Abstract
The purpose of the present study was to investigate the influence of molecular weights on the chemical, biophysical, and biological properties of bioreducible oligoethylenimine conjugates. The cationic conjugates were synthesized by polyaddition between branched oligoethylenimine 800 Da (OEI) and the disulfide bond containing N,N′-cystamine bisacrylamide (CBA) linker. A correlation between the copolymer molecular weights and the polyplex transfection properties was found. The OEI–CBA copolymers differing in molecular weights (from 8.6 to 37 kDa) showed good plasmid DNA binding ability resulting in compact 90- to 150-nm-sized polyplexes. Colloidal stability of the polyplexes was lost in reductive environment. A low concentration of dithiothreitol of 5 µM was sufficient to render polyplexes unstable in size. Reducing conditions at physiological salt concentration triggered polyplex dissociation. The bioreducible polymers displayed much lower cytotoxicity (IC50 ∼ 100 μg/mL in cell culture) than branched polyethylenimine 25 kDa (BPEI) and linear polyethylenimine 22 kDa (LPEI). Reporter gene transfection experiments were done with CHO-K1 and B16-F10 cells. The largest (37 kDa) copolymer HC-6-8 demonstrated highest transfection levels among all the bioreducible copolymers, which was comparable with LPEI and much more effective than BPEI.
Key words: bioreducible, gene delivery, PEI, polyplexes, synthetic vectors
INTRODUCTION
For the success of gene therapy, appropriate carrier formulations are essential which can protect therapeutic nucleic acids from enzyme degradation and efficiently deliver them into the desired cell population. Among the various synthetic polymers used for non-viral gene delivery (1), high-molecular weight (HMW) polyethylenimines (linear polyethylenimine 22 kDa, LPEI, or branched polyethylenimine 25 kDa, BPEI) can deliver plasmid DNA (pDNA) into many types of cells with high efficacy in vitro and in vivo.(2,3). This mostly depends on their inherent “proton sponge” effect to facilitate the endosomal release of the polyplexes (4,5). However, the non-degradable PEIs are known to induce significantly cytotoxic side effects. The strategy of preparing HMW polycations from low-molecular weight (LMW) oligocations via biodegradable linkages has been proven to reduce toxicity while retaining the gene transfer ability of the polyplexes. Up to now, several examples had been reported (6–21). Hydrolysable PEIs were developed by cross-linking oligoethylenimine 800 Da (OEI) with ester bond bearing monomers or polyesters (6–12). Biocleavable PEIs were prepared by thiolation of LMW PEI (13,14) or by cross-linking of OEI with disulfide bond containing linkers, including dimethyl 3, 3′-dithiobis(propionimidate) and dithiobis (succinimidyl propionate) (6,15,16). Bioreducible copolymers from N,N′-cystamine bisacrylamide (CBA) linker and different primary amines or oligoamines were explored too (17–21). The polyplexes from these hydrolysable or bioreducible polycations were designed to keep stable in extracellular environment while dissociate efficiently upon intracellular delivery when exposed to reducing intracellular condition (22–24).
Although encouraging results about bioreducible polycations have already been described, only few examples discuss the influence of polymer synthesis and purification conditions on the chemical, biophysical, and biological properties of the resulting polycations. In the current study, we synthesized a set of bioreducible BPEI analogs with different molecular weights starting from OEI and CBA via the Michael addition reaction (17,20,21). The synthesis and purification procedures were optimized by evaluating different OEI/cross-linker feed ratios, monomer concentrations, and molecular weight cutoffs (MWCO) of dialysis membranes to obtain polycations with high transfection efficiency but low toxicity. The resulting compounds were structurally characterized by 1H NMR spectra and gel permeation chromatograph (GPC) measurement. Their biophysical properties were investigated by pDNA binding and cytotoxicity assay. The dissociation kinetics of the reducible polyplexes was thoroughly examined by particle size measurement and gel electrophoreses assay. Furthermore, the gene transfer properties of the copolymers were tested in B16-F10 and CHO-K1 cells. The results suggest that the better defined copolymers with high molecular weights and narrow molecular weight polydispersity were able to transfer pDNA efficiently in both cell lines with low cytotoxicity. The transfection efficiency is comparable to that of LPEI, while much higher than that of the BPEI control.
MATERIALS AND METHODS
Chemicals and Reagents
Dimethylsulfoxide (DMSO, water-free) was obtained from Fluka (Fluka Chemie GmbH, Deisenhofen, Germany). CBA was purchased from Polysciences Europe Gmbh (Eppelheim, Germany). LPEI was purchased from Polyplus Transfections (Strasburg, France). BPEI, OEI, DTT, ethidium bromide (EtBr), and 2,5-diphenyl-3-(4, 5- dimethyl-2-thiazolyl) tetrazolium bromide (MTT) were all obtained from Sigma-Aldrich (Munich, Germany). Cellulose dialysis bags with different MWCO (6–8 and 3.5 kDa) were obtained from Spectrum Medical Industries, Inc. (CA, USA). Cell culture media, antibiotics, and fetal calf serum (FCS) were all purchased from Life Technologies (Karlsruhe, Germany). Cell culture lysis buffer and d-luciferin sodium salt were obtained from Promega (Mannheim, Germany). EGFP-N1 and pCMV-Luc [Photinus pyralis luciferase under control of the CMV enhancer/promoter, described by Plank et al. (25)] plasmid DNAs were purified with the EndoFree plasmid kit from Qiagen (Hilden, Germany). Water was used as purified Millipore water (MQ).
Synthesis and Purification of the Bioreducible PEI Analogs
In a typical synthetic procedure, CBA and OEI stock solutions (both 1.0 M in DMSO) were mixed at certain molar ratios. The reactions were continued for 2 days at 40°C. Afterwards, the reaction mixtures were neutralized to pH value of 6–7 by 1.0 M hydrogen chloride solution and dialysed against MQ water using MWCO membranes of either 6–8 or 3.5 kDa. The final products were obtained after freeze drying.
Chemical Structure and Molecular Weight Characterization
1H NMR spectra of the products was recorded on an Eclipse 500 spectrometer from JOEL (Tokyo, Japan) in D2O at 500 MHz. The copolymer molecular weights [weight average (Mw) and number average (Mn) molecular weights] and the coresponding polydispersity indexes (PDI, Mw/Mn) were determined by GPC measurement (Agilent Technologies, Waldbronn, Germany). The GPC was equipped with multi-detectors (refractive index, UV and viscosity detectors), a NOVEMA 10-mm pre-column and a NOVEMA 300 analytical column (PSS, Mainz, Germany). An acidic mobile phase [0.1% (v/v) HCOOH and 0.1 M NaCl in MQ, pH 2.8] at a flow rate of 1.0 mL/min were selected. Pullulans with different molecular weights were used as standards for calibration curve determination (12).
DNA Binding Ability Determined by EtBr Exclusion Assay
Aliquots of OEI–CBA copolymer, LPEI, or BPEI stock solution was added to 20 μg/mL pCMVLuc pDNA solution in HBG [20 mM HEPES and 5% (w/v) glucose, pH 7.4] containing 400 ng/mL EtBr. The decrease of fluorescence intensity was measured by a Varian Cary Eclipse fluorescence spectrophotometer at λex = 510 nm and λem = 590 nm (Varian Eclipse, Mulgrave, Australia). The fluorescence intensity of EtBr/DNA complexes was set as 100% prior to polycation addition (9).
Polyplex Particle Size and Zeta Potential Measurements
Polyplexes were prepared by adding polymer solutions to 10 µg/ml pDNA solution at various polymer to pDNA weight ratios (c/p ratios). All polymer and pDNA solutions were prepared in HBG. The polyplex mixture was incubated for at least 20 min at room temperature before the measurements. The particle sizes and zeta potentials of the polyplexes were determined by a Malvern Zetasizer ZS (Malvern Instruments, Worcestershire, UK) (26).
Stability of the Polyplexes Under Reducing Conditions
Polyplex dissociation in the presence of DTT was studied by particle size measurement and gel electrophoresis assay. Polyplexes were prepared in HBG solution and pre-incubated for 1 h at 37°C water bath. Oxygen-free HBG buffer was used where oxygen had been removed by argon bubbling for 30 min. Polyplexes were treated either with 5 mM DTT or kept untreated; particle sizes were then measured after additional 1-h incubation. Two different DTT final concentrations of 5 µM and 5 mM were selected to investigate the kinetics of polyplex dissociation. The polyplex particle size was recorded every 2 min for at least 40 min.
Cytotoxicity Assay
The metabolic activity of the cells incubated with polymer solutions was determined by MTT assay. B16-F10 (murine melanoma cells) and CHO-K1 (Chinese hamster ovary cells) cells were seeded in 96-well tissue culture plates (TPP, Transdingen, Switzerland) at a density of 5,000 cells per well in 100 μL of cell culture medium (penicillin 100 U/mL, streptomycin 100 μg/mL, and FCS 10%). Dulbecco's modified Eagle's medium (DMEM) or DMEM/HAMS-F12 (1:1) was used for culturing of B16-F10 or CHO-K1 cell, respectively. Defined amounts of polymer solution at different final concentrations were added when cells reached a confluency of 60–80%. The cell culture medium was exchanged to a fresh one after 4-h incubation. MTT solution (5.0 mg/ml in phosphate-buffered saline, 10 μL/well) was added 24 h later. The medium was replaced by 100 µL of DMSO after an additional 2-h incubation. The optical absorbance was measured at 590 nm (reference wavelength 630 nm) by a microplate reader (Spectraflour Plus, Tecan Austria GmbH, Austria) (9). The metabolic activity of the polymer treated cells was expressed as a relative to untreated cell controls taken as 100% metabolic activity.
Gene Transfection
Gene transfer properties of the synthesized copolymers were investigated in B16-F10 and CHO-K1 cell lines. Cells were seeded in 96-well TPP plates at a density of 5,000 cells per well in 100 μL of 10% FCS containing cell culture medium. The polyplexes in HBG were added at indicated polymer to pDNA weight ratios 24 h later after cell seeding. Unless indicated otherwise, the pDNA concentration was 200 ng per well and the medium was replaced by 100 μL of fresh medium after 4-h polyplex incubation. The luciferase reporter gene expression levels were determined by luciferase assay (12). The luciferase activity was expressed as relative light units per well (RLU/well). In parallel, the cellular metabolic activity after transfection was evaluated by MTT assay. Enhanced green fluorescent protein (EGFP) transfections were studied using an inverted fluorescence microscope (Axiovert 200, Carl Zeiss, Jena, Germany) and flow cytometry (Cyan MLE flow cytometer, Dako, Copenhagen, Denmark). The EGFP transfection results were shown as percentage of positive cells compared to untreated cell controls. The optimized c/p ratio of 0.8 was selected for BPEI and LPEI polyplex mediated transfection. All data represent the mean out of three independent measurements and each measurement was performed in triplicate.
RESULTS AND DISCUSSION
Preparation and Characterization of the OEI–CBA Copolymers
HMW polyethylenimines (BPEI and LPEI) are considered to be “golden standard” for non-viral gene delivery due to their high transfection efficiency in vitro and in vivo (27). However, both BPEI and LPEI are non-degradable and display significant toxicities, limiting their clinical application in gene therapy (28,29). In this study, a set of bioreducible analogs of PEI, the copolymers OEI–CBA, were prepared via the Michael addition reaction (Fig. 1). It has been reported that not only the monomer molar ratios but also the reaction conditions in Michael-type polyaddition reactions can display important roles regarding the molecular weights and chemical structure of the resulting copolymers (30, 31). According to the literature, all reactions were proceeded at 40°C to produce preferably copolymers with linear structure (see Fig. 1) (31). Three different CBA to OEI molar ratios of A (1:1), B (1.2:1), and C (1.4:1) were tested to optimize the synthesis conditions. Precipitation occurred in the case of the molar ratio of 1.4:1 already after 24-h reaction due to the formation of highly cross-linked copolymers. Neither gelation nor precipitation was obseved for A- and B-type copolymers with lower monomer molar ratios. The resulting products were both well water-soluble. In order to investigate the influences of the purification degree of the copolymers on both cytotoxicity and transfection efficiency, dialysis bags with different MWCOs of 3.5 and 6–8 kDa were used. Preliminary cytotoxicity studies demonstrated a lower toxicity of products purified with dialysis membranes of 3.5 kDa compared to polymers purified at a MWCO of 6–8 kDa. This is consistent with the presence of more LMW copolymers. Furthermore, B-type copolymers (CBA/OEI = 1.2:1) demonstrated higher toxicity than A (CBA/OEI = 1:1), presumably due to their higher molecular weights (data not shown).
Fig. 1.
Scheme for synthesis of bioreducible BPEI analogs from OEI and CBA
Hence, the CBA to OEI molar ratio of 1:1 was found as the best synthesis condition and selected for further optimization. Two different OEI concentrations of 1.0 and 0.5 M were applied. The resulting polymers labeled as “HC” (high concentration) and “LC” (low concentration) were compared. Their molecular weights and corresponding PDIs were shown in Table I. It was found that different reaction concentrations and purification conditions resulted in copolymers with different molecular weights. The HC condition led to products with much higher Mw than that of the LC copolymers. For example, copolymer HC-6-8 had Mw of 37 kDa, which was significantly higher than that of copolymer LC-6-8, which had 8.7 kDa. Moreover, the selected MWCOs of dialysis membranes in the purification procedure also influenced the copolymer molecular weights and, more obviously, their PDIs. For example, HC-3.5, which was purified at a MWCO of 3.5 kDa, had a PDI of 13. In contrast, HC-6-8 (MWCO 6–8 kDa) showed a much lower PDI of 5.5 due to the removal of the lower molecular weight fraction during dialysis.
Table I.
Synthesis Conditions and Molecular Parameters for OEI–CBA Copolymers
| Polymersa | Monomer concentration (M) | Molecular weightsb | OEI to CBA molar ratio | |||
|---|---|---|---|---|---|---|
| M w/103 | M n/103 | PDI | Feed ratio | Actual ratioc | ||
| HC-6-8 | 1.0 | 37 | 6.6 | 5.5 | 1.0 | 0.80 |
| HC-3.5 | – | 25 | 1.9 | 13 | – | 0.72 |
| LC-6-8 | 0.5 | 8.7 | 4.1 | 2.1 | – | 0.90 |
| LC-3.5 | – | 8.6 | 1.6 | 5.5 | – | 0.72 |
| LPEI | 59 | 27 | 2.2 | |||
| BPEI | 36 | 9.7 | 3.6 | |||
| OEI | 0.33 | 0.15 | 2.0 | |||
aThe copolymers prepared at the same monomer concentration were distinguished by the MWCOs of the dialysis bags used for purification. b Copolymer molecular weights and their polydispersity index (PDI, M w/M n) determined by GPC. c OEI to CBA molar ratios in the copolymers determined by 1H NMR spectra
Biophysical Characterization
DNA Binding Ability
An EtBr exclusion assay was firstly performed to test the pDNA binding kinetics of the copolymers (9). As shown in Table II, copolymers HC-6-8 and HC-3.5 had EB50 of 0.28 and 0.35, whereas it increased to 0.38 and 0.50 for LC-6-8 and LC-3.5, respectively. For more detailed studies, the pDNA binding ability of the copolymers was investigated by gel electrophoresis assay. It was found that BPEI and LPEI completely retard pDNA already at c/p ratios of 0.5 and 1.0. There was no free pDNA found for the HC copolymer polyplexes at a c/p ratio of 1. The LC copolymers, however, required a c/p ratio of 2 or higher to completely bind and retard pDNA. Similar results were also found for OEI (see Table II). We also found that copolymer HC-6-8 possesses the same PDI of 5.5 as LC-3.5, but it displayed much better pDNA binding ability than the latter. Taken together, all these data clearly demonstrated that the copolymer HC-6-8 with higher molecular weight and narrower PDI than other copolymers exhibited the highest pDNA binding ability. The molecular weight was obviously the dominating parameter for an effective pDNA binding.
Table II.
EB50 and Required Polymer to pDNA Weight Ratios (c/p) for Effective pDNA Binding and Compacting Mediated by BPEI, LPEI, OEI, and OEI–CBA Copolymers
| Polymers | BPEI | LPEI | OEI | HC-6-8 | HC-3.5 | LC-6-8 | LC-3.5 |
|---|---|---|---|---|---|---|---|
| EBa50 | 0.26 | 0.30 | 0.44 | 0.28 | 0.35 | 0.38 | 0.50 |
| Full Retardationb | 0.50 | 0.80 | 2.0 | 1.0 | 1.0 | 2.0 | 2.0 |
aEB50 was calculated as polymer to pDNA weight ratio (c/p) when the relative fluorescence intensity was reduced to 50% comparing to free pDNA. b c/p ratios when the pDNA movement in the agarose gel was fully retarded
Particle Size and Zeta Potential Measurement
Dynamic light scattering (DLS) measurements were performed to examine the hydrodynamic particle sizes of the polyplexes. As demonstrated in Fig. 2a, all tested copolymers, LPEI and BPEI, were generally able to compact pDNA to polyplexes with particle sizes ranging from 80 to 200 nm. In contrast, the complexes formed by OEI and pDNA tend to aggregate and showed hydrodynamic diameters over 1,000 nm even at a c/p ratio of 8 (data not shown). Both copolymers of HC-6-8 and HC-3.5 resulted in polyplexes about 120 nm at all tested c/p ratios ranging from 2 to 8, which was similar to that of LPEI and BPEI polyplexes. For copolymers LC-3.5 and LC-6-8 with lower molecular weights, the measured particle size was over 180 nm at a c/p ratios of 2. Increasing the c/p ratio, the polyplexes became smaller. Among all the copolymers, the HC compounds always resulted in much smaller particles than the LC ones at the same c/p ratios. It can be concluded that the copolymers with higher molecular weights could bind and compact pDNA more effectively due to their higher OEI contents (see Table I). All these results demonstrated the good pDNA binding and compacting properties of the OEI–CBA copolymers.
Fig. 2.
Particle size and zeta potential of the polyplexes. a Particle size of the polyplexes measured by DLS. b Zeta potential of the polyplexes. All the samples were prepared in HBG buffer and the measurements were performed in triplicates (n = 3)
The surface charges of the polyplexes were determined by zeta potential measurement. As shown in Fig. 2b, all polyplexes were positively charged, while the surface charges increased with raised c/p ratios. The polyplexes from HC copolymers generally had higher surface charges than those from LC ones at the same c/p ratios. This can be explained by the lower pDNA compacting ability of LC copolymers, resulting in less tightened particles and diffused surface charges.
Polyplex Stability Under Reducing Conditions
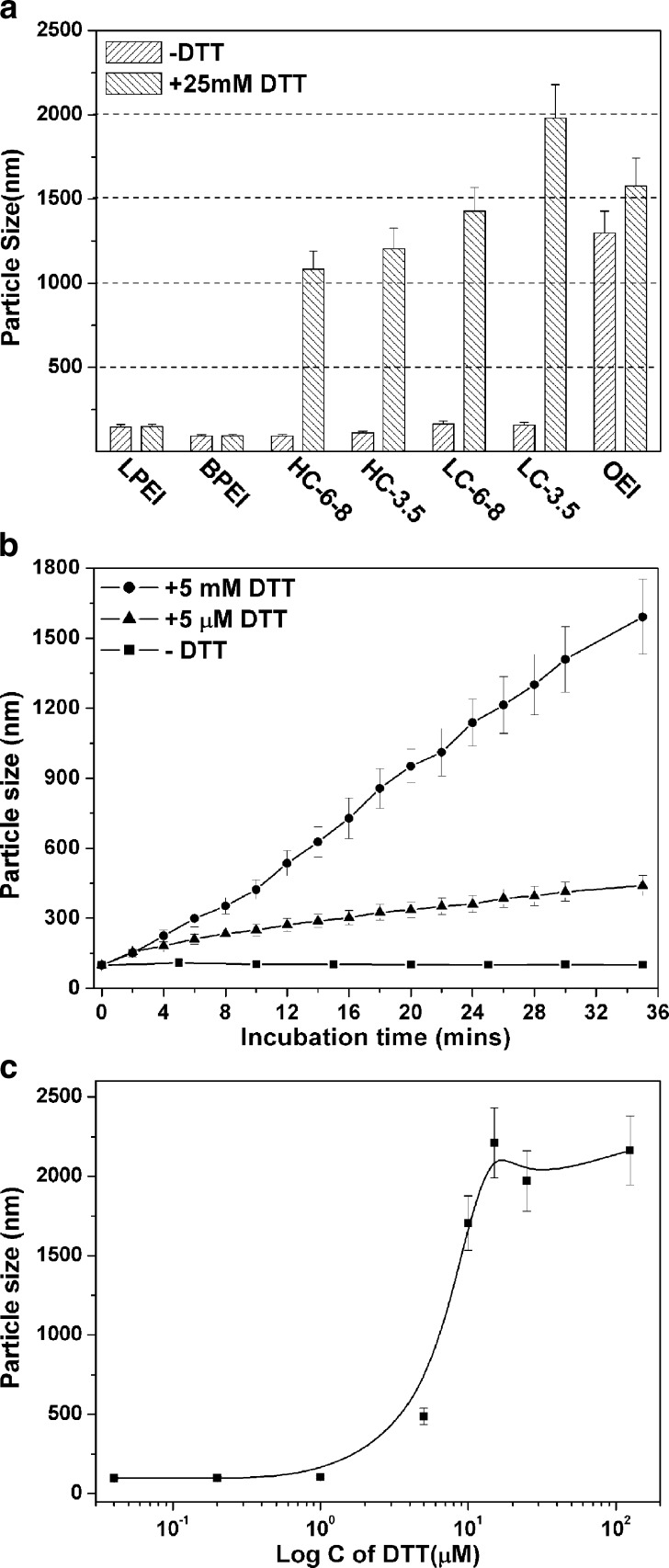
The polyplex formulations should possess several functions for efficient gene delivery, including protection of the nucleic acid payloads from extracellular polyanion exchange and enzyme degradation, facilitating cellular uptake, and delivery of the payloads and polyplex disassociation before or after nuclear entry. In the case of disulfide bond bearing bioreducible polyplexes, it is hypothesized that the polyplexes are kept stable in the extracellular environment while dissociate easily under intracellular reducing conditions due to disulfide cleavage (32,33). In the present study, the polyplex stability was thoroughly investigated by particle size measurement and gel electrophoresis assay in the presence of DTT. Firstly, both non-degradable and bioreducible PEI polyplexes were prepared and exposed to 25 mM DTT, a large excess to the calculated amount of the disulfide bonds in the polyplexes (Fig. 3a). The experiments were performed in HBG buffer with low ionic strength to avoid salt-induced polyplex aggregation. In the absence of DTT, all polyplexes except the OEI ones were stable without any obvious change in particle size. Importantly, the hydrodynamic diameters of the polyplexes could even be kept at around 100 nm for more than 48 h at 37°C water bath. However, addition of 25 mM DTT changed the particle sizes of different polyplexes to different extents. After 1-h incubation, the particle size of LPEI and BPEI polyplexes kept almost constant, indicating the good stability of these polyplexes. At the same time, all the copolymer polyplexes swelled from about 100 nm to over 1 μm due to disulfide cleavage in the bioreducible polyplexes.
Fig. 3.
Reduction with DTT together with physiological salt mediates polyplex disassociation and pDNA release. a–c Particle size measurements of polyplexes. The polyplexes were prepared in HBG (low ionic strength) and incubated at 37°C applying various DTT concentrations. a Particle sizes of indicated polyplexes (c/p 4) in the absence or presence of 25 mM DTT. b Expansion kinetics of the HC-6-8 polyplex (c/p 4) at 37°C in the presence of 5 µM or 5 mM DTT. c DTT concentration-dependent HC-6-8 polyplex (c/p 4) dissociation presented as particle sizes at 1 h after DTT incubation. d pDNA release from the polyplexes detected by gel shift assay. Polyplexes were incubated at indicated sodium chloride concentrations (0, 0.15, and 0.30 M) in the absence or presence of 25 mM DTT; same c/p ratio of 1 was selected for HC-6-8 and OEI polyplexes
HC-6-8 polyplexes were then selected to detect the dissociation kinetics of the bioreducible polyplexes against reduction induced by DTT (see Fig. 3b). With 5-mM DTT treatment, the polyplexes swelled gradually to a final size of over 1.5 μm after 30 min. But even the 1,000-fold lower DTT concentration of 5 µM resulted in significant polyplex swelling to about 400 nm, which indicated partial cleavage of the disulfide bonds. To further confirm these findings, HC-6-8 polyplexes were then treated by DTT at several concentrations for 1 h (see Fig. 3c). When the DTT concentration was in the range of 0 to 1 μM, the polyplexes remained stable with diameters around 100 nm. Increasing the DTT concentration to 5 μM, particle size of about 500 nm was found. Adding more DTT to a concentration of 15 μM, the polyplexes swelled dramatically to about 2 μm. Adding more DTT over 100 μM, the particle size would not increase anymore due to the complete cleavage of the disulfide bonds. In summary, swelling of the polyplexes (resulting from partial dissociation) was mediated by disulfide cleavage with DTT concentration-dependent kinetics. To our knowledge, this is the first time to evaluated the dissociation kinetics of disulfide bond containing polyplexes with reducing treatment. It had been reported that the total concentration of reducing agents, such as glutathione, in the extracellular environment is typically in the range of 1–5 μM, but in the cytosol, it is higher than millimolar levels (22). We therefore assume that our bioreducible polyplexes could effectively dissociate in intracellular environment while retaining their stability at least during the first half an hour circulation. When long-term circulation of small polyplexes is required, polyplex swelling resulting from disulfide bond cleavage might be a factor affecting the biodistribution.
Polyplex stability also depends on the ionic buffer conditions. Figure 3d shows the percentage of pDNA released from the polyplexes as determined by agarose gel electrophoresis. The release was quantified in the form of supercoiled pDNA (SC-pDNA) compared to SC-pDNA in the naked pDNA control. For HC-6-8 polyplexes (c/p 1), in the absence of DTT, no free SC-pDNA could be detected even with 0.3 M sodium chloride addition. Similar observations were made in the case of BPEI polyplexes. It suggested the good stability of the HC-6-8 polyplexes. In the presence of DTT, however, with salt adding at physiological concentration of 0.15 M, over 20% of SC-pDNA was released from the HC-6-8 polyplexes due to the reductive degradation of the copolymer. Enhancing the salt concentration further to 0.3 M, the detected pDNA release was 65%, practically identical to the level of OEI complexes with 66% pDNA release. This highlights the characteristic of the copolymers to facilitate intracellular pDNA release triggered by reducing conditions. It is noteworthy that DTT in the absence of physiological salt destabilizes the polyplexes (Fig. 3a–c), but this is not sufficient for pDNA release (see Fig. 3d).
Biological Characterization
Cytotoxicity of Free Copolymers
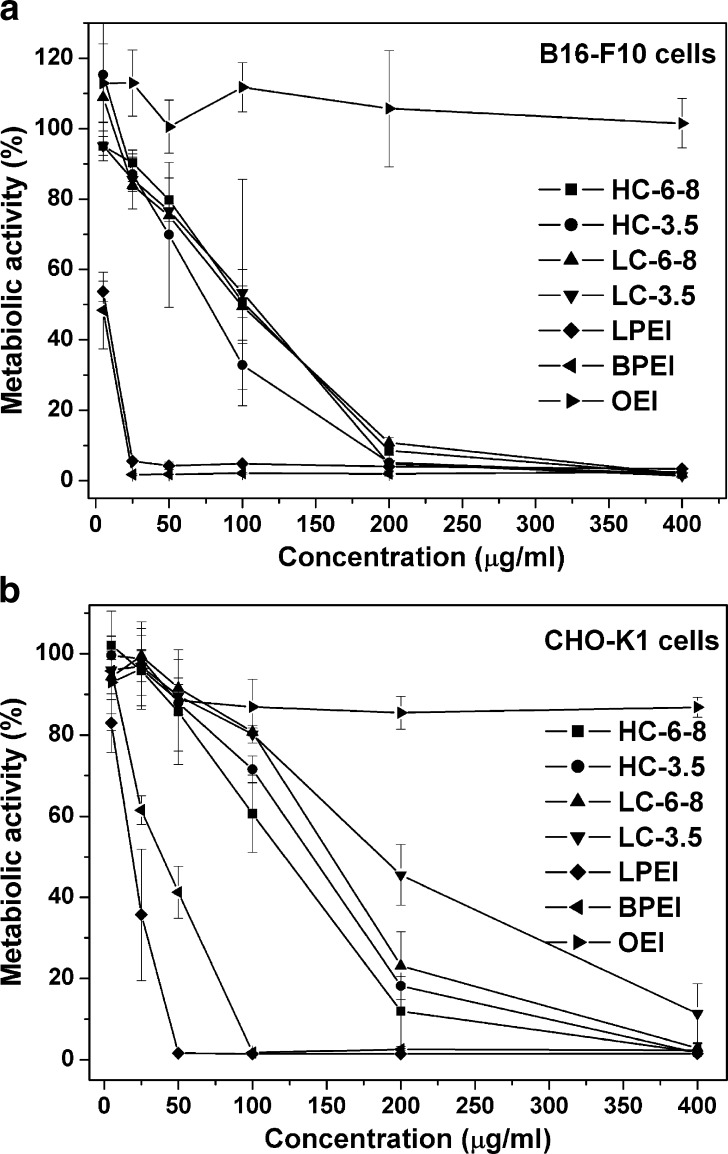
The cytotoxicity of the copolymers in B16-F10 and CHO-K1 cells was tested by MTT assay. As shown in Fig. 4a, the metabolic activity of B16-F10 cells treated with LPEI or BPEI at 50 μg/mL is lower than 5%, indicating high toxicity of BPEI and LPEI, whereas cells treated with OEI–CBA copolymers at the same concentration retained at least 70% of their metabolic activity. The same trend was also found for CHO-K1 cells (see Fig. 4b). The IC50 values of the tested compounds were shown in Table III. The IC50 values ranged for the reducible copolymers from 77 to 102 μg/mL, i.e., about 15- to 20-fold above that of the PEIs in B16-F10 cells, and in CHO-K1 cells from 122 to 186 μg/mL, i.e., about three to tenfold above the PEI standards. In the selected concentration ranges from 1 to 400 μg/mL, OEI never displayed any measurable toxic effect in both cell lines. As demonstrated in both cell lines, the HC copolymers with higher molecular weight displayed lower IC50 than the LC ones. Difference in IC50 was also found between the copolymers prepared at the same reaction concentration with different dialysis membranes used in the purification. For example, the copolymers HC-6-8 and LC-6-8 always resulted in lower IC50 values than HC-3.5 and LC-3.5. Our finding was consistent with the literature reports that cytotoxic properties of polycations are usually correlated with their molecular weights; those with lower molecular weights usually result in reduced cytotoxicity (32,34,35). Taken all together, the preliminary toxicity data encouraged to evaluate these bioreducible OEI–CBA copolymers as candidates for gene delivery.
Fig. 4.
Cytotoxicity assays of the plain OEI–CBA copolymers, OEI, LPEI, and BPEI in a B16-F10 and b CHO-K1 cells. The polymers were incubated with cells for 4 h (n = 3)
Table III.
IC50 Data of BPEI, LPEI, and OEI–CBA Copolymers in B16-F10 and CHO-K1 Cells
| Polymers | BPEI | LPEI | HC-6-8 | HC-3.5 | LC-6-8 | LC-C3.5 | |
|---|---|---|---|---|---|---|---|
| ICa50 (μg/ml) | B16-F10 | 4.9 | 6.4 | 77 | 99 | 87 | 102 |
| CHO-K1 | 39 | 19 | 122 | 140 | 153 | 186 | |
aIC50 was defined as the polymer concentration with 50% cell survival
Transfection Efficiency and Cytotoxicy of Polyplexes
Both B16-F10 and CHO-K1 cells were transfected with polyplexes prepared at various c/p ratios with 4-h incubation. The luciferase reporter gene expression was determined 24 h after polyplex addition. LPEI and BPEI were most commonly used as polyplex formulations; therefore, we included these polymers in optimized formulations. LPEI was previously found to show particularly high transfection activity and presents the “golden standard” (12,36). In CHO-K1 cells, as shown in Fig. 5a, all copolymer polyplexes achieved their maximal transfection efficiencies at a c/p ratio of 4. Out of all copolymer polyplexes, those from HC-6-8 resulted in the highest reporter gene expression level comparable to that of LPEI polyplexes and about 150-fold higher than that of BPEI ones. In B16-F10 cells, HC-6-8 was also the most efficient among the copolymers at an optimized c/p ratio of 2. For all the other copolymers, higher c/p ratios were required to achieve their highest luciferase gene expression yields (Fig. 5a, b). The metabolic activity of the cells treated by the polyplexes was determined by MTT assay. The results were shown in Fig. 5c, d. At optimized c/p ratios leading to the highest transfection yields, B16-F10 and CHO-K1 cells treated by the copolymer polyplexes exhibited metabolic activity higher than 80%, which was much higher than that treated by LPEI polyplexes.
Fig. 5.
Luciferase reporter gene transfer efficiency of the polymer/pDNA polyplexes and metabolic activity of the transfected CHO-K1 and B16-F10 cells. All the polyplexes were prepared in HBG buffer; 200 ng of pDNA/well was used for 96-well tissue culture plate. The cell culture medium was changed 4 h after polyplexes addition; the relative luciferase intensity (a, c) and metabolic activity of the polyplexes treated cells (b, d) was measured 24 h later. Presented data show the average of experiments performed three times as triplicates (n = 3 × 3)
Effects of Polyplex Concentrations on the Transfection Efficiency
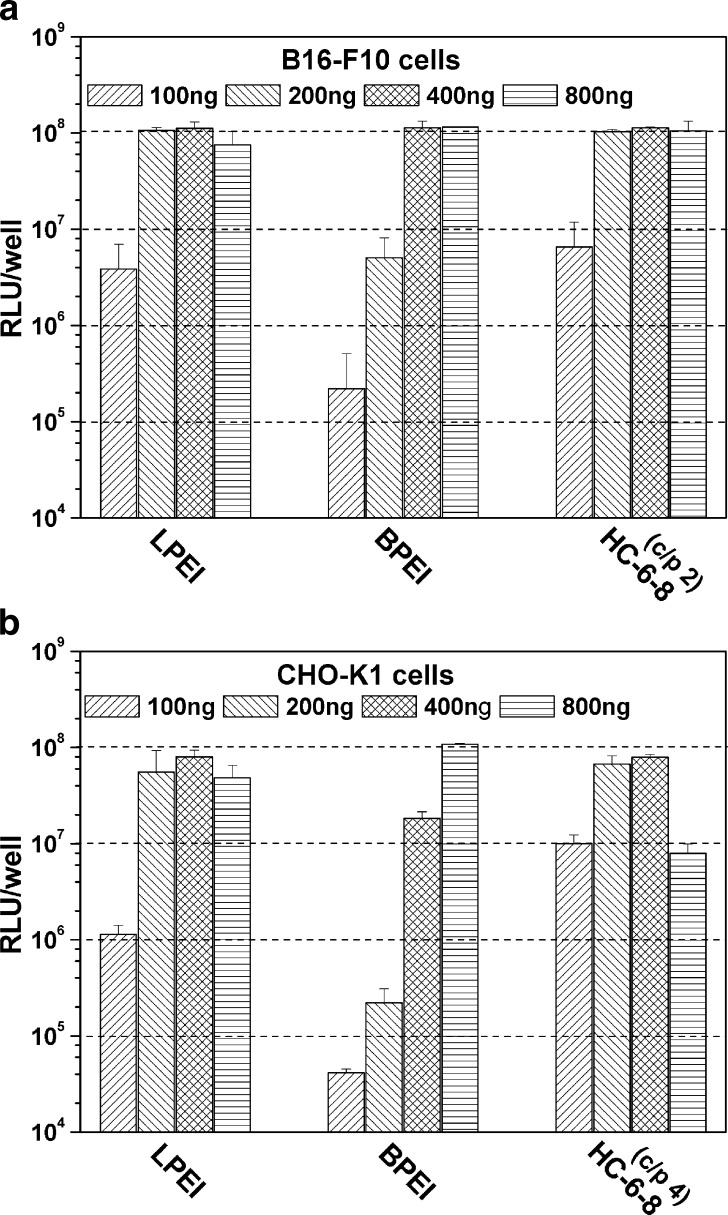
B16-F10 and CHO-K1 cells were transfected with HC-6-8 polyplexes at optimized c/p ratios of 2 and 4, respectively. As shown in Fig. 6a, in B16-F10 cells, at a lower pDNA concentration of 100 ng per well, all three polymers, including HC-6-8, LPEI, and BPEI, produced only moderate reporter gene expression. Increasing the pDNA amount to 200 ng per well, both HC-6-8 and LPEI reached their comparable highest RLU levels. A much higher pDNA concentration of 400 ng per well was required by BPEI polyplexes to reach the highest RLU level. Similar trends were also found in CHO-K1 cells (Fig. 6b). Here, HC-6-8 and LPEI needed a pDNA concentration of 400 ng per well to reach the highest transfection yields. However, BPEI need much higher pDNA concentration of 800 ng per well to efficiently transfect CHO-K1 cells.
Fig. 6.
Gene transfer efficiency of the polyplexes built on HC-6-8, BPEI, and LPEI with different pDNA concentrations (n = 3 × 3)
Effects of Polyplex Incubation Time on the Transfection Efficiency
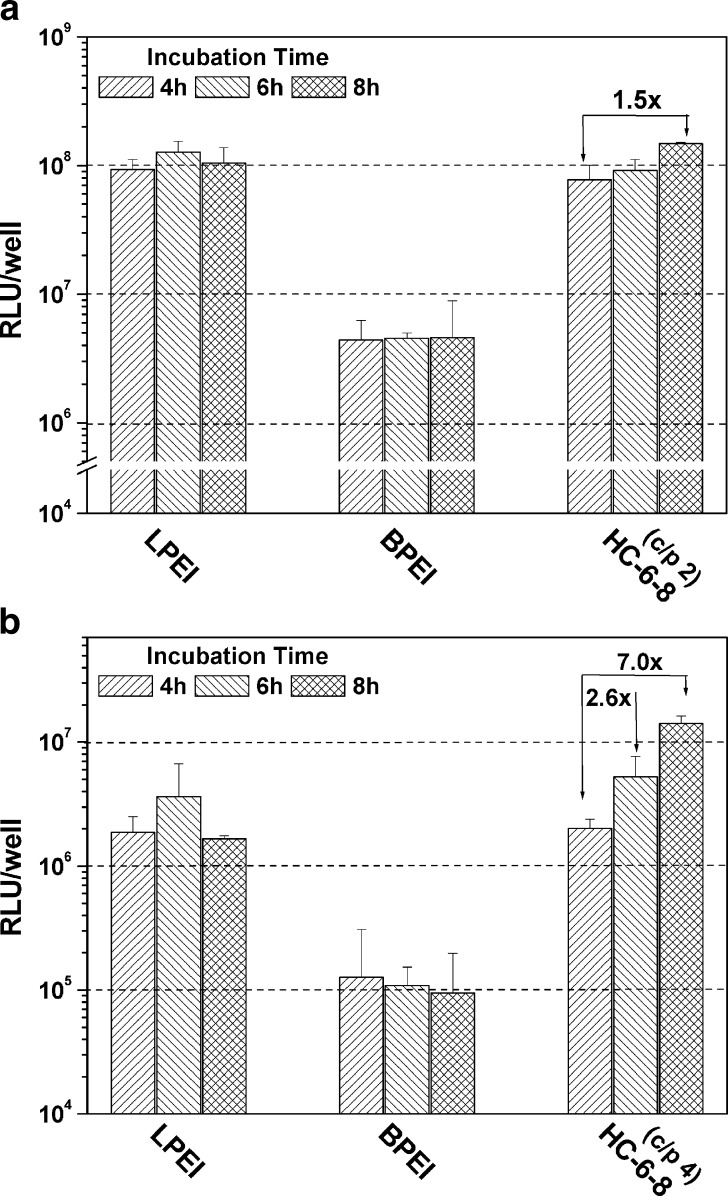
CHO-K1 cell line had been well investigated for membrane protein associated disulfide cleavage (22,37–39). The cleavage began in the first hour of cell membrane binding and continued for more than 6 h (38). In order to examine the influence of disulfide cleavage on the transfection property of the polyplexes, different polyplex incubation durations were tested. As demonstrated in Fig. 7a, the reporter gene expression level for HC-6-8 polyplexes was improved by 2.6-fold in CHO-K1 cells when the incubation time was increased from 4 to 6 h. Further extending the incubation time to 8 h, even a sevenfold enhancement was achieved compared to polyplex treatment for 4 h. Similar trends were found by extending incubation durations in B16-F10 cells, while the results were less pronounced (see Fig. 7b). In both cell lines, no significant effects of different incubation times were found for LPEI and BPEI polyplexes.
Fig. 7.
Gene transfer efficiency of the polyplexes built on HC-6-8, BPEI, and LPEI with different incubation durations (n = 3 × 3)
EGFP Transfection
In addition to luciferase gene expression studies, pDNA encoding an EGFP-N1 reporter gene was also used to examine the transfection ability of the copolymers in the two selected cell lines. The EGFP pDNA transfected cells were observed by fluorescence microscopy. Forty-eight hours later after polyplex addition, copolymer HC-6-8 displayed excellent EGFP transfection results in both cell lines (see Fig. 8a). The percentages of EGFP-positive cells as quantified by flow cytometry measurements were shown in Fig. 8b. Copolymer HC-6-8 resulted in 68% of EGFP positively expressing B16-F10 cells. It was comparable to LPEI (62%) and even 20-fold more efficient than BPEI. In CHO-K1 cells, copolymer HC-6-8 transfected 30% of cell population, which was about tenfold higher than that of BPEI, but lower than that of LPEI (57%). Generally, the EGFP transfection results at optimized c/p ratios were found to go quite well along with the luciferase transfection results (see Fig. 5), indicating a high transfection efficiency at low cytotoxicity of the bioreducible copolymers.
Fig. 8.
EGFP reporter gene transfer results. a EGFP transfection effects in B16-F10 and CHO-K1 cells. The polyplexes were prepared in HBG at optimized c/p ratios and incubated with cells for 4 h. All the pictures were taken 48 h later after polyplex addition. b Flow cytometry measurement results for percentages of EFGP expressing cells. The cells were measured immediately after photograph taken (n = 3)
CONCLUSION
In this study, a set of bioreducible PEI analogues, OEI–CBA copolymers were prepared by a polyaddition reaction. The influence of synthesis conditions on the chemical, biophysical, and biological properties of the OEI–CBA copolymers was investigated. The polyplexes built on these copolymers can be kept stable in salt-free HBG buffer, but are destabilized upon treatment with the reducing agent DTT. Copolymer HC-6-8 with a molecular weight of 37 kDa can efficiently deliver pDNA both into B16-F10 and CHO-K1 cells. The transfection efficiencies are comparable to that of LPEI, while much higher than that of BPEI. Given the higher cytotoxicity and non-degradability of LPEI and BPEI, and low gene transfection ability of the latter, bioreducible copolymer HC-6-8 is much superior and promising for polymer-based gene delivery.
Acknowledgment
We thank Ms Olga Brück for skilful assistance in preparing the manuscript, Ms. Terese Magnusson and Dr. Michael Günther for their assistance in FACS measurement. We acknowledge the financial support by DFG SPP1230, excellence cluster NIM and EC project GIANT.
Abbreviations
- BPEI
Branched PEI 25 kDa
- CBA
N,N′-cystamine bisacrylamide
- DLS
Dynamic light scattering
- DTT
Dithiothreitol
- GPC
Gel permeation chromatograph
- HC
High concentration
- HMW
High molecular weight
- LC
Low concentration
- LMW
Low molecular weight
- LPEI
Linear PEI 22 kDa
- MWCO
Molecular weight cutoff
- OEI
Oligoethylenimine 800 Da
- pDNA
Plasmid DNA
References
- 1.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari S, Moro E, Pettenazzo A, Behr JP, Zacchello F, Scarpa M. ExGen 500 is an efficient vector for gene delivery to lung epithelial cells in vitro and in vivo. Gene Ther. 1997;4:1100–1106. doi: 10.1038/sj.gt.3300503. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet ME, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm Res. 2008;25:2972–2982. doi: 10.1007/s11095-008-9693-1. [DOI] [PubMed] [Google Scholar]
- 4.Kichler A, Leborgne C, Coeytaux E, Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J Gene Med. 2001;3:135–144. doi: 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- 5.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine–DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 6.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. Eur J Pharm Sci. 2006;29:414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Petersen H, Merdan T, Kunath K, Fischer D, Kissel T. Poly(ethylenimine-co-l-lactamide-co-succinamide): a biodegradable polyethylenimine derivative with an advantageous pH-dependent hydrolytic degradation for gene delivery. Bioconjug Chem. 2002;13:812–821. doi: 10.1021/bc0255135. [DOI] [PubMed] [Google Scholar]
- 8.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003;14:934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 9.Kloeckner J, Bruzzano S, Ogris M, Wagner E. Gene carriers based on hexanediol diacrylate linked oligoethylenimine: effect of chemical structure of polymer on biological properties. Bioconjug Chem. 2006;17:1339–1345. doi: 10.1021/bc060133v. [DOI] [PubMed] [Google Scholar]
- 10.Ahn CH, Chae SY, Bae YH, Kim SW. Biodegradable poly(ethylenimine) for plasmid DNA delivery. J Control Release. 2002;80:273–282. doi: 10.1016/S0168-3659(01)00547-8. [DOI] [PubMed] [Google Scholar]
- 11.Arote R, Kim TH, Kim YK, et al. A biodegradable poly(ester amine) based on polycaprolactone and polyethylenimine as a gene carrier. Biomaterials. 2007;28:735–744. doi: 10.1016/j.biomaterials.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 12.Russ V, Elfberg H, Thoma C, Kloeckner J, Ogris M, Wagner E. Novel degradable oligoethylenimine acrylate ester-based pseudodendrimers for in vitro and in vivo gene transfer. Gene Ther. 2008;15:18–29. doi: 10.1038/sj.gt.3303046. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Mo H, Koo H, et al. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjug Chem. 2007;18:13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 14.Peng Q, Zhong Z, Zhuo R. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjug Chem. 2008;19:499–506. doi: 10.1021/bc7003236. [DOI] [PubMed] [Google Scholar]
- 15.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Chen P, Shen J. The development and characterization of a glutathione-sensitive cross-linked polyethylenimine gene vector. Biomaterials. 2006;27:5292–5298. doi: 10.1016/j.biomaterials.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 17.Lin C, Zhong Z, Lok MC, et al. Novel bioreducible poly(amido amine) s for highly efficient gene delivery. Bioconjug Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 18.Zhong Z, Song Y, Engbersen JF, Lok MC, Hennink WE, Feijen J. A versatile family of degradable non-viral gene carriers based on hyperbranched poly(ester amine) s. J Control Release. 2005;109:317–329. doi: 10.1016/j.jconrel.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 19.Hoon JJ, Christensen LV, Yockman JW, et al. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28:1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Lin C, Blaauboer CJ, Timoneda MM, et al. Bioreducible poly(amido amine) s with oligoamine side chains: synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126:166–174. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Sun YX, Zeng X, Meng QF, Zhang XZ, Cheng SX, Zhuo RX. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29:4356–4365. doi: 10.1016/j.biomaterials.2008.07.045. [DOI] [PubMed] [Google Scholar]
- 22.Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv Drug Deliv Rev. 2003;55:199–215. doi: 10.1016/S0169-409X(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen CP, Kim JS, Steenblock E, Liu D, Rice KG. Gene transfer with poly-melittin peptides. Bioconjug Chem. 2006;17:1057–1062. doi: 10.1021/bc060028l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blacklock J, Handa H, Soundara MD, Mao G, Mukhopadhyay A, Oupicky D. Disassembly of layer-by-layer films of plasmid DNA and reducible TAT polypeptide. Biomaterials. 2007;28:117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Plank C, Zatloukal K, Cotten M, Mechtler K, Wagner E. Gene transfer into hepatocytes using asialoglycoprotein receptor mediated endocytosis of DNA complexed with an artificial tetra-antennary galactose ligand. Bioconjug Chem. 1992;3:533–539. doi: 10.1021/bc00018a012. [DOI] [PubMed] [Google Scholar]
- 26.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 27.Zou SM, Erbacher P, Remy JS, Behr JP. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. J Gene Med. 2000;2:128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Chollet P, Favrot MC, Hurbin A, Coll JL. Side-effects of a systemic injection of linear polyethylenimine–DNA complexes. J Gene Med. 2002;4:84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- 29.Wagner E. The silent (R) evolution of polymeric nucleic acid therapeutics. Pharm Res. 2008;25:2920–2923. doi: 10.1007/s11095-008-9689-x. [DOI] [PubMed] [Google Scholar]
- 30.Lynn DM, Langer R. Degradable poly(ß-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. doi: 10.1021/ja0015388. [DOI] [Google Scholar]
- 31.Hong CY, You YZ, Wu DC, Liu Y, Pan CY. Thermal control over the topology of cleavable polymers: from linear to hyperbranched structures. J Am Chem Soc. 2007;129:5354–5355. doi: 10.1021/ja070871+. [DOI] [PubMed] [Google Scholar]
- 32.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read ML, Singh S, Ahmed Z, et al. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33:e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim YB, Kim SM, Suh H, Park JS. Biodegradable, endosome disruptive, and cationic network-type polymer as a highly efficient and nontoxic gene delivery carrier. Bioconjug Chem. 2002;13:952–957. doi: 10.1021/bc025541n. [DOI] [PubMed] [Google Scholar]
- 35.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 36.Thomas M, Lu JJ, Ge Q, Zhang C, Chen J, Klibanov AM. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci U S A. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci U S A. 1993;90(9):4112–4116. doi: 10.1073/pnas.90.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feener EP, Shen WC, Ryser HJ. Cleavage of disulfide bonds in endocytosed macromolecules. A processing not associated with lysosomes or endosomes. J Biol Chem. 1990;265:18780–18785. [PubMed] [Google Scholar]
- 39.Neu M, Germershaus O, Mao S, Voigt KH, Behe M, Kissel T. Crosslinked nanocarriers based upon poly(ethylene imine) for systemic plasmid delivery: in vitro characterization and in vivo studies in mice. J Control Release. 2007;118:370–380. doi: 10.1016/j.jconrel.2007.01.007. [DOI] [PubMed] [Google Scholar]