SUMMARY
6S RNA is a small, noncoding RNA that interacts with σ70-RNA polymerase and down-regulates transcription at many promoters during stationary phase. When bound to σ70-RNA polymerase, 6S RNA is engaged in the active site of σ70-RNA polymerase in a manner similar enough to promoter DNA that the RNA can serve as a template for RNA synthesis. It has been proposed that 6S RNA mimics the conformation of DNA during transcription initiation, suggesting contacts between RNA polymerase and 6S RNA or DNA may be similar. Here we demonstrate that region 4.2 of σ70 is critical for the interaction between 6S RNA and RNA polymerase. We define an expanded binding surface that encompasses positively charged residues throughout the recognition helix of the helix-turn-helix motif in region 4.2, in contrast to DNA binding that is largely focused on the N-terminal region of this helix. Furthermore, negatively charged residues in region 4.2 weaken binding to 6S RNA but do not similarly affect DNA binding. We propose that the binding sites for promoter DNA and 6S RNA on region 4.2 of σ70 are overlapping but distinct, raising interesting possibilities for how core promoter elements contribute to defining promoters that are sensitive to 6S RNA regulation.
INTRODUCTION
6S RNA is an untranslated, small RNA that was first discovered in E. coli as a highly abundant RNA [Hindley, 1967]. Although its cellular function remained elusive for many years, it is now known that 6S RNA regulates transcription through direct interaction with RNA polymerase (RNAP)[reviewed in Wassarman, 2007; Willkomm and Hartmann, 2005]. Bacterial RNAP contains a multi-subunit core enzyme (β, β′, ω and two αsubunits) and a specificity subunit (σ) that together form the holoenzyme (Eσ). Although core RNAP can carry out transcription elongation, the holoenzyme form is required for DNA promoter recognition and transcription initiation [reviewed in Gross et al, 1998; Murakami and Darst, 2003; Paget and Helmann, 2003]. E. coli has seven σ subunits: the housekeeping σ70 in addition to six alternative σ factors important during growth in suboptimal environments [Gruber et al, 2003]. 6S RNA binds specifically and tightly to the Eσ70 form of RNAP and it accumulates to high levels in late stationary phase where the vast majority of Eσ70 is bound by 6S RNA [Trotochaud and Wassarman, 2005; Wassarman and Storz, 2000]. 6S RNA interactions with Eσ70 result in down-regulation of transcription at hundreds of σ70-dependent promoters, making 6S RNA a global regulator of transcription in stationary phase [Cavanagh et al, 2008; Trotochaud and Wassarman, 2005; Wassarman and Storz, 2000]. In addition, cells lacking 6S RNA are altered in cell survival, particularly during competitive growth in stationary phase and growth at high pH [Trotochaud and Wassarman, 2004; 2006].
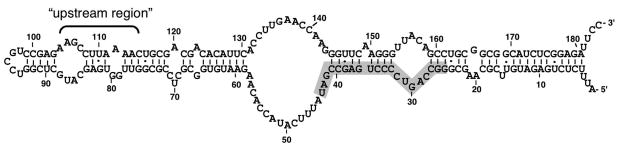
A highly conserved secondary structure is required for 6S RNA binding to Eσ70 [Trotochaud and Wassarman, 2005][Fig 1]. The RNA is primarily double-stranded with a large central bulge [Barrick, et al 2005; Trotochaud and Wassarman, 2005] and is reminiscent of the conformation of DNA within the “open complex” formed during transcription initiation when the DNA surrounding the start site of transcription is melted. Similar to promoter DNA, 6S RNA resides within the active site of RNAP and can be used as a template by Eσ70 to synthesize product RNAs (pRNA) [Gildehaus et al, 2007; Wassarman and Saecker, 2006]. pRNA synthesis results in the release of 6S RNA from RNAP, which appears to be a mechanism to liberate Eσ70 from 6S RNA regulation upon outgrowth from stationary phase [Wassarman and Saecker, 2006]. However, during stationary phase the 6S RNA:Eσ70 complex is stabilized, potentially due to the inability of pRNA synthesis to occur under low nucleotide concentrations [Murray et al, 2003; Wassarman and Saecker, 2006; Wassarman and Storz, 2000]. The presence of 6S RNA within the active site of Eσ70 blocks promoter DNA from binding to RNAP in vitro [Wassarman and Saecker, 2006]. In addition, the similarity in overall structure of 6S RNA and promoter DNA, as well as the fact that both reside in the active site of Eσ70, suggest that 6S RNA-dependent inhibition of transcription results from direct competition between 6S RNA and promoter DNA for binding to Eσ70.
Figure 1. E. coli 6S RNA.
Schematic of 6S RNA in a secondary structure supported by phylogenetic and experimental analyses [see Barrick et al, 2005; Trotochaud and Wassarman, 2005]. The sequence in 6S RNA used as a template for synthesis of an RNA product (pRNA) is highlighted in gray. The “upstream region” designates a general region predicted to contain the interaction site for σ70 region 4.2 based on the location of the active site of Eσ70 [Wassarman and Saecker, 2006] and by analogy to promoter DNA.
σ70-dependent promoters contain two core sequences called the -35 element (consensus TTGACA) and -10 element (consensus TATAAT) based on their approximate distances from the start site of transcription at position +1 [reviewed in Gross et al, 1998; Paget and Helmann, 2003]. Some promoters contain an “extended -10 element” that is defined by a conserved TGn immediately upstream of the -10 element (consensus TGnTATAAT) [Bown et al, 1997; Voskuil, et al, 1995]. Promoter recognition by Eσ70 is mediated primarily through interactions with σ70: region 4 of σ70 contacts the -35 element, region 2 contacts the -10 element, region 3.0 contacts the extended -10 element, and region 1.2 interacts with the discriminator region between the -10 element and the start site of transcription [Baldwin and Dombroski, 2001; Barne et al, 1997; Campbell et al, 2002; Dombroski et al, 1992; Gross et al, 1998; Haugen et al, 2006; Haugen et al, 2008; Lonetto et al, 1992; Murakami et al, 2002A; Paget and Helmann, 2003; Sanderson et al, 2003]. The relative strength of each of these contacts and their contribution to transcription initiation varies at different promoters. For example, promoters with extended -10 elements do not require a -35 element or region 4 of σ70 to retain Eσ70 recognition and subsequent transcription initiation [Baxter et al, 2006; Kumar et al, 1993; Minakhin and Severinov, 2003; Nechaev and Geiduschek, 2006].
Region 4.2 of σ70 uses a canonical helix-turn-helix motif to directly bind the -35 element, and many of the residues that are important for DNA binding are near the N-terminus of the second helix (i.e. the “recognition helix”) [Campbell et al, 2002; Gardella et al, 1989; Kuldell and Hochschild 1994; Siegele et al, 1989]. The C-terminal portion of this recognition helix, in addition to residues beyond the helix referred to as the “C-tail”, contributes to regulation of transcription initiation through contact with trans-acting transcription factors and/or the αsubunit of RNAP [Campbell et al, 2008; Chen et al, 2003; Dove et al, 2003; Ross et al, 2003]. Additional contacts are made between region 4 and the C-tail of σ70 with a flexible element in the β subunit called the β-flap. It has been suggested that this interaction is critical for promoter binding due to its proposed role in positioning region 4.2 relative to other regions of RNAP that allows appropriate recognition of -35 and -10 elements simultaneously [Geszvain et al., 2004; Kuznedelov et al., 2002; Murakami et al., 2002A; 2002B; Vassylyev et al, 2002].
Intriguingly, 6S RNA regulation of transcription in vivo is promoter specific, even during late stationary phase when the vast majority of Eσ70 is bound by 6S RNA [Cavanagh et al, 2008; Trotochaud and Wassarman, 2004; Wassarman and Storz, 2000]. We have shown that two promoter features, a weak -35 element and an extended -10 element, contribute independently to 6S RNA-sensitivity of σ70-dependent promoters. In contrast, neither overall promoter affinity for Eσ70 nor the strength of the -10 element contributes to 6S RNA sensitivity [Cavanagh et al, 2008]. These observations suggest that the mechanism underlying promoter specific regulation by 6S RNA is likely to be more complicated than simple sequestration of Eσ70 by 6S RNA such that only strong promoters can compete efficiently for the reduced pool of free Eσ70.
We have found that the C-terminal region of σ70 (region 4.2 and the C-tail) is required for 6S RNA to form stable complexes with Eσ70 [Cavanagh et al, 2008]. Given that region 4.2 of σ70 mediates binding to the -35 element in promoter DNA as well as 6S RNA, together with the observation that the -35 element strength is one critical determinant for 6S RNA sensitive promoters, we have proposed that competition for binding Eσ70 by 6S RNA and promoter DNA may be mediated through competition for binding region 4.2 of σ70 [Cavanagh et al, 2008]. As shown here, we investigated the role of region 4.2 of σ70 in 6S RNA binding in more detail by examining the contribution of individual residues throughout this region. Several residues that are important for binding the -35 element in promoter DNA also are important for binding to 6S RNA, although their relative contributions to DNA and RNA binding may be different. We also found that additional residues in σ70 region 4.2, specifically residues in the C-terminus of the second helix, contribute significantly to 6S RNA binding but not to binding of a minimal promoter, raising interesting possibilities for further competition between 6S RNA and transcription factors with Eσ70. Our data demonstrate that many positively charged residues within the second helix of σ70 region 4.2 define a 6S RNA-binding surface that overlaps with, but is distinct from, the DNA-binding surface.
RESULTS
Previously, we demonstrated that region 4.2 of σ70 was required for 6S RNA binding to Eσ70 as indicated by the observation that an Eσ70 variant lacking this region does not bind 6S RNA [Eσ70(1–565); Cavanagh et al, 2008]. To identify the relative importance of individual residues within σ70 region 4.2 for 6S RNA binding, here we test 6S RNA binding to Eσ70 variants containing single amino acid substitutions throughout this region [see Fig 2A]. All purified σ70 variants described here were able to form similar levels of Eσ70 holoenzyme when a 5 to 8-fold excess of σ70 over core protein was used in reconstitution reactions as measured by co-immunoprecipitation (data not shown, see Experimental procedures) in agreement with previous studies on many of these proteins [Chen et al, 2003; Lonetto et al, 1998; Ross et al, 2003]. We were able to determine the specific activity of Eσ70 variants with changes in region 4.2 of σ70 by measuring binding to DNA containing an extended -10 promoter [Pext-10] (see Experimental procedures), since extended -10 promoters do not require an active region 4.2 of σ70 for binding [Baxter et al, 2006; Kumar et al, 1993; Minakhin and Severinov, 2003; Nechaev and Geiduschek, 2006]. Reconstituted holoenzymes had similar specific activities (data not shown, see Experimental procedures), and Eσ70 levels were normalized in all RNA and DNA binding studies below to include equal concentrations of active holoenzyme. Note that all work here is using E. coli RNAP, and therefore we will refer to specific residues in σ70 by the E. coli numbering, even when referring to crystal structure data from T. aquaticus or T. thermophilus σA (the σ70 homologs in these species).
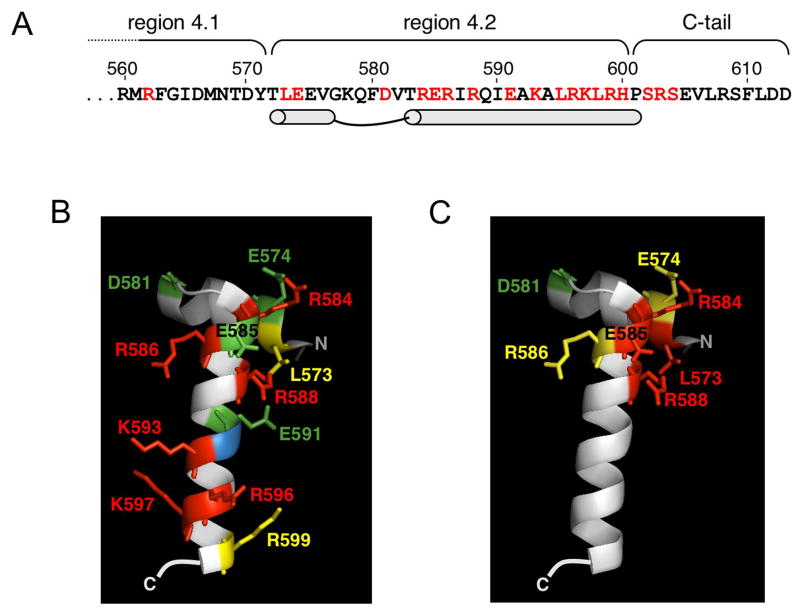
Figure 2. The C-terminal region of σ70.
(A). The amino acid sequence of the C- terminal end of σ70 with regions 4.1, 4.2 and the C-tail indicated [Lonetto et al, 1992]. The location of the helix-turn-helix motif is designated by schematic and the location of alanine substitutions examined here are shown in red. (B and C). The conformation of region 4.2 of T. aquaticusσA bound to DNA as observed by crystallography [Campbell et al, 2002] positioned to show the second helix of the helix-turn-helix motif (N-C is top- bottom). The -35 element DNA (not shown) would be oriented with the upstream region to the left, and the downstream region to the right. T. aquaticusσA is very similar to E. coli σ70 in this region, except for residues beyond R599 and thus the C-terminal region of the structure is not shown. Numbers are based on homologous positions in the E. coli protein. The relative effects on 6S RNA binding (B) or P-35-10 DNA binding (C) for alanine substitutions are indicated by color: Red, strong defect in binding; yellow, moderate defect in binding; and green, increased binding. Position 592 is blue; this residue is an alanine in wild type E. coli σ70.
Many residues important for DNA binding also are important for 6S RNA binding
Given the model that 6S RNA may mimic DNA interactions with σ70, we first examined whether alanine substitutions at residues important for binding the -35 element in promoter DNA also influence 6S RNA binding. The association of 6S RNA with Eσ70 variants was monitored by native gel electrophoresis [see Fig 3 and 4A]. Binding was done at 18° C to slow association to more manageable time frames similar to previous binding studies of Eσ70 and variants to a consensus extended -10 promoter [Schroeder et al, 2008]. Similar trends in changes in association of 6S RNA to variant Eσ70 proteins compared to wild type were observed at both 30° C and 18° C (data not shown). We also monitored DNA binding of our reconstituted Eσ70 proteins to a promoter with consensus -35 and -10 σ70 elements [P-35-10] that lacks an extended -10 element; thus efficient E binding to this promoter is dependent on -35 element contacts within σ70 region 4.2.
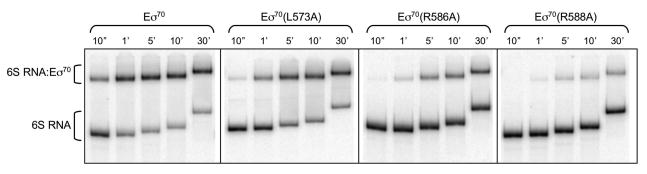
Figure 3. 6S RNA association with wild type or variant Eσ70.
0.5 nM 6S RNA and 2.2 nM active Eσ70 (wild type or variant) was incubated at 18° C. At times indicated (10 sec to 30 min), aliquots were removed and heparin was added to 100 μg/ml final concentration, samples were incubated for an additional two minutes at 4° C, and samples were loaded onto native gels. Gels were run continuously through incubation times and samples were loaded as they were generated; therefore the migration distance for 6S RNA and complexes are different for different time points. Representative gels are shown; all experiments were repeated at least three times to generate data shown in Figs 4–7. Fraction bound was calculated as the signal intensity in the 6S RNA:Eσ70 complex divided by the sum of the signal intensity in free 6S RNA and 6S RNA:Eσ70 complexes.
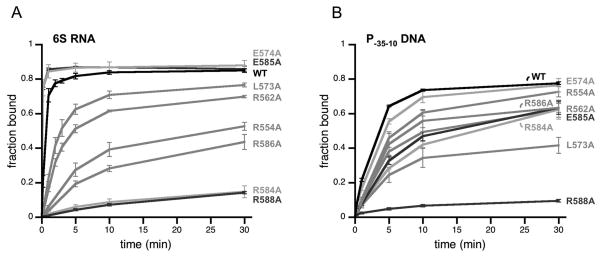
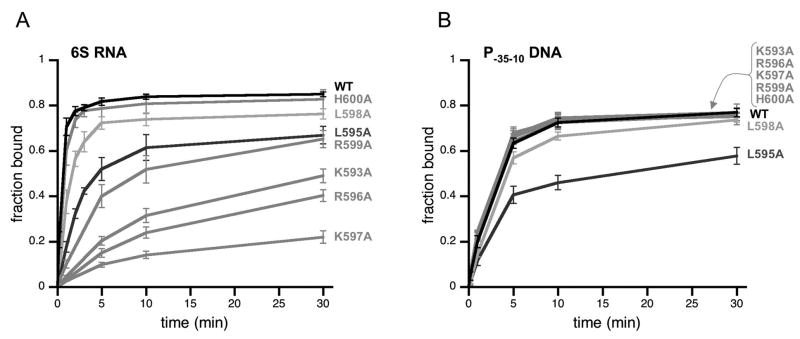
Figure 4. Many residues important for DNA binding to Eσ70 also are important for 6S RNA binding.
(A) Fraction of 6S RNA or (B) fraction of P-35-10 DNA bound to Eσ70 or Eσ70 variants with alanine substitutions in σ70 at positions indicated as a function of time as assayed by native gel electrophoresis in experiments similar to those in Fig 3. Data shown are an average of at least 3 experiments, and error bars correspond to ± standard deviation of the average.
As expected from previous work, Eσ70(L573A), Eσ70(R584A), Eσ70(E585A), Eσ70(R586A), and Eσ70(R588A) had decreased binding to DNA containing a -35 element-dependent promoter [P-35-10] compared to wild type Eσ70 [Fig 4B; Table S1], presumably due to decreased -35 element contacts [Campbell et al, 2002; Gardella et al, 1989; Gregory et al, 2005; Kuldell and Hochschild, 1994; Siegele et al, 1989]. With the exception of Eσ70(E585A), the Eσ70 variants also had decreased 6S RNA binding activity [Fig 4A; Table S1] suggesting that these residues contribute to both 6S RNA binding and -35 element-dependent DNA binding in the wild type protein. For instance, Eσ70(R588A) was the most defective in binding either RNA or DNA. However, some residues had differential effects on 6S RNA or DNA binding. For example, while Eσ70(R584A) is very defective for RNA binding relative to wild type Eσ70, it has less of an impact on DNA binding, and Eσ70(L573A) is more defective in DNA binding than RNA binding. Eσ70(E585A) is decreased for DNA binding, yet is not defective for RNA binding and appears to associate with 6S RNA faster than wild type (see below). Note that DNA binding was performed at 30° C as association of the P-35-10 promoter lacking an extended -10 element used here was sufficiently slow to measure at this temperature and conditions (2.2 nM active Eσ70 and 0.5 nM DNA), in contrast to previous reports using an extended -10 promoter and higher Eσ70 and DNA concentrations (50 nM Eσ70 and 10 nM DNA) [Schroeder et al, 2008]. As a consequence of the difference in temperature for NA and DNA binding experiments (18 C and 30 C, respectively), the differences in actual observed rate constants between wild type and Eσ70 variants can only be compared for either RNA or DNA binding, and not for a comparison of association rates of RNA to DNA directly.
Eσ70(E574A) was only minimally, if at all, defective for DNA binding in our assays, in contrast to our expectations based on the apparent contribution of this residue to DNA binding inferred from crystal structures [Campbell et al, 2002; Jain et al, 2004]. Eσ70(E574A) was not defective for 6S RNA binding, but instead exhibited faster binding to 6S RNA than wild type Eσ70 (see below).
In addition to region 4.2, other residues of σ70 were observed to be in close proximity to the -35 element in promoter DNA in crystal structures, including R562 and R554 [Campbell et al, 2002; Jain et al, 2004]. We found that Eσ70(R562A) and Eσ70(R554A) were moderately defective in RNA and DNA binding. However, once again the magnitude of change in 6S RNA binding and promoter DNA binding relative to wild type Eσ70 was not the same: Eσ70(R554A) was more defective in 6S RNA binding than Eσ70(R562A), while the opposite was observed for DNA binding.
Additional residues in σ70-region 4.2 are important for 6S RNA binding
Although many of the same residues appear to contribute to both RNA and DNA binding as observed above [see Fig 4], several residues appear to contribute differentially to RNA and DNA binding. In addition, the region of 6S RNA predicted to interact with region 4.2 of σ70 does not appear to resemble a -35 element in sequence or structure (see Discussion), suggesting that recognition of 6S RNA may be quite different than DNA. Therefore, we expanded our studies to test other residues in region 4.2 of σ70 and found that many residues more C-terminal in the second helix of region 4.2 were necessary for efficient binding to 6S RNA [see Fig 5A; Table S2]. For example, Eσ70(K597A) was particularly defective for 6S RNA-binding, with levels almost as low as Eσ70(R588A). However, in contrast to Eσ70(R588A), Eσ70(K597A) bound to promoter DNA [P-35-10] similarly to wild type Eσ70 [Fig 5B; Table S2]. Likewise, Eσ70(K593A), Eσ70(R596A), and Eσ70(R599A) also had decreased binding to 6S RNA but wild type levels of binding to promoter DNA [Fig 5; Table S2], indicating that residues in σ70 region 4.2 important for 6S RNA binding extend beyond residues that contribute to -35 element binding.
Figure 5. Residues in the C-terminal portion of the second helix in region 4.2 of σ70 are important for 6S RNA binding to Eσ70 but not for promoter DNA binding.
(A) The fraction of 6S RNA or (B) the fraction of P-35-10 DNA bound to Eσ70 and Eσ70 variants with alanine substitutions in σ70 at positions indicated as a function of time as assayed by native gel assays similar to those shown in Fig 3. Data shown are an average of at least 3 experiments, and error bars correspond to ± standard deviation of the average.
Eσ70(L595A) and Eσ70(L598A) were moderately or slightly defective for 6S RNA binding but also showed similar defects in DNA binding. Since these residues are not in close proximity to the -35 element within promoter DNA and have not been implicated in direct DNA binding [Campbell et al, 2002], it is possible L595 and L598 contribute to general positioning or folding of region 4.2 within Eσ70. Consistent with this model, L595 and L598 are on the face of the helix oriented towards the rest of σ70 and the β-subunit in holoenzyme crystal structures [Murakami et al, 2002A; 2002B; Vassylyev et al, 2002] and σ70(L598A) is decreased for competitive binding to core RNAP compared to wild type σ70 [Sharp et al, 1999], although our noncompetitive reconstitution conditions yielded similar levels of Eσ70(L598A).
Alanine substitutions at H600, S602, and S604 had little or no effect on 6S RNA association [Fig 5A and data not shown] suggesting these C-tail residues may not be important for 6S RNA binding. However, other residues within the C-tail might still contribute to 6S RNA binding or the relative positioning of region 4.2 of σ70 to allow efficient binding, as suggested by the observation that deletion of the last 5 amino acids of σ70 resulted in a moderate binding defect [Cavanagh et al, 2008].
A positively charged surface is key for 6S RNA binding
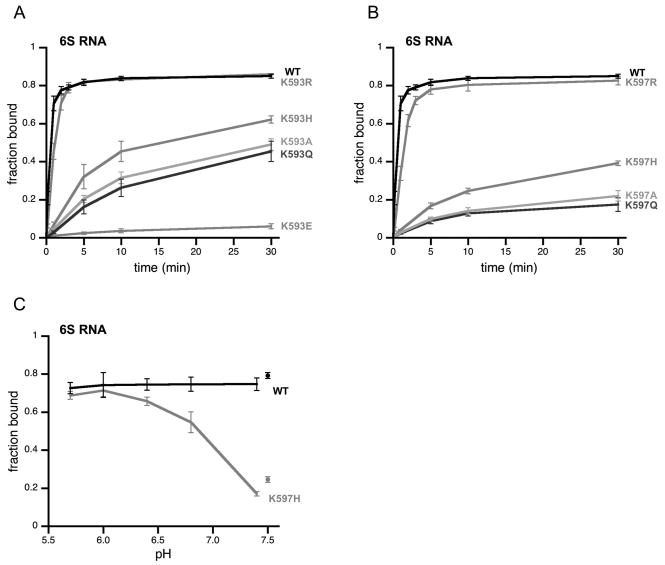
Most of the residues within region 4.2 of σ70 that we have shown are important for 6S RNA binding are positively charged arginine or lysine residues. To specifically address the role of charge contributions to 6S RNA binding, we made additional amino acid substitutions at two lysine residues (K593, K597) [see Fig 6]. We found that neutral glutamine substitutions were defective for 6S RNA binding at a level similar to the alanine substitutions [Eσ70(K593Q), Eσ70(K597Q)], while incorporation of a negatively charged residue essentially eliminated binding [Eσ70(K593E)]. Positively charged arginine substitutions maintained near wild type binding [Eσ70(K593R), Eσ70(K597R)], but histidine substitutions only slightly improved binding compared to alanine substitutions at K593 or K597 under standard binding conditions [Eσ70(K593H), Eσ70(K597H)](see also below). K593 and K597 do not contribute to binding the -35 element in promoter DNA as suggested by crystal structures [Campbell et al, 2002; Jain et al, 2004]; likewise substitutions to arginine, histidine, glutamine or glutamic acid did not significantly alter DNA binding at the P-35-10 promoter (data not shown), indicating observed changes in 6S RNA binding reflect changes in this defined activity.
Figure 6. Positively charged residues are critical for 6S RNA association with Eσ70.
(A and B) The fraction of 6S RNA bound to Eσ70 and Eσ70 variants with single amino acid substitutions in σ70 at positions indicated as a function of time as assayed by native gel assays similar to those shown in Fig 3. (C) 6S RNA binding to Eσ70 and Eσ70(K597H) was measured as a function of pH (50 mM sodium cacodylate) as assayed by native gel assays similar to those shown in Fig 3. For comparison, 6S RNA binding to Eσ70 and Eσ70(597H) under standard conditions (20 mM Hepes pH 7.5) are shown as single points. Data shown are an average of at least 3 experiments, and error bars correspond to ± standard deviation of the average.
To further address the potential role of positively charged residues for 6S RNA binding, we tested if 6S RNA binding to Eσ70(K597H) would be sensitive to pH, since the likelihood that the histidine R group would be protonated, and thus positively charged, increases with decreasing pH (pKa ~ 6.0 in free histidine). We found that Eσ70(K597H) binding to 6S RNA did increase with decreasing pH, reaching near wild type levels of binding at pH 6 [Fig 6C], further supporting the importance of positively charged side residues for 6S RNA binding. Wild type Eσ70 binding to 6S RNA was insensitive to pH in the range tested [Fig 6C].
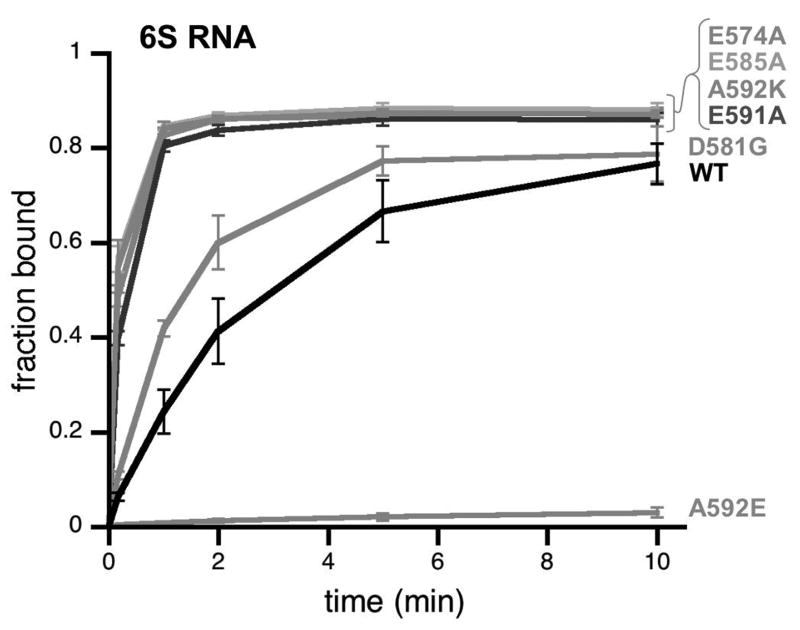
In addition to decreased 6S RNA binding when positively charged residues were neutralized, we consistently observed that removal of negatively charged residues appeared to result in faster 6S RNA binding [i.e. see Eσ70(E574A) and Eσ70(E585A) in Fig 4A]. Reducing the binding temperature to 9° C slowed the association of 6S RNA to Eσ70 enough to clearly demonstrate that Eσ70(E574A) and Eσ70(E585A) bind 6S RNA faster than wild type Eσ70 [Fig 7; Table S3]. Substitutions at other negatively charged residues within region 4.2 also enhanced 6S RNA binding [see Eσ70(E591A) and Eσ70(D581G)].
Figure 7. Negatively charged residues within wild type σ70 region 4.2 detract from 6S RNA binding to Eσ70.
The fraction of 6S RNA bound to Eσ70 and Eσ70 variants with amino acid substitutions that neutralize negative charges in σ70 at positions indicated as a function of time assayed by native gel assays similar to those shown in Fig 3 except that binding assays were done at 9° C. Data shown are an average of at least 3 experiments, and error bars correspond to ± standard deviation of the average.
Next, we substituted a naturally occurring alanine residue at position 592 with a basic (arginine) or acidic (glutamic acid) residue. Consistent with the importance of positive charges for 6S RNA binding, Eσ70(A592K) bound 6S RNA more rapidly than wild type Eσ70, while Eσ70(A592E) did not bind 6S RNA [Fig 7]. Replacement of a positively charged residue with a negatively charged residue at 596 or 593 [Eσ70(R596E), Eσ70(K593E)] resulted in a further reduction in binding compared to the neutral alanine substitution [Eσ70(R596A), Eσ70(K593A)][Fig 6A and data not shown]. The effects of these charge changes appear to be specific for 6S RNA binding as similar trends were not seen for DNA binding [P-35-10], except for Eσ70(D581G), which bound both 6S RNA and DNA [P-35-10] slightly faster than wild type Eσ70 (data not shown).
DISCUSSION
Here we have identified residues within region 4.2 of σ70 that contribute to 6S RNA association with Eσ70 [see Fig 2]. These residues define a positively charged surface on σ70 that is essential for 6S RNA binding. We have found that several residues important for recognition and binding of the -35 element in promoter DNA also contribute to 6S RNA binding (e.g. R584, R588). However, the relative importance of each residue for binding DNA and 6S RNA differs greatly, suggesting that 6S RNA contacts with region 4.2 of σ70 are not the same, or do not contribute to overall binding similarly to DNA binding. Although the approach used here cannot differentiate residues that directly interact with 6S RNA from those that are required to maintain a specific structure or orientation of σ70 region 4.2 relative to the rest of RNAP, the fact that different residues contribute to RNA and DNA binding differently, either through direct or indirect effects, supports our conclusion that 6S RNA interacts with σ70 region 4.2 in a manner distinct from DNA binding. Moreover, residues important for 6S RNA binding to Eσ70 extend further C-terminal on σ70 than those required for DNA binding, and are likely to encompass contacts throughout the second helix of the helix-turn-helix motif within region 4.2 (e.g. K593, R596, K597)[see Fig 2]. Therefore, although region 4.2 of σ70 is key for recognition of both DNA and 6S RNA, the manner of recognition and binding to each is distinct, which may contribute to the nature of the competition between 6S RNA and DNA for binding Eσ70 and, thus, the promoter specificity of 6S RNA regulation of transcription.
Does RNA mimic DNA structure to bind nucleic acid binding proteins?
Our results strongly support a model where 6S RNA recognition by region 4.2 of σ70 is different than DNA recognition. For DNA binding, the recognition helix within region 4.2 of σ70 interacts exclusively within the major groove of the double-stranded DNA [Campbell et al, 2002]. Double-stranded RNA usually adopts an A-form helix with a narrow major groove that would not allow similar contacts to be made, perhaps hinting that RNA binding would require different contacts. However, RNA can adopt a B-form helix structure that mimics a DNA major groove as observed for an RNA aptamer that binds NF-κB [Cassiday and Maher, 2002; Huang et al, 2003; Reiter et al, 2008]. A crystal structure of the aptamer RNA:NF-κB complex revealed that the RNA mimics a DNA major groove to such an extent that protein-nucleic acid contacts are largely conserved. An NMR solution structure demonstrated that the RNA alone adopts this unusual RNA structure, using non-canonical basepairing and stacking interactions within an asymmetrical loop. Therefore, the fact that 6S is an RNA did not preclude the possibility that it could directly mimic DNA interactions with σ70 region 4.2.
We can predict the general region of 6S RNA that is likely to interact with σ70 region 4.2 based on the location of the active site of Eσ70 in 6S RNA:Eσ70 complexes [Gildehaus et al, 2007; Wassarman and Saecker, 2006] in comparison to the location of the active site and region 4.2 of σ70 in DNA:Eσ70 complexes (“upstream region”, see Fig 1). The predicted secondary structures of 6S RNA in the upstream region indicate it does not form a continuous double-helix of Watson-Crick basepairs [Barrick et al, 2005; Trotochaud and Wassarman, 2005]. Instead, this region is fairly open with asymmetric loops lacking significant basepairing potential, which raised the possibility that it could form an unusual structure, perhaps even as a direct mimic of B-form DNA similar to the NF-κB RNA aptamer that also has asymmetric loops in simple secondary structure predictions (in contrast to the actual structure described above)[Reiter et al, 2008]. However, given that 6S RNA utilizes different contacts than DNA for binding σ70 region 4.2, it would appear that a direct mimic of DNA in this region is unlikely, although the possibility of an unusual RNA structure in this region is still quite possible.
Another RNA aptamer (FC) and the murine B2 RNA have been found to interact with eukaryotic RNA polymerase II and inhibit transcription [Espinoza et al, 2004; Goodrich and Kugel, 2009; Thomas et al, 1997], perhaps by competing with DNA for binding RNAP in a manner resembling 6S RNA inhibition of transcription. A variation of the FC aptamer, FC* RNA, binds in the downstream channel of RNAP where B-form double-stranded DNA normally resides, but in this case the FC* RNA maintains an A-form helix structure [Kettenberger et al, 2006]. The FC* RNA and promoter DNA binding sites are overlapping but not identical, as indicated by a subset of protein residues that are important for both RNA and DNA binding, and similar to our conclusions about σ70 region 4.2 binding to 6S RNA and promoter DNA.
It is important to note that although FC* and 6S RNA both bind RNAP, the two sites of interaction that have been studied in detail (FC* in the downstream channel and 6S RNA interactions with region 4.2 of σ70) are not the same as they occur in distant regions of RNAP. Although FC* does not interact with regions of RNAP normally bound by upstream DNA [Kettenberger et al, 2006], it is possible that 6S RNA makes contacts with E. coli RNAP in a manner similar to FC* in the downstream channel, especially given the location of 6S RNA near the active site and the fact that it is used as a template for RNA synthesis, suggesting that the “downstream region” of 6S RNA is likely to reside in the downstream channel of RNAP [Gildehaus et al, 2007; Wassarman and Saecker, 2006]. Any such interactions would be in addition to the contacts with σ70 we have defined here, and we suggest it is likely that there are multiple interactions between 6S RNA and Eσ70. Further experiments will be necessary to fully define the 6S RNA:Eσ70 interactions and to understand the contribution of each to the strength and specificity of 6S RNA binding to Eσ70.
Region 4.2 of σ70 is a target for transcriptional regulation
Region 4.2 of σ70, particularly the C-terminal portion of the second helix, is a target for binding of many transcription factors [reviewed in Campbell et al, 2008; Dove et al, 2003]. These proteins often activate transcription by binding specific DNA sequences in promoters and helping recruit RNAP through interactions with region 4.2 of σ70, the αsubunit of RNAP, or both. For example, region 4.2 of σ70 is required for CRP regulation at some promoters as indicated by decreased CRP-sensitivity of Eσ70 variants containing alanine substitutions at K593, K597 or R599 [Lonetto et al, 1998]. In contrast, other proteins interact with region 4.2 of σ70 to regulate transcription by preventing or altering σ70 activity without directly binding DNA [see Campbell et al, 2008; Dove et al, 2003]. For example, Rsd, an anti-sigma factor of σ70, binds to region 4.2 of σ70 (in addition to other regions), and prevents σ70 association with core RNAP [Jishage and Ishihama, 1998; Jishage et al, 2001; Patikoglou et al, 2007; Westblade et al, 2004, Yuan et al, 2008].
Both classic and nonclassic transcription factors interact with basic residues in region 4.2 (as well as in the C-tail); however, the identity of which residues are necessary for binding defines a pattern of recognition specific for each protein [Lonetto et al, 1998]. We propose that the 6S RNA interaction with σ70 also utilizes a specific pattern that is likely to be dependent on the structure/shape of the upstream region of 6S RNA. In addition, we suggest that the interaction with region 4.2 of σ70 is critical for the initial recognition and/or overall affinity for 6S RNA binding to Eσ70, based on our observations that the association of 6S RNA with Eσ70 appears to be particularly sensitive to changes in region 4.2 of σ70 as exemplified by the severe loss of binding of variants such as Eσ70(R588A) and Eσ70(K597A).
6S RNA and promoter DNA bind to Eσ70 in similar, if not identical, locations at the active site and σ70 region 4.2, thereby supporting the model that 6S RNA regulates transcription through direct competition with DNA for binding Eσ70. It is intriguing that the 6S RNA binding to σ70 region 4.2 includes residues that also mediate interactions with other proteins such as transcription factors and the α-subunit of RNAP. It is possible the expanded interaction between 6S RNA and Eσ70 reduces the potential for transacting factors to gain access to the C-terminal portion of region 4.2 and influence 6S RNA binding. Alternatively, if 6S RNA acts by displacing Eσ70 from DNA in vivo, the requirement for residues within 593-599 of σ70 for 6S RNA binding may prevent 6S RNA from displacing Eσ70 from DNA when transcription factors also are bound. Further details of the dynamics of exchange between 6S RNA and promoter DNA on Eσ70, with and without other factors, is required before we can fully understand the molecular details underlying 6S RNA regulation of σ70-dependent transcription.
How is 6S RNA recognized by region 4.2 of σ70?
We have mapped a highly basic region of σ70 region 4.2 that is important for the binding of 6S RNA to Eσ70. Given that the phosphate backbone of RNA is negatively charged, it is likely that these charge-charge interactions contribute to the high affinity of 6S RNA for Eσ70 as has been observed for many RNA:protein interactions [Bahadur et al, 2008]. The location of the basic “patch” is likely to define a path for the RNA that allows the central region of 6S RNA to access the active site. One might speculate that the larger region of 4.2 of σ70 that contributes to RNA binding may reflect flexibility in positioning the RNA, and that it primarily provides strength to the interaction. However, we propose that region 4.2 of σ70 contributes a high degree of specificity to 6S RNA recognition, in addition to affinity. First, there are specific residues that have quite dramatic effects on 6S RNA binding that are on different faces and ends of the helix (e.g. R584 and K597), suggesting their contribution may be a direct interaction rather than a general contribution to a positively charged surface. Second, the upstream region of E. coli 6S RNA that we believe contacts region 4.2 of σ70 is particularly sensitive to single nucleotide substitutions, in contrast to the rest of the molecule where similar changes often have no effect (KMW, unpublished data). This sensitivity to single nucleotide mutations may indicate these nucleotides are sites for direct contact with σ70, or that they are required to maintain a precise structure for recognition. If structure is the key, as might be suggested by the low conservation of sequence in this region [Barrick et al, 2005; Trotochaud and Wassarman, 2005], we suggest the RNA forms an unusual structure since many of the single nucleotide substitutions would not dramatically alter current secondary structure models. A better understanding of the tertiary structure of the upstream region of 6S RNA, alone or in complex with σ70 region 4.2, will be needed to differentiate such possibilities as well as to distinguish the residues mediating direct contact with 6S RNA from those potentially important for maintaining a precise protein structure or orientation.
In summary, we have found that 6S RNA interactions with Eσ70 include a strong interaction with region 4.2 of σ70, mediated largely through charge-charge interactions. Although this region of σ70 also is important for binding to DNA, we propose the RNA binding site is not coincident with the DNA binding site. Instead, we suggest the RNA and DNA binding sites overlap, which results in competition between 6S RNA and promoter DNA as the mechanism underlying 6S RNA down-regulation of σ70-dependent transcription. In agreement with this model, the strength of the -35 element is one critical factor in determining 6S RNA sensitive promoters. Although it is not yet clear how an extended -10 element also determines 6S RNA sensitivity, observations that σ70 region 4.2 interacts with extended -10 promoters differently than with nonextended -10 promoters [Minakhin and Severinov, 2003] may suggest that competition for region 4.2 of σ70 also is important at these promoters [Cavanagh et al, 2008]. In any event, the details of how 6S RNA regulates transcription and interacts with RNAP continue to reveal surprising results, such as the finding that 6S RNA binds Eσ70 in a manner that allows it to be used as a template for RNA synthesis and that it binds through contacts to σ70 region 4.2, but not entirely as a molecular mimic of promoter DNA.
EXPERIMENTAL PROCEDURES
Plasmids and DNA
pLA4 [Anthony et al, 2003] and derivatives (see below) were used to express σ70 proteins without additional sequences or tags. Most rpoD derivatives were generated by replacing the C-terminal region of rpoD in pLA4σ70(1–565) [Cavanagh et al, 2008] with the C-terminal region from pGEXσ70 and derivatives [Lonetto et al, 1998] or pET-σ70(K593A), pET-σ70(L595A), and pET-σ70(K597A), [Ross et al, 2003; W. Ross, unpublished] to regenerate full length rpoD genes with appropriate substitutions. Other rpoD derivatives were generated by site directed mutagenesis (Quikchange, Stratagene). All rpoD mutations were confirmed by sequencing. Oligonucleotide sequences are available upon request. pT3–6S was used to generate 32P-6S RNA as previously described [Trotochaud and Wassarman, 2005]. Promoter DNA duplexes were generated by annealing oligonucleotides in 25 mM Tris pH 7.5, 50 mM NaCl as previously described [deHaseth and Tsujikawa, 2003; Schroeder and deHaseth, 2005]. The Pext-10 promoter is as described by Schroeder et al, 2005 (“duplex” promoter), and the P-35-10 promoter is the same except for a G to C substitution at position -14 to remove the extended -10 element. CGCACGGTGTTTGACATTTATCCCTTGCTTTCGTATAATTAACGTATGAGCACAAAAAAGA GGCC is the sequence of P-35-10 with the -35 and -10 elements in bold and the -14 position in italics.
RNA polymerase
σ70 proteins were purified from inclusion bodies after overproduction, followed by refolding as previously described [Arthur and Burgess, 1998; Arthur et al, 2000]. Core RNAP preparations were generous gifts from R.A. Saecker and M.T. Record or T. Gaal and R.L. Gourse.
Eσ70 and variants were reconstituted from core RNAP and σ70 proteins by incubation in 40 mM Hepes pH 7.5, 240 mM KCl, 2.5 mM DTT, and 28.3% glycerol for 30 minutes at 37° C, except for experiments testing the effects of pH on 6S RNA binding [Fig 6C], which used 20 mM instead of 40 mM Hepes. The efficiency of σ70 binding to core RNAP was measured as the relative co-immunoprecipitation of σ70 with core specific antibodies as previously described [Cavanagh et al, 2008]. Except forσ70(R562A), a 5-fold excess of σ70 over core saturated input core and resulted in similar concentrations of reconstituted Eσ70. For σ70(R562A) 5-fold excess was not sufficient, in agreement with reports that σ70(R562A) did not bind core as efficiently as wild typeσ70 in a competitive binding assay [Sharp et al, 1999]. However, when σ70(R562A) was at 8-fold excess over core, the level of reconstituted Eσ70(R562A) was comparable to wild type and therefore 8-fold excess of σ70(R562A) over core was used in experiments done here while all other σ70 variants were at 5-fold excess relative to core.
The specific activity of reconstituted Eσ70 and variants was estimated from the minimum protein concentration required to obtain maximum binding to DNA, as described in Roe et al, 1984. DNA with an extended -10 promoter was used since region 4.2 of σ70 is not required for Eσ70 binding [Kumar et al, 1993], and therefore all our Eσ70 variants were able to bind this promoter. See below for DNA binding conditions. Most reconstituted Eσ70 preparations were very similar in specific activity (~25% active), but Eσ70(L573A), Eσ70(E585A) and Eσ70(R588A) were somewhat lower (~18% active). Similar results were observed for at least two independent σ preparations, suggesting the lower activity is inherent in the protein identities.
6S RNA binding assay
6S RNA binding to Eσ70 and variants was done essentially as described [Trotochaud and Wassarman, 2005; Wassarman and Saecker, 2006]. Specifically, reconstituted Eσ70 (2.2 nM active) and 32P-6S RNA (0.5 nM) were incubated in 20 mM HEPES pH 7.5, 120 mM KCl, 14.1% glycerol, and 1.25 mM DTT for 10 sec to 30 min at 18° C [Fig 3–6] or 9° C [Fig 7], except for experiments examining pH effects [Fig 6C] which were incubated in 50 mM sodium cacodylate, 120 mM KCl, 1.25 mM DTT, 14.1% glycerol for 10 min at 18° C at pH 5.7 to 7.4 as indicated.
DNA binding assay
Promoter DNA binding to Eσ70 and variants was done essentially as described [Wassarman and Saecker, 2006]. Specifically, for binding to P-35-10 as shown in Fig 4 and 5, 0.5 nM labeled promoter DNA was incubated with 2.2 nM active Eσ70 at 30° C for times indicated. For binding to Pext-10 for specific activity determination, 10 nM labeled promoter DNA was incubated with Eσ70 (~2 nM to 20 nM active) at 18° C for 30 min. All DNA binding was done in 20 mM Hepes pH 7.5, 120 mM KCl, 14.1% glycerol, and 1.25 mM DTT.
Native gel electrophoresis
Native gels were as previously described [Trotochaud and Wassarman, 2005; Wassarman and Saecker, 2006]. All samples from RNA and DNA binding assays were incubated an additional 2 min at 4° C after addition of heparin (100 μg/ml final concentration), diluted 1:2 with 50% glycerol containing xylene cyanol and bromophenol blue, and immediately loaded onto running native polyacrylamide gels [5% polyacrylamide (37.5:1)/5% glycerol/0.5x TBE]. Gels were run at 200V at 4 C for 2 hours after loading of the first time point. Gels were visualized on a Typhoon Phosphorimager and quantified using Imagequant software. The fraction of bound RNA or DNA was calculated and the averages from at least three independent experiments were plotted as a function of time [See Fig 4–7]. Observed pseudo first-order rate constants (kobs) were determined from the equation y = ymax(1 − e−kobst) ymax is the maximal level of complex formation using SigmaPlot [Ross and Gourse, 2009; Schroeder et al, 2008], followed by a correction for proteins that did not reach the same level of maximal binding as wild type Eσ70, presumably due to reversibility of the reaction, as described by Saecker et al, 2002 [see Tables S1–S3].
Supplementary Material
Acknowledgments
We thank R. Gourse, W. Ross, T. Gaal, R. Saecker, A. Cavanagh and K. Forest for helpful discussions throughout this work. We thank R. Saecker, M.T. Record, T. Gaal and R. Gourse for core RNAP preparations; R. Gourse and W. Ross for plasmids; W. Ross for help with determination of kobs; and A. Cavanagh and E. Bruger for technical assistance. This work was supported by the National Institutes of Health (GM67955).
References
- Anthony LC, Foley KM, Thompson NE, Burgess RR. Expression, purification of, and monoclonal antibodies to σ factors from Escherichia coli. Methods Enzymol. 2003;370:181–192. doi: 10.1016/S0076-6879(03)70016-0. [DOI] [PubMed] [Google Scholar]
- Arthur TM, Anthony LC, Burgess RR. Mutational analysis of β′ 260–309, a σ70 binding site located on Escherichia coli core RNA polymerase. J Biol Chem. 2000;275:23113–23119. doi: 10.1074/jbc.M002040200. [DOI] [PubMed] [Google Scholar]
- Arthur TM, Burgess RR. Localization of a σ70 binding site on the N terminus of the Escherichia coli RNA polymerase β′ subunit. J Biol Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin NE, Dombroski AJ. Isolation and characterization of mutations in region 1.2 of Escherichia coli σ70. Mol Microbiol. 2001;42:427–437. doi: 10.1046/j.1365-2958.2001.02642.x. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter K, Lee J, Minakhin L, Severinov K, Hinton DM. Mutational analysis of σ70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA. J Mol Biol. 2006;363:931–944. doi: 10.1016/j.jmb.2006.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown JA, Barne KA, Minchin SD, Busby SJW. Extended -10 promoters. Nucleic Acids Mol Biol. 1997;11:41–52. [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, et al. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Klocko AD, Liu X, Wassarman KM. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol Microbiol. 2008;67:1242–1256. doi: 10.1111/j.1365-2958.2008.06117.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Tang H, Ebright RH. Functional interaction between RNA polymerase αsubunit C-terminal domain and σ70 in UP-element- and activator-dependent transcription. Mol Cell. 2003;11:1621–1633. doi: 10.1016/s1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- deHaseth PL, Tsujikawa L. Probing the role of region 2 of Escherichia coli σ70 in nucleation and maintenance of the single-stranded DNA bubble in RNA polymerase-promoter open complexes. Methods Enzymol. 2003;370:553–567. doi: 10.1016/S0076-6879(03)70047-0. [DOI] [PubMed] [Google Scholar]
- Dove SL, Darst SA, Hochschild A. Region 4 of σ as a target for transcription regulation. Mol Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Record MT, Jr, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;9 :822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Gardella T, Moyle H, Susskind MM. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates σ70 region 4 function. J Mol Biol. 2004;343:569–587. doi: 10.1016/j.jmb.2004.08.063. [DOI] [PubMed] [Google Scholar]
- Gildehaus N, Neuser T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–1896. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: the expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Nickels BE, Darst SA, Hochschild A. An altered-specificity DNA-binding mutant of Escherichia coli σ70 facilitates the analysis of σ70 function in vivo. Mol Microbiol. 2005;56:1208–1219. doi: 10.1111/j.1365-2958.2005.04624.x. [DOI] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Manrique M, Gourse RL. Fine structure of the promoter-σ region 1.2 interaction. Proc Natl Acad Sci U S A. 2008;105:3292–3297. doi: 10.1073/pnas.0709513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley J. Fractionation of 32P-labelled ribonucleic acids on polyacrylamide gels and their characterization by fingerprinting. J Mol Biol. 1967;30:125–136. doi: 10.1016/0022-2836(67)90248-3. [DOI] [PubMed] [Google Scholar]
- Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd, Ghosh G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci U S A. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Nickels BE, Sun L, Hochschild A, Darst SA. Structure of a ternary transcription activation complex. Mol Cell. 2004;13:45–53. doi: 10.1016/s1097-2765(03)00483-0. [DOI] [PubMed] [Google Scholar]
- Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc Natl Acad Sci U S A. 1998;95 :4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Dasgupta D, Ishihama A. Mapping of the Rsd contact site on the sigma 70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 2001;183:2952–2956. doi: 10.1128/JB.183.9.2952-2956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- Kuldell N, Hochschild A. Amino acid substitutions in the -35 recognition motif of σ70 that result in defects in phage λ repressor-stimulated transcription. J Bacteriol. 1994;176:2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, et al. A role for interaction of the RNA polymerase flap domain with the σ subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The σ70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase σ70 region 4.2 and α-subunit C-terminal domains in promoter complex formation on the extended -10 galP1 promoter. J Biol Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002A;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002B;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Geiduschek EP. The role of an upstream promoter interaction in initiation of bacterial transcription. EMBO J. 2006;25:1700–1709. doi: 10.1038/sj.emboj.7601069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The σ70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patikoglou GA, Westblade LF, Campbell EA, Lamour V, Lane WJ, Darst SA. Crystal structure of the Escherichia coli regulator of σ70, Rsd, in complex with σ70 domain 4. J Mol Biol. 2007;372:649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Maher LJ, 3rd, Butcher SE. DNA mimicry by a high-affinity anti-NF-κB RNA aptamer. Nucleic Acids Res. 2008;36:1227–1236. doi: 10.1093/nar/gkm1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JH, Burgess RR, Record MT., Jr Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the λPR promoter. J Mol Biol. 1994;176:495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- Ross W, Gourse RL. Analysis of RNA polymerase-promoter complex formation. Methods. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the αC-terminal domain and σ region 4. Genes Dev. 2003;17:1293–1307. doi: 10.1101/gad.1079403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Record MT., Jr Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: large scale conformational changes in forming the kinetically significant intermediates. J Mol Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- Sanderson A, Mitchell JE, Minchin SD, Busby SJ. Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended -10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- Schroeder LA, deHaseth PL. Mechanistic differences in promoter DNA melting by Thermus aquaticus and Escherichia coli RNA polymerases. J Biol Chem. 2005;280:17422–17429. doi: 10.1074/jbc.M501281200. [DOI] [PubMed] [Google Scholar]
- Schroeder LA, Karpen ME, deHaseth PL. Threonine 429 of Escherichia coli σ70 is a key participant in promoter DNA melting by RNA polymerase. J Biol Chem. 2008;376:153–165. doi: 10.1016/j.jmb.2007.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, et al. The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J Biol Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J Bacteriol. 2004;186:4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J Bacteriol. 2006;188:3936–3943. doi: 10.1128/JB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Voskuil MI, Voepel K, Chambliss GH. The -16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol Microbio. 1995;17 :271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a regulator of transcription. Mol Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Westblade LF, Ilag LL, Powell AK, Kolb A, Robinson CV, Busby SJ. Studies of the Escherichia coli Rsd-σ70 complex. J Mol Biol. 2004;335:685–692. doi: 10.1016/j.jmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Willkomm DK, Hartmann RK. 6S RNA - an ancient regulator of bacterial RNA polymerase rediscovered. Biol Chem. 2005;386:1273–1277. doi: 10.1515/BC.2005.144. [DOI] [PubMed] [Google Scholar]
- Yuan AH, Gregory BD, Sharp JS, McCleary KD, Dove SL, Hochschild A. Rsd family proteins make simultaneous interactions with regions 2 and 4 of the primary sigma factor. Mol Microbiol. 2008;70:1136–1151. doi: 10.1111/j.1365-2958.2008.06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.