Abstract
UDP glucuronosyltransferases (UGTs) are an important class of Phase II enzymes involved in the metabolism and detoxification of numerous xenobiotics including therapeutic drugs and endogenous compounds (e.g. bilirubin). To date, there are 21 human UGT genes identified, and most of them contain single-nucleotide polymorphisms (SNPs). Non-synonymous SNPs (nsSNPs) of the human UGT genes may cause absent or reduced enzyme activity and polymorphisms of UGT have been found to be closely related to altered drug clearance and/or drug response, hyperbilirubinemia, Gilbert’s syndrome, and Crigler-Najjar syndrome. However, it is unlikely to study the functional impact of all identified nsSNPs in humans using laboratory approach due to its giant number. We have investigated the potential for bioinformatics approach for the prediction of phenotype based on known nsSNPs. We have identified a total of 248 nsSNPs from human UGT genes. The two algorithms tools, sorting intolerant from tolerant (SIFT) and polymorphism phenotyping (PolyPhen), were used to predict the impact of these nsSNPs on protein function. SIFT classified 35.5% of the UGT nsSNPs as “deleterious”; while PolyPhen identified 46.0% of the UGT nsSNPs as “potentially damaging” and “damaging”. The results from the two algorithms were highly associated. Among 63 functionally characterized nsSNPs in the UGTs, 24 showed altered enzyme expression/activities and 45 were associated with disease susceptibility. SIFT and Polyphen had a correct prediction rate of 57.1% and 66.7%, respectively. These findings demonstrate the potential use of bioinformatics techniques to predict genotype–phenotype relationships which may constitute the basis for future functional studies.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-009-9126-z) contains supplementary material, which is available to authorized users.
Key words: phenotype, PolyPhen, SIFT, SNP, UGT
INTRODUCTION
The UDP glucuronosyltransferases (UGTs) represent a superfamily of endoplasmic-reticulum-bound enzymes that catalyze glucuronidation, a process that increases the polarity of xenobiotics and some endogenous compounds to facilitate their excretion via the bile or urine (1). Glucuronidation accounts for ~35% of all drugs metabolized by Phase II drug-metabolizing enzymes and therefore plays an important role in the detoxification and excretion of drugs and/or their metabolites (1,2). Additionally, UGTs play a critical role in the disposition of several important endogenous substrates, including bilirubin, bile acids, steroids, thyroxine, biogenic amines, and fat-soluble vitamins (3,4).
Up to date, there are 21 human UGTs identified (http://www.ugtalleles.ulaval.ca, access date: 11 June 2009). Based on sequence identities, human UGTs comprise UGT1, 2, 3, and 8 families (3,5). The UGT1 locus in humans has been mapped to chromosome 2q37 (3,6). All UGT1 genes contain unique first exons, and they are subsequently spliced into common exons 2 through 5, leading to different N-terminal halves but identical C-terminal halves of the gene products (3). Unlike the UGT1 family, the human UGT2 mRNAs are transcribed from separate genes and are divided into UGT2A and 2B subfamilies (4,7). In humans, the genes encoding several UGT2 enzymes form a cluster on chromosome 4 (UGT2B7-UGT2B4-UGT2B15; 4). UGT3 has two members, namely UGT3A1 and 3A2, and UGT8 family has one member.
In the human genome, the most frequent type of DNA variation is single base change, namely single-nucleotide polymorphisms (SNPs) (8,9). Currently, there are a total of 14,708,752 SNPs in human genome identified and deposited to the NCBI dbSNP (http://www.ncbi.nlm.nih.gov/sites/entrez, Build 129). A non-synonymous (nsSNPs) or missense variant is a single base change in a coding region that leads to amino acid substitution in the corresponding protein (8,10,11). Many nsSNPs do not cause any change in the corresponding protein and are regarded as tolerated nsSNPs (9,12). On the other hand, when mutations occur and nsSNPs do cause clinical phenotypic consequences in individuals (e.g. disease or biochemical abnormality), they are termed deleterious nsSNPs (9,12).
In the case of UGTs, deleterious nsSNPs of UGT have shown to alter bilirubin metabolism, drug disposition, and predisposition to diseases in individuals carrying the polymorphism (13,14). Variation in UGT1A coding sequences may lead to deficiency of UGT1A1 activity, often resulting in high level of unconjugated bilirubin which is closely related to diseases such as Crigler-Najjar syndrome (CN; 15) and Gilbert’s syndrome (16). Nonsense mutations in the UGT1 gene resulting in a premature termination codon and absent UGT1A1 activity have been reported in CN patients (17,18). Furthermore, some genetic variations in the UGT1A1 promoter (*28) is associated with increased drug toxicity and hyperbilirubinemia in cancer patients receiving irinotecan-based chemotherapy (19–22), and reports also show increased risk of tranilast-induced hyperbilirubinemia (23). UGT1A7*3 leading to a low-enzyme activity has been linked to increased risk of the cancer of the colon (24), mouth (25,26), esophagus (26), stomach (26), and the liver (27). A number of UGT2 genetic variants have been shown to relate to altered drug metabolism and disease risk (1,28,29). For example, the UGT2B7 promoter variant is associated with significantly reduced glucuronidation of morphine in sickle cell disease and contributes to the variability in hepatic clearance of morphine in these patients (28). A UGT2B15 variant has been shown to relate to prostate cancer (30).
The identification of deleterious nsSNPs in the human genome is important. This process relates human phenotypes to variation at the DNA level and links genetic and phenotypic differences in individuals; it is believed that nsSNPs and SNPs in regulatory regions together have the highest impact on phenotype (8). The identification and characterization of SNPs of major drug-metabolizing enzymes such as UGTs is crucial for understanding individual differences in drug metabolism, therapeutic efficacy, inherited diseases and also for the predisposition toward diseases such as cancer (13). However, humans have vast numbers of SNPs and presently it is unlikely to investigate their functional impact of these SNPs by experiments. Alternatively, bioinformatics approaches have gained increased use in the prediction of the functional effect of SNPs. Two algorithms sorting intolerant from tolerant (SIFT) and polymorphism phenotyping (PolyPhen) represent powerful bioinformatic tools, they enable high-throughput prediction of the potential impact of nsSNPs and large-scale polymorphism analyses. This process has the prospective to reduce the number of SNPs that may have clinical implications and needs to be evaluated in future clinical studies.
The elementary theory of SIFT have been developed and illustrated by Ng and Henikoff (9,31,32); it is used to predict the effect of an amino acid substitution on protein function according to sequence homology and the physical properties of amino acids. SIFT examines sequence similarity among related genes and does not require the knowledge of protein structural or functional information; therefore, it can be applied to a much larger number of proteins (9). The PolyPhen algorithm is a structure-sequence-based amino acid substitution prediction method; it utilizes the data available in Swiss-Prot (http://au.expasy.org/sprot/userman.html) and predicts the possible impact of amino acid substitutions on the structure and function of a human protein (8). Mapping of an amino acid replacement to a known 3D structure reveals whether the replacement is likely to destroy the hydrophobic core of a protein, electrostatic interactions, interactions with ligands or other important features of a protein (8,33). Several programs including TMHMM algorithm, Coils2 program, SignalIP programs, and PHAT transmembrane-specific matrix scores are also used to predict the possible functional effect of nsSNPs (8,34–37). Multiple alignment-based profile scores provide the major contribution to the prediction therefore reliable prediction can be reasonably achieved even if the proteins has no known 3D structure (8). In this study, we have investigated the potential effect of known human UGT nsSNPs on protein function using both SIFT and PolyPhen algorithms.
MATERIALS AND METHODS
Gene Nomenclature and Dataset
The human UGT genes examined in this study were named in agreement with the UGT Nomenclature Committee (http://www.ugtalleles.ulaval.ca/, access date: 11 June 2009). The data on human UGT genes were collected from Entrez Gene on NCBI Website (http://www.ncbi.nlm.nih.gov/sites/enterz, access date: 11 June 2009). Expired and merged gene names were excluded from the study. The majority of the variants included in this analysis were identified during the screening of 21 human UGT genes from MutDB and PolyDoms. MutDB integrates genetic variants from Swiss-Prot and dbSNP and links to functional disruption prediction scores. PolyDoms incorporate the results of multiple algorithmic procedures and functional criteria applied to the entire Entrez dbSNP dataset. Information including gene symbol, gene name, mRNA accession number (NM), protein accession number (NP), SNP ID, amino acid residue 1 (wild-type), amino acid position, and amino acid residue 2 (missense) were collected. Most of the data and information describing the genes and variants, including Entrez Gene ID are available at http://www.mutdb.org and http://www.polydoms.cchmc.org/polydoms. Supplementary variants were identified from Entrez Gene on NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez, access date: 11 June 2009) and added to the dataset after cross-examination. The information on the effect of the nsSNPs on enzyme activity and the correlation between the nsSNPs and disease were extracted from in vivo and in vitro experiments (e.g. site-directed mutagenesis analysis) according to literature.
Prediction of the Phenotype of nsSNPs in Human UGT Genes
Identified UGT nsSNPs and prediction effect of the variant amino acid substitution on protein function was performed using SIFT (http://blocks.fhcrc.org/sift/SIFT.html) and Polyphen (http://genetics.bwh.harvard.edu/pph/), listed in Supplementary Table 1.
In this study, SIFT version 3 was used. This algorithm uses a query sequence to search for similar sequences that may share similar function, generates the alignment of the chosen sequences, and predicts the probability score of the impact of an amino acid substitution on protein function effects. SIFT scores ranges from 0 to 1, outcome scores from 0.00–0.05 are elected as intolerant, 0.051–0.10 as potentially tolerant, 0.101–0.20 as borderline, and 0.201–1.00 as tolerant (31,38). Information such as dbSNP ID and GI number of an amino acid substitution can be employed to predict the effect on protein functions in SIFT. The algorithm and instructions for analysis of amino acid substitutions are available at http://www.blocks.fhcrc.org/sift/SIFT.html. The data employed in the SIFT analysis were the UGT gene sequences available in the NCBI non-redundant database (http://www.ncbi.nlm.nih.gov, access date: 11 June 2009), orthologous sequences were used as paralogous sequences confounds predictions (32).
PolyPhen uses empirically derived rules based on previous research in protein structure, interaction, and evolution that automatically predict whether a replacement is likely to be deleterious for the protein on the basis of three-dimensional structure and multiple alignments of homologous sequences (8,33). PolyPhen input is a protein amino acid sequence or the SWALL database (http://srs.ebi.ac.uk/srs6bin/cgi-bin/wgetz?-page+LibInfo+-newId+-lib+SWALL) ID or accession number together with sequence position and two amino acid variants characterizing the polymorphism (8). PolyPhen scores range from 0 ≤ 2 ≤ X, outcome scores of 0.00–0.99 are classified as benign, 1.00–1.24 as borderline, 1.25–1.19 as potentially damaging, 1.50–1.99 as possibly damaging, and ≥2 as damaging (38). Additional details of the algorithm and instructions for analysis of amino acid substitutions are available at http://www.bork.embl-heidelberg.de/PolyPhen/.
Validation of the Prediction Results
nsSNPs with experimental evidences of changing enzyme activity or disease association were regarded as deleterious. The phenotypic data are from both in vivo and in vitro studies, in which analysis of site-directed mutagenesis or enzymatic changes often provide direct evidence indicating the functional impact of nsSNPs. Prediction accuracy was analyzed according to these positive findings from these experiments.
As a test for the ability of both SIFT and PolyPhen algorithms to identify substitutions impacting enzymatic activity of UGT proteins, scores were obtained and compared for the collected nsSNPs of human UGT genes related to loss of enzyme activity and disease based on experimental and clinical studies.
Statistical Analysis
Given that SIFT and PolyPhen employ different approaches and also different datasets as foundations for their analysis, it is important to find the concordance of the two prediction tools on functional consequences of each nsSNP prediction. Concordance analysis of each nsSNP predicted by SIFT and PolyPhen were assessed using Spearman’s rank correlation coefficient ρ using SPSS 15. Prediction scores of each nsSNP were plotted on scatter graphs and analyzed using linear trend lines.
RESULTS
Validated nsSNPs of Human UGT Genes
Two hundred and forty eight amino acid substitution variants were identified in the systemic screening of 21 human UGT genes for the analysis of the potential impact of all nsSNPs in human UGT genes. With development and updated data in bioinformatics, some previously reported SNPs in dbSNP have been identified as invalid by later studies due to wrong sequencing and alignment. These incorrect SNPs have either been terminated or merged into other SNPs. We have cross-examined the databases and removed those invalid SNPs. Number of variants from four UGT subfamilies (UGT1, UGT2, UGT3, and UGT8) are shown in Table I. Three SNPs were identified in the screening of UGT2B17 but none were non-synonymous, therefore not included in this study.
Table I.
A Summary of Allelic Variants of Human UGT Genes
| Gene symbol | No. nsSNPs | No. of nsSNPs scored by SIFT | No. of nsSNPs scored by PolyPhen | No. of nsSNPs scored by both algorithms |
|---|---|---|---|---|
| UGT1A1 | 51 | 51 | 50 | 50 |
| UGT1A3 | 20 | 20 | 20 | 20 |
| UGT1A4 | 12 | 12 | 12 | 12 |
| UGT1A5 | 25 | 25 | 25 | 25 |
| UGT1A6 | 9 | 9 | 9 | 9 |
| UGT1A7 | 14 | 14 | 14 | 14 |
| UGT1A8 | 29 | 29 | 29 | 29 |
| UGT1A9 | 11 | 11 | 11 | 11 |
| UGT1A10 | 25 | 25 | 25 | 25 |
| UGT2A1 | 5 | 5 | 5 | 5 |
| UGT2A2 | 3 | 3 | 3 | 3 |
| UGT2A3 | 1 | 1 | 1 | 1 |
| UGT2B4 | 9 | 9 | 9 | 9 |
| UGT2B7 | 10 | 10 | 10 | 10 |
| UGT2B10 | 6 | 6 | 6 | 6 |
| UGT2B11 | 6 | 6 | 6 | 6 |
| UGT2B15 | 2 | 2 | 2 | 2 |
| UGT2B17 | 0 | 0 | 0 | 0 |
| UGT2B28 | 5 | 5 | 5 | 5 |
| UGT3A1 | 1 | 1 | 1 | 1 |
| UGT3A2 | 2 | 2 | 2 | 2 |
| UGT8 | 2 | 2 | 2 | 2 |
| Total | 248 | 248 | 247 | 247 |
Prediction of Functional Effect of nsSNPs of Human UGT Genes
SIFT scores for all 248 nsSNPs identified in this study was found, Polyphen classified UGT1A1 (L15R) as benign but did not provide a prediction score, therefore 247 nsSNPs in this study were scored by both SIFT and PolyPhen, these nsSNPs were used in the statistical concordance test.
As shown in Table II, 88 of 248 or 35.5% of identified UGT nsSNPs exhibited SIFT scores of <0.05 and are classified as “Intolerant” variants by SIFT. One hundred fourteen of 248 or 46.0% of identified UGT nsSNPs had prediction scores of ≥1.5 and are classified as “probably damaging” by PolyPhen. SIFT predicted a few nsSNPs of UGT genes to have no deleterious nsSNPs causing functional effects on protein function of UGT2A3, of 2B15, of 2B28, 3A1, and 8 (Table III), including Ala496Thr, Asp84Tyr, Lys522Thr, Leu364His, Asn441Ser, Cys120Gly, and Pro225Leu. PolyPhen also predicted some nsSNPs of UGT genes to have no deleterious nsSNPs, these included UGT2A3, 2B15, and 8 (Table III). UGT2A3, 2B15, and 8 did not have any deleterious nsSNPs predicted by both SIFT and PolyPhen (Table III), which are Ala496Thr, Asp84Tyr, Lys522Thr, and Ile367Met.
Table II.
Prediction Results of nsSNPs of Human UGT Genes
| Prediction result | SIFTa | Polyphenb | ||
|---|---|---|---|---|
| Number of nsSNPs | Percent | Number of nsSNPs | Percent | |
| Tolerated | 160 | 35.5 | 114 | 46 |
| Deleterious | 88 | 64.5 | 133 | 53.6 |
| Not scored | 0 | 1 | 0.4 | |
| Total | 248 | 100 | 248 | 100 |
aSee website: SIFT (http://blocks.fhcrc.org/sift/SIFT.html); positions with normalized probabilities <0.05 are predicted to be deleterious, those ≥0.05 are predicted to be tolerated
bSee website: polyphen (http://genetics.bwh.harvard.edu/pph/); positions with normalized probabilities ≥1.5 are predicted to be deleterious, those <1.5 are predicted to be tolerated
Table III.
Distribution of Deleterious nsSNPs in Human UGT Genes Predicted by SIFT and PolyPhen
| UGT gene | Number of deleterious nsSNPs predicted by SIFT | UGT gene | Number of deleterious nsSNPs predicted by PolyPhen | UGT gene | Number of deleterious nsSNPs predicted by SIFT or PolyPhen | Number of deleterious nsSNPs predicted by SIFT and PolyPhen |
|---|---|---|---|---|---|---|
| UGT1A1 | 32 | UGT1A1 | 38 | UGT1A1 | 38 | 32 |
| UGT1A3 | 8 | UGT1A3 | 10 | UGT1A3 | 11 | 7 |
| UGT1A4 | 3 | UGT1A4 | 7 | UGT1A4 | 7 | 3 |
| UGT1A5 | 5 | UGT1A5 | 6 | UGT1A5 | 7 | 4 |
| UGT1A6 | 3 | UGT1A6 | 4 | UGT1A6 | 5 | 4 |
| UGT1A7 | 2 | UGT1A7 | 3 | UGT1A7 | 3 | 2 |
| UGT1A8 | 8 | UGT1A8 | 10 | UGT1A8 | 10 | 8 |
| UGT1A9 | 6 | UGT1A9 | 7 | UGT1A9 | 7 | 6 |
| UGT1A10 | 7 | UGT1A10 | 8 | UGT1A10 | 9 | 6 |
| UGT2A1 | 1 | UGT2A1 | 1 | UGT2A1 | 1 | 1 |
| UGT2A2 | 1 | UGT2A2 | 1 | UGT2A2 | 1 | 1 |
| UGT2A3 | 0 | UGT2A3 | 0 | UGT2A3 | 0 | 0 |
| UGT2B4 | 4 | UGT2B4 | 3 | UGT2B4 | 4 | 3 |
| UGT2B7 | 2 | UGT2B7 | 3 | UGT2B7 | 3 | 2 |
| UGT2B10 | 2 | UGT2B10 | 4 | UGT2B10 | 4 | 2 |
| UGT2B11 | 3 | UGT2B11 | 3 | UGT2B11 | 3 | 3 |
| UGT2B15 | 0 | UGT2B15 | 0 | UGT2B15 | 0 | 0 |
| UGT2B28 | 0 | UGT2B28 | 2 | UGT2B28 | 2 | 0 |
| UGT3A1 | 0 | UGT3A1 | 1 | UGT3A1 | 1 | 0 |
| UGT3A2 | 1 | UGT3A2 | 2 | UGT3A2 | 2 | 1 |
| UGT8 | 0 | UGT8 | 0 | UGT8 | 0 | 0 |
| Total | 88 | 113 | 118 | 85 |
Potential Effect of nsSNPs of UGTs on Amino Acid Change
Representative deleterious nsSNPs and the corresponding amino acid substitution of various UGT genes are listed in Table IV. As many as 31 UGT1A1 nsSNPs of 80 UGT nsSNPs were predicted as deleterious by both algorithms. For UGT1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, and 1A10, deleterious nsSNPs predicted by both SIFT and PolyPhen includes Arg45Trp, Ile322Thr, Leu60His, Ser69Tyr, Ile318Thr, Thr202Ala, Val264Glu, and Ile211Thr. Thirteen UGT nsSNPs (Gly307Arg, Gly309Arg, Phe396Leu, Asp457Glu, Leu46Pro, Ser299Phe, Ala381Thr, Leu497Pro, Cys155Arg, Pro288Leu, and Lys367Ile) predicted deleterious by both SIFT and PolyPhen algorithms were from the UGT2 family, including UGT2A1, 2A2, 2B4, 2B7, 2B10, and 2B11. For UGT3A2, Ala343Thr was predicted as deleterious, respectively, by both SIFT and PolyPhen algorithms.
Table IV.
Deleterious nsSNPs Predicted by Both SIFT and PolyPhen
| Gene symbol | SNP ID | Amino acid change | Gene symbol | SNP ID | Amino acid change |
|---|---|---|---|---|---|
| UGT1A1 | VAR_026134 | Pro34Gln | UGT1A5 | rs17851756 | Ile322Thr |
| UGT1A1 | VAR_026135 | His39Asp | UGT1A5 | rs28934877 | Asn400Asp |
| UGT1A1 | – | Gln185Pro | UGT1A6 | rs1042708 | Ser69Tyr |
| UGT1A1 | VAR_007698 | Arg209Trp | UGT1A6 | rs17851756 | Ile320Thr |
| UGT1A1 | VAR_007699 | Gly276Arg | UGT1A6 | rs28934877 | Asn398Asp |
| UGT1A1 | VAR_026138 | Glu291Val | UGT1A6 | rs34993780 | Tyr485Asp |
| UGT1A1 | VAR_026139 | Ile294Thr | UGT1A6 | rs1042709 | Ala509Pro |
| UGT1A1 | VAR_007701 | Gly308Glu | UGT1A7 | rs17851756 | Ile318Thr |
| UGT1A1 | rs17851756 | Ile322Thr | UGT1A7 | rs28934877 | Asn396Asp |
| UGT1A1 | VAR_007702 | Gln331Arg | UGT1A8 | rs45504099 | His53Asn |
| UGT1A1 | VAR_026140 | Arg336Leu | UGT1A8 | VAR_015546 | Thr202Ala |
| UGT1A1 | VAR_026142 | Arg336Trp | UGT1A8 | rs17863762 | Cys276Tyr |
| UGT1A1 | VAR_026143 | Trp354Arg | UGT1A8 | rs17851756 | Ile318Thr |
| UGT1A1 | VAR_007703 | Gln357Arg | UGT1A8 | rs34946978 | Pro361Leu |
| UGT1A1 | – | Pro364Leu | UGT1A8 | rs55750087 | Arg364Gly |
| UGT1A1 | VAR_012283 | Arg367Gly | UGT1A8 | rs28934877 | Asn396Asp |
| UGT1A1 | VAR_007704 | Ala368Thr | UGT1A8 | rs34993780 | Tyr483Asp |
| UGT1A1 | VAR_007705 | Ser375Phe | UGT1A9 | rs4663870 | Val264Glu |
| UGT1A1 | VAR_026144 | His376Arg | UGT1A9 | rs17851756 | Ile318Thr |
| UGT1A1 | VAR_026145 | Gly377Val | UGT1A9 | rs34946978 | Pro361Leu |
| UGT1A1 | VAR_007706 | Ser381Arg | UGT1A9 | rs55750087 | Arg364Gly |
| UGT1A1 | VAR_026146 | Pro387Ser | UGT1A9 | rs28934877 | Asn396Asp |
| UGT1A1 | VAR_026147 | Gly395Val | UGT1A9 | rs34993780 | Tyr483Asp |
| UGT1A1 | VAR_019412 | Asn400Asp | UGT1A10 | – | Ile211Thr |
| UGT1A1 | VAR_007707 | Ala401Pro | UGT1A10 | rs17851756 | Ile318Thr |
| UGT1A1 | VAR_026148 | Arg403Cys | UGT1A10 | rs34946978 | Pro361Leu |
| UGT1A1 | VAR_026149 | Trp461Arg | UGT1A10 | rs55750087 | Arg364Gly |
| UGT1A1 | – | Glu463Ala | UGT1A10 | rs28934877 | Asn396Asp |
| UGT1A1 | VAR_026150 | Ala478Asp | UGT1A10 | rs34993780 | Tyr483Asp |
| UGT1A1 | VAR_007709 | Tyr486Asp | UGT2A1 | rs4148301 | Gly307Arg |
| UGT1A1 | – | Leu496Asn | UGT2A2 | rs4148301 | Gly309Arg |
| UGT1A3 | rs45625338 | Arg45Trp | UGT2B4 | VAR_011329 | Phe396Leu |
| UGT1A3 | rs45595237 | Arg49Trp | UGT2B4 | rs13119049 | Asp457Glu |
| UGT1A3 | rs17851756 | Ile322Thr | UGT2B4 | rs41298245 | Cys511Arg |
| UGT1A3 | rs34946978 | Pro365Leu | UGT2B7 | rs61361928 | Leu46Pro |
| UGT1A3 | rs55750087 | Arg368Gly | UGT2B7 | rs34620993 | Ser299Phe |
| UGT1A3 | rs28934877 | Asn400Asp | UGT2B10 | rs4095564 | Ala381Thr |
| UGT1A3 | rs34993780 | Tyr487Asp | UGT2B10 | rs3208727 | Leu497Pro |
| UGT1A4 | rs17851756 | Ile322Thr | UGT2B11 | rs7697037 | Cys155Arg |
| UGT1A4 | rs28934877 | Asn400Asp | UGT2B11 | rs3890590 | Pro288Leu |
| UGT1A4 | VAR_009507 | Tyr487Asp | UGT2B11 | rs11936018 | Lys367Ile |
| UGT1A5 | rs28898602 | Leu60His | UGT3A2 | rs2591714 | Ala343Thr |
| UGT1A5 | rs41270755 | Arg49Trp |
Concordance Analysis of Predicted Results by SIFT and PolyPhen
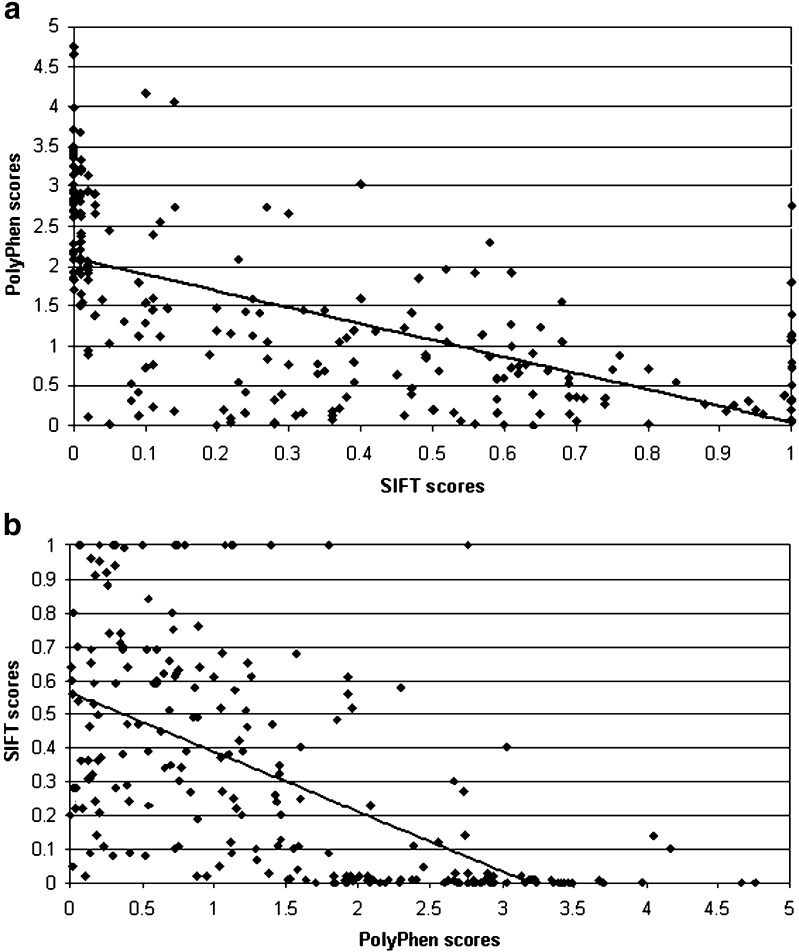
Table V shows the concordance analysis between the functional consequences for 247 nsSNPs predicted by SIFT and PolyPhen. Raw scores rather than arbitrarily defined categories were used for the correlation analysis. The row percentage of PolyPhen scores that falls into a SIFT category was calculated. Remarkably, 85.7% of “benign” predictions from PolyPhen falls into SIFT “tolerant” predictions and 84.2% of “probably damaging” predictions from PolyPhen falls into SIFT “intolerant” predictions. Scatter graphs plotted using 247 prediction scores from SIFT and PolyPhen showed negative correlation (Fig. 1). Spearman’s rank correlation coefficient ρ = −0.709 (P ≤ 0.01) illustrate the significant concordance between the prediction scores from SIFT and PolyPhen algorithms.
Table V.
Concordance Analysis Between the Functional Consequences of each nsSNP Predicted by SIFT and Polyphen
| SIFT prediction | ||||||
|---|---|---|---|---|---|---|
| Tolerant | Borderline | Potentially intolerant | Intolerant | Total | ||
| 1.000 ~ 0.201 | 0.200 ~ 0.101 | 0.100 ~ 0.050 | 0.049 ~ 0.000 | |||
| Polyphen prediction | ||||||
| Benign | 0.00 ~ 0.99 | 84 (85.7%) | 5 (5.1%) | 6 (6.1%) | 3 (3.1%) | 98 |
| Borderline | 1.00 ~ 1.24 | 17 (81%) | 2 (9.5%) | 2 (9.5%) | 0 | 21 |
| Potentially damaging | 1.25 ~ 1.49 | 7 (53.8%) | 3 (23.1%) | 2 (15.4%) | 1 (7.7%) | 13 |
| Possibly damaging | 1.50 ~ 1.99 | 8 (24.2%) | 8 (24.2%) | 2 (6.1%) | 15 (45.5%) | 33 |
| Probably damaging | ≥2.00 | 6 (7.3%) | 5 (6.1%) | 2 (2.4%) | 69 (84.2%) | 82 |
| Total | 122 | 23 | 14 | 88 | 247 | |
Fig. 1.
Scatter graph displaying the negative correlation between prediction scores for nsSNPs in human UGT genes using SIFT and PolyPhen programs a and b
Validation of the Prediction Results
The confirmed phenotype of nsSNPs manifests as alteration of enzyme activity and susceptibility to diseases such as CN I, CN II and Gilbert’s syndrome. Up to date, a total of 63 nsSNPs of human UGT genes are identified to demonstrate a relation to decreased activity, loss of enzyme activity, or susceptibility to diseases based on experimental and clinical studies. Using positive findings from the experiments, if the variants were predicted to be deleterious, it is considered a correct prediction. An incorrect/error prediction was considered when such nsSNPs were predicted as tolerant.
The confirmed variants were collected from results derived from site-directed mutagenesis studies of the enzyme using biochemical characterization (18,39–47) or clinical data from family-based and association studies (48–56). The biochemical/in vitro and in vivo UGT variants and the predictions for their functional impact scores by SIFT and PolyPhen are displayed in Table VI. If a confirmed UGT nsSNP was predicted as “intolerant” by SIFT and “probably damaging” by PolyPhen, then it is termed “true positive finding”. If a confirmed UGT nsSNP was predicted as “tolerated” by SIFT and “benign” by PolyPhen then it is termed “false negative finding”. According to these criteria, approximately 57% and 67% of characterized UGT nsSNPs were correctly predicted as deleterious and are “true positive predictions” by SIFT and PolyPhen, respectively (Table VII). SIFT predicted 27 confirmed UGT alleles as “tolerated” and Polyphen predicted 21 confirmed UGT alleles as “benign”, based on this finding, false negative error for SIFT and PolyPhen was 33% and 43%, respectively (Table VII). Ninety-seven percent of confirmed allelic variants of human UGTs were distributed in the UGT1A family, with two additional variants from the UGT2B family. Even more interesting is UGT1A1 alone contributes 74.6% of the confirmed allelic variants of human UGTs, this correlates with the fact that UGT1A1 was the top UGT gene with most frequent deleterious nsSNPs predicted by both SIFT and PolyPhen.
Table VI.
Confirmed Allelic Variants of Human UGT Genes using In Vivo and In Vitro Studies
| Allele naming | Residue 1 | Position | Residue 2 | SNP Id | Phenotype | Enzyme activity | Site directed mutagenesis | SIFT prediction score | PolyPhen prediction score | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| In vitro | In vivo | ||||||||||
| UGT1A1*3 | Ser | 375 | Phe | VAR_007705 | CN1 | Inactive | Inactive | Yes | 0.02 | 1.949 | (18,43) |
| UGT1A1*6 | Gly | 71 | Arg | rs4148323 | Reduced | Reduced | Yes | 0.1 | 1.285 | (45,57) | |
| UGT1A1*7 | Tyr | 486 | Asp | VAR_007709 | CN2 | Reduced | Reduced | Yes | 0 | 3.39 | (45,57,59) |
| UGT1A1*8 | Arg | 209 | Trp | VAR_007698 | CN2 | 4.40% | Reduced | Yes | 0 | 3.408 | (58,70) |
| UGT1A1*9 | Gln | 331 | Arg | VAR_007702 | CN2 | Reduced | Reduced | Yes | 0 | 2.814 | (17) |
| UGT1A1*11 | Gly | 308 | Glu | VAR_007701 | CN1 | Inactive | Absent | Yes | 0 | 2.946 | (43) |
| UGT1A1*12 | Leu | 175 | Gln | VAR_019411 | CN2 | 38.40% | Reduced | Yes | 0.1 | 1.547 | (46) |
| UGT1A1*14 | Gly | 276 | Arg | VAR_007699 | CN1 | Inactive | Inactive | Yes | 0 | 2.6 | (46) |
| UGT1A1*15 | Cys | 177 | Arg | VAR_007697 | CN1 | Inactive | Inactive | Yes | 0.14 | 2.739 | (46) |
| UGT1A1*16 | Gln | 357 | Arg | VAR_007703 | CN1 | Absent | Absent | No | 0.01 | 2.105 | (44) |
| UGT1A1*17 | Ser | 381 | Arg | VAR_007706 | CN1 | Absent | Absent | No | 0.01 | 1.507 | (44) |
| UGT1A1*18 | Ala | 401 | Pro | VAR_007707 | CN1 | Absent | Absent | No | 0.02 | 1.92 | (44) |
| UGT1A1*20 | Ala | 368 | Thr | VAR_007704 | CN1 | Absent | Absent | No | 0.01 | 1.957 | (44) |
| UGT1A1*22 | Ala | 291 | Val | VAR_026138 | CN1 | Absent | Absent | No | 0 | 2.766 | (44) |
| UGT1A1*23 | Lys | 428 | Glu | VAR_007708 | CN1 | Absent | Absent | No | 0.09 | 1.124 | (44) |
| UGT1A1*27 | Pro | 229 | Gln | VAR_009505 | Gilbert | Reduced | Reduced | No | 0.3 | 0.759 | (39) |
| UGT1A1*29 | Arg | 367 | Gly | VAR_012283 | Gilbert | Reduced | Reduced | No | 0 | 2.94 | (39) |
| UGT1A1*30 | Leu | 15 | Arg | VAR_019410 | CN2 | Reduced | Reduced | Yes | 0.16 | No score | (47) |
| UGT1A1*31 | Pro | 387 | Arg | VAR_026146 | CN1 | Absent | Absent | Yes | 0 | 3.158 | (41) |
| UGT1A1*32 | Arg | 336 | Trp | VAR_026142 | CN1 | 0–10% | Absent | Yes | 0 | 3.235 | (40) |
| UGT1A1*33 | Ile | 294 | Thr | VAR_026139 | CN2 | 40–55% | Yes | 0 | 1.839 | (40) | |
| UGT1A1*34 | Met | 310 | Val | – | CN2 | 26–51% | Yes | 0.52 | 1.956 | (42) | |
| UGT1A1*351 | Ile | 431 | Thr | – | CN2 | 61–81% | Yes | 0.26 | 1.42 | (42) | |
| UGT1A1*38 | Asn | 400 | Asp | VAR_019412 | CN2 | No | 0.01 | 2.623 | (52) | ||
| UGT1A1*42 | Glu | 463 | Ala | – | CN2 | No | 0 | 2.702 | (48) | ||
| UGT1A1*44 | His | 39 | Asp | VAR_026135 | CN1 | Yes | 0.01 | 3.673 | (51) | ||
| UGT1A1*51 | His | 376 | Arg | VAR_026144 | CN2 | Yes | 0.01 | 2.669 | (51) | ||
| UGT1A1*52 | Gly | 377 | Val | VAR_026145 | CN2 | Yes | 0 | 2.847 | (51) | ||
| UGT1A1*55 | Leu | 496 | Asn | – | CN1 | Yes | 0.01 | 2.375 | (51) | ||
| UGT1A1*62 | Phe | 83 | Leu | VAR_026136 | Gilbert | Yes | 0.58 | 0.863 | (56) | ||
| UGT1A1*63 | Pro | 364 | Leu | – | – | Reduced | – | No | 0.01 | 3.219 | (50) |
| UGT1A1*69 | Ile | 159 | Thr | – | Gilbert | – | – | Yes | 0.2 | 1.46 | (49) |
| UGT1A1*70 | Ala | 321 | Gly | – | Gilbert | – | – | Yes | 0.37 | 1.051 | (49) |
| UGT1A1*72 | Asp | 359 | Asn | – | Gilbert | – | – | Yes | 0.05 | 2.45 | (49) |
| UGT1A1*73 | Pro | 364 | Leu | – | Gilbert | – | – | Yes | 0.01 | 3.219 | (49) |
| UGT1A1*94 | Trp | 461 | Arg | VAR_026149 | CN1 | – | No detectable activity | Yes | 0 | 4.657 | (53) |
| UGT1A1*95 | Arg | 336 | Gln | VAR_026141 | CN1 | – | – | Yes | 0.11 | 1.445 | (55) |
| UGT1A1*96 | Arg | 336 | Leu | VAR_026140 | CN1 | – | – | Yes | 0 | 3.03 | (55) |
| UGT1A1*97 | Gly | 395 | Val | VAR_026147 | CN1 | – | – | Yes | 0.01 | 2.292 | (55) |
| UGT1A1*98 | Pro | 387 | Ser | VAR_026146 | CN1 | – | – | Yes | 0 | 3.158 | (55) |
| UGT1A1*101 | Trp | 354 | Arg | VAR_026143 | CN2 | – | – | Yes | 0 | 4.753 | (55) |
| UGT1A1*102 | Pro | 34 | Gln | VAR_026134 | CN2 | – | – | Yes | 0 | 2.803 | (55) |
| UGT1A1*103 | Arg | 403 | Cys | VAR_026148 | CN2 | – | – | Yes | 0 | 2.92 | (55) |
| UGT1A1*106 | Ala | 478 | Asp | VAR_026150 | CN2 | – | – | Yes | 0 | 2.149 | (55) |
| UGT1A1*107 | Trp | 40 | Arg | – | CN2 | – | – | Yes | 0.14 | 4.05 | (54) |
| UGT1A1*109 | Gln | 18 | Pro | – | CN2 | – | – | Yes | 0.03 | 1.535 | (54) |
| UGT1A1*113 | Gly | 493 | Arg | – | CN2 | – | – | No | 0.23 | 2.086 | (71) |
| UGT1A3*4a | Arg | 45 | Trp | – | Low | Yes | 0.01 | 2.078 | (72) | ||
| UGT1A3*7a | Phe | 110 | Ile | – | Intermediate | Yes | 0.39 | 0.804 | (73) | ||
| UGT1A3*8a | Ala | 158 | Val | – | Low | Yes | 0.54 | 1.223 | (73) | ||
| UGT1A3*10a | Val | 47 | Ala | rs6431625 | Intermediate | Yes | 0.1 | 0.726 | (73) | ||
| UGT1A4*2 | Pro | 24 | Thr | – | Substrate dependent | No | 0.58 | 2.3 | (74) | ||
| UGT1A4*3b | Leu | 48 | Val | – | Low activity | Yes | 1 | 0.072 | (75) | ||
| UGT1A7*4 | Trp | 208 | Arg | rs11692021 | Low | Yes | 0.39 | 1.199 | (76) | ||
| UGT1A7*5 | Gly | 115 | Ser | – | Low | Yes | 0.74 | 0.262 | (77) | ||
| UGT1A7*6 | Glu | 139 | Asp | – | High | Yes | 0.59 | 0.318 | (77) | ||
| UGT1A8*3 | Cys | 277 | Tyr | rs17863762 | Severely reduced | Yes | 0 | 3.076 | (78) | ||
| UGT1A9*3a | Met | 33 | Thr | – | Substrate-dependent impact | Yes | 1 | 1.074 | (77) | ||
| UGT1A9*5 | Asp | 256 | Asn | – | Decreased activity | Yes | 0.61 | 0.994 | (79) | ||
| UGT1A10*6 | Thr | 202 | Ile | VAR_018355 | Reduced by 50% | Yes | 0.03 | 1.377 | (79,80) | ||
| UGT1A10*7 | Ile | 211 | Thr | No activity | Yes | 0.02 | 2.003 | (81) | |||
| UGT2B10*2 | Asp | 67 | Tyr | – | Associated with lower level of nicotine and cotinine glucuronidation in HLM | No | 0.08 | 0.524 | (60) | ||
| UGT2B15*2 | Asp | 85 | Tyr | rs1902023 | Increased activity on androgens | No | 0.08 | 0.299 | (61) | ||
Table VII.
Evaluation of Predicting Effect of SIFT and PolyPhen on Confirmed Human UGT nsSNPs
| Number | % | |
|---|---|---|
| SIFT prediction | ||
| True positive predictions | 36 | 57 |
| False negative predictions | 27 | 43 |
| Total | 63 | |
| PolyPhen prediction | ||
| True positive predictions | 42 | 67 |
| False negative predictions | 21 | 33 |
| Total | 63 | |
Based on the results of in vivo and in vitro research, among 63 functionally characterized nsSNPs in the UGTs, there were 24 showing altered enzyme expression/activities and 45 associated with disease susceptibility. In agreement with earlier studies, reduced levels of UGT enzyme activities were observed in CN II and Gilbert’s syndrome, whilst absence of UGT enzyme activities were observed only in CN I patients. Additional deleterious nsSNPs of human UGT were predicted by SIFT and PolyPhen but the phenotypic prediction of these nsSNPs has not yet been confirmed using experiments.
More than two-thirds of confirmed variants were UGT1A1 variants (Table VI). In the confirmed variants of UGT1A1, the associated phenotypes were CN I, CN II (15), and Gilbert’s syndrome (16) which are all closely related to bilirubin levels. To date, 47 mutant UGT1A1 alleles have been identified (Table VI; 17,18,39,42–51,53–59). Many of these SNPs were predicted to have phenotypical effects by the algorithms and the correct prediction rates were 68% and 81% by SIFT and PolyPhen, respectively.
A number of the confirmed allelic variants of UGT1A1 associated with Gilbert’s syndrome, including UGT1A1*27 (Pro229Gln), *29 (Arg367Gly), *62 (Phe83Leu), *69 (Ile159Thr), *70 (Ala321Gly), *72 (Asp359Asn), and *73 (Pro364Leu). Both SIFT and PolyPhen correctly predicted the phenotype of Por364Leu. In addition, UGT1A1*7 (Tyr486Asp) and *9 (Gln331Arg) closely associated with Crigler-Najjar II syndrome (3,18,45,59), were predicted as probably damaging with high PolyPhen scores. Furthermore, another UGT allelic variant UGT1A1*11 (Gly308Glu) identified in Crigler-Najjar I syndrome (3,43), was also correctly predicted by both SIFT and PolyPhen algorithms.
Only two variant alleles of the human UGT2 mRNAs have been confirmed in the UGT2 gene family, which are Asp67Tyr and Asp85Tyr (60,61). Hyperbilirubinemia can be caused by a number of reasons including liver disease and hemolysis (62). Rate of production of bilirubin also needs to be considered when bilirubin level is abnormal, e.g. in inherited hemolytic syndromes such as glucose 6-phosphate dehydrogenase deficiency and sickle cell anemia. UGT2B7 promoter variant 840G>A is associated with significantly reduced glucuronidation of morphine in sickle cell disease and contributes to the variability in hepatic clearance of morphine in these patients (28). UGT2B15 variant (Asp85Tyr) have been shown to relate to prostate cancer risk (30,63).
DISCUSSION
Humans have vast numbers of SNPs and presently there are more than a million SNPs in dbSNP that can be screened for association with diseases. Prediction tool SIFT was tested against unbiased experimental datasets in which mutagenesis was performed throughout the entire protein, and both wild-type and negative phenotypes were assayed, only three datasets fit the criteria and the scarcity of unbiased data sets indicates how difficult characterization of mutant proteins on a large scale can be (31). Therefore, determining disease causing SNPs via site-directed mutagenesis experiments and gene knockout/knockin experiments is complex, lengthy, and unrealistic taking into consideration the mass amount of SNPs (64). Using bioinformatic tools to predict nsSNPs most likely to be damaging, this process acts as a filter and reduces the number of SNPs required to be screened for association with disease to those that most likely alter gene function (8,9). By predicting nsSNPs of UGT, we are able to distinguish mutations that are unlikely to affect protein function (9), reducing the number of nsSNPs for experimentation thus saving time and resources. Based on prediction results of deleterious nsSNPs, on a much smaller scale we then via experiments and clinical studies are able to screen for polymorphisms of UGT that may potentially cause disease and increase drug toxicity. Moreover, the detection of individual single-nucleotide polymorphisms that alter enzyme function can be a useful tool for the identification of disease risk or to personalize drug regimens (65,66).
A total number of 248 nsSNPs were identified through the screening of 21 human UGT genes from NCBI dbSNP, Mutdb, and Polydoms. However, according to published results from in vivo and in vitro studies, only about a quarter of nsSNPs in the dataset of validated human UGT genes were found to attribute to alteration of enzyme activity and correlation with diseases. These confirmed phenotypes of nsSNPs were related to reduced or absence of enzyme activity, and correlated to susceptibility to diseases such as CN, Gilbert’s syndrome and hyperbilirubinemia related to certain therapeutic drugs.
There are 49 non-synonymous SNPs in exons 1–5 of UGT1A1 (Supplementary Table 1). To date, 47 mutant UGT1A1 alleles have been identified (17,18,39,42–51,53–59). Deleterious nsSNPs of UGT1A1 such as Ser375Phe, Gly308Glu, Ala291Val, and His39Asp which are phenotypically presented as CN I were all correctly predicted by both SIFT and PolyPhen. The Tyr486Asp (UGT1A1*7) mutation in exon 1 and is the most abundant mutation in CN II in Japanese patients (67), was correctly predicted as deleterious nsSNP by both SIFT and PolyPhen. Variant UGT1A1*27 (Pro229Gln) identified in CN II with reduced UGT enzyme activities in vitro and in vivo was not predicted to be deleterious by SIFT and PolyPhen. In seven confirmed alleles associated with Gilbert’s syndrome (UGT1A1*27, *29, *62, *69, *70, *72, and *73), only Arg367Gly and Pro364Leu were correctly predicted as deleterious nsSNPs of UGT1A1. Other examples of incorrect predictions includes UGT1A1*6 (Gly71Arg) and UGT1A1*15 (Cys177Arg). Overall, SIFT and PolyPhen predicted UGT1A1 SNPs to have phenotypical effects with correct prediction rates of 68% and 81%, respectively.
SIFT and PolyPhen are in silico algorithm tools that use protein sequence alignment, physicochemical differences, mapping to know protein 3-D structures to predict the functional impact of nsSNPs on protein structure and activities (8,9,31,33). Even though SIFT and PolyPhen employ different approaches and types of reference data for their predictions and different scales for scoring, there was significant concordance observed on functional consequences of each nsSNP prediction on human UGTs (Spearman’s ρ = –0.709, P ≤ 0.01). SIFT and PolyPhen can discriminate diseased variants from neutral variants, results from this study are consistent with reports by Ng and Henikoff (9) who predicted 757 of 3,084 or 25% nsSNPs from dbSNP to be damaging and having protein activity impacts. Ramensky et al. (8) correctly predicted 27.6% of nsSNPs to affect protein function in the human genome variation database. Xi et al. (38) reported 30–50% of over 500 amino acid substitution variants identified in DNA repair genes were predicted to exhibit reduced activity. Zhang et al. (68) studied variants in H+/peptide cotransporter (PEPT1) involved in drug transportation, SIFT correctly predicted the SNP that reduced transport capacity to “affect protein function”, confirming the accuracy of SIFT on prediction of individual protein predictions. Our recent study on phenotype prediction of 791 validated nsSNPs in human cytochrome P450s using SIFT and PolyPhen, found that 70% of nsSNPs were correctly predicted as damaging (69).
Furthermore, the scatter graph plotted using functional consequence predictions from SIFT and PolyPhen showed negative correlation between the two sets of scores. Low scores from SIFT indicate an “intolerant” prediction, correlated with high scores from PolyPhen which indicates a “probably damaging” prediction. High scores from SIFT indicate a “tolerant” prediction, correlated with low scores from PolyPhen indicating a “benign” prediction. This correlation was further established by row percentage calculated between scores for each one of 247 nsSNPs predicted by SIFT and PolyPhen. The two algorithms show agreements at two extremes of the scores, a high percentage (85.7%) of “benign” predictions from PolyPhen was predicted “tolerant” by SIFT and a high percentage (84.2%) of “probably damaging” predictions from PolyPhen was predicted “intolerant” by SIFT. There were slight correlations for scores in between, showing a lack of agreement crossing different prediction categories of the two algorithms, demonstrating a strong correlation in their predictions on potential effect of known human UGT nsSNPs on protein function. The concordance between SIFT and PolyPhen prediction scores suggest they can be used in combination in the future to improve accuracy of prediction studies.
Although bioinformatics tools show their potential in reducing the number of nsSNPs for disease association studies by filtering nsSNPs that are most likely to be disease related, error predictions do occur. In this study, based on collected data from 63 in vitro and in vivo studies, the false negative predictions for SIFT and PolyPhen is 43% and 33%, respectively. There are several aspects affecting the prediction accuracy for prediction tools like SIFT and PolyPhen algorithms. Firstly, SIFT and PolyPhen rely on several different databases for SNP information, polluted databases with erroneous SNP reports and bias of the data towards disease-related allelic variants are likely to lead to an over prediction of the number of deleterious nsSNPs (8). For example, Ramensky et al. (8) compared a fraction of nsSNPs predicted to be damaging for HGV-base entries and found that the overall prediction rate for the category “Suspected” nsSNPs was 31.4%, for the category “Proven” nsSNPs was 28.9% and for “Proven” nsSNPs from systematic studies on healthy individual was 27.6%. Furthermore, programs identifying SNPs may detect base differences between the functional gene and a pseudogene and erroneously report these differences as SNPs in the function protein. Including nsSNPs erroneously mapped from pseudogenes in the SNP database will affect prediction accuracy on prediction tools using SNP information from these databases (9). The growth of public SNP data and improvements of SNP database data will assist in acquiring correct information on SNPs to assist in improving prediction accuracy of bioinformatics tools. Secondly, SIFT has a weighted false positive error of 19% and PolyPhen 9%, indicating that if all of the nsSNPs from dbSNP were functionally neutral, 19% or 9% would have been predicted as damaging by SIFT and PolyPhen, respectively (9). Additionally, there are factors overlooked when the predictions occurs, SIFT predicts on amino acid substitution in the protein product and does not take into account mutations that affect transcription, translation, splicing, and other possible pretranslational alterations (9), erroneous predictions may then arise. Furthermore, the prediction might appear incorrect based on the lack of association with obvious altered phenotypes. SIFT and PolyPhen may be sensitive to a mutation and predict it to be damaging to protein function, but if the phenotype is undiagnosed, or has not yet been assayed for therefore condemned an “error prediction”. On the other hand, SIFT and PolyPhen may predict a deleterious mutation to be “tolerated” and because it is not an obvious phenotype, we believe that it is a correct prediction whereas in actual fact it is a deleterious mutation that is recessive or undiagnosed. Therefore it is important to identify the association of SNPs with various phenotype/diseases. Identifying deleterious nsSNPs using bioinformatics tools like SIFT and PolyPhen may lay the foundation and initiate the process of identifying SNPs that has clinical implications. There lies the ability to reduce the number and/or the field of possible deleterious nsSNPs that needs to be evaluated in clinical studies.
In summary, in silico analysis predicted that 35.3–46% of over 200 amino acid substitution variants currently identified in the human UGT genes might cause reduced enzyme activity and/or elevated bilirubin levels. The identification of these variants using bioinformatic tools such as SIFT and PolyPhen narrows down study fields and is the first step of the progress to evaluate their possible phenotypic importance in costly clinical studies.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
(XLS 158 kb)
Acknowledgements
The authors appreciate the technical assistance of Dr Lin-Lin Wang of Institute of Reproductive and Child Health, Peking University, Beijing, China. Ms. Yuan Ming Di is a holder of RMIT University PhD Scholarship. The authors appreciate the support of RMIT Health Innovations Research Institute, RMIT University, Bundoora, Victoria, Australia. We also would like to thank Associate Professor Clifford Da Costa (Department of Mathematics and Statistics, RMIT University, Melbourne, Australia) for his assistance in the statistical analysis of the data in this paper.
Abbreviations
- UGTs
UDP glucuronosyltransferase
- nsSNP
non-synonymous single-nucleotide polymorphism
References
- 1.Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3:136–158. doi: 10.1038/sj.tpj.6500171. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie PI, Owens IS, Burchell B, et al. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7:255–269. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 4.King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab. 2000;1:143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 5.Mackenzie PI, Bock KW, Burchell B, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 6.Harding D, Jeremiah SJ, Povey S, Burchell B. Chromosomal mapping of a human phenol UDP-glucuronosyltransferase, GNT1. Ann Hum Genet. 1990;54:17–21. doi: 10.1111/j.1469-1809.1990.tb00356.x. [DOI] [PubMed] [Google Scholar]
- 7.Turgeon D, Carrier JS, Levesque E, Hum DW, Belanger A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/en.142.2.778. [DOI] [PubMed] [Google Scholar]
- 8.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 11.Herrgard S, Cammer SA, Hoffman BT, et al. Prediction of deleterious functional effects of amino acid mutations using a library of structure-based function descriptors. Proteins. 2003;53:806–816. doi: 10.1002/prot.10458. [DOI] [PubMed] [Google Scholar]
- 12.Dobson RJ, Munroe PB, Caulfield MJ, Saqi MA. Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC Bioinformatics. 2006;7:217. doi: 10.1186/1471-2105-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehmer U, Vogel A, Schutte JK, Krone B, Manns MP, Strassburg CP. Variation of hepatic glucuronidation: novel functional polymorphisms of the UDP-glucuronosyltransferase UGT1A4. Hepatology. 2004;39:970–977. doi: 10.1002/hep.20131. [DOI] [PubMed] [Google Scholar]
- 14.Guillemette C, Millikan RC, Newman B, Housman DE. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60:950–956. [PubMed] [Google Scholar]
- 15.Crigler JF, Jr, Najjar VA. Congenital familial nonhemolytic jaundice with kernicterus. Pediatrics. 1952;10:169–180. [PubMed] [Google Scholar]
- 16.Black M, Billing BH. Hepatic bilirubin udp-glucuronyl transferase activity in liver disease and Gilbert’s syndrome. N Engl J Med. 1969;280:1266–1271. doi: 10.1056/NEJM196906052802303. [DOI] [PubMed] [Google Scholar]
- 17.Moghrabi N, Clarke DJ, Boxer M, Burchell B. Identification of an A-to-G missense mutation in exon 2 of the UGT1 gene complex that causes Crigler-Najjar syndrome type 2. Genomics. 1993;18:171–173. doi: 10.1006/geno.1993.1451. [DOI] [PubMed] [Google Scholar]
- 18.Bosma PJ, Chowdhury JR, Huang TJ, et al. Mechanisms of inherited deficiencies of multiple UDP-glucuronosyltransferase isoforms in two patients with Crigler-Najjar syndrome, type I. Faseb J. 1992;6:2859–2863. doi: 10.1096/fasebj.6.10.1634050. [DOI] [PubMed] [Google Scholar]
- 19.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–47. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 20.Ando Y, Saka H, Ando M, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 2000;60:6921–6926. [PubMed] [Google Scholar]
- 21.Wasserman E, Myara A, Lokiec F, et al. Severe CPT-11 toxicity in patients with Gilbert’s syndrome: two case reports. Ann Oncol. 1997;8:1049–1051. doi: 10.1023/A:1008261821434. [DOI] [PubMed] [Google Scholar]
- 22.Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33:458–465. doi: 10.1124/dmd.104.001800. [DOI] [PubMed] [Google Scholar]
- 23.Danoff TM, Campbell DA, McCarthy LC, et al. A Gilbert’s syndrome UGT1A1 variant confers susceptibility to tranilast-induced hyperbilirubinemia. Pharmacogenomics J. 2004;4:49–53. doi: 10.1038/sj.tpj.6500221. [DOI] [PubMed] [Google Scholar]
- 24.Strassburg CP, Vogel A, Kneip S, Tukey RH, Manns MP. Polymorphisms of the human UDP-glucuronosyltransferase (UGT) 1A7 gene in colorectal cancer. Gut. 2002;50:851–856. doi: 10.1136/gut.50.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Z, Park JY, Guillemette C, Schantz SP, Lazarus P. Tobacco carcinogen-detoxifying enzyme UGT1A7 and its association with orolaryngeal cancer risk. J Natl Cancer Inst. 2001;93:1411–1418. doi: 10.1093/jnci/93.18.1411. [DOI] [PubMed] [Google Scholar]
- 26.Vogel A, Ockenga J, Ehmer U, et al. Polymorphisms of the carcinogen detoxifying UDP-glucuronosyltransferase UGT1A7 in proximal digestive tract cancer. Z Gastroenterol. 2002;40:497–502. doi: 10.1055/s-2002-32805. [DOI] [PubMed] [Google Scholar]
- 27.Vogel A, Kneip S, Barut A, et al. Genetic link of hepatocellular carcinoma with polymorphisms of the UDP-glucuronosyltransferase UGT1A7 gene. Gastroenterology. 2001;121:1136–1144. doi: 10.1053/gast.2001.28655. [DOI] [PubMed] [Google Scholar]
- 28.Darbari DS, van Schaik RH, Capparelli EV, Rana S, McCarter R, van den Anker J. UGT2B7 promoter variant -840G>A contributes to the variability in hepatic clearance of morphine in patients with sickle cell disease. Am J Hematol. 2008;83:200–202. doi: 10.1002/ajh.21051. [DOI] [PubMed] [Google Scholar]
- 29.Holthe M, Rakvag TN, Klepstad P, et al. Sequence variations in the UDP-glucuronosyltransferase 2B7 (UGT2B7) gene: identification of 10 novel single nucleotide polymorphisms (SNPs) and analysis of their relevance to morphine glucuronidation in cancer patients. Pharmacogenomics J. 2003;3:17–26. doi: 10.1038/sj.tpj.6500139. [DOI] [PubMed] [Google Scholar]
- 30.MacLeod SL, Nowell S, Plaxco J, Lang NP. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7:777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- 31.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 34.Ng PC, Henikoff JG, Henikoff S. PHAT: a transmembrane-specific substitution matrix. Predicted hydrophobic and transmembrane. Bioinformatics. 2000;16:760–766. doi: 10.1093/bioinformatics/16.9.760. [DOI] [PubMed] [Google Scholar]
- 35.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 39.Aono S, Adachi Y, Uyama E, et al. Analysis of genes for bilirubin UDP-glucuronosyltransferase in Gilbert’s syndrome. Lancet. 1995;345:958–959. doi: 10.1016/S0140-6736(95)90702-5. [DOI] [PubMed] [Google Scholar]
- 40.Ciotti M, Chen F, Rubaltelli FF, Owens IS. Coding defect and a TATA box mutation at the bilirubin UDP-glucuronosyltransferase gene cause Crigler-Najjar type I disease. Biochim Biophys Acta. 1998;1407:40–50. doi: 10.1016/s0925-4439(98)00030-1. [DOI] [PubMed] [Google Scholar]
- 41.Ciotti M, Obaray R, Martin MG, Owens IS. Genetic defects at the UGT1 locus associated with Crigler-Najjar type I disease, including a prenatal diagnosis. Am J Med Genet. 1997;68:173–178. doi: 10.1002/(SICI)1096-8628(19970120)68:2<173::AID-AJMG10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Ciotti M, Werlin SL, Owens IS. Delayed response to phenobarbital treatment of a Crigler-Najjar type II patient with partially inactivating missense mutations in the bilirubin UDP-glucuronosyltransferase gene. J Pediatr Gastroenterol Nutr. 1999;28:210–213. doi: 10.1097/00005176-199902000-00024. [DOI] [PubMed] [Google Scholar]
- 43.Erps LT, Ritter JK, Hersh JH, Blossom D, Martin NC, Owens IS. Identification of two single base substitutions in the UGT1 gene locus which abolish bilirubin uridine diphosphate glucuronosyltransferase activity in vitro. J Clin Invest. 1994;93:564–570. doi: 10.1172/JCI117008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labrune P, Myara A, Hadchouel M, et al. Genetic heterogeneity of Crigler-Najjar syndrome type I: a study of 14 cases. Hum Genet. 1994;94:693–697. doi: 10.1007/BF00206965. [DOI] [PubMed] [Google Scholar]
- 45.Maruo Y, Nishizawa K, Sato H, Sawa H, Shimada M. Prolonged unconjugated hyperbilirubinemia associated with breast milk and mutations of the bilirubin uridine diphosphate-glucuronosyltransferase gene. Pediatrics. 2000;106:E59. doi: 10.1542/peds.106.5.e59. [DOI] [PubMed] [Google Scholar]
- 46.Seppen J, Bosma PJ, Goldhoorn BG, et al. Discrimination between Crigler-Najjar type I and II by expression of mutant bilirubin uridine diphosphate-glucuronosyltransferase. J Clin Invest. 1994;94:2385–2391. doi: 10.1172/JCI117604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seppen J, Steenken E, Lindhout D, Bosma PJ, Elferink RP. A mutation which disrupts the hydrophobic core of the signal peptide of bilirubin UDP-glucuronosyltransferase, an endoplasmic reticulum membrane protein, causes Crigler-Najjar type II. FEBS Lett. 1996;390:294–298. doi: 10.1016/0014-5793(96)00677-1. [DOI] [PubMed] [Google Scholar]
- 48.Chalasani N, Chowdhury NR, Chowdhury JR, Boyer TD. Kernicterus in an adult who is heterozygous for Crigler-Najjar syndrome and homozygous for Gilbert-type genetic defect. Gastroenterology. 1997;112:2099–2103. doi: 10.1053/gast.1997.v112.pm9178703. [DOI] [PubMed] [Google Scholar]
- 49.Farheen S, Sengupta S, Santra A, et al. Gilbert’s syndrome: high frequency of the (TA)7 TAA allele in India and its interaction with a novel CAT insertion in promoter of the gene for bilirubin UDP-glucuronosyltransferase 1 gene. World J Gastroenterol. 2006;12:2269–2275. doi: 10.3748/wjg.v12.i14.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang CS, Luo GA, Huang ML, Yu SC, Yang SS. Variations of the bilirubin uridine-diphosphoglucuronosyl transferase 1A1 gene in healthy Taiwanese. Pharmacogenetics. 2000;10:539–544. doi: 10.1097/00008571-200008000-00007. [DOI] [PubMed] [Google Scholar]
- 51.Kadakol A, Ghosh SS, Sappal BS, Sharma G, Chowdhury JR, Chowdhury NR. Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler-Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutat. 2000;16:297–306. doi: 10.1002/1098-1004(200010)16:4<297::AID-HUMU2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 52.Labrune P, Myara A, Chalas J, Le Bihan B, Capel L, Francoual J. Association of a homozygous (TA)8 promoter polymorphism and a N400D mutation of UGT1A1 in a child with Crigler-Najjar type II syndrome. Hum Mutat. 2002;20:399–401. doi: 10.1002/humu.10122. [DOI] [PubMed] [Google Scholar]
- 53.Maruo Y, Serdaroglu E, Iwai M, et al. A novel missense mutation of the bilirubin UDP-glucuronosyltransferase gene in a Turkish patient with Crigler-Najjar syndrome type 1. J Pediatr Gastroenterol Nutr. 2003;37:627–630. doi: 10.1097/00005176-200311000-00024. [DOI] [PubMed] [Google Scholar]
- 54.Petit F, Gajdos V, Capel L, et al. Crigler-Najjar type II syndrome may result from several types and combinations of mutations in the UGT1A1 gene. Clin Genet. 2006;69:525–527. doi: 10.1111/j.1399-0004.2006.00616.x. [DOI] [PubMed] [Google Scholar]
- 55.Servedio V, d’apolito M, Maiorano N, et al. Spectrum of UGT1A1 mutations in Crigler-Najjar (CN) syndrome patients: identification of twelve novel alleles and genotype-phenotype correlation. Hum Mutat. 2005;25:325. doi: 10.1002/humu.9322. [DOI] [PubMed] [Google Scholar]
- 56.Sutomo R, Laosombat V, Sadewa AH, et al. Novel missense mutation of the UGT1A1 gene in Thai siblings with Gilbert’s syndrome. Pediatr Int. 2002;44:427–432. [PubMed] [Google Scholar]
- 57.Aono S, Yamada Y, Keino H, et al. Identification of defect in the genes for bilirubin UDP-glucuronosyl-transferase in a patient with Crigler-Najjar syndrome type II. Biochem Biophys Res Commun. 1993;197:1239–1244. doi: 10.1006/bbrc.1993.2610. [DOI] [PubMed] [Google Scholar]
- 58.Huang CS, Luo GA, Huang MJ, Chen ES, Young TH, Chao YC. A novel compound heterozygous variation of the uridine-diphosphoglucuronosyl transferase 1A1 gene that causes Crigler-Najjar syndrome type II. Pharmacogenetics. 2001;11:639–642. doi: 10.1097/00008571-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 59.Udomuksorn W, Elliot DJ, Lewis BC, Mackenzie PI, Yoovathaworn K, Miners JO. Influence of mutations associated with Gilbert and Crigler-Najjar type II syndromes on the glucuronidation kinetics of bilirubin and other UDP-glucuronosyltransferase 1A substrates. Pharmacogenet Genomics. 2007;17:1017–1029. doi: 10.1097/FPC.0b013e328256b1b6. [DOI] [PubMed] [Google Scholar]
- 60.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 2007;67:9024–9029. doi: 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- 61.Levesque E, Beaulieu M, Green MD, Tephly TR, Belanger A, Hum DW. Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1997;7:317–325. doi: 10.1097/00008571-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 62.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 63.Gsur A, Preyer M, Haidinger G, et al. A polymorphism in the UDP-Glucuronosyltransferase 2B15 gene (D85Y) is not associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:497–498. [PubMed] [Google Scholar]
- 64.Li Y, Wang Y, Li Y, Yang L. Prediction of the deleterious nsSNPs in ABCB transporters. FEBS Lett. 2006;580:6800–6806. doi: 10.1016/j.febslet.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 65.Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- 66.Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: an opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999;20:342–349. doi: 10.1016/S0165-6147(99)01363-2. [DOI] [PubMed] [Google Scholar]
- 67.Yamamoto K, Soeda Y, Kamisako T, et al. Analysis of bilirubin uridine 5′-diphosphate (UDP)-glucuronosyltransferase gene mutations in seven patients with Crigler-Najjar syndrome type II. J Hum Genet. 1998;43:111–114. doi: 10.1007/s100380050050. [DOI] [PubMed] [Google Scholar]
- 68.Zhang EY, Fu DJ, Pak YA, et al. Genetic polymorphisms in human proton-dependent dipeptide transporter PEPT1: implications for the functional role of Pro586. J Pharmacol Exp Ther. 2004;310:437–445. doi: 10.1124/jpet.104.065912. [DOI] [PubMed] [Google Scholar]
- 69.Wang L. A bioinformatics approach for the phenotype prediction of non-synonymous single nucleotide polymorphisms in human cytochrome P450s. Drug Metab Dispos. 2009;37:977–91. [DOI] [PubMed]
- 70.Bosma PJ, Goldhoorn B, Oude Elferink RP, Sinaasappel M, Oostra BA, Jansen PL. A mutation in bilirubin uridine 5′-diphosphate-glucuronosyltransferase isoform 1 causing Crigler-Najjar syndrome type II. Gastroenterology. 1993;105:216–220. doi: 10.1016/0016-5085(93)90029-c. [DOI] [PubMed] [Google Scholar]
- 71.Yusoff S, Van Rostenberghe H, Yusoff NM, et al. Frequencies of A(TA)7TAA, G71R, and G493R mutations of the UGT1A1 gene in the Malaysian population. Biol Neonate. 2006;89:171–176. doi: 10.1159/000088844. [DOI] [PubMed] [Google Scholar]
- 72.Iwai M, Maruo Y, Ito M, Yamamoto K, Sato H, Takeuchi Y. Six novel UDP-glucuronosyltransferase (UGT1A3) polymorphisms with varying activity. J Hum Genet. 2004;49:123–128. doi: 10.1007/s10038-003-0119-y. [DOI] [PubMed] [Google Scholar]
- 73.Caillier B, Lepine J, Tojcic J, et al. A pharmacogenomics study of the human estrogen glucuronosyltransferase UGT1A3. Pharmacogenet Genomics. 2007;17:481–495. doi: 10.1097/FPC.0b013e32806d87a4. [DOI] [PubMed] [Google Scholar]
- 74.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–1196. doi: 10.1158/0008-5472.CAN-03-3219. [DOI] [PubMed] [Google Scholar]
- 75.Saeki M, Saito Y, Jinno H, et al. Genetic variations and haplotypes of UGT1A4 in a Japanese population. Drug Metab Pharmacokinet. 2005;20:144–151. doi: 10.2133/dmpk.20.144. [DOI] [PubMed] [Google Scholar]
- 76.Guillemette C, Ritter JK, Auyeung DJ, Kessler FK, Housman DE. Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene. Pharmacogenetics. 2000;10:629–644. doi: 10.1097/00008571-200010000-00006. [DOI] [PubMed] [Google Scholar]
- 77.Villeneuve L, Girard H, Fortier LC, Gagne JF, Guillemette C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther. 2003;307:117–128. doi: 10.1124/jpet.103.054072. [DOI] [PubMed] [Google Scholar]
- 78.Huang YH, Galijatovic A, Nguyen N, et al. Identification and functional characterization of UDP-glucuronosyltransferases UGT1A8*1, UGT1A8*2 and UGT1A8*3. Pharmacogenetics. 2002;12:287–297. doi: 10.1097/00008571-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 79.Jinno H, Saeki M, Saito Y, et al. Functional characterization of human UDP-glucuronosyltransferase 1A9 variant, D256N, found in Japanese cancer patients. J Pharmacol Exp Ther. 2003;306:688–693. doi: 10.1124/jpet.103.051250. [DOI] [PubMed] [Google Scholar]
- 80.Saeki M, Ozawa S, Saito Y, et al. Three novel single nucleotide polymorphisms in UGT1A10. Drug Metab Pharmacokinet. 2002;17:488–490. doi: 10.2133/dmpk.17.488. [DOI] [PubMed] [Google Scholar]
- 81.Martineau I, Tchernof A, Belanger A. Amino acid residue ILE211 is essential for the enzymatic activity of human UDP-glucuronosyltransferase 1A10 (UGT1A10) Drug Metab Dispos. 2004;32:455–459. doi: 10.1124/dmd.32.4.455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 158 kb)