Abstract
The design of effective pH responsive MRI contrast agents is a key goal in the development of new diagnostic methods for conditions such as kidney disease and cancer. A key factor determining the effectiveness of an agent is the difference between the relaxivity of the “on” state compared to that of the “off” state. In this paper, we demonstrate that it is possible to improve the pH-responsive action of a low molecular weight agent by conjugating it to a macromolecular construct. The synthesis of a bifunctional pH responsive agent is reported. As part of that synthetic pathway we examine the Ing–Manske reaction, identifying an undesirable by-product and establishing effective conditions for promoting a clean and effective reaction. Reaction of the bifunctional pH responsive agent with a G5-PAMAM dendrimer yielded a product with an average of 96 chelates per dendrimer. The relaxivity of the dendrimer conjugate rises from 10.8 mm−1 s−1 (pH 9) to 24.0 mm−1 s−1 (pH 6) per Gd3+ ion. This more than doubles the relaxivity pH response, Δr1, of our agent from just 51% for the original low molecular weight chelate to 122% for the dendrimer.
Keywords: dendrimers, imaging agents, lanthanide complexes, MRI contrast agents
Introduction
Diseases, such as kidney disorders and cancer, often result in a significant reduction in the extracelluar pH of these tissues.[1–3] Thus, imaging tissue pH could be quite useful in clinical diagnose of these and other conditions. Current methods for assessing tissue pH involve relatively invasive procedures and typically can assess pH only from a limited number of locations.[2–4] Less invasive techniques such as 31P NMR NMR spectroscopy[4] can provide a direct measure of pH but the concentrations of the endogenous phosphorus metabolites that respond to tissue pH are relatively low so pH measurements can only be taken from relatively large volumes of tissue. pH sensitive fluorescence dyes are quite sensitive but applications of optical techniques in human imaging is limited to tissues near the surface of skin where sufficient light penetration can be achieved.[4] A pH responsive T1-shortening contrast agent could offer a minimally invasive and highly practical approach to mapping of tissue pH so it is hardly surprising that a large number of pH sensitive MRI contrast agents have been reported.[5–13]
Typically, Gd3+-based complexes used to relax bulk water have relatively low T1 relaxivities. Relaxivity is defined as the increase in water proton relaxation rate per unit concentration of contrast agent and is a measure of the effectiveness of a contrast agent. At typical magnetic fields used for clinical imaging, the relaxivity of a Gd3+ complex is primarily determined by three factors: q, the number of water molecules in the inner coordination sphere of Gd3+; τM, the residence lifetime of these water molecule on Gd3+; and τR, the rotational correlation time, or how fast the complex tumbles in solution.[14–18] Examples of contrast agents that respond to pH through changes in each of these parameters have been reported,[5] but the only agent actually used to image tissue pH to date is Gd1 (see below).[12, 13] Although Gd1 exhibits a relatively small a change in relaxivity over a physiologically relevant pH range, it has been used successfully in vivo to generate pH maps of kidneys and tumors in small animals.[19–21] The mechanism by which Gd1 operates as a pH responsive agent is unique among responsive Gd3+ agents and stems from the presence of the phosphonate groups of the pendant arms.[12] The sole inner-sphere water molecule of Gd1 is in slow exchange with the bulk solvent which, ordinarily, would limit relaxivity. However, the phosphonate groups of Gd1 are able to catalyse the exchange of the protons of this single Gd3+-bound water molecule with bulk solvent protons. The effectiveness of the phosphonates at catalysing proton exchange is dependent upon their protonation state so as the four phosphonates become successively protonated at the pH falls below ≈8.5, the rate of proton exchange increases and the paramagnetic relaxation effects of Gd3+ are transferred to the bulk solvent protons. In addition the phosphonates can organise a number of other water molecules into a second hydration sphere through hydrogen-bonding interactions.[22, 23] The extent, and proton residence lifetime, of this second hydration sphere is also likely to fluctuate with the protonation state of the phosphonate groups. The increase in relaxivity of Gd1 on passing from pH 9 to 6 was found to be the result of a combination of these two effects.[12]
Despite the demonstrated utility of Gd1, the maximum increase in relaxivity with changing pH, Δr1, is relatively modest, rising from 3.5 mm−1 s−1 at pH 9.5 to 5.3 mm−1 s−1 at pH 6.3; a Δr1 of just 51%.[12] The relaxivity arising from an exchanging inner-sphere water molecule, r1 i.s., is given by Equation (1), where τM is the lifetime of the inner-sphere bound water molecule and T1M is the T1 of the inner-sphere water protons. The relaxivity arising from water molecules in the second hydration sphere follows a similar relationship. From Equation (1) it can be seen that relaxivity will be higher if T1M is shorter, but the rapid rotational dynamics of Gd1 cause T1M to be long thereby limiting relaxivity.
| (1) |
Slowing molecular rotation is most easily achieved by conjugating the contrast agent to a macromolecule, such as a dendrimer. However, improving relaxivity alone will not improve the function of Gd1 as a pH responsive contrast agent. To do that it is necessary to accentuate the difference between the high and low relaxivity regimes. We hypothesized that T1M could be reduced by conjugating Gd1 to a dendrimer which would render the system more sensitive to changes in τM. At higher pH values, the relaxivity would be expect to be limited by slow exchange whereas at lower pH values, catalysis of inner-sphere proton exchange by the phosphonate groups should lift the limiting effect of exchange upon relaxivity (T1M > τM). Overall, coupling Gd1 to the dendrimer should improve both the absolute relaxivity, r1, and the change in relaxivity, Δr1, on going from low to high pH. We therefore set out to conjugate Gd1 to a larger macromolecule, in this case an ethylenediamine core G5-PAMAM dendrimer, in order to reduce the rate of rotation and improve the pH responsive characteristics of the complex.
Results and Discussion
Synthesis
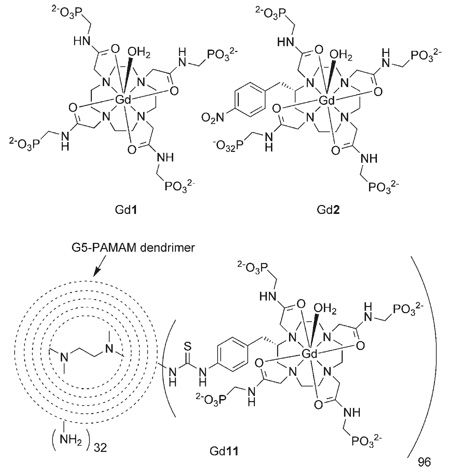
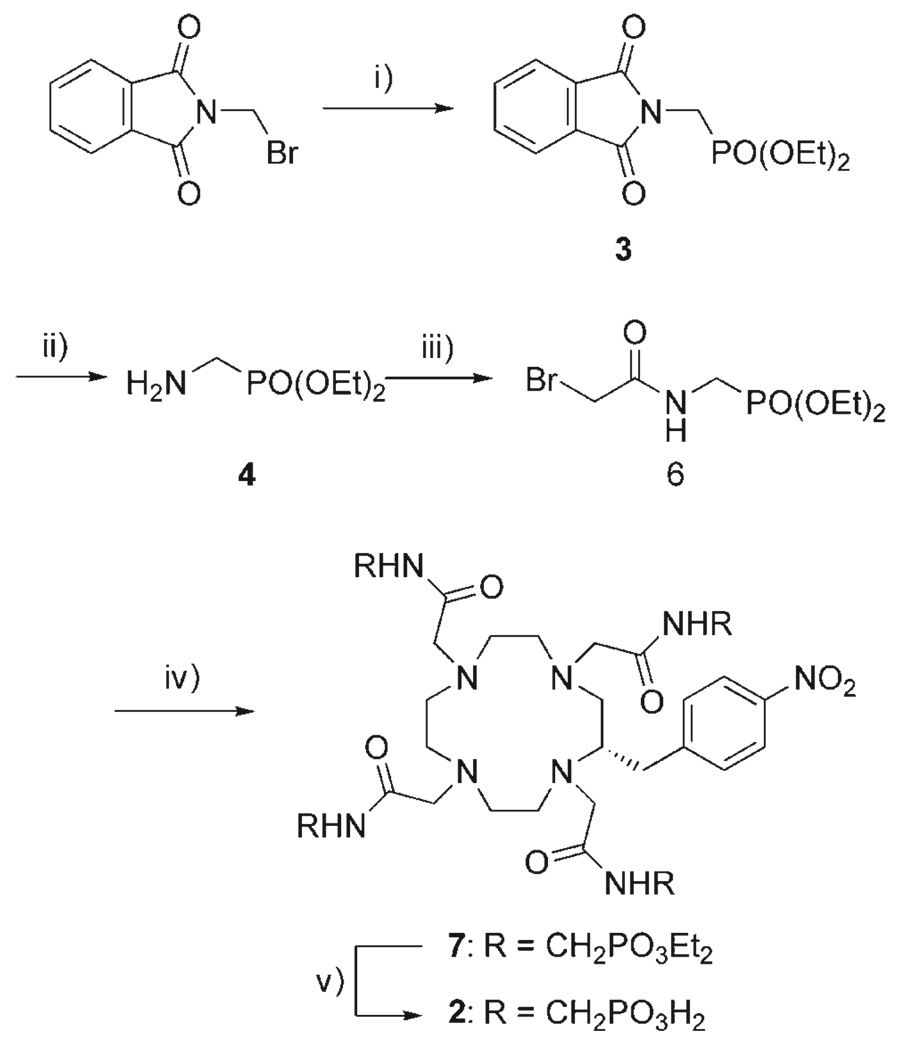
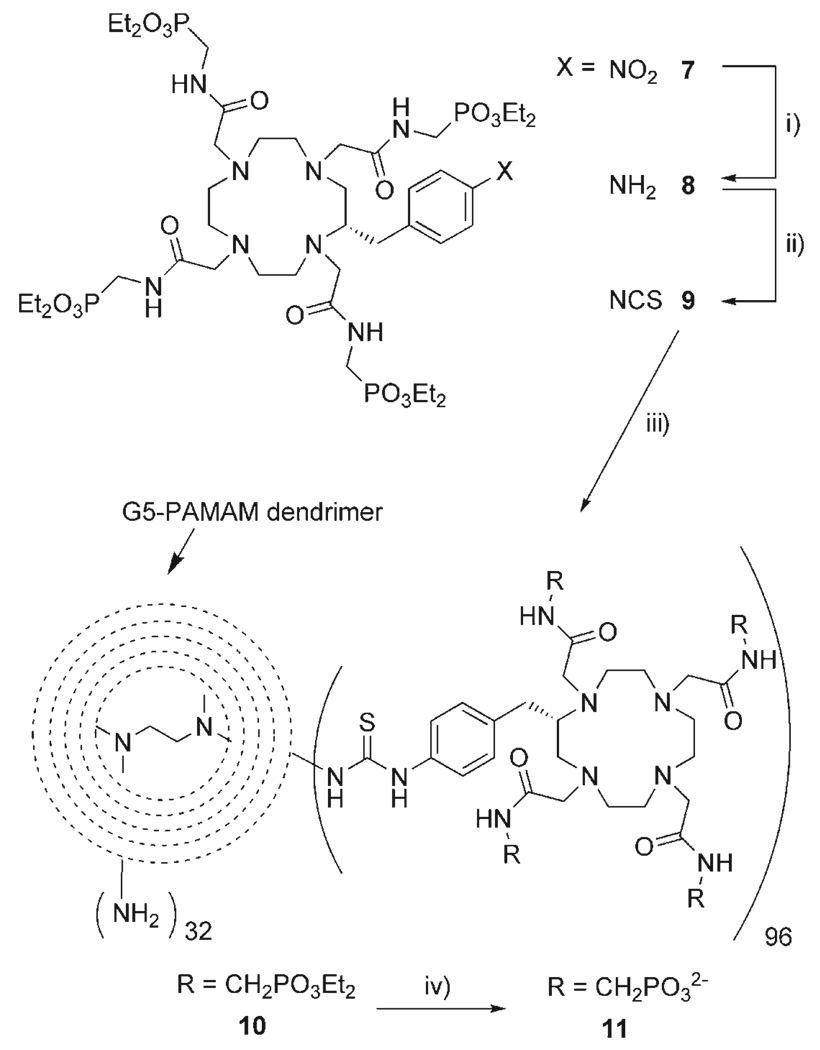
As the phosphonate groups of the pendant arms were found to be responsible for the pH responsive nature of Gd1,[12] it was important to maintain this structural feature when the complex was modified to facilitate attachment to the dendrimer. Accordingly, the complex was modified by incorporating a functionalized benzyl group onto the macro-cyclic backbone of the complex, leaving the four phosphonate groups intact. The functionalized complex Gd2 was prepared following the same synthetic procedure used for the preparation of Gd1[12] simply substituting (S)-2-(p-nitrobenzyl) cyclen for cyclen (Scheme 1).
Scheme 1.
Synthesis of a functionalised pH responsive contrast agent. i) P(OEt)3/Δ ii) H2NNH2/EtOH; iii) BrCH2COBr/K2CO3/CH2Cl2; iv) (S)-2-(p-nitrobenzyl) cyclen/K2CO3/MeCN/60°C; v) 30% HBr in AcOH.
Compound 3 was prepared by the Michaelis–Arbuzov reaction of triethylphosphite with bromomethyl phthalimide. The key intermediate 4 was then obtained by removal of the phthalimide protecting group by an Ing–Manske[24] reaction with 1.2 equivalents of hydrazine in ethanol. Normally this type of deprotection reaction proceeds in good yields;[24] however, in our case the yields were moderate at best.[12] Furthermore, the presence of significant quantities of a reaction by-product meant that column chromatography was necessary to purify amine 4, an oil at room temperature. Amines can often be purified by an acid/base extraction but 4 shows good water solubility, even at high pH, and this precluded purification by extraction. Low yielding reactions that require complicated purification procedures are unsuitable for the scale-up necessary for the production of large quantities of an MRI contrast agent. We therefore undertook an investigation into the reasons for the low reaction yield of this step.
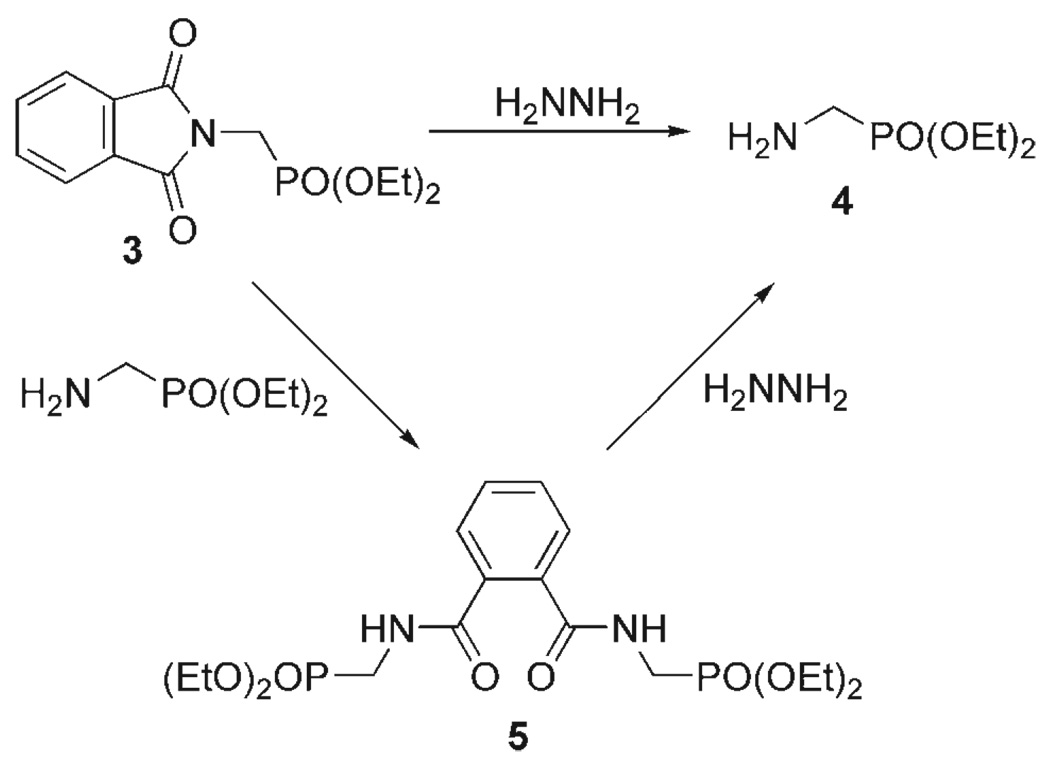
1H NMR analysis of the crude reaction mixture indicated that the primary contaminant of the product 4 was a single reaction by-product. This compound was isolated by column chromatography and characterized. Rather surprisingly, this by-product was identified as compound 5, the product of a reaction between the starting material 3 and the intended reaction product 4 (Scheme 2). A significant effort has been applied to understanding the mechanism and intermediates of the Ing–Manske reaction,[25–28] however, this particular reaction pathway is rarely included in these discussions.[29] The quantities of 5 produced suggest that this side reaction may, on occasion, be more important than generally described. The mechanism of the Ing–Manske reaction is quite complex[26–29] but it is initiated by nucleophilic attack of hydrazine at one of the carbonyls of the phthalimide. The effectiveness of an amine at removing phthalimide protecting groups has been shown to relate to the protonation constant of the attacking amine; the more basic the amine, the more effective the reaction.[27] Thus, if the product amine is significantly more basic than hydrazine, it is likely to compete with hydrazine in attacking the phthalimide 3 to yield significant quantities of compound 5.
Scheme 2.
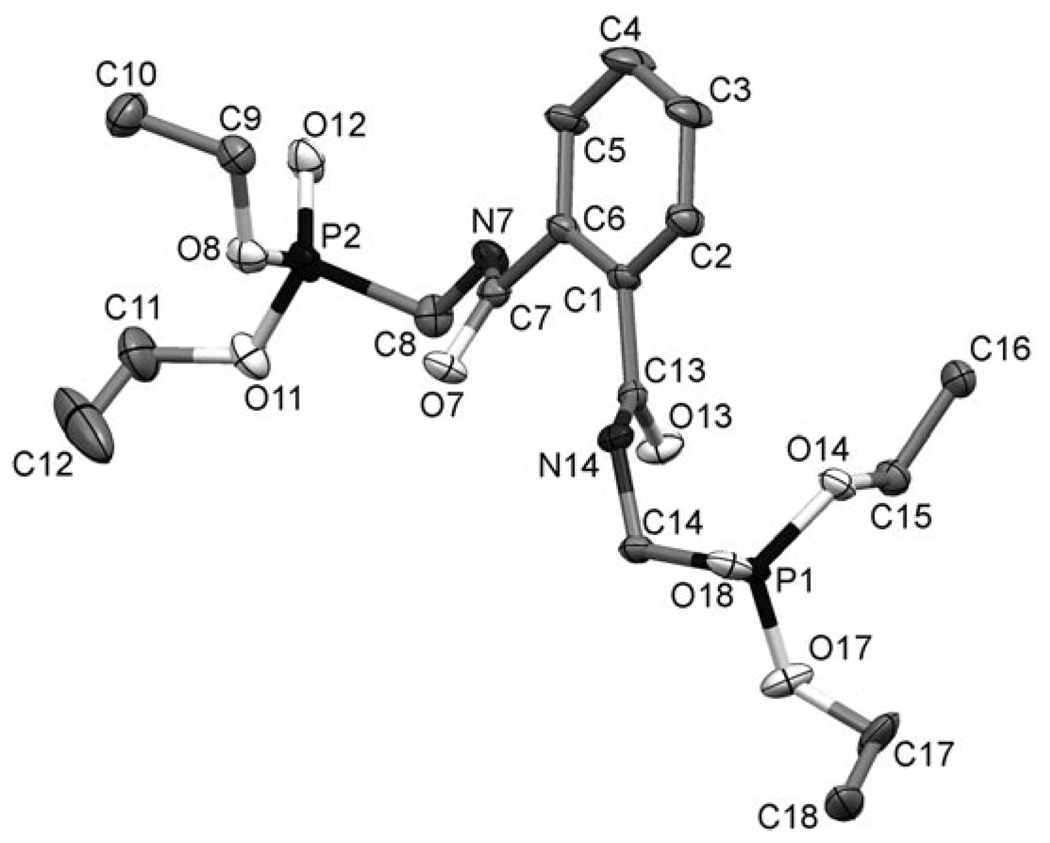
The identity of compound 5 was established by NMR spectroscopy and mass spectrometry and, although attempts to remove compound 5 from the crude reaction mixture by crystallisation were unsuccessful, compound 5 readily crystallises once purified. X-ray quality crystals could be grown at room temperature from a solution of 5 in dichloromethane by addition of diethyl ether and hexanes. This allowed the structure of compound 5 to be confirmed by X-ray crystallography (Figure 1). The production of compound 5 during the deprotection of the phthalimide 3 is doubly detrimental to the yield of 4 since two molecules of 4 are taken up into one molecule of 5; this is in addition to the need for chromatography. Thus, eliminating compound 5 from the reaction would both improve yield and simplify purification since the other reaction by-product, the insoluble phthalhydrazide, can be removed by filtration. A number of improvements to the Ing–Manske reaction have been suggested, most notably raising the reaction pH through addition of sodium hydroxide.[26, 27] However, concerns over the lability of the phosphonate diester moiety towards base-catalysed hydrolysis precluded this as a solution to this problem. We investigated a number of the other Ing–Manske reaction conditions to assess whether the reaction conditions could be improved.
Figure 1.
ORTEP rendering of the crystal structure of 5 showing 50% ellipsoids. Hydrogen atoms have been omitted for clarity.
Compounds 4 and 5 are easily distinguished by 1H NMR spectroscopy; a shift difference of almost 1 ppm is observed for the methylene resonance α to the phosphorous. This resonance appears as a doublet at 2.9 ppm (coupled to phosphorous) in 4 and as a doublet of doublets at 3.8 ppm (coupled to phosphorous and the amide proton) in 5. A series of experiments were performed in which various reaction conditions were altered and the ratio of the two reaction products in the crude reaction mixture determined by 1H NMR spectroscopy. Neither increasing the reaction temperature from room temperature to reflux nor increasing the reaction time from 6 h to 5 d was found to have a significant effect upon the distribution of reaction products. However, when the amount of hydrazine used in the reaction was increased the amount of 4 obtained from the reaction was greatly increased at the expense of 5. Increasing the amount of hydrazine from one to four equivalents greatly reduced the amount of 5 produced in the reaction. At six equivalents of hydrazine the yield of 5 was < 5%, and ten equivalents of hydrazine eliminated all traces of 5 from the reaction 1H NMR spectrum. This observation does not indicate that 5 is not produced during the reaction. Indeed, when an purified sample of 5 was treated with ten equivalents of hydrazine it was found to undergo a quantitative conversion to 4, as determined by 1H NMR (Scheme 2). Thus, 5 may still be produced in the reaction but, importantly, it is reactive under these conditions and does not persist as a reaction product.
Changing the conditions of the Ing–Manske reaction by increasing to ten the equivalents of hydrazine allowed purification to be simplified from column chromatography to filtration to remove the phthalhydrazide. It also improved the reaction yield to 97% and rendered this process suitable for scale-up. Condensation of the amine 4 with bromoacetyl bromide afforded the bromoacetamide 6 which was used to alkylate (S)-2-(p-nitrobenzyl) cyclen in acetonitrile with K2CO3 as base (Scheme 1). Subsequent deprotection of the phosphonate esters of 7 with HBr in AcOH afforded the ligand 2 a functionalized analogue of 1 that preserved the integrity of the four phosphonate groups.
It was later found that conjugation of the bifunctional ligand to the PAMAM dendrimer was more efficient if the protected ester ligand 7 was used instead of the free acid. Reduction of the nitro group with hydrogen and palladium catalyst afforded the corresponding amine 8 in 72% yield (Scheme 3). The amine was then converted to the isothiocyanate 9 by reaction with thiophosgene in a biphasic reaction at pH 2. The isothiocyanate group is ideal for conjugation with primary amines, such as those that decorate the surface of the PAMAM dendrimer, under mild conditions. The phosphonate ester was preferred for this conjugation reaction in order to minimize non-reactive salt formation between the amines of the dendrimer and phosphonic acids and repulsion between conjugated and incoming phosphonates. The ethylenediamine core G5-PAMAM dendrimer selected as the basis of our macromolecular construct has 128 primary amine groups on its surface. The dendrimer was reacted with 256 equivalents of 9 for 24 h at 40 °C followed by a further 256 equivalents for 48 h. The reaction pH was maintained at 9 throughout by addition of a 1m solution of NaOH. The reaction was analysed by HPLC using a Phenomenex BIOSEP SEC S-3000 size exclusion column (5–700 kD, PBS buffer, pH 7.4). The chelate–dendrimer conjugate was purified by repeated diafiltration using a Centricon C-30 membrane with a 30 kD cut-off (Millipore) until no low molecular weight materials could be detected by HPLC. HPLC analysis of the resulting dendrimer indicated that an average hydrodynamic volume equating to a molecular weight of about 140 kD was achieved. This corresponds to an average of 75% coverage or 96 ligands per dendrimer. A ligand/dendrimer coverage ratio of 97:1 was confirmed by elemental analysis of the carbon and sulfur content of the conjugate. Similar loading values were obtained by 1H NMR analysis of the aromatic and alkyl protons, however, the reproducibility of loading values determined by this method was poorer than SEC and combustion analysis. The conjugation reaction was performed in H2O, DMSO and mixtures of the two, the extent of ligand/dendrimer ratio was found to be unaffected by the choice of solvent. The phosphonate esters of the conjugate 10 were finally removed under identical conditions to those used in the preparation of the ligand 2, HBr and AcOH, to afford the conjugate 11.
Scheme 3.
Synthesis of the dendrimer-based pH responsive contrast agent Gd11. i) H2/Pd on C/H2O; ii) SCCl2/CHCl3/H2O pH 2; iii) G-5 PAMAM dendrimer/H2O pH 8; iv) 30% HBr in AcOH.
Formation of the gadolinium complexes of 2 and 11 also requires special attention. With most DOTA–tetraamide ligands, this step is relatively simple; however, we have recently reported that the phosphonate groups of 1 can interfere with these complexation reactions[12] so care was taken to perform the complexation reactions of both 2 and 11 at pH 9 in order to ensure that the gadolinium ion was bound by the macrocyclic coordination cage. In the case of ligand 2 equimolar amounts of ligand and gadolinium chloride hexahydrate were reacted together at pH 9 and 60 °C in aqueous solution. No further purification was undertaken. However, 1.2 equivalents of gadolinium chloride were used in the reaction with 11 to ensure complete reaction of the ligands. After 48 h at 40 °C and pH 9 in aqueous solution the excess gadolinium was removed by addition of EDTA followed by dialysis in water (12 kD molecular weight cut-off, Sigma Aldrich). The conjugate Gd11 was then further purified by diafiltration with Centricon C-10 (10 kD cut-off) in water at pH 7.4. Although addition of the Gd3+ ion into each ligand of the conjugate would lead to a substantial increase in molecular weight, this increase would be expect to have little or no effect upon the hydrodynamic volume of the conjugate. Thus, size exclusion HPLC was used to verify that the apparent molecular weight remained near 140 kD.
Relaxometric studies
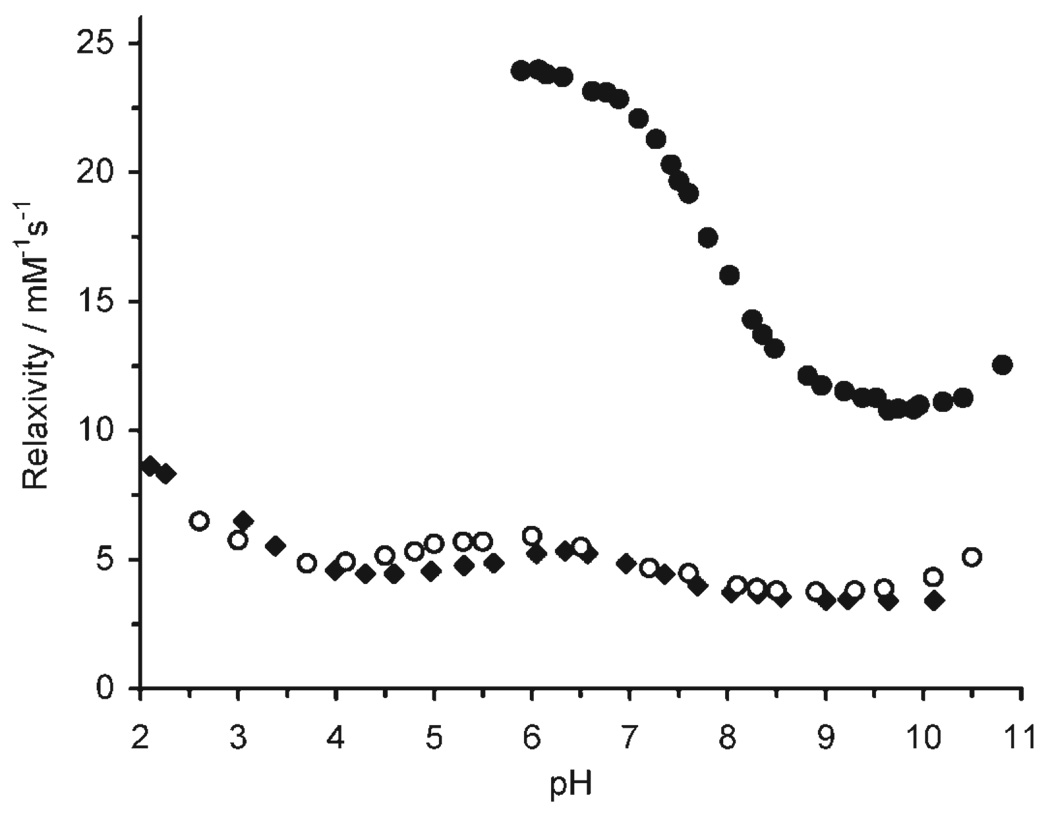
The relaxivity pH profile of Gd2 was recorded and compared with that of Gd1 to ensure that the introduction of the benzylic function on the macrocyclic ring did not negatively impact the pH responsive properties of the complex. The relaxivity pH profiles of Gd1 and Gd2 (Figure 2) are comparable, rising and falling to approximately the same relaxivity at approximately the same pH. The only slight difference between the two profiles is that the relaxivity of Gd2 is slightly higher than that of Gd1 between pH 4 and 6. This may be a reflection of slight changes in the protonation constants of the phosphonates, but nonetheless indicates that introduction of the nitrobenzyl substituent does not negatively impact the behaviour of Gd2. Like Gd1 the relaxivity of Gd2 changes over a physiologically relevant pH range. The relaxivity pH profile of Gd11 is also shown (Figure 2); here the advantage of conjugating the low molecular weight chelate to a dendrimer is immediately apparent. The relaxivity of Gd11 changes over the same physiologically relevant pH range as that of Gd1 and Gd2 but is much higher on a per Gd3+ basis, rising from 10.8 mm−1 s−1 at pH 9.5 to 24.0 mm−1 s−1 at pH 6. (On a per molecule basis relaxivity rises from 1037 mm−1 s−1 to 2304 mm−1 s−1.) This equates to a relaxivity pH response, Δr1, of 122% on passing from pH 9.5 to 6.0, more than doubling the Δr1 of Gd1 and Gd2, Δr1=51 and 59%, respectively, over the same pH range. Although the pH profiles of Gd1 and Gd2 exhibit a drop in relaxivity on passing below pH 6 the profile of Gd11 cannot be measured below this pH since the dendrimer conjugate precipitates from solution immediately below pH 5.9. This is presumably the result of the high molecular weight conjugate suddenly reaching its isoelectric point.
Figure 2.
Relaxivity pH profiles of Gd1 (◆)[12] Gd2 (○) and Gd11 (●) recorded at 20 MHz and 298 K. Relaxivity is expressed per Gd3+ ion.
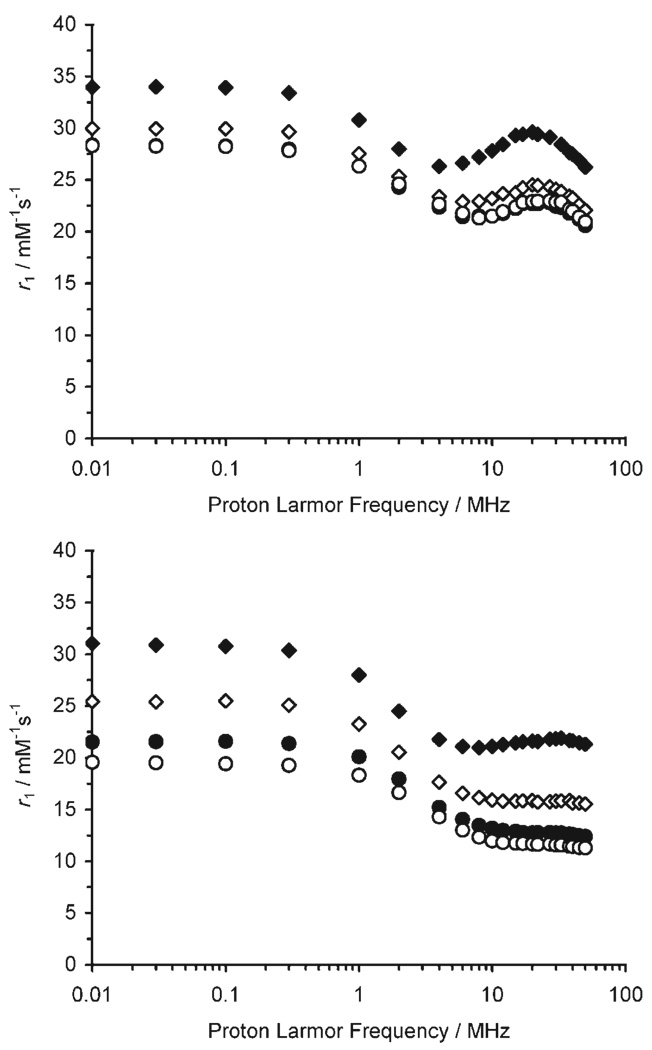
The difference in relaxivity between the “on” (pH 6) and “off” (pH 9.6) states of the dendrimer-based pH responsive agent was improved by more than a factor of 2 by slowing molecular rotation. This should render the dendrimer-conjugate, Gd11, a more effective pH responsive agent for imaging tissue pH. In order to assess the origins of this improvement in pH responsive behaviour, nuclear magnetic resonance dispersion (NMRD) profiles of Gd11 were recorded at high and low pH values (Figure 3). At both pH 6.5 and at pH 9.3, the relaxivity increased at lower temperatures indicating that the observed relaxivities are not limited by slow water proton exchange between complex and bulk water. Comparing the high field (1–100 MHz) regions of NMRD profiles recorded at the same temperature we can see that at higher pH (9.3) the curve is flatter and has a lower magnitude than at lower pH (6.5). This indicates that at pH 9.3 the effective correlation time, τC, which is responsible for modulating relaxation, is shorter than it is at pH 6.5.
Figure 3.
NMRD profiles of Gd11 recorded at pH 6.5 (top) and pH 9.3 (bottom); ◆: 5, ◇: 15, ●: 25, ○: 35°C.
Owing to the large number of ionizable phosphonate and dendrimer amine groups on the dendrimer, multiple protonated species must exist at each pH value. This complicates any attempt to “fit” these NMRD profiles in a quantitative, meaningful way. Nonetheless, it is useful examine the parameters that influence relaxivity in a qualitative sense in order to probe which factors are responsible for the observed behaviour. One may assume that each of the 96 Gd3+ chelates on the dendrimer surface has one water molecule in its inner coordination sphere. In addition, the phosphonate groups of each pendant arm may form hydrogen-bonding interactions with nearby water molecules forming a second hydration sphere. The extent of this second hydration sphere is likely to vary as the phosphonates are protonated or deprotonated with changing pH. Over a certain pH range it has also been shown that these phosphonates can catalyse exchange of protons from the coordinated water molecule to the bulk solvent.[12] Finally there is an outer-sphere contribution to relaxivity resulting from the diffusion water molecules of the bulk solvent close by the slowly tumbling Gd3+ chelates. This outer-sphere effect depends primarily on the rate of diffusion of water and is insensitive to changes in pH.
Conjugating Gd3+ complexes to dendrimers is a common strategy for slowing the rotational dynamics of paramagnetic complexes.[30–37] In order to quantitatively describe the rotational dynamics of these dendrimer systems, the Lipari–Szabo approach is typically used.[38–43] This model employs two correlation times; a long correlation time that defines the global motion of the entire dendrimer conjugate, τg, and a second shorter correlation time, τl, that reflects the local motion of the metal complex about its point of attachment to the dendrimer. This fast local motion is superimposed upon the slower global motion of the dendrimer. For systems conjugated to G5-PAMAM dendrimers the global correlation time, τg, has been reported on the order of 4–5 ns.[42, 44] The correlation time describing the local motion, τl, of the complex range from 0.07–0.76 ns.[41–44] The relative weighting of these two correlation times is given by an order parameter, S2, that can range from 0 (where local motion is dominant) to 1 (where isotropic, global motion is dominant). For dendrimers modified with Gd3+ chelates S2 was found to range from 0.28–0.5.
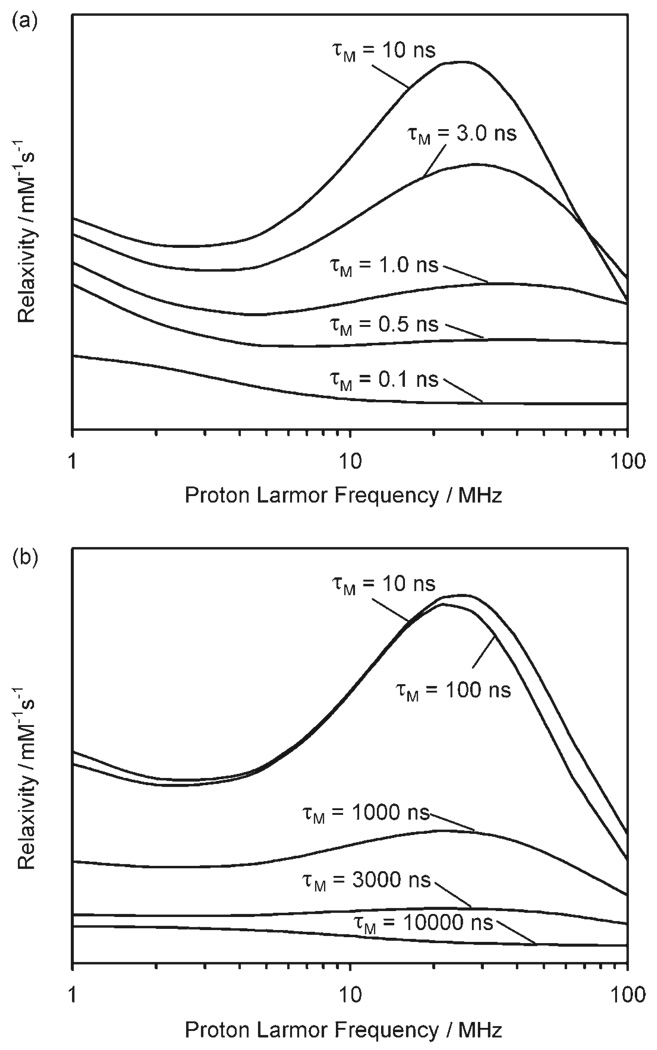
We assumed that similar rotational dynamics applied to Gd11 and these parameters to simulate the high field region (1–100 MHz) of an NMRD profile (Figure 4). Electron-spin relaxation parameters were fixed at values similar to those used elsewhere.[40–44] When the rate of proton exchange between water molecules associated with the complex (either 1st or 2nd hydration sphere) and the bulk solvent is extremely fast (short τM) the high field NMRD profile is flat and relaxivity low (Figure 4a). This is because these protons have a low probability of being relaxed before they exchange back into the bulk solvent. At the other extreme, very slow exchange that is, very long τM, the profile is also flat and relaxivity low (Figure 4b) because these protons, once relaxed by Gd3+, remain on the complex preventing others from being relaxed. So in both cases relaxation is not effectively transferred to the bulk solvent and the T1 of the bulk remains long. Between these two extremes, where 1 ns < τM < 1000 ns, the profile is characterized by higher relaxivities and a “hump” between 10 and 60 MHz (Figure 4). The magnitude of the relaxivity in the profiles will depend on these exchange kinetics, but also on the number of exchangeable protons and their distance from Gd3+. However, these latter two parameters will not affect the shape of the NMRD profile.
Figure 4.
Effect of proton residence lifetime (τM) on the relaxivity of Gd3+-G5 PAMAM dendrimer conjugates in the fast (top) and slow (bottom) exchange regimes. The simulated profiles are plotted at same scale using parameters: τg=4.5 ns, τl=0.15 ns, S2=0.5, τv=20 ps, Δ=2×109 s.
In order to mimic the flat, field independent behaviour observed for Gd11 at pH 9.3, the proton residence lifetime, τM, must be either very short (<1 ns) or very long (>1 µs). The rate of water proton exchange is temperature dependent; as the temperature is lowered τM becomes longer. This increase in τM applies to both water molecules in the inner and second hydration spheres; however, it has an opposing effect on the relaxivity of each. The water protons of the inner-sphere water molecule of Gd1 were found to be in slow exchange with the bulk,[12] and so these protons contribute poorly to the overall relaxivity. In contrast, water protons of a second hydration sphere are known to undergo very rapid exchange that also limits their contribution to relaxivity.[22] Whereas making τM of the inner sphere longer (lower temperature) will not result in an increase in relaxivity, making τM of the second sphere longer could bring these protons into a range where they are able to contribute more substantially to relaxivity (Figure 4a). Inspecting the NMRD profile of Gd11 recorded at pH 9.3 and 35 °C, the high field region is flat suggesting that exchange of inner-sphere water protons is too slow and exchange of second sphere water protons too fast for a relaxivity enhancement “hump” to be observed at high field. As the temperature is taken down to 5 °C the profile begins to take on the appearance of a small “hump” at high field. Clearly exchange from the inner-sphere water protons will continue to be too slow to provide a contribution to relaxivity at lower temperatures. The small increase in relaxivity must therefore be the result of slowing the exchange rate of second-sphere water protons into a range that allows some contribution to relaxivity. This second-sphere contribution to relaxivity is not observed for either Gd1 or Gd2 because their rotational dynamics are too rapid. Given the small size of the high field relaxivity hump, exchange of second-sphere water molecules is likely to be in the range of τM=0.5–5 ns.
Over a certain pH range, the phosphonate groups of the pendant arms of the low molecular weight complexes Gd1 and Gd2, catalyse exchange of inner-sphere water protons with the bulk solvent.[12] The fast rotational dynamics (τg ≈0.1 ns) of these low molecular weight chelates limits their relaxivity but conjugation to the dendrimer lifts this restriction in Gd11 and so the observed relaxivity is higher by a factor of almost 5. At pH 6.5 and 35 °C the NMRD profile of Gd11 already has a slight high field “hump” which becomes more pronounced as the temperature is lowered. From studies on Gd1, a contribution to this high field relaxivity from the inner-sphere proton exchange is expected. However, a contribution from protons in the second hydration sphere is also apparent. This is most clearly seen when the temperature is lowered. The τM of inner-sphere water protons of Gd1 was found to be on the order of microseconds[12] and so as the temperature is lowered the inner-sphere relaxivity should decrease as exchange becomes increasingly limited (cf. Figure 4b). The observed relaxivity increases with decreasing temperature indicating that a substantial second-sphere component must be present (Figure 4a). The fact that the increase in high field relaxivity with decreasing temperature is larger at pH 6.5 than it is at pH 9.3 suggests that either the second hydration sphere is larger or it is more ordered, leading to longer τM values, at pH 6.3 than at 9.3. Gd11 would seem to be a rare example of a system in which relaxivity is limited both by prototropic exchange in the 2nd-sphere that is too fast and by water exchange in the inner-sphere that is too slow.
It is worth noting that a third factor may also play a role in improving the relaxivity pH response of Gd11. Gd3+ complexes that exhibit no pH response have been found to behave as pH responsive agents once conjugated to PAMAM dendrimers.[44] The origin of this phenomenon is thought to be changes in the internal motion of the dendrimer itself as the pH changes. Protonation of amines within the body of the dendrimer is believed to make the dendrimer more rigid making τR longer at lower pH. Thus the relaxivity of agents conjugated to these dendrimers has been found to increase as the pH drops. It is likely that, in addition to the interplay of inner-sphere and second-sphere water proton exchange rates, a third contribution to the pH responsive behaviour of Gd11 arises from changes in the rigidity of the dendrimer with changes in pH.
Conclusions
It is possible to take advantage of the interplay between molecular reorientation and water proton exchange kinetics to enhance the response behaviour of complexes that exhibit changes in relaxivity arising from changes in water or proton exchange kinetics. Our initial hypothesis was that coupling the pH-responsive agent Gd1 to a dendrimer would increase both the overall relaxivity, r1, the responsiveness of relaxivity to pH, Δr1. This was achieved by slowing rotation via conjugation to a dendrimer, increasing Δr1 by more than a factor of 2. This enhancement should enable Gd11 to serve as an effective pH responsive MRI contrast agent. Furthermore, this dendrimer system has provided further insights into the mechanism by which this class if pH responsive agent, Gd1, Gd2 and Gd11, operates. The pH response is the result of a complex interplay between the rate of proton exchange between the bulk solvent and water molecules in the inner and second hydration spheres. The overall relaxivity is ultimately limited by the slow exchange kinetics of protons in the inner hydration sphere and rapid exchange kinetics of protons in the second hydration sphere. From an imaging point of view the substantial improvements in both r1 and Δr1 afforded by Gd11 should allow improved determination of in vivo pH by MRI. However, it is worth noting that improving Δr1 through increased molecular weight may also negatively impact the effectiveness of such agents. Large molecules, such as dendrimers, remain in vasculature longer than discrete agents, such as Gd1, which are better able to diffuse into all extracellular space. Furthermore, large molecules, such as Gd11, tend to clear more slowly from the body as a result of increased liver uptake. This extends the retention time of Gd3+ in the body. Further studies into the in vivo behaviour of dendrimer-based MRI contrast media will be required to establish if this approach, which is successful for increasing both r1 and Δr1, will yield agents that can actually be applied in vivo.
Experimental Section
General remarks
All reagents and solvents were purchased from commercial sources and used as received unless otherwise stated. The ethylene core G5-PAMAM dendrimer from Dendritech was purchased through Sigma-Aldrich as a 5% solution in methanol. Prior to use the solvents were removed under vacuum and the dendrimer redissolved in the reaction solvent. 1H NMR spectra were recorded on a JOEL Eclipse 270 spectrometer operating at 270.17 MHz, a Varian Mercury 300 spectrometer operating at 299.95 MHz and a Bruker Avance III spectrometer operating at 400.13 MHz. 13C NMR spectra were obtained using a Bruker Avance III spectrometer operating at 100.61 MHz. Longitudinal relaxation times were measured using the inversion recovery method on a MRS-6 NMR analyzer from the Institut “Jožef Stefan”, Ljubljana, Slovenjia operating at 20 MHz. The pH of samples for relaxivity measurements was adjusted by addition of either lithium hydroxide monohydrate or p-toluenesulfonic acid monohydrate in order to avoid dilution. Melting points were determined on a Fisher-Johns melting point apparatus and are uncorrected. NMRD profiles between 0.01 and 50 MHz were recorded using the field cycling relaxometer at New York Medical Collge, Valhala NY.
Synthesis
The synthesis of diethyl phthalimidomethylphosphonate (3) and diethyl bromoacetamidomethylphosphonate (6) have been reported previously.[12]
Diethyl aminomethylphosphonate (4)
Hydrazine (81.6 mL, 168.2 mmol) was added to a solution of the phthalimide 3 (50.0 g, 1.68 mol) in dry ethanol (500 mL). The reaction was stirred at room temperature for 18 h. The solvents were removed under reduced pressure and the residue placed under high vacuum to remove as much excess hydrazine as possible. To the residue was added diethyl ether (750 mL) which was then filtered to remove the precipitate. The precipitate was washed with diethyl ether (2000 mL). The solvents were removed from the filtrate under reduced pressure to afford the title compound as a colourless oil (41.9 g, 97%). Characterisation data is identical to that reported previously.[12]
N′,N″-Bis-(diethyl)methylphosphonate phthalamide (5)
The title compound was isolated as a by-product of the reaction used to synthesise 4.[12] X-ray quality crystals were grown by dissolving 5 (0.5 g) in a minimum of dichloromethane in a 20 mL scintillation vial. Diethyl ether (8 mL) was added and the components mixed thoroughly. Hexanes (9 mL) were then added slowly such that a layer of hexanes lay on top of the solution of 5. The vial was sealed and the layers were allowed to diffuse together at room temperature, affording high quality crystals of 5. M.p. 119–120°C; 1H NMR (400 MHz, CDCl3): δ=1.21 (t, 12H, 3J(H,H)=7 Hz, CH3), 3.73 (dd, 4H, 2J(H,P)=12 Hz, 3J(H,H)=6 Hz, CH2P), 3.98 (q, 4H, 3J(H,H)=7 Hz, OCH2), 4.00 (q, 4H, 3J(H,H)=7 Hz, OCH2), 7.36 (dd, 2H, 3J(H,H)=6 Hz, 4J(H,H)=3 Hz, Ar), 7.48 (dd, 2H, 3J(H,H)=6 Hz, 4J(H,H)=3 Hz, Ar), 7.70 ppm (dd, 2H, 3J(H,H)=6 Hz, NH); 13C NMR (100 MHz, CDCl3): δ=16.3 (3J(C,P)=6 Hz, CH3), 35.2 (1J(C,P)=156 Hz, CH2P), 62.4 (2J(C,P)=6 Hz, OCH2), 128.4 (3-Ar), 130.1 (2-Ar), 134.5 (1-Ar), 168.7 (3J(C,P)=5 Hz, C=O); IR (KBr disc): vmax = 3251 (NH), 3063, 2984, 2931, 2910, 2236 (PO), 1661 (CO) 1595, 1539, 1479, 1444, 1369, 1319, 1217 (PO), 1163, 1098, 1023 (PO), 973, 786, 730 cm−1; ESMS-: m/z (%): 463 (100) [M–H+]−; elemental analysis calcd (%) for C18H30N2O8P2 : C 46.6, H 6.5, N 6.0; found: C 46.1, H 6.4, N 5.9.
(S)-2-(p-Nitrobenzyl)-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetamidomethylene-(diethyl)phosphonate (7)
(S)-2-(p-Nitrobenzyl) cyclen (0.48 g, 1.56 mmol) was dissolved in acetonitrile (20 mL). Potassium carbonate (1.45 g, 11.0 mmol) and bromoacetamide 6 (2 g, 6.94 mmol) were added and the reaction mixture stirred for 72 h at 60 °C. The reaction mixture was filtered and the solvents removed under reduced pressure. The residue was purified by column chromatography over silica gel eluting with 10% methanol in dichloromethane to afford the title compound as pale yellow solid (0.53 g, 30%). ESI+: m/z (%): 1136 (35) [M+H]+, 1158 (100) [M+Na]+, 1174 (60) [M+K]+.
(S)-2-(p-Aminobenzyl)-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetamidomethylene-(diethyl)phosphonate (8)
Nitro compound 7 (0.50 g, 0.44 mmol) was dissolved in water (15 mL) and 10% palladium on carbon (0.12 g) was added. The reaction mixture was shaken on a Parr hydrogenation apparatus for 12 h under H2 (25 psi). The catalyst was removed by filtration and the solvents removed by lyophilization to afford the title compound as a colourless solid (0.35 g, 72%). ESI+: m/z (%): 1106 (65) [M+H]+, 1128 (100) [M+Na]+.
(S)-2-(p-Isothiocyanatobenzyl)-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetamidomethylene-(diethyl)phosphonate (9)
Amine 8 (0.50 g, 0.45 mmol) was dissolved in water (5 mL) and the pH of the resulting solution adjusted to 2 by addition of a dilute HCl solution. Chloroform (15 mL) was added to the reaction which was then stirred vigorously at room temperature. Thiophosgene (0.052 mg, 0.45 mmol) was added to the reaction which was then stoppered and stirred vigorously for 18 h at room temperature. The reaction mixture was then transferred to a separatory funnel and the chloroform layer was allowed to run off. The aqueous layer was then washed with chloroform (2×20 mL). The aqueous layer was then collected and the solvents removed under reduced pressure to afford the title compound as a colourless solid (0.48 g, 93%). ESI+: m/z (%): 1148 (100) [M+H]+, 1170 (78) [M+Na]+.
(S)-2-(p-Nitrobenzyl)-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacet-amidomethylene phosphonic acid (2)
Octaethyl ester 7 (0.75 g, 0.66 mmol) was dissolved in a 30% solution of HBr in acetic acid (8 mL). The resulting solution was stirred at room temperature for 18 h. The solvents were removed under reduced pressure and the residue taken up in EtOH (20 mL), the solvents were again removed under reduced pressure. The solid residue was then taken up into MeOH (10 mL) and the title compound precipitated by dropwise addition of Et2O. The title compound was isolated by filtration, dissolved in water and the solvents removed by lypholization to afford a tan solid (0.49 g, 81.4%). ESI+: m/z (%): 511 (100) [H4L+5Na]2+.
G5-PAMAM dendrimer–ligand phosphonate diethyl ester conjugate (10)
G5-PAMAM dendrimer (40 mg, 1.4 µmol) was dissolved in DMSO/water 1:1 (4 mL). Isothiocyanate 9 (0.41 g, 0.36 mmol) was added and the pH of the resulting solution raised to 9 by addition of 1m NaOH solution. The reaction was then stirred at 40 °C for 24 h with the pH maintained at 9 by addition of NaOH before a solution of 9 (0.41 g, 0.36 mmol) in DMSO (1 mL) was added. The reaction was stirred for a further 48 h at pH 9. The reaction mixture was then transferred to a Centircon C-30 diafiltration cell with a 30 kD molecular weight cut-off. Diafiltration was repeated until SEC-HPLC revealed that no further low molecular weight material was present, the solvents were then removed by lyophylisation to afford a colourless solid (0.11 g). Anal found C 28.63, S 1.34%, or 56.975 carbon atoms per sulfur atom. The ligand formula C44H81N9O16P4S equates to 1 ligand per 12.975 carbon atoms of the dendrimer. The dendrimer formula C1262H2528N506O252 affords a ratio 1262/12.975 or 97.26 ligands per dendrimer.
G5-PAMAM dendrimer–ligand phosphonic acid conjugate (11)
Conjugate 10 (0.11 g) was dissolved in a 30% solution of HBr/glacial acetic acid (3 mL) under an argon atmosphere. The reaction was stirred at room temperature for 18 h. The solvents were then removed under reduced pressure and residue taken up into methanol (10 mL) which was then also removed under reduced pressure. Dissolution and evaporation of methanol was performed a further three times to removes as much excess acetic acid as possible. The residue was then washed with diethyl and vacuum dried to afford the title compound as a colourless solid (0.09 g).
SEC-HPLC analysis
Size-exclusion HPLC analysis of 10 and Gd11 were performed on a phenomenex BIOSEP SEC S-3000 size exclusion column (5–700 kD) eluting with PBS buffer at pH 7.4. The column was standardised using commercially available protein (globular) molecular weight standards: cytochrome C (12.4 kD), carbonic anhydrase (29 kD), bovine serum albumin (66 kD), γ-globulins (160 kD), apoferritin (480 kD), dextran blue (2000 kD).
Crystal structure determination of 5
Data collection
A colourless irregular block with approximate orthogonal dimensions 0.46×0.41×0.25 mm was placed and optically centered on the Bruker SMART1000 CCD system (Bruker, SMART, Version 5.054 (2004) and SAINT, Version 7.23 A, Bruker AXS Inc.) at −183 °C. The initial unit cell was indexed using a least-squares analysis of a random set of reflections collected from three series of 0.3° wide ω scans, 10 seconds per frame, and 25 frames per series that were well distributed in reciprocal space. Five ω-scan data frame series were collected (MoKa) with 0.3° wide scans, 20 seconds per frame and 606 frames collected per series at varying ϕ angles (ϕ=0, 72, 144, 216, 288°). The crystal to detector distance was 4.123 cm, thus providing a complete sphere of data to 2θmax= 60.48°.
Structural determination and refinement
A total of 88185 reflections were collected and corrected for Lorentz and polarization effects and absorption using Blessing’s method[45] as incorporated into the program SADABS (Sheldrick, G.M., SADABS, Version 2.10 (2003), “Siemens Area Detector Absorption Correction” Universität Göttingen, Göttingen, Germany) with 13581 unique. The SHELXTL (G. M. Sheldrick, SHELXTL, Version 6.1, 2002, Bruker AXS Inc.) program package was implemented to determine the probable space group and set up the initial files. System symmetry, systematic absences, and intensity statistics indicated the non-centrosymmetric orthorhombic space group Pna21 (no. 33). The structure was determined by direct methods with the successful location of a majority of the molecule within the asymmetric unit using the program XS (G. M. Sheldrick, SHELXS97 and SHELXL97, 1997), which was also used to refine the structure. The data collected were merged based upon identical indices yielding 53260 data [R(int)=0.0267] that were truncated to 2θmax=60.00° resulting in 51 060 data that were further merged during least-squares refinement to 13 322 unique data [R(int)=0.0393]. A series of least-squares difference-Fourier cycles were required to locate the remaining non-hydrogen atoms and optimize the various disorders present for the two molecules in the asymmetric unit. All full occupancy non-hydrogen atoms were refined anisotropically. Hydrogen atoms were idealized throughout the final refinement stages. The final structure was refined to convergence with R(F)=6.55%, wR(F2)=12.39%, GOF=1.118 for all 13 322 unique reflections [R(F)=5.72%, wR(F2)=12.02% for those 11 744 data with Fo > 4σ(Fo)]. The final difference-Fourier map was featureless indicating that the structure is both correct and complete. An empirical correction for extinction was also attempted but found to be negative and therefore not applied. The absolute structure parameter, Flack(x),[46] was refined and found to be 0.38(9) indicating that racemic twinning is present.
CCDC 680275 (5) contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
Supplementary Material
Acknowledgements
The authors thank the National Institutes of Health (RR-02584, CA-126608 and CA-115531), the Robert A. Welch Foundation (AT-584) for financial assistance.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.200800402.
References
- 1.Wike-Hooley JL, Haveman J, Reinhold JS. Radiother. Oncol. 1984;2:343. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- 2.Tannock IF, Rotin D. Cancer Res. 1989;49:4373. [PubMed] [Google Scholar]
- 3.Vaupel P, Kallinowski F, Okunieff P. Cancer Res. 1989;49:6449. [PubMed] [Google Scholar]
- 4.Khramtsov VV. Curr. Org. Chem. 2005;9:909. [Google Scholar]
- 5.Woods M, Zhang S, Kovacs Z, Sherry AD. In: Advances in Supra-molecular Chemistry. Gokel G, editor. Vol. 9. Amsterdam: Elsevier; 2003. p. 1. and references therein. [Google Scholar]
- 6.Aime S, Castelli DD, Terreno E. Angew. Chem. 2002;114:4510. [Google Scholar]; Angew. Chem. Int. Ed. 2002;41:4334. [Google Scholar]
- 7.Aime S, Barge A, Castelli DD, Fedeli F, Mortillaro A, Nielsen FU, Terreno E. Magn. Reson. Med. 2002;47:639. doi: 10.1002/mrm.10106. [DOI] [PubMed] [Google Scholar]
- 8.Woods M, Zhang S, Von Howard E, Sherry AD. Chem. Eur. J. 2003;9:4634. doi: 10.1002/chem.200305159. [DOI] [PubMed] [Google Scholar]
- 9.Terreno E, Castelli Daniela D, Cravotto G, Milone L, Aime S. Invest. Radiol. 2004;39:235. doi: 10.1097/01.rli.0000116607.26372.d0. [DOI] [PubMed] [Google Scholar]
- 10.Woods M, Kiefer GE, Bott S, Castillo-Muzquiz A, Eshelbrenner C, Michaudet L, McMillan K, Mudigunda SDK, Ogrin D, Tircso G, Zhang S, Zhao P, Sherry AD. J. Am. Chem. Soc. 2004;126:9248. doi: 10.1021/ja048299z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aime S, Fedeli F, Sanino A, Terreno E. J. Am. Chem. Soc. 2006;128:11326. doi: 10.1021/ja062387x. [DOI] [PubMed] [Google Scholar]
- 12.Kalman FK, Woods M, Caravan P, Jurek P, Spiller M, Tircso G, Kiraly R, Brucher E, Sherry AD. Inorg. Chem. 2007;46:5260. doi: 10.1021/ic0702926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Wu K, Sherry AD. Angew. Chem. 1999;111:3382. [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 1999;38:3192. [Google Scholar]
- 14.Bloembergen N. J. Chem. Phys. 1957;27:572. [Google Scholar]
- 15.Bloembergen N, Morgan LO. J. Chem. Phys. 1961;34:842. [Google Scholar]
- 16.Bloembergen N, Purcell EM, Pound RV. Phys. Rev. 1948;73:679. [Google Scholar]
- 17.Solomon I. Phys. Rev. 1955;99:559. [Google Scholar]
- 18.Solomon I, Bloembergen N. J. Chem. Phys. 1956;25:261. [Google Scholar]
- 19.Garcia-Martin ML, Martinez GV, Raghunand N, Sherry AD, Zhang S, Gillies RJ. Magn. Reson. Med. 2006;55:309. doi: 10.1002/mrm.20773. [DOI] [PubMed] [Google Scholar]
- 20.Raghunand N, Howison C, Sherry AD, Zhang S, Gillies RJ. Magn. Reson. Med. 2003;49:249. doi: 10.1002/mrm.10347. [DOI] [PubMed] [Google Scholar]
- 21.Raghunand N, Zhang S, Sherry AD, Gillies RJ. Acad. Radiol. 2002;9 Suppl 2:S481. doi: 10.1016/s1076-6332(03)80270-2. [DOI] [PubMed] [Google Scholar]
- 22.Borel A, Helm L, Merbach AE. Chem. Eur. J. 2001;7:600. doi: 10.1002/1521-3765(20010202)7:3<600::aid-chem600>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 23.Botta M. Eur. J. Inorg. Chem. 2000:399. [Google Scholar]
- 24.Ing HR, Manske RHF. J. Chem. Soc. 1926:2348. [Google Scholar]
- 25.Khan MN, Ohayagha JE. React. Kinet. Catal. Lett. 1996;42:97. [Google Scholar]
- 26.Khan MN. J. Org. Chem. 1996;61:8063. doi: 10.1021/jo961172a. [DOI] [PubMed] [Google Scholar]
- 27.Ariffin A, Khan MN, Lan LC, May FY, Yun CS. Synth. Commun. 2004;34:4439. [Google Scholar]
- 28.Khan MN. J. Org. Chem. 1995;60:4536. [Google Scholar]
- 29.Curley OMS, McCormick JE, McElhinney RS, McMurry TBH. ARKIVOC. 2003:180. [Google Scholar]
- 30.Wang SJ, Brechbiel M, Wiener EC. Invest. Radiol. 2003;38:662. doi: 10.1097/01.rli.0000084887.47427.75. [DOI] [PubMed] [Google Scholar]
- 31.Wiener E, Narayanan VV. Adv. Dendritic Macromol. 2002;5:129. [Google Scholar]
- 32.Konda SD, Aref M, Brechbiel M, Wiener EC. Invest. Radiol. 2000;35:50. doi: 10.1097/00004424-200001000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Wiener EC, Konda S, Shadron A, Brechbiel M, Gansow O. Invest. Radiol. 1997;32:748. doi: 10.1097/00004424-199712000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Wiener EC, Brechbiel MW, Gansow OA, Foley G, Lauterbur PC. Polym. Mater. Sci. Eng. 1997;77:193. [Google Scholar]
- 35.Wiener EC, Auteri FP, Chen JW, Brechbiel MW, Gansow OA, Schneider DS, Belford RL, Clarkson RB, Lauterbur PC. J. Am. Chem. Soc. 1996;118:7774. [Google Scholar]
- 36.Wiener EC, Brechbiel MW, Brothers H, Magin RL, Gansow OA, Tomalia DA, Lauterbur PC. Magn. Reson. Med. 1994;31:1. doi: 10.1002/mrm.1910310102. [DOI] [PubMed] [Google Scholar]
- 37.Bryant LH, Jr, Brechbiel MW, Wu C, Bulte JW, Herynek V, Frank JA. J. Magn. Reson. Imag. 1999;9:348. doi: 10.1002/(sici)1522-2586(199902)9:2<348::aid-jmri30>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 38.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546. [Google Scholar]
- 39.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4559. [Google Scholar]
- 40.Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ. Inorg. Chem. 2007;46:6632. doi: 10.1021/ic700686k. [DOI] [PubMed] [Google Scholar]
- 41.Jaszberenyi Z, Moriggi L, Schmidt P, Weidensteiner C, Kneuer R, Merbach AE, Helm L, Toth E. J. Biol. Inorg. Chem. 2007;12:406. doi: 10.1007/s00775-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 42.Lebduskova P, Sour A, Helm L, Toth E, Kotek J, Lukes I, Merbach AE. Dalton Trans. 2006:3399. doi: 10.1039/b517847a. [DOI] [PubMed] [Google Scholar]
- 43.Nicolle GM, Toth E, Schmitt-Willich H, Raduchel B, Merbach AE. Chem. Eur. J. 2002;8:1040. doi: 10.1002/1521-3765(20020301)8:5<1040::aid-chem1040>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 44.Laus S, Sour A, Ruloff R, Toth E, Merbach AE. Chem. Eur. J. 2005;11:3064. doi: 10.1002/chem.200401326. [DOI] [PubMed] [Google Scholar]
- 45.Blessing RH. Acta Crystallogr. Sect. A. 1995;51:33. doi: 10.1107/s0108767394005726. [DOI] [PubMed] [Google Scholar]
- 46.Flack HD. Acta Crystallogr. Sect. A. 1983;39:876. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.