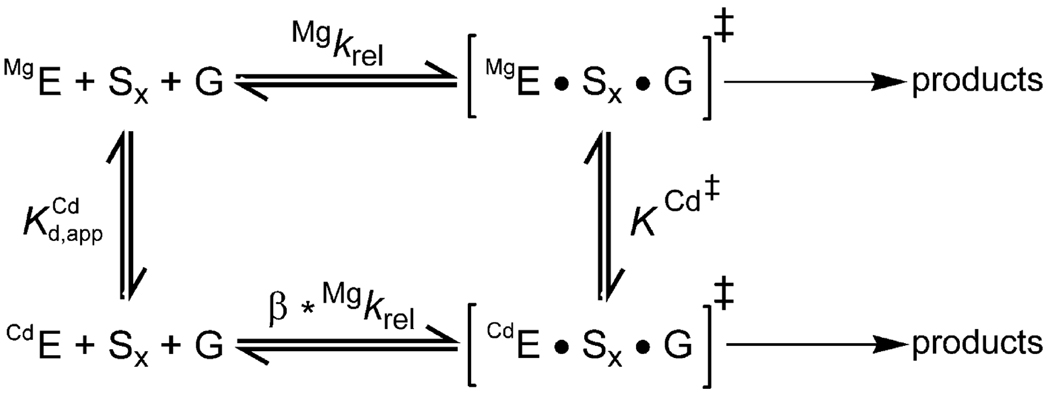Abstract
Many enzymes use metal ions within their active sites to achieve enormous rate acceleration. Understanding how metal ions mediate catalysis requires elucidation of metal ion interactions with both the enzyme and the substrate(s). The three-dimensional arrangement determined by X-ray crystallography provides a powerful starting point for identifying ground state interactions, but only functional studies can establish and interrogate transition state interactions. The Tetrahymena group I ribozyme is a paradigm for the study of RNA catalysis, and previous work using atomic mutagenesis and quantitative analysis of metal ion rescue behavior identified catalytic metal ions making five contacts with the substrate atoms. Here, we have combined atomic mutagenesis with site-specific phosphorothioate substitutions in the ribozyme backbone to establish transition state ligands on the ribozyme for one of the catalytic metal ions, referred to as MA. We identified the pro-S P oxygen atoms at nucleotides C208, A304, and A306 as ground state ligands for Ma, verifying interactions suggested by the Azoarcus crystal structures. We further established that these interactions are present in the chemical transition state, a conclusion that requires functional studies, such as those carried out herein. Elucidating these active site connections is a crucial step toward an in-depth understanding of how specific structural features of the group I intron lead to catalysis.
Phosphoryl transfer is a ubiquitous reaction in biology, yet it is extremely slow in solution (1, 2). To accelerate this class of reactions to an extent compatible with life, many enzymes have evolved active sites that contain metal ions. To understand how enzymes utilize the catalytic power of metal ions, we must identify individual metal ions, define their coordination environment, and establish the network of interactions that position the coordinating groups. X-ray crystallography has been successfully used in many cases to determine the location of metal ions and nearby residues, providing powerful starting points to unveil the network of catalytic interactions and the effects of perturbation of these interactions on catalysis and the enzyme’s reaction cycle. Nevertheless, X-ray crystallography provides ground state structures, and uncertainties about transition state interactions remain in the absence of functional data.
Metal ion rescue experiments provide a powerful means to identify specific metal ion interactions. As first developed by Cohn and Eckstein (3, 4), a decrease in reactivity upon single-atom replacements of an oxygen atom by sulfur or nitrogen in Mg2+, which interacts weakly with sulfur, and subsequent rescue of activity upon addition of softer cations, such as Mn2+ or Cd2+, which interact more strongly with sulfur, can provide evidence of direct metal ion coordination during catalysis. This technique has revealed catalytic metal ion interactions with enzyme substrates in many protein enzymes, especially for metal ion interactions within active site ATP • Mg2+ complexes (ref 5 and references cited therein; see also Supporting Information).
In contrast to the relative ease of introducing single-atom substitutions in substrates, it remains a formidable challenge to introduce single-atom substitutions in proteins (6), pre-cluding the use of site-specific single-atom substitutions to identify metal ion binding sites in most protein backbones. In contrast, RNA enzymes, or ribozymes, are readily amenable to single-atom substitutions (7). Given these favorable properties and the broad interest in understanding RNAs catalytic abilities and mechanisms, ribozymes have been attractive systems for investigation of the role of metal ions in enzymatic catalysis (ref 5 and references cited therein; see also Supporting Information).
The Tetrahymena ribozyme catalyzes a nucleotidyl transfer reaction from an oligonucleotide substrate (S) that mimics the natural 5′-splice site to an exogenous guanosine (G), which serves as the nucleophile in a reaction analogous to the first step of group I intron self-splicing (eq 1) (8, 9).
| (1) |
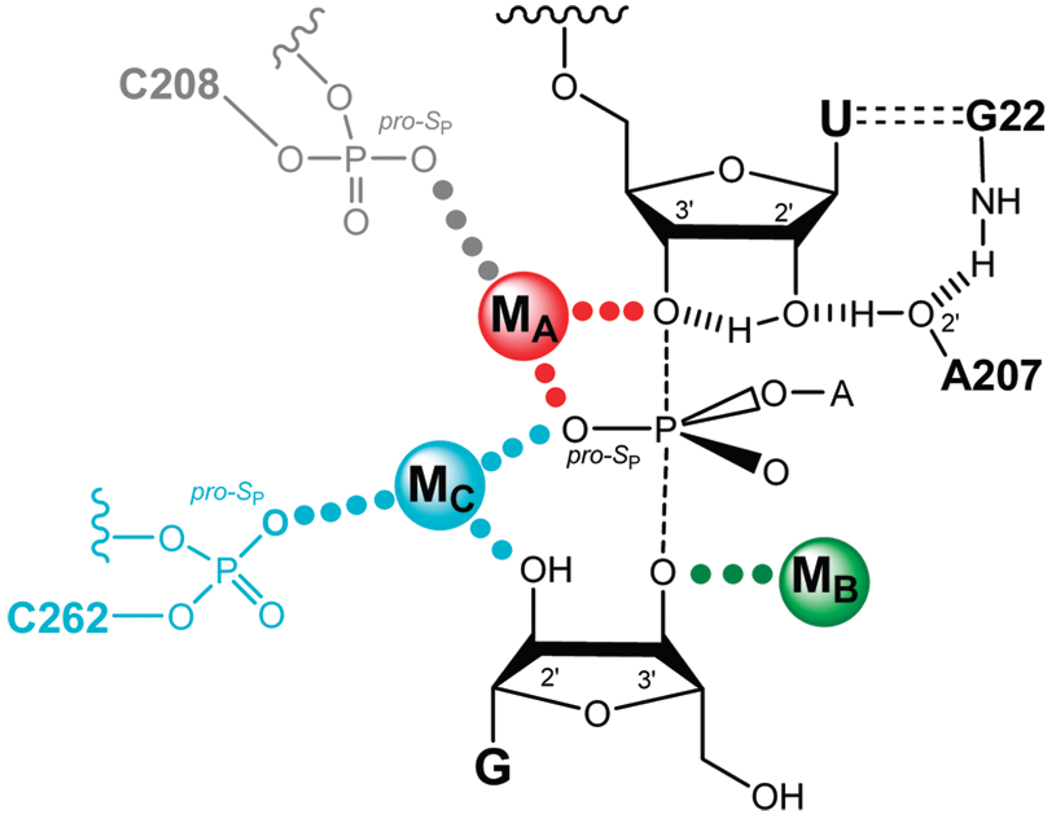
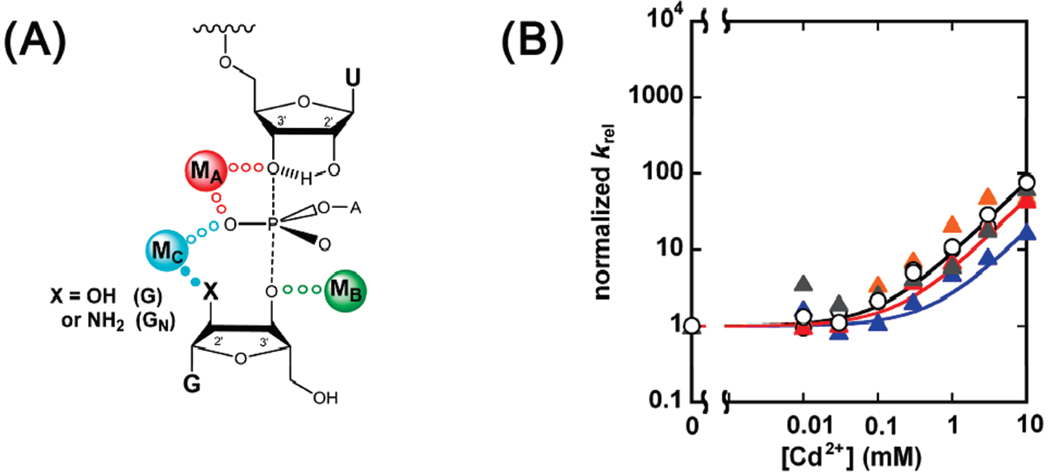
Early metal ion rescue experiments identified four transition state contacts between metal ions and atoms on the substrates through use of thio- or amino-substituted substrates and provided evidence against metal ion interactions at other positions (ref 10 and references cited therein). To determine whether one or several distinct metal ions mediate these interactions, Shan et al. developed “thermodynamic finger-print analysis” (TFA),1 a quantitative approach that determines whether the same or different metal ions give rescue at different positions (11). TFA and related analyses (11, 12) provided functional evidence for a network of transition state interactions within the Tetrahymena ribozyme active site, involving five substrate atoms interacting with three metal ions and additional hydrogen-bonding interactions (Figure 1). According to the resultant model, metal ions coordinate to the 3′-oxygen of the leaving group (MA), the 3′-oxygen of the G nucleophile (MB), and the 2′-hydroxyl of the G nucleophile (MC); MA and MC also contact the pro-S P oxygen of the scissile phosphate (Figure 1). These metal ions contribute to catalysis in a number of ways (ref 10 and references cited therein). MA stabilizes the developing negative charge on the 3′-leaving group oxygen in the transition state, along with an intramolecular hydrogen bond from the neighboring 2′-hydroxyl group, and may contribute to electrostatic destabilization of the ground state. MA, in conjunction with MC, may help to stabilize negative charge developing on the pro-S P oxygen in the transition state. MB is likely to activate the nucleophile. Finally, MC may also contribute to the correct positioning of the two substrates, one relative to the other.
FIGURE 1.
Proposed model of the Tetrahymena ribozyme transition state from functional data (ref 10 and references cited therein). Bonds forming and breaking in the transition state are represented by dashed lines. Hydrogen bonds are represented by hashed lines. Active site metal ions (MA, MB, and MC) are color coded, as are putative ligand interactions, in this and subsequent figures. Contacts between metal ions and their ligands supported by quantitative functional data are represented by solid dots of the same color of the metal ion; contacts proposed by qualitative analysis are represented by gray dots. Charges are omitted for clarity.
Four crystal structures of group I introns have been published in the past four years (13–16), providing an unprecedented atomic view of the group I intron. Despite the similar architecture, the four structures display significant variations in the number of active site metal ions and in the specific interactions made between the metal ions and the RNA backbone. In addition, one of the active site metal ions, MB, was not detected by X-ray crystallography. Instead, in the second Azoarcus crystal structure (16), MC (or M2) appeared to make contact with both the 2′- and the 3′-oxygens of the guanosine nucleophile on the basis of the observed distances between electron densities. The possible reasons for this difference have been discussed in the literature (10, 13, 16) and are not the subject of this report. Here we determine to what extent the observed ground state ribozyme environment around the catalytic metal ion, MA, is supported by functional data and can be extended to the fleeting transition state.
To functionally test potential ligands of catalytic MA, we introduced site-specific phosphorothioate substitutions on the ribozyme and applied TFA (5, 17). We confirmed the pro-S P oxygen atom of C208 as a ligand for MA, as previously proposed on the basis of a qualitative analysis (18), and identified two additional ligands for this metal ion, the pro-S P oxygen atoms of residues A304 and A306. Our results establish three ground state ligands for MA, consistent with results from the Azoarcus crystal structures (13, 16), and further indicate that these contacts are present in the transition state.
MATERIALS AND METHODS
Materials
Wild-type in vitro transcribed Tetrahymena L-21 ScaI ribozyme was prepared as described previously (9). 2′-Aminoguanosine (GN) was prepared as described (19, 20). Standard oligonucleotide substrates were purchased from Dharmacon Inc. (Lafayette, CO) and 5′-32P-end-labeled using standard methods (9). Oligonucleotide substrates with phosphorothiolate linkage (S3′S and CUCG3′SA) were prepared as described (21). Diastereoisomers of the oligonucleotides corresponding to nucleotides 204–211 of the ribozyme, containing single phosphorothioate substitution at position C208, and to nucleotides 297–311, containing single phosphorothioate substitutions at positions A304, A306, and U307, were separated by anion-exchange HPLC (0–370 mM NaCl over 5 min, then 370–470 mM NaCl over 35 min in a background of 25 mM Tris, pH 7.4) and desalted by Sep-Pak (Waters, Milford, MA). Stereochemistry was assigned by elution under conditions in which the R P diastereoisomer elutes first (5, 22).
Ribozyme Preparation
Variant ribozymes were constructed semisynthetically using a single-step three-piece ligation (7). Constructs corresponding to nucleotides 22–203, 22–296, 212–409, and 312–409 of the Tetrahymena ribozyme were transcribed using a DNA template produced by PCR truncation of the plasmid-encoded ribozyme sequence, with excess GMP present in the transcription of the 3′-constructs (212–409 and 312–409) to yield a 5′-monophosphate. The 5′-construct contained a 3′-flanking hammerhead cassette to ensure homogeneous 3′-ends; the terminal 2′,3′-cyclic phosphate was removed after gel purification by treatment with T4 polynucleotide kinase (23). The transcripts were ligated to the HPLC-purified synthetic oligonucleotides via a single-step ligation with T4 DNA ligase and a DNA splint to yield full-length ribozyme containing a single phosphorothioate mutation at the desired site. Yields were ~10% in purified, fully ligated ribozyme. The ligated ribozymes were >90% active, as indicated by virtually monophasic kinetics under conditions in which oligonucleotide substrate cleavage occurs faster than oligonucleotide substrate dissociation (data not shown).
General Kinetic Methods
All cleavage reactions were single turnover, with ribozyme in excess of the radiolabeled 5′-splice site analogue (*S) or 3′-splice site analogue (*P), which was always present in trace quantities. Reactions were carried out at 30 °C in 50 mM buffer, 50 mM MgCl2, and varying concentrations of CdCl2. The buffers used were NaMES (pH 5.6–6.7), NaMOPS (pH 6.5–7.7), and Na-HEPES (pH 6.9–8.1). Reaction mixtures containing 10 mM MgCl2 and all components except CdCl2 and *S (or *P) were preincubated at 50 °C for 30 min to renature the ribozyme. Additional components were added, and reactions were allowed to equilibrate at 30 °C for 15 min before the addition of *S (or *P). Reactions were followed by gel electrophoresis and analyzed as described previously (8, 24).
Determination of Rate and Equilibrium Constants
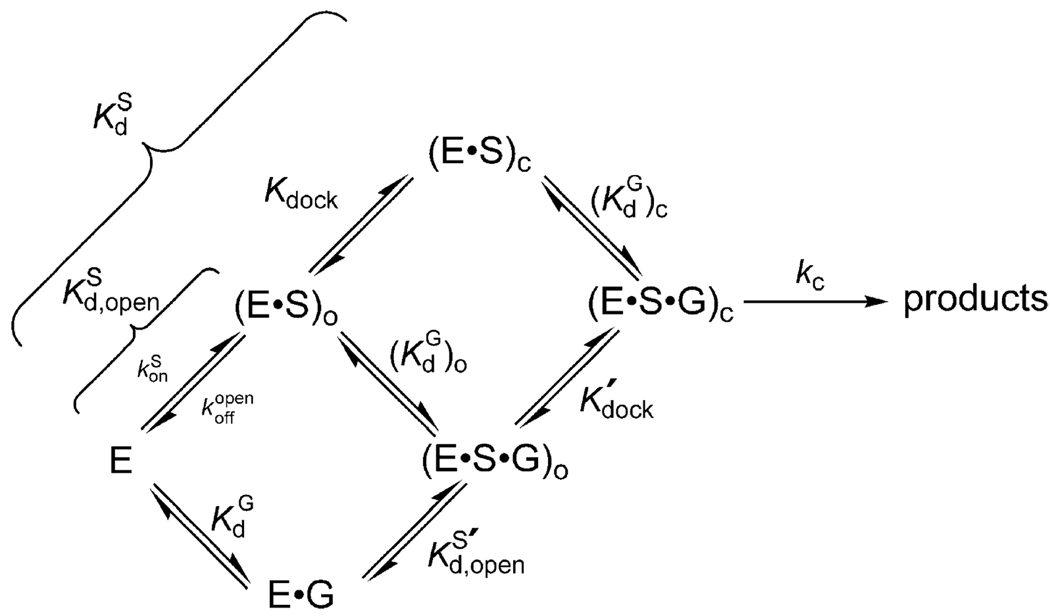
Table 1 describes substrates used and steps monitored. Values of k c, which represents the first-order rate constant for the reaction (E•S•xG)c → products (S = CCCUCdUA5, with “d” representing a 2′-deoxynucleotide; xG = G or UCG), and kopen, which represents the first-order rate constant for the reaction (E•S•xG)o → products (S = CCCd(UCU)A5, with “d” representing a 2′-deoxynucleotide; xG = G or UCG), were determined at pH 6.9 with ribozyme saturating with respect to the oligonucleotide substrate (50 nM E) and with saturating guanosine nucleophile (xG; 2 mM G or 0.5 mM UCG). The subscripts “c” and “o” refer respectively to the closed and open complex, in which the duplex between the ribozyme’s internal guide sequence (IGS) and S is either docked into tertiary contacts with the ribozyme’s core or undocked and not making additional contacts (8, 25, 26). Atomic level substitutions on S allowed control over which complex is formed (8, 24).
Table 1.
Reaction Steps Monitored and Oligonucleotide Substrates Used in This Work
| step monitoreda | measured rate constantb | S (or P)c | abbreviation used in text for S or P | |
|---|---|---|---|---|
| (E•S•xG)c → P | kc | CCCUCdUA5 | −1d,rSA5 | |
| (E•S3′O • xG)o → P | kopen | CCCd(UCU)A5 | S3′O | |
| (E•S3′S • xG)o → P |
|
m(CCC)UCdU3′SA | S3′S | |
| (E•S)o + G → P | (kc/KM)G | d(CCCUC)UA5 | −1r,dSA5 | |
| (E•S)o + GN → P | (kc/KM)GN | d(CCCUC)UA5 | −1r,dSA5 | |
| (E•S•GN)o → P |
|
d(CCCUC)UA5 | −1r,dSA5 | |
| (E•P) + CUCGA → S | (kc/KM)CUCGA3′SA | d(CCCUC)U | −1r,dP | |
| (E•P) + CUCG3′SA → S | (kc/KM)CUCGA | d(CCCUC)U | −1r,dP | |
| E + S → (E•S) | kon | CCCUCdUA5 | −1d,rSA5 | |
| (E•S) → E + S | koff | CCCUCdUA5 | −1d,rSA5 |
xG = G or UCG.
Rate constants are defined according to the reaction framework in Scheme 3.
3′S refers to a 3′-phosphorothiolate linkage; m = 2′-OCH3; d = 2′-H.
, which represents the first-order rate constant for the reaction (E•S3′S•xG)o → products (S3′S = m(CCC)UCdU3′SA, with “d” representing a 2′-deoxynucleotide, “m” representing a 2′-methoxynucleotide, and 3′S representing a 3′-phosphorothiolate linkage; xG = G or UCG), was determined at pH 6.9 with ribozyme saturating with respect to the oligonucleotide substrate (50 nM E) and with saturating xG (2 mM G or 0.5 mM UCG).
(kc/KM)G and (kc/KM)GN are the second-order rate constants for the reaction E•S + G (or GN) → products, where G is guanosine and GN is 2′-aminoguanosine. In these experiments, we used the oligonucleotide substrate −1r,dSA5, which binds to the wt and modified ribozymes in the open complex (Table 1, ref 5, and data not shown). Values of (k c/K M)G and (kc/KM)GN were determined at pH 6.9, with ribozyme saturated with respect to the oligonucleotide substrate (50 nM E) and with subsaturating G or GN (30 µM, which is at least 4-fold below saturation for all of the ribozymes; data not shown).
(kc/KM)CUCG3′X (X = O or S) is the second-order rate constant for the reaction (E•P) + CUCG3′XA → products, where P = d(CCCUC)U and the 3′-X subscript refers to the atom present at the 3′-position of the G nucleoside, either an oxygen (3′O) or a sulfur (3′S) atom. Values of were determined at pH 5.5 with trace amounts of radiolabeled CUCG3′XA and (E•P) subsaturating with respect to CUCG3′XA (20 nM E for reactions with CUCG3′OA, 200 nM E for reactions of CUCG3′SA).
Association (k on) and dissociation (k off) rate constants were measured by gel mobility shift assay using pulse–chase experiments, as described previously (5, 8). In Table 3, k off values for the wt and A306S P ribozymes represent the averages from at least three independent measurements; values for the C208S P, A304S P, and U307S P ribozymes were determined in side-by-side experiments in one of these measurements.
Table 3.
Rate Constants for Dissociation of the Oligonucleotide Substrate −1d,rSA5 in the wt and Modified Ribozymes
| ribozyme | ratio modified/wt | ||
|---|---|---|---|
| wt | 0.0063 ± 0.0015 | (1) | |
| C208SP | 0.10 ± 0.02 | 16 | |
| A304SP | 0.13 ± 0.03 | 21 | |
| A306SP | 0.32 ± 0.12 | 51 | |
| U307SP | 0.013 ± 0.001 | 2 |
EXPERIMENTAL APPROACH AND RESULTS
Metal Ion Rescue and Functional Detection of Ribozyme Ligands of Specific Catalytic Metal Ions
As noted in the introduction, metal ion rescue experiments provide a powerful means to identify functional interactions and have been used extensively for protein and RNA enzymes. This approach has been extended to provide quantitative information that allows determination of whether substrate atoms are contacted by the same or distinct metal ions, using TFA (11, 12). In a further extension, coupled manipulation of both substrate and potential ribozyme ligands can determine whether such groups are indeed ligands for particular metal ions (5, 17, 27). Possible complications that may arise in these analyses, but that do not apply to the Tetrahymena ribozyme, are described in the Supporting Information.
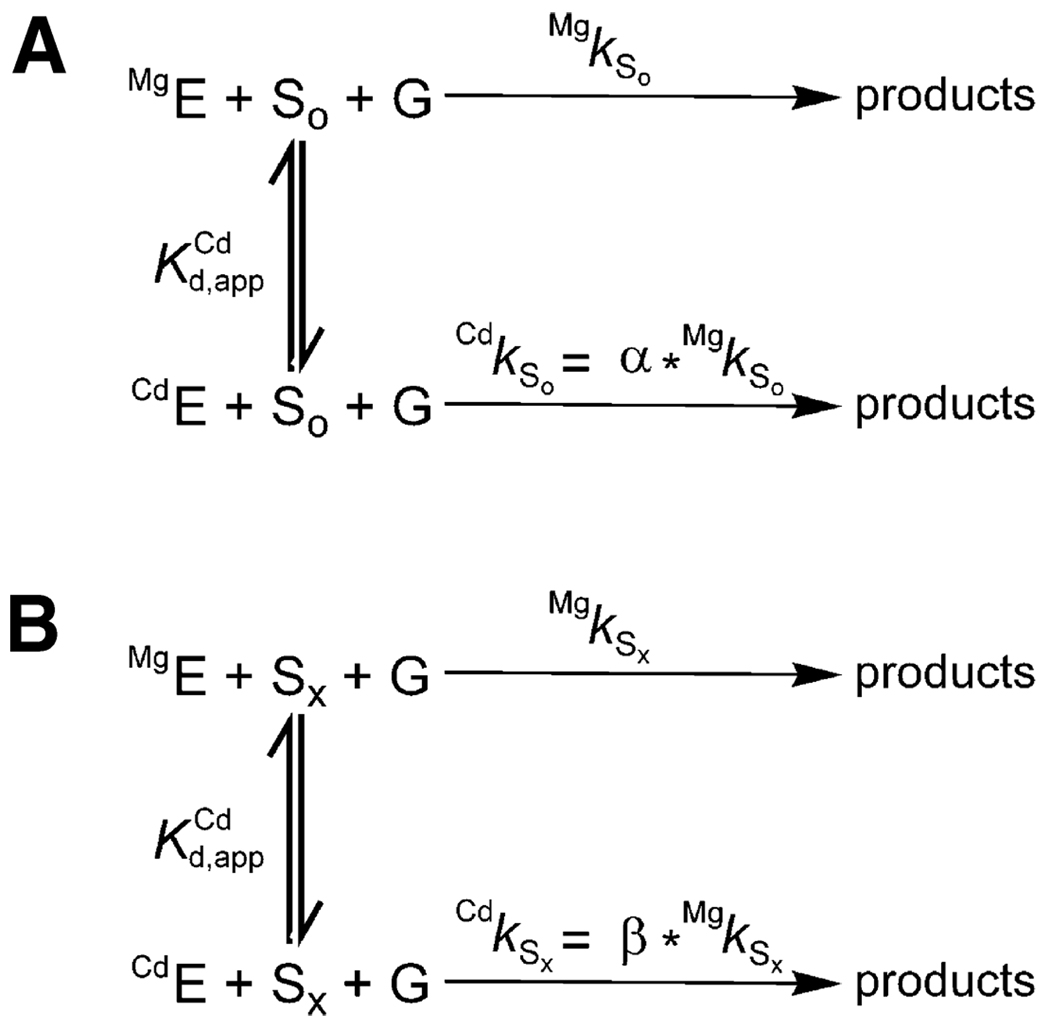
Consider two substrates, So and Sx, corresponding to the unmodified and sulfur- or amino-modified substrates, respectively. Schemes 1A and 1B show simplified reaction frameworks for the unmodified (or cognate) and modified substrates, respectively. We use Cd2+ as the rescuing metal ion for simplicity, but the same consideration can be applied to metal ions that are softer than the native Mg2+, such as Mn2+ and Zn2+. We show the reaction for the Tetrahymena ribozyme and use the symbols and abbreviations used to describe the experiments herein to simplify the presentation. In this reaction S is the oligonucleotide substrate that is an intermolecular analogue of the 5'-splice site and G is the guanosine nucleophile. In Scheme 1, the reaction rate reflects the fraction of ribozyme that has Cd2+ bound, with a rate enhancement with saturating Cd2+ given by the parameters α and β for So and Sx, respectively. The Cd2+ concentration dependence of the observed rate constants gives the dissociation constant for Cd2+, as shown in eq 2a,eq 2b.
| (2a) |
| (2b) |
Scheme 1.
There are two critical features of this analysis. First, represents binding of Cd2+ to the reaction’s ground state, even though it is determined by following the reaction rate that represents transient attainment of the reaction’s transition state. Second, for standard TFA the reaction’s ground state is the free ribozyme and free substrates so that the substrate modification has no effect on the affinity of Cd2+ for the ribozyme. Thus, the value of provides a “fingerprint” for the binding to the free ribozyme of the metal ion that rescues this particular substrate modification, and this value can be compared with the value of obtained for rescue at other positions to determine if the same or distinct metal ions give rescue. In practice, experiments are sometimes carried out with certain substrates bound, provided that control experiments have determined that the association of those substrates is not coupled to the binding of the rescuing metal ion; i.e., the binding of such substrates does not affect the dissociation constant for the rescuing metal ion (5, 10, 11).
In particular with RNA enzymes, metal ions other than the one involved in rescue can bind and affect activity. In many cases, high concentrations of thiophilic metal ions cause inhibition of the normal reaction, apparently by binding at one or more alternate sites. There can also be stimulatory effects from occupancy of other sites. If these effects are the same with the normal and the modified substrate, they can be readily eliminated by using a relative rate constant, k rel, which is simply the ratio of rate constants for the modified and unmodified substrates at each Cd2+ concentration. This is often the case, and this approach has been used on multiple occasions (11, 12, 27–29). Further, experiments are usually carried out in a background of excess Mg2+ to minimize the effects from binding of the soft metal ions at other sites. The resultant obtained by plotting k rel versus Cd2+ concentration and fitting to eq 3 can be used in TFA just as described above.
| (3) |
Macromolecular structural models derived from X-ray crystallographic data provide a breadth of structural detail that would be impossible to match through functional studies alone. Nevertheless, there is no guarantee that interactions observed in ground state structures obtained crystallographically or by other means reflect the functional interactions made in the reaction’s transition state, as exemplified in the case of another RNA enzyme, the hammerhead ribozyme (30, 31). Thus, it is critical to “connect” X-ray structures to functional data, and the knowledge obtained is typically synergistic.
For group I ribozymes, a wealth of functional data predated the recent X-ray structures, and the vast majority of the interactions detected by functional studies were reflected in the X-ray structures (10). In total, of 19 interactions identified by functional data at or near the active site, 17 were reproduced in the second Azoarcus crystal structure (16). Despite this wealth of information, contacts between active site metal ions and the ribozyme’s backbone remained less explored functionally, and only the contact between MC and the pro-S P phosphoryl oxygen of C262 was functionally probed in the reaction’s transition state (5). As introduced above, MA (Figure 1) plays a pivotal role in group I intron catalysis by providing electrostatic stabilization of negative charges in the transition state and possibly electrostatic destabilization of the ground state. We have functionally probed the coordination sphere of MA proposed from crystallographic data (13, 14, 16). In this section, we describe the extension of thermodynamic fingerprint analysis that allows such identification and then present the relevant data. The basic approach of identifying metal ion ligands from the ribozyme is to determine which “fingerprint” is perturbed to give stronger binding upon thio substitution at potential ligand sites.
Consider two ribozymes, the wild type (wt) and a modified ribozyme in which one phosphoryl oxygen atom is replaced by a sulfur atom. For the wt ribozyme, addition of Cd2+ does not usually affect reactions of the unmodified substrate, So, except for inhibition at high concentration that can be accounted for by using k rel. Therefore, we can treat the parameter α in Scheme 1A as being close to unity; α refers to the reactivity of the unmodified substrates in the presence of Cd2+ relative to the reactivity in the absence of Cd2+. This situation may or may not hold for the modified ribozyme. In a case in which a contact between the sulfur atom and a metal ion is not present in the reaction’s ground state but must be formed in the transition state, the simplest expectation is that this modified ribozyme would show an increase in the observed rate constant as the Cd2+ concentration is increased; in this case, we expect α > 1. Conversely, if there is a ground state contact between the sulfur atom and a metal ion that needs to be broken in going to the transition state, a decrease in the observed rate constant is expected as the Cd2+ concentration is increased; i.e., α < 1. However, a lack of dependence on Cd2+ concentration is not conclusive, as it can arise from the absence of a contact between the sulfur atom and a metal ion or, alternatively, a contact that is fully formed in both the ground state and the transition state with either Cd2+ or Mg2+.
To establish the identity of ligands for metal ions, let us first extend Scheme 1 by using Scheme 2, which explicitly contains both the ground state () and the transition state (K Cd‡) dissociation constants of the rescuing metal ion. These two dissociation constants are linked by the thermodynamic cycle in Scheme 2. TFA can now be applied to both the wt and the modified ribozymes, and can be determined for both ribozymes from their Cd2+ dependencies of k rel; similarly, K Cd‡ can be determined by considerations from the observed rate constants and the thermodynamic cycle in Scheme 2. Because of the high affinity of Cd2+ for sulfur, the simplest prediction is that a sulfur atom introduced in the modified ribozyme that contacts the rescuing metal ion in both the ground state and transition state will lower the dissociation constants (ground and transition states) of this metal ion compared to the wt ribozyme. This effect would shift the rescue profile for the modified ribozyme toward lower Cd2+ values. If there is no contact between the rescuing metal ion and the sulfur atom on the ribozyme, the simplest expectation is that the profiles for the wt and modified ribozymes will coincide, and and K Cd‡ will be unaffected. More complex situations in which the metal ion/ligand interaction is made only in the ground state or the transition state can be distinguished and were described above.
Scheme 2.
The cases described above are ideal situations, in which the affinity of the rescuing metal ion is affected only by direct effects. In practice, indirect effects from altered charge distributions, steric clashes, and structural rearrangements upon substitution of oxygen for sulfur can result in changes in the value of the dissociation constants. On the other hand, direct contacts will likely induce larger changes in the dissociation constants compared to indirect effects. Thus, it is crucial to also determine the magnitude of effects arising from sulfur substitutions at different positions. In the following section, we describe how these comparisons have allowed the identification of three ligands for MA, one of the catalytic metal ions in the Tetrahymena ribozyme. These results unify the structural and functional models for this metal ion and its interactions.
Metal A and Its Environment from Crystal Structures and Previous Functional Data
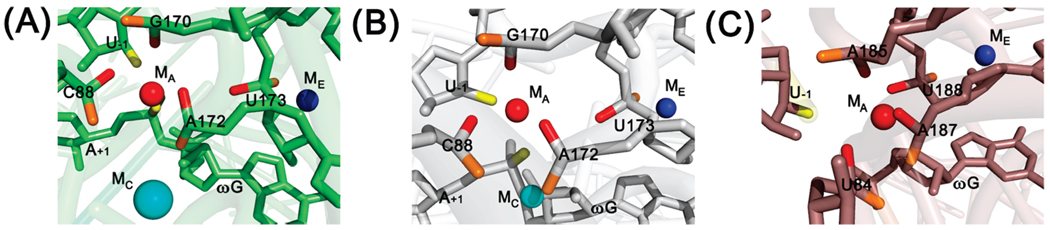
Functional data detected a metal ion, MA, that interacts with both the 3′-oxygen of the leaving group and the pro-SP oxygen of the transferred phosphoryl group (32, 33). A metal ion that appears to satisfy these contacts is also present in the Twort (14) and the two Azoarcus (13, 16) crystal structures (Figure 2), but this metal ion was absent in the structure of the truncated Tetrahymena intron (15). In the Twort crystal structure model (Table 2 and Figure 2C), the pro-SP oxygen atoms of residues U84, A185, A187, and U188 are respectively 3.8, 2.9, 3.3, and 2.8 Å from MA, and these residues were proposed to make direct contacts with the metal ion (14, 34). In both Azoarcus crystal structures, the pro-SP oxygens of the corresponding residues, C88, G170, A172, and U173, are also in the vicinity of a metal ion, M1, that was proposed to be equivalent to MA (Table 2 and Figure 2A,B); however, the oxygen of residue U173 was 4.4 Å from the metal ion, although in the right geometry to complete the octahedral shell for the magnesium ion. This residue was thus proposed to make an outer sphere interaction with MA through an intervening water molecule not seen in the X-ray structure. These residues correspond to residues C208, A304, A306, and U307, respectively, in the Tetrahymena ribozyme, and we use the numbering from Tetrahymena throughout for simplicity.
FIGURE 2.
Location of MA in the different group I intron crystal structures. (A) First Azoarcus crystal structure, PDB accession number 1U6B (13). (B) Second Azoarcus crystal structure, PDB accession number 1ZZN (16). (C) Twort crystal structure, PDB accession number 1Y0Q (14). MA and its putative ribozyme ligands are in red, and MA ligands on the substrates are in yellow; MC (cyan) and ME (blue) are shown to aid orientation. MC is larger in panel A than in panel B because K+ and Mg2+ ions are present, respectively. Residues in orange are phosphoryl oxygen atoms with stereochemistry opposite to that of the putative ribozyme MA ligands. Residues are indicated according to their original numbering; see Table 2 for the equivalent residues in the Tetrahymena ribozyme. Selected residues are shown as sticks; the other parts of the ribozyme are shown as a transparent cartoon in the three panels. The figure was prepared using PyMol (DeLano Scientific, Palo Alto, CA; http://www.pymol.org).
Table 2.
Distances (Å) from MA to Selected Residues in Azoarcus (Azo1 and Azo2, PDB Accession Numbers 1U6B and 1ZZN, Respectively) and Twort (Two, PBD Accession Number 1Y0Q) Crystal Structures
| residue (Tetrahymena numbering)a |
corresponding residue (Azo) |
distance in Azo1 |
distance in Azo2 |
corresponding residue (Two) |
distance in Two |
|---|---|---|---|---|---|
| C208SP | C88 | 2.3 | 2.2 | U84 | 3.8 |
| C208RP | C88 | 4.4 | 3.9 | U84 | 4.8 |
| A304SP | G170 | 2.5 | 2.1 | A185 | 2.9 |
| A304RP | G170 | 4.4 | 4.2 | A185 | 5.2 |
| A306SP | A172 | 2.1 | 2.2 | A187 | 2.8 |
| A306RP | A172 | 2.8 | 3.8 | A187 | 3.3 |
| U307SP | U173 | 4.4 | 4.4 | U188 | 2.8 |
| U307RP | U173 | 6.9 | 6.8 | U188 | 4.9 |
Residues in bold represent putative MA ligands.
To test these proposals using the “extended TFA” described above, we prepared four ribozymes containing a regio- and stereospecific sulfur substitution in place of the proposed oxygen ligands (C208SP, A304SP, A306SP, and U307SP). In addition, we prepared four control ribozymes containing the sulfur modification at the same residue, but with the opposite stereochemistry (C208RP, A304RP, A306RP, and U307RP). The pro-RP oxygens of residues C208 and A304RP are not proposed to make contacts with metal ions, although previous qualitative data from nucleotide analogue interference mapping (NAIM) experiments (35, 36) suggest a role for these atoms. The pro-RP oxygen of residue A306 is proposed to make a contact with MC on the basis of the Azoarcus (13, 16) and Tetrahymena (15) crystal structures. Finally, the pro-RP oxygen of residues U307 was functionally linked to a peripheral metal ion, referred to as ME (22), consistent with a metal ion proposed to be present in the Azoarcus and Twort crystal structures (13, 14, 16) but not observed in the Tetrahymena structure (15).
The reactivity of six of these modified ribozymes (C208SP, C208RP, A304SP, A304RP, A306SP, and A306RP) was partially characterized in previous work (18). In that report the effect of phosphorothioate substitution was evaluated by determining the reactivity of the modified ribozymes with an all-RNA 5′-splice site mimic, CCCUCUA5 (rSA5). Among the different ribozymes studied in that paper, the C208SP, A306SP, and A306RP ribozymes were found to react 30–100-fold slower than the wt ribozyme. Addition of 1 mM Mn2+ or 0.5 mM Zn2+ increased the observed rate constant for the C208SP ribozyme by ~20-fold, but this rate stimulation was not observed for the other modified ribozymes. Addition of 0.2 mM Cd2+ did not have any effect on any of these ribozymes. The enhancement in reactivity upon Mn2+ or Zn2+ addition was taken as support for a direct contact between the pro-SP oxygen of C208 and a metal ion. This metal ion was suggested to be MA by using a substrate containing a modification at the sulfur modification at the 3′-oxygen of the oligonucleotide substrate, because this substrate exhibited an ~1000-fold rate acceleration upon addition of Cd2+ for the modified ribozyme compared to the wt ribozyme.
For the comparisons between modified and wt ribozymes to be interpretable, it is crucial to monitor the same individual reaction steps. The reaction catalyzed by the Tetrahymena ribozyme involves multiple steps, and a kinetic and thermodynamic reaction framework has been determined and is summarized in Scheme 3 (ref 10 and references cited therein). The oligonucleotide substrate (S), which mimics the 5′-splice site of the normal self-splicing reaction, binds to the ribozyme to form the so-called P1 duplex in an “open complex”, indicated with the subscript “o” [(E • S)o and (E • S•G)o]. The P1 duplex can then dock into the ribozyme’s core, forming tertiary interactions and generating the “closed complex”, denoted with the subscript “c” [(E • S)c and (E•S•G)c]. Guanosine (G) can bind at any time along this framework, and there is thermodynamic coupling between guanosine binding and P1 docking (24, 37), resulting in increased affinity of guanosine for the closed complex relative to the open complex. Short oligonucleotides with 3′-guanosine residues, such as UCG, can form base pairings with the ribozyme adjacent to the G-binding site (38–40) and thus can bind stronger and be used in place of G to ensure saturation of the nucleophilic species (24, 38). When the 3′-hydroxyl of G (or UCG) is deprotonated and S and G (or UCG) are aligned in the ribozyme’s active site, the reaction’s chemical step can proceed, in which the deprotonated 3′-oxygen atom of guanosine attacks the phosphoryl center in a transition state stabilized by the catalytic metal ions and other interactions (Figure 1) (ref 10 and references cited therein). Because only a limit (pKa ≥ 10) has been established for deprotonation of the attacking guanosine nucleophile (24), this step is included with the actual chemical step in Scheme 3 and in the rate constants reported here and elsewhere (ref 10 and references cited therein).
Scheme 3.
The oligonucleotide substrate rSA5, used in the previous report aimed at probing possible metal ion ligands contributed from the ribozyme (18), binds and reacts from the closed complex in the wt ribozyme (26). However, no characterization of the ground state for the modified ribozymes was carried out. To determine whether the wt and modified ribozymes react from the same ground state complex, we measured the off-rate for the oligonucleotide substrate CCCUCdUA5 (−1d,rSA5). The observed off-rate () depends on two factors: the stability of the duplex between the ribozyme and the oligonucleotide substrate, which is expected to be the same for the modified and wt ribozymes, and the tertiary contacts between the ribozyme and the P1 duplex in the closed complex (ref 10 and references cited therein). For the wt ribozyme, an oligonucleotide substrate that binds predominantly in the open complex, i.e., the open complex is the ground state, gives (Scheme 3). An oligonucleotide substrate that binds predominantly in the closed complex dissociates slower, with the value of Kdock is 22 for the standard oligonucleotide substrate containing all ribose residues, rSA5, and for its slower reacting analogue, −1d,rSA5 (41). We found that oligonucleotides that bind to the wt ribozyme in the closed complex dissociate ~20-fold faster when bound to the C208SP, A304SP, and A306SP ribozymes (Table 3). As expected, these modified ribozymes did not show any difference in the stability of the open complex, measured with oligonucleotides that bind predominantly in the open complex (data not shown; see also refs 5 and 22 for related examples). These results indicate that the closed complex is destabilized in the C208SP, A304SP, and A306SP ribozymes. This amount of destabilization is similar to, or greater than, the docking equilibrium in the wt ribozyme [Kdock = 22 (41)], suggesting that these modified ribozymes may react from the open complex. Given this complexity and the possible resultant ambiguities, we included the previously studied C208SP ribozyme in our tests of coordination to MA and assessed rescue starting from the open complex rather than the closed complex, carrying out additional controls as described below.
Functional Tests Detect Three Transition State Contacts between MA and the Ribozyme
To monitor Cd2+ binding at the metal ion site A for each of the thio-substituted ribozymes, we used the approach described in the previous section, following the effect of increasing Cd2+ concentration on reactivity of an oligonucleotide substrate containing a 3′-thiophosphoryl linkage at the cleavage site, S3′S, relative to the unmodified substrate, S3′O (Table 1). Previous work showed that both substrates bind and react from the wt ribozyme in the open complex (11), thereby not perturbing the ground state dissociation constant for MA, as compared to the free ribozyme. In addition, the log linearity of the pH–rate dependencies of the reactions of both the S3′O and −1r,dSA5 substrates in the presence of saturated amounts of G or UCG, in the range between pH 5.5 and pH 7.5 (data not shown), strongly suggests that the chemical step is rate-limiting, by analogy to the wt ribozyme (42–45). We first established the reactivity of the modified ribozymes using the unmodified substrate, S3′O, in the absence of Cd2+ (Table 4). All of the modified ribozymes displayed a reduced reactivity compared to the wt ribozyme, by values ranging from 6- to 270-fold. These results are consistent with the previous functional data (18, 36), suggesting an important role for each of the substituted oxygen atoms. However, it remained to be established whether this role is to coordinate a metal ion.
Table 4.
Observed Rate Constants for the Reaction (E•S3′O • xG)o → Products (kopen; 50 mM MgCl2, pH 6.9) with Different Ribozymes without and with 1 mM CdCl2 a
| ribozyme |
Mgkopen (min−1) × 10−2 |
fold down relative to wt |
Cdkopen (min−1) × 10−2 |
Cdkopen/ Mgkopen |
|---|---|---|---|---|
| wt | 2.6 | (1) | 1.8 | 0.69 |
| C208SP | 0.046 | 57 | 0.069 | 1.5 |
| C208RP | 0.066 | 39 | 0.057 | 0.86 |
| A304SP | 0.018 | 140 | 0.032 | 1.8 |
| A304RP | 0.44 | 6 | 1.1 | 2.5 |
| A306SP | 0.042 | 62 | 0.058 | 1.4 |
| A306RP | 0.0095 | 270 | 0.0058 | 0.61 |
| U307SP | 0.26 | 10 | 0.27 | 1.0 |
| U307RP | 0.25 | 10 | 2.0 | 8.0 |
S3′O is defined in the text (xG = G or UCG).
We next tested the effect of addition of 1 mM Cd2+ on the reactivity of S3′O in the wt and modified ribozymes (Table 4 and Figure 3). With the exception of the A304RP and the U307RP ribozymes, Cd2+ affected the reactivity of the ribozymes less than 3-fold. The stimulatory effect on the U307RP ribozyme was previously investigated and ascribed to a peripheral metal ion, ME, that interacts with the introduced sulfur atom on the ribozyme and modulates several reaction steps (22). There was also a small stimulatory effect on the A304RP ribozyme (2.5-fold), even though the substituted oxygen atom was not implicated in functional contacts with metal ions from structural data. The small magnitude of this rate enhancement and the small decrease in reactivity of the A304RP ribozyme compared to the wt ribozyme (6-fold) suggest that the stimulatory Cd2+ effect for this ribozyme may arise from indirect effects rather than direct coordination to a metal ion (see Discussion).
FIGURE 3.
Effect of Cd2+ (1 mM) on the reactivity of the oligonucleotide substrate S3′O with modified ribozymes, normalized for the effect observed in the wt ribozyme. Data are from Table 4.
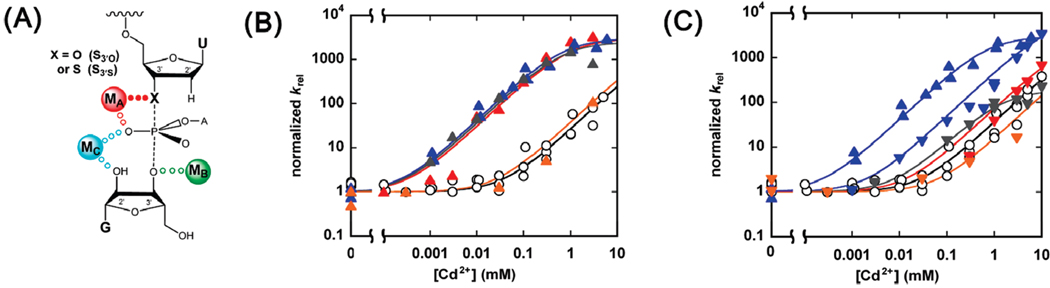
Finally, we monitored the reactivity of S3′S relative to S3′O as a function of Cd2+ concentration (Figure 4) to establish the complete rescue profiles for the wt and modified ribozymes. To allow direct comparison of the rescue profiles, we normalized the data so that Mg krel = 1 for all the ribozymes; the raw data are presented as Supporting Information. As shown in Figure 4B, the ribozymes containing a sulfur atom in a position proposed to be direct ligands of MA, namely, the C208SP, A304SP, and A306SP ribozymes, displayed rescue profiles significantly shifted toward lower Cd2+ concentrations, with values of ~ 0.4–0.7 mM for the three ribozymes (Table 5); this shift is in sharp contrast to the lack of saturation observed with the wt and U307SP ribozymes ( ≥ 10 mM). Following the analyses described above and the additional control reactions described below, these results strongly suggest that the pro-SP oxygen atoms of residues C208, A304, and A306 are both ground state and transition state ligands for MA.
FIGURE 4.
MA rescue of S3′S cleavage for wild-type and modified ribozymes. Cd2+ rescue reactions were carried out as indicated in the text and in Materials and Methods. (A) The metal ion-substrate contact being probed is indicated by solid dots in the model for the Tetrahymena ribozyme transition state. X represents an oxygen or sulfur atom in the oligonucleotide substrates S3′O and S3′S, respectively. (B) Rescue profiles for the wt (○), C208SP (red △), A304SP (blue △), A306SP (gray △), and U307SP (orange △) ribozymes. (C) Rescue profiles for the wt (○), C208RP (red ▽), A304RP (blue ▽), A306RP (gray ▽), and U307RP (orange ▽) ribozymes. The rescue profile of the A304SP ribozyme (blue △) is repeated from panel B to aid comparison. krel represents the observed rate constant for cleavage of S3′S relative to S3′O and was normalized to equal 1 in the absence Cd2+.
Table 5.
Values of the Parameters in Scheme 3 for the Rescue of the S3′S Substrate with Different Ribozymes
| ribozyme | β | (mM)c | KCd‡ (mM)d × 10−2 |
KCd‡ relative to wt |
|---|---|---|---|---|
| wt | (3000)a | (120 ± 10) | 4.0 | (1) |
| C208 SP | 3000 b | 0.36 ± 0.10 | 0.012 | 0.0030 |
| C208RP | (3000)a | (51 ± 7) | 1.7 | 0.44 |
| A304SP | 3000 b | 0.47 ± 0.15 | 0.016 | 0.0040 |
| A304RP | 7000 | (14 ± 2) | 0.20 | 0.050 |
| A306SP | 2500 b | 0.77 ± 0.10 | 0.031 | 0.0078 |
| A306RP | 260 ± 70b | 3.0 ± 1.8 | 1.2 | 0.30 |
| U307SP | (3000)a | 80 ± 2 | 2.7 | 0.66 |
| U307RP | (>3000)a | (200 ± 20) | >6.7 | >1.7 |
Inferred from the saturation observed in the C208SP, A304SP, and A306SP ribozymes.
Derived from experimental measurements, assuming the same maximal rate for wt and modified ribozymes not giving enhanced rescue as for the preferentially rescued modified ribozymes.
Represents the best fit of the experimental data (Figure 4B,C) to eq 2b using values of β indicated in this table.
Derived from Scheme 3 using the values of β and indicated in this table.
The four control ribozymes, C208RP, A304RP, A306RP, and U307RP, showed different behavior (Figure 4C, inverted triangles). Despite the decrease in reactivity observed using the S3′O substrate (see Table 4), the rescue profile for C208RP (inverted red triangles) was virtually identical to that of the wt ribozyme, suggesting a lack of contacts between MA and the sulfur substitution. U307RP, which was identified to be a ligand for ME, a peripheral metal ion (22), showed a rescue profile slightly shifted to higher Cd2+ concentrations, suggesting an increased value of compared to the wt ribozyme. This result is consistent with a subtle effect introduced by the perturbation of ME binding site that affects the environment within the active site, as discussed previously (22).
The A304RP ribozyme (inverted blue triangles) displayed a shift in the rescue profile compared to the wt ribozyme, although this shift is less pronounced than that observed in the modified ribozymes whose sulfur modification is proposed to directly interact with MA. Indeed, the A304RP ribozyme reacts with S3′O within 6-fold of the wt ribozyme in the absence of Cd2+, indicating that the modified ribozyme is not substantially impaired compared to the wt.
Finally, the A306RP ribozyme displayed saturation with ~ 3 mM, but also a lower plateau compared to the other ribozymes. The possible reasons for this result are summarized in the Discussion. Regardless, the effects on the RP modified ribozymes are far less dramatic than those on the putative MA ligands (Figure 4C; compare inverted triangles and normal triangles).
The Putative MA Ligands Are Not Ligands for MB and MC
The results presented in the previous section strongly suggest the presence of ground state and transition state contacts between the sulfur atoms in the C208SP, A304SP, and A306SP ribozymes and CdA. To further test this model and the conclusions arising, we tested whether these atoms interact with the other catalytic metal ions, MB and MC. We also included U307SP, because of its proposed outer-sphere coordination to MA and its vicinity to the 3′-oxygen of the guanosine nucleophile in the Twort crystal structure, suggesting a possible role of the pro-SP oxygen of U307 in coordination of MB (14).
To follow Cd2+ binding to the MB site, we monitored Cd2+ rescue of the reactivity of CUCG3′SA relative to CUCGA in the reverse reaction with E•P (eq 4 and Figure 5A). In previously published experiments (5, 11, 12), this test was carried out using the oligonucleotide substrate CCCUCdT (−1d,rP), which binds and reacts from the closed complex. However, as described above, docking is significantly weakened in the C208SP, A304SP, and A306SP ribozymes. In addition, the flat pH–rate profile for the reaction of CUCGA and −1d,rP using the A304SP and A306SP ribozymes (data not shown) suggests a nonchemical rate-limiting step for these reactions. Given these complications, we decided to follow the reaction using the oligonucleotide substrate d(CCCUC)U (eq 4), which docks weaker than the −1d,rP substrate (24) used in the previously published MB tests and whose pH–rate profile is linear up to pH 6.5 (data not shown). Additional controls showed that using d(CCCUC)U instead of CCCUCdU does not change the rescue profile for CUCG3′SA (Supporting Information Figure S4).
| (4) |
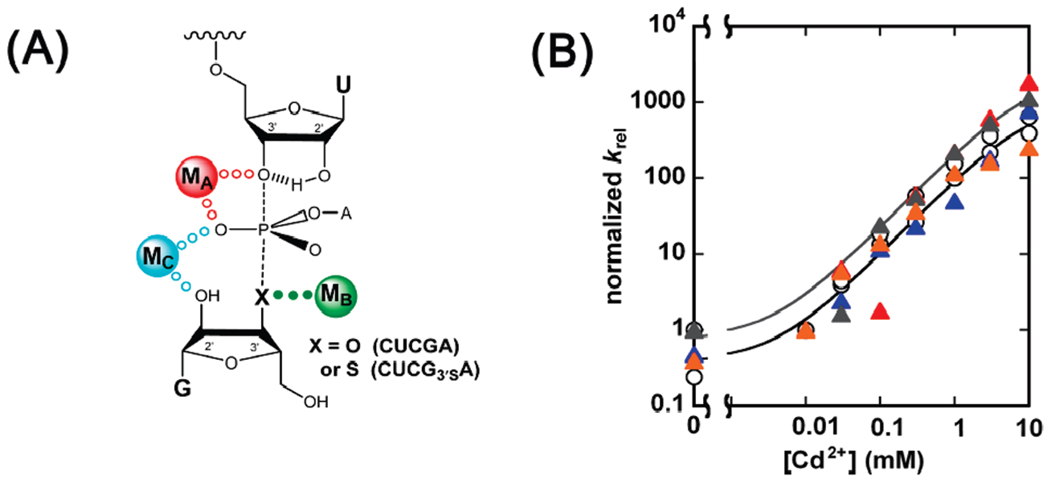
FIGURE 5.
MB rescue of CUCG3′SA cleavage for wild-type and modified ribozymes. Rescue reactions were carried out as indicated in the text and in Materials and Methods. (A) The metal ion-substrate contact being probed is indicated by green solid dots in the model for the Tetrahymena ribozyme transition state. X represents an oxygen or sulfur atom in the guanosine analogues CUCGA and CUCG3′SA, respectively. (B) Rescue profiles for the wt (○), C208SP (red △), A304SP (blue △), A306SP (gray △), and U307SP (orange △) ribozymes. krel represents the observed rate constant for cleavage of CUCG3′SA relative to CUCGA and was normalized to equal 1 in the absence of Cd2+.
In reactions catalyzed by the wt and modified ribozymes, Cd2+ specifically stimulates the reactivity of CUCG3′SA, relative to CUCGA, by more than 800-fold (Figure 5B and Supporting Information Table S1). This rescue fits well to a model in which a single Cd2+ ion binds to the MB site and rescues the reaction (Figure 5B). The modified ribozymes with phosphorothioate substitutions at the SP positions of residues C208, A304, A306, and U307 gave nearly identical rescue profiles (Figure 5B and Supporting Information Table S1). These results strongly suggest that the pro-SP phosphoryl oxygens of residues C208, A304, A306, and U307 are not ground state or transition state ligands for MB.
To follow Cd2+ binding to the MC site, we monitored Cd2+ rescue of oligonucleotide substrate cleavage by 2′-aminoguanosine (GN) relative to G (Figure 6A) (5, 11, 46, 47). Previous work showed that saturating conditions of G or GN give a larger shift in the MC rescue profile for a MC ligand, compared to subsaturating G or GN (5). This approach alters the ground state dissociation constant for MC, with respect to a situation with unbound G or GN, and therefore the resultant does not provide a fingerprint for MC. However, our goal was to determine whether the introduction of a sulfur atom on the ribozyme affected the dissociation constant of the rescuing metal ion, regardless of whether this dissociation constant was already altered by the modification on the substrate.
FIGURE 6.
MC rescue of −1r,dSA5 cleavage with saturating GN by wild-type and modified ribozymes. Rescue reactions were carried out as indicated in the text and in Materials and Methods. (A) The metal ion–substrate contact being probed is indicated by cyan solid dots in the model for the Tetrahymena ribozyme transition state. X represents the −OH or −NH2 group of guanosine and 2′-aminoguanosine, respectively. (B) Rescue profiles for the wt (○), C208SP (red △), sA304SP (blue △), A306SP (gray △), and U307SP (orange △) ribozymes. krel represents the observed rate constant for −1r,dSA5 cleavage by 2′-aminoguanosine relative to that by guanosine and was normalized to equal 1 in the absence of Cd2+.
The rescue profiles for C208SP, A306SP, and U307SP ribozymes were nearly identical to that of the wt ribozyme, showing lack of saturation in the monitored range of Cd2+ concentration (Figure 6B and Supporting Information Table S2). The A304SP ribozyme showed a slightly different rescue profile, but within 4-fold from that of the wt ribozyme. Further, the small shift observed in the rescue profile was toward higher concentrations of Cd2+ and thus in the opposite direction of what is predicted for a direct contact between the sulfur atom and MC. Stronger binding was observed in the same test with the C262SP ribozyme (5), consistent with a ground state and transition state contact between MC and the sulfur atom at position C262SP. The phosphorothioate incorporation into the ribozymes gave small or no effect on the MC Cd2+ rescue profiles, strongly suggesting that the pro-SP phosphoryl oxygen atoms of residues C208, A304, A306, and U307 are not ground state or transition state ligands for MC.
DISCUSSION
Metalloenzymes play multiple roles in biology, serving as kinases, phosphatases, polymerases, and nucleases, among other functions. To understand how enzymes use metal ions to provide rate acceleration, the contacts made by these metal ions at the reaction’s transition state must be defined. The results presented provide a functional snapshot of the environment surrounding a catalytic metal ion in the Tetrahymena group I ribozyme in the reaction’s ground state and transition state, a necessary step toward unraveling contributions to the overall rate enhancement.
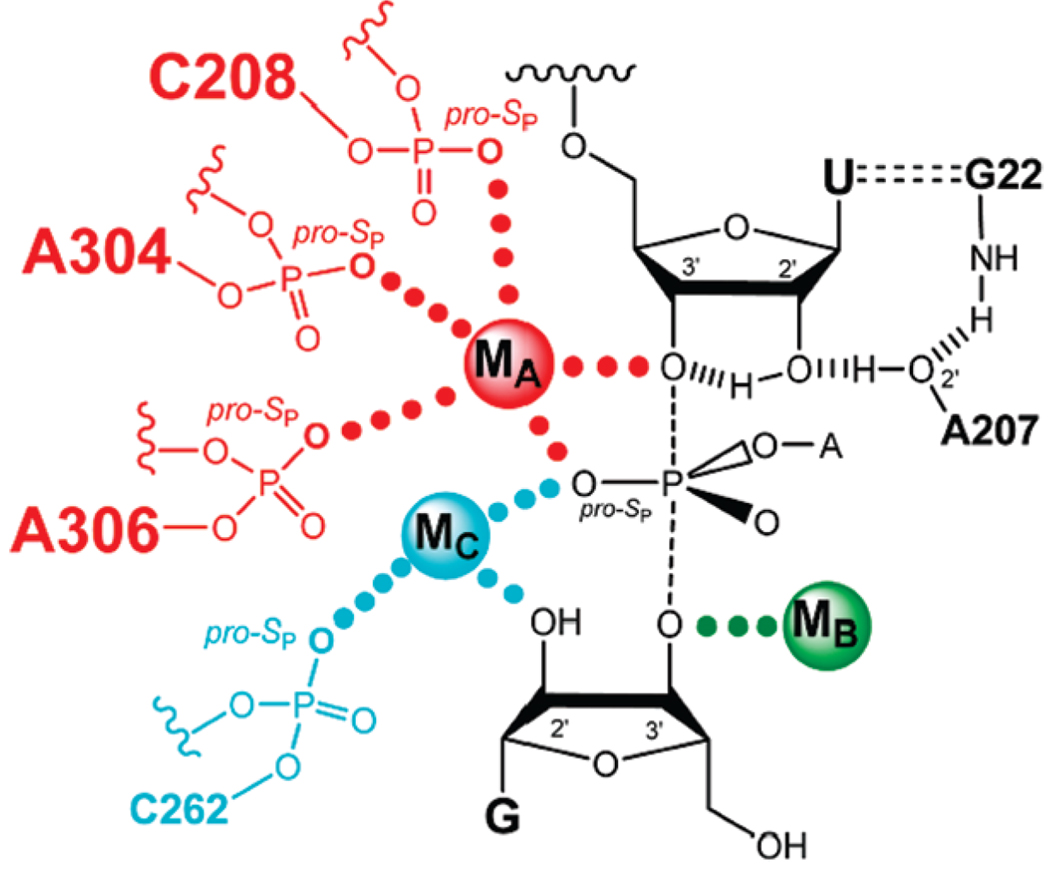
The analysis of simultaneous atomic substitutions within the Tetrahymena ribozyme core and its substrates, under conditions that allow rigorous thermodynamic comparisons, provides strong evidence that the pro-SP oxygen atoms of residues C208, A304, and A306 serve as ligands for the catalytic metal ion MA (Figure 7). A single phosphorothioate substitution at these sites dramatically decreases the apparent dissociation constant for CdA (Figure 4B and Table 5) in both the ground state and the transition state. These results are consistent with proposals from the Azoarcus and Twort crystal structures (13, 14, 16). The lack of a shift in the rescue profile for the U307SP ribozyme suggests that there is no direct contact between the sulfur at position 307SP and MA, in contrast to the proposal from the Twort crystal data (14).
FIGURE 7.
Proposed model of the Tetrahymena ribozyme transition state from functional data (ref 10 and references cited therein and this work). Bonds made and broken at the transition state are represented by dashed lines. Hydrogen bonds are represented by hashed lines. Functional contacts between metal ions and their ligands are represented by solid dots of the same color of the metal ion; contacts from this work are in larger font.
Coordination of CdA to the modified ribozymes does not stimulate reaction with the normal substrates, as shown by lack of Cd2+ dependence of the reaction of So and G (Table 4). These results suggest that site A is fully occupied by Mg2+ in the absence of Cd2+ in the modified ribozyme and that the contacts between MA and its transition state ligands on the ribozyme (the pro-SP oxygen atoms of residues C208, A304, and A306) are fully formed in the ground state of the reaction and do not provide additional transition state stabilization. In contrast, there is a modest stimulation for two other modified ribozymes, U307RP and A308RP, in which the introduced sulfur atom coordinates a peripheral metal ion, ME, apparently due to subtle local rearrangements (22).
In contrast to the other proposed MA ligands, phosphorothioate substitution at position U307SP gives no change in the rescue profile, relative to the wt ribozyme. This result suggests that if a contact is formed between MA and the pro-SP oxygen of U307, it does not occur through direct coordination. The ~10-fold reduced reactivity of the U307SP ribozyme indicates a role that is consistent with the proposal of an outer-sphere coordination of the pro-SP oxygen of U307 to MA (13, 16), although our experiments do not probe this coordination. Regardless, our results suggest that introduction of a phosphorothioate substitution in a position ~4 Å from a metal ion is not sufficient to induce direct coordination. While possible in principle, the prior proposal that outersphere metal ligands can lead to rescue (48) was based on an incomplete thermodynamic analysis (J. K. Frederiksen and J. A. Piccirilli, unpublished) and is not substantiated by the results herein.
The ribozymes carrying a phosphorothioate modification with stereochemistry opposite to the proposed MA ligands showed more heterogeneous behavior. Virtually all of these ribozymes showed a decrease in overall reactivity compared to the wt ribozyme (Table 4), including the C208RP and A304RP ribozymes, where the sulfur substitution is not predicted to interact with any metal ion. While it is possible that these atoms interact with a metal ion, the C208RP ribozyme rescue profile for the oligonucleotide substrate S3′S overlaps with that of the wt ribozyme (Figure 4C), strongly suggesting that there are no ground state or transition state contacts between the sulfur atom and MA. The detrimental effect upon introduction of a sulfur atom in the C208RP ribozyme presumably results from indirect effects, such as disruption of a network of interactions that does not involve a metal ion, introduction of a steric clash, or redistribution of charge within the phosphodiester linkage. In the second Azoarcus crystal structure, the pro-RP oxygen of the residue corresponding to C208 is 2.5 Å from the 2′-OH of A306 and 3.1 Å from the pro-RP oxygen atom of the same residue. These atoms have been proposed to coordinate metal ions (16), and it is possible that the introduced sulfur atom affects their positioning.
For the A304RP ribozyme, the rescue profile for S3′S is shifted toward lower Cd2+ concentrations compared to the wt ribozyme. However, this shift is less pronounced than those for the putative MA ligands, suggesting differences between this group of ribozymes and the A304RP ribozyme. In addition, functional data already support five transition state ligands for MA (the pro-SP phosphoryl oxygen atoms of residues C208, A304, and A306, the pro-SP oxygen on the scissile phosphate, and the 3′-bridging oxygen of the leaving group), with only one ligand missing to satisfy an octahedral geometry. These five ligands support the spatial environment observed in the Azoarcus crystal structures (13, 16). Given this agreement of function and structure in the environment surrounding MA, including the functionally tested contact between the pro-RP oxygen of U307 (22), it is reasonable for the sixth MA ligand to be in the vicinity of the pro-SP oxygen of U307, as proposed from the Azoarcus crystal structures (13, 16). In these structures, the pro-RP oxygen of A304 approaches the metal ion site from a distinct orientation than the pro-SP oxygen of U307 [Figure 2A,B; cf. the red oxygen atom for Azoarcus residues U173 (U307) and the orange oxygen atom for Azoarcus residue G170 (A304)] and therefore seems unlikely to represent the missing MA ligand. This analysis supports the model that the effects observed upon introduction of a sulfur atom at the RP position of A304 are due to a rearrangement of the active site upon thio substitution, as described in other systems (22, 49–51). One interesting possibility for this rearrangement is that a direct coordination of MA to the sulfur atom is induced in a conformation distinct from the predominant active conformer of the wt ribozyme that still allows catalytic interactions with MA. There is evidence for such an effect for the hammerhead ribozyme (ref 52 and S. Wang, K. Karbstein, and D. Herschlag, unpublished results).
Phosphorothioate substitution at position U307RP, a position implicated in binding a peripheral metal ion referred to as ME, shifts the rescue profile to higher Cd2+ concentrations. Possible reasons for this effect have been discussed in previous work (22).
The last ribozyme studied, the A306RP ribozyme, shows a significant but modest decrease in the ground state dissociation constant for MA, (2.5 mM vs ≥10 mM for the wt ribozyme). It is possible that this residue indeed interacts with MA, and its distance from MA was calculated as 2.8 and 3.8 Å in the first and subsequent Azoarcus structures, respectively (13, 16). The longer distance and the unfavorable angle of approach observed in the second structure, which appears to better mimic the active ribozyme, the structural and functional evidence that the pro-SP oxygen atom of the residues is a MA ligand, and the modest change in MA affinity raise questions about whether the pro-RP oxygen atom of A306 is normally a MA ligand. We propose instead that this atom, when replaced by sulfur, may take advantage of the strong sulfur/Cd2+ affinity to make a noncognate interaction, as has been observed with the hammerhead ribozyme (52). Further, the rescue profiles of Figure 4C show that the pro-RP sulfur substitution at A306 does not increase reactivity and, indeed, decreases reactivity at saturating Cd2+, consistent with formation of a nonproductive, inhibitory interaction. Finally, this phosphoryl oxygen atom has been proposed to be a ligand for MC (13, 15, 16), and initial functional tests support this proposal (M. Forconi, J. Lee, J. L. Hougland, J. A. Piccirilli, and D. Herschlag, unpublished).
In contrast to the large shift of MA rescue profiles, the C208SP, A304SP, and A306SP ribozymes did not show significantly altered rescue profiles for substrates that probe Cd2+ occupancy at the MB and MC sites (CUCG3′SA and GN, respectively; Figure 5B and Figure 6B). These observations strongly suggest that none of the introduced sulfur atoms interacts with the metal ions responsible for rescue with these substrates in either the ground state or the transition state. The lack of altered rescue profiles in MB and MC tests underscores the power of the approach used here, as contacts with different metal ions can be distinguished on the basis of the simultaneous introduction of substitutions on the substrates and on the ribozyme.
Our results on MA coordination are in full agreement with all the previous functional data and clarify, minimally for the Tetrahymena ribozyme, the metal ion coordination for the ribozyme, distinguishing between different coordination schemes suggested from different X-ray structures (13, 14, 16). Combining the considerable functional work from sulfur substitution on the ribozyme backbone with the structural data suggests that these substitutions generally do not alter the metal ion contacts. We also have provided evidence for one instance of such change, based on careful dissection of functional data. These results, in combination with previous functional data linking the pro-SP oxygen of C262 to MC in both the ground state and the transition state of the reaction (5), suggest that the second Azoarcus crystal structure (16) most closely represents the ground state interactions that occur in solution. As noted above, the main difference between transition state models based on functional or structural data is the presence or absence of a third metal ion, MB, and this question is not answered herein. Nevertheless, the extensive functional data exploring catalytic interactions provide a portrait of catalysis by this ribozyme in nearly full agreement with one of the proposals based on these X-ray data (16), as described previously and summarized in Figure 7 (10).
The results presented herein emphasize the enormous power of the interplay of structural and functional data. Structural data, derived from different group I intron crystal structures, provided important initial clues regarding the environment surrounding catalytic metal ions in the group I intron. From this, functional assays were used to probe the ground state and transition state coordination around a catalytic metal ion, MA, in the Tetrahymena ribozyme-catalyzed reaction. Using site-specific phosphorothioate substitutions in the ribozyme, coupled to atomic substitutions on the substrates under conditions that allow valid thermodynamic comparison between the wild type and modified ribozymes, we have identified three ligands for MA. Our results also suggest that the ribozyme–MA ligands remain bound to MA over the reaction cycle, prior to association with substrates, in the E•S complexes and in the reaction’s transition state. These results provide the basis for dissection of the role of this metal ion in the ribozyme reaction cycle and, in conjunction with the information from crystallography, for understanding of the structural features that allow construction of positioned, catalytic metal ion sites.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Alexander Kravchuk for preliminary experiments with the modified ribozymes and members of the Herschlag and Piccirilli laboratories for helpful discussion and comments on the manuscript.
Footnotes
This work was supported by a grant from the NIH (GM 49243) to D.H. and by a grant from the Howard Hughes Medical Institute to J.A.P.
Abbreviations: TFA, thermodynamic fingerprint analysis; GN = 2′-aminoguanosine; IGS) internal guide sequence. Refer to Table 1 for the list of oligonucleotide substrates used and their abbreviations.
SUPPORTING INFORMATION AVAILABLE
Supporting text with references to metal ion rescue experiments in protein and RNA enzymes and discussion of possible complications in TFA analysis; Table S1, providing the values of the parameters in Scheme 3 for the MB rescue of CUCG3′SA; Table S2, providing the values of the parameters in Scheme 3 for the MC rescue of GN; Figures S1, S2, S3, and S5, providing the data prior to normalization associated with Figure 4B,C, Figure 5B, and Figure 6B, respectively; and Figure S4, providing the comparison of MB rescue of CUCG3′SA using two different substrates, −1r,dP and −1d,rP, for the wt and U307SP ribozymes. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Lad C, Williams NH, Wolfenden R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroeder GK, Lad C, Wyman P, Williams NH, Wolfenden R. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn M, Shih N, Nick J. Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ATP in the arginine kinase reaction. Structure of the metal-nucleoside triphosphate substrate. J. Biol. Chem. 1982;257:7646–7649. [PubMed] [Google Scholar]
- 4.Eckstein F. Phosphorothioate analogs of nucleotides–Tools for the investigation of biochemical processes. Angew. Chem., Int. Ed. 1983;22:423–439. [Google Scholar]
- 5.Hougland JL, Kravchuk AV, Herschlag D, Piccirilli JA. Functional identification of catalytic metal ion binding sites within RNA. PLoS Biol. 2005;3:1536–1548. doi: 10.1371/journal.pbio.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: Chasing a protein’s tail. Curr. Opin. Chem. Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA–The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 8.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 9.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozymes– Enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 10.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. How the group I intron works: A case study of RNA structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 133–206. [Google Scholar]
- 11.Shan S, Yoshida A, Piccirilli JA, Herschlag D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12299–12304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan S, Kravchuk AV, Piccirilli JA, Herschlag D. Defining the catalytic metal ions interactions in the Tetrahymena ribozyme reaction. Biochemistry. 2001;40:5161–5171. doi: 10.1021/bi002887h. [DOI] [PubMed] [Google Scholar]
- 13.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 14.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 15.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell. 2005;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 18.Szewczak AA, Kosek AB, Piccirilli JA, Strobel SA. Identification of an active site ligand for a group I ribozyme catalytic metal ion. Biochemistry. 2002;41:2516–2525. doi: 10.1021/bi011973u. [DOI] [PubMed] [Google Scholar]
- 19.Dai Q, Deb SK, Hougland JL, Piccirilli JA. Improved synthesis of 2′-amino-2′-deoxyguanosine and its phosphoramidite. Bioorg. Med. Chem. 2006;14:705–713. doi: 10.1016/j.bmc.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 20.Benseler F, Williams DM, Eckstein F. Synthesis of suitably protected phosphoramidites of 2′-fluoro-2′-deoxyguanosine and 2′-amino-2′-deoxyguanosine for incorporation into oligoribonucleotides. Nucleosides Nucleotides. 1992;11:1333–1351. [Google Scholar]
- 21.Sun SG, Yoshida A, Piccirilli JA. Synthesis of 3′-thioribonucleosides and their incorporation into oligoribonucleotides via phosphoramidite chemistry. RNA. 1997;3:1352–1363. [PMC free article] [PubMed] [Google Scholar]
- 22.Forconi M, Piccirilli JA, Herschlag D. Modulation of individual steps in group I intron catalysis by a peripheral metal ion. RNA. 2007;13:1656–1667. doi: 10.1261/rna.632007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morl M, Lizano E, Willkomm DK, Hartmann RK. Production of RNAs with homogeneous 5′and 3′ends. In: Hartmann RK, editor. Handbook of RNA Biochemistry. Weinheim: Wiley-VCH Verlag GmbH & Co.; 2005. [Google Scholar]
- 24.Karbstein K, Carroll KS, Herschlag D. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry. 2002;41:11171–11183. doi: 10.1021/bi0202631. [DOI] [PubMed] [Google Scholar]
- 25.Bevilacqua PC, Turner DH. Comparison of binding of mixed ribose deoxyribose analogs of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: Evidence for ribozyme specific interactions with 2′-OH groups. Biochemistry. 1991;30:10632–10640. doi: 10.1021/bi00108a005. [DOI] [PubMed] [Google Scholar]
- 26.Herschlag D. Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry. 1992;31:1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 27.Gordon PM, Piccirilli JA. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol. 2001;8:893–898. doi: 10.1038/nsb1001-893. [DOI] [PubMed] [Google Scholar]
- 28.Peracchi A, Beigelman L, Scott EC, Uhlenbeck OC, Herschlag D. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- 29.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13:1505–1515. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 31.Nelson JA, Uhlenbeck OC. Hammerhead redux: Does the new structure fit the old biochemical data? RNA. 2007;14:605–6015. doi: 10.1261/rna.912608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piccirilli JA, Vyle JS, Cartuhers MH, Cech TR. Metal-ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida A, Sun SG, Piccirilli JA. A new metal ion interaction in the Tetrahymena ribozyme reaction revealed by double sulfur substitution. Nat. Struct. Mol. Biol. 1999;6:318–321. doi: 10.1038/7551. [DOI] [PubMed] [Google Scholar]
- 34.Stahley MR, Strobel SA. RNA splicing: group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr. Opin. Struct. Biol. 2006;16:319–326. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Christian EL, Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 36.Strauss-Soukup JK, Strobel SA. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 37.McConnell TS, Cech TR, Herschlag D. Guanosine binding to the Tetrahymena ribozyme: Thermodynamic coupling with oligonucleotide binding. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran S, Kierzek R, Turner DH. Binding of guanosine and 3′ splice site analogues to a group I ribozyme: Interactions with functional groups of guanosine and with additional nucleotides. Biochemistry. 1993;32:5247–5256. doi: 10.1021/bi00070a037. [DOI] [PubMed] [Google Scholar]
- 39.Russell R, Herschlag D. Specifity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanner NK, Cech TR. Guanosine binding required for cyclization of the self-splicing intervening sequence ribonucleic acid from Tetrahymena thermophila. Biochemistry. 1987;26:3330–3340. doi: 10.1021/bi00386a013. [DOI] [PubMed] [Google Scholar]
- 41.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 42.Herschlag D, Koshla M. Comparison of pH dependencies of the Tetrahymena ribozyme reactions with RNA 2′-substituted and phosphorothioates substrates reveal a rate-limiting conformational step. Biochemistry. 1994;33:5291–5297. doi: 10.1021/bi00183a036. [DOI] [PubMed] [Google Scholar]
- 43.Karbstein K, Herschlag D. Extraordinary slow binding of guanosine to the Tetrahymena group I ribozyme: Implications for RNA preorganization and function. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2300–2305. doi: 10.1073/pnas.252749799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knitt DS, Herschlag D. pH dependencies of the Tetrahymena ribozyme reveal an unconventional origin of an apparent pKa. Biochemistry. 1996;35:1560–1570. doi: 10.1021/bi9521147. [DOI] [PubMed] [Google Scholar]
- 45.Narlikar GJ, Bartley LE, Koshla M, Herschlag D. Characterization of a local folding event of the Tetrahymena group I ribozyme: Effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps. Biochemistry. 1999;38:14192–14204. doi: 10.1021/bi9914309. [DOI] [PubMed] [Google Scholar]
- 46.Shan S, Herschlag D. Probing the role of metal ions in RNA catalysis: Kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry. 1999;38:10958–10975. doi: 10.1021/bi990388e. [DOI] [PubMed] [Google Scholar]
- 47.Sjogren AJ, Petterson E, Sjoberg BM, Stromberg R. Metal ion interaction with cosubstrate in self-splicing group I introns. Nucleic Acids Res. 1997;25:648–653. doi: 10.1093/nar/25.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu S, Strobel SA. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4-P6 domain. RNA. 1999;5:1399–1407. doi: 10.1017/s135583829999115x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brannvall M, Mikkelsen NE, Kirsebom LA. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 2001;29:1426–1432. doi: 10.1093/nar/29.7.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maderia M, Horton TE, DeRose VJ. Metal interactions with a GAAA RNA tetraloop characterized by 31P NMR and phosphorothioate substitutions. Biochemistry. 2000;39:8193–8200. doi: 10.1021/bi000140l. [DOI] [PubMed] [Google Scholar]
- 51.Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- 52.Maderia M, Hunsicker LM, DeRose VJ. Metal-phosphate interactions in the hammerhead ribozyme observed by 31P NMR and phosphorothioate substitutions. Biochemistry. 2000;39:12113–12120. doi: 10.1021/bi001249w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.