. [DOI] [PubMed] [Google Scholar]
- 44.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 45.Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput. Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 47.Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozowsky JS, Newburger D, Sayward F, Wu J, Jordan G, et al. The DART classification of unannotated transcription within the ENCODE regions: associating transcription with known and novel loci. Genome Res. 2007;17:732–745. doi: 10.1101/gr.5696007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schadt EE, Lum PY. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J. Lipid Res. 2006;47:2601–2613. doi: 10.1194/jlr.R600026-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 53.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 54.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 55.Stein AD, Lumey LH. The relationship between maternal and offspring birth weights after maternal prenatal famine exposure: the Dutch Famine Birth Cohort Study. Hum. Biol. 2000;72:641–654. [PubMed] [Google Scholar]
- 56.Takahashi K, Mizuarai S, Araki H, Mashiko S, Ishihara A, et al. Adiposity elevates plasma MCP-1 levels leading to the increased CD11b-positive monocytes in mice. J. Biol. Chem. 2003;278:46654–46660. doi: 10.1074/jbc.M309895200. [DOI] [PubMed] [Google Scholar]
- 57.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. USA. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 60.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 61.Vickers MH, Gluckman PD, Coveny AH, Hofman PL, Cutfield WS, et al. Neonatal leptin treatment reverses developmental programming. Endocrinology. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- 62.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 63.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 66.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 67.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]