Abstract
Human CYP1A2 is one of the major CYPs in human liver and metabolizes a variety of clinically important drugs (e.g., clozapine, tacrine, tizanidine, and theophylline), a number of procarcinogens (e.g. benzo[a]pyrene and aflatoxin B1), and several important endogenous compounds (e.g. steroids and arachidonic acids). Like many of other CYPs, CYP1A2 is subject to induction and inhibition by a number of compounds, which may provide an explanation for some drug interactions observed in clinical practice. A large interindividual variability in the expression and activity of CYP1A2 and elimination of drugs that are mainly metabolized by CYP1A2 has been observed, which is largely caused by genetic (e.g., SNPs) and epigenetic (e.g., DNA methylation) and environmental factors (e.g., smoking and comedication). CYP1A2 is primarily regulated by the aromatic hydrocarbon receptor (AhR) and CYP1A2 is induced through AhR-mediated transactivation following ligand binding and nuclear translocation. To date, more than 15 variant alleles and a series of subvariants of the CYP1A2 gene have been identified and some of they have been associated with altered drug clearance and response to drug therapy. For example, lack of response to clozapine therapy due to low plasma drug levels has been reported in smokers harboring the −163A/A genotype; there is an association between CYP1A2*1F (−163C>A) allele and the risk for leflunomide-induced host toxicity. The *1F allele is associated with increased enzyme inducibility whereas *1C causes reduced inducibility. Further studies are warranted to explore the clinical and toxicological significance of altered CYP1A2 expression and activity caused by genetic, epigenetic, and environmental factors.
Key words: CYP1A2, inducer, inhibitor, single nuclear polymorphism, substrate
INTRODUCTION
The CYP1A gene cluster has been mapped to chromosome 15, with a close link between CYP1A1 and 1A2 sharing a common 5′-flanking region (1). The CYP1A1 and 1A2 genes are separated by a 23-kb segment that contains no open-reading frames (1). Between CYP1A2 and CYP1A1, exons 2, 4, 6, and especially 5 are strikingly conserved in both nucleotides and total number of bases (2). Jaiswal et al. (3) were the first to isolate a cDNA corresponding to the CYP1A2 gene located on human chromosome 15. The human CYP1A2 gene spans almost 7.8 kb comprising seven exons and six introns (2), and the first exon is a 55-bp-long non-coding exon. CYP1A2 is a 515-residue protein with a molecular mass of 58,294 Da (2).
CYP1 family comprises three members, namely CYP1A1, 1A2, and 1B1, with CYP1A2 being one of the major CYPs in human liver (∼13–15%) (4). A large interindividual variability in the elimination of drugs that are metabolized by CYP1A2 has been observed, which has been ascribed to both genetic mutations and environmental factors (5–8). This review updates our current knowledge on the substrate specificity, regulation, structural features, and polymorphisms of human CYP1A2. To retrieve relevant data, the authors have searched through computer-based literatures by full text search in Medline (via Pubmed), ScienceDirect, Genetics Abstracts (CSA), SCOPUS, Chemical Abstracts, Current Contents Connect (ISI), Cochrane Library, CINAHL (EBSCO), CrossRef Search, and Embase (all from inception to 12 June 2009). Keyword search terms used included cytochrome P450, CYP1A2, substrate, inhibitor, inducer, structure, mutation, single nucleotide polymorphism (SNP), mutagenesis, genotype, phenotype, and allele, with combination terms pharmacokinetics, response, clearance, adverse reaction, drug interaction, toxicity, response, and human.
STRUCTURAL FEATURES AND FUNCTIONAL RELEVANCE OF CYP1A2
Homology modeling studies have suggested that most of the CYP1A2 substrates are hydrophobic with high LogP values, suggesting that hydrophobic interactions play an important role in their binding to CYP1A2 (9). Our modeling studies have indicated that the common features of ligands for CYP1A2 are of one to two hydrophobic regions, an aromatic ring and a hydrogen bond acceptor. A number of mutagenesis studies (10,11) have demonstrated that a series of residues in the substrate recognition sequence (SRS) regions of CYP1A2 (e.g., Arg108, Thr124, Thr223, Glu225, Phe226, Lys250, Arg251, Lys253, Asn312, Asp313, Glu318, Thr319, Asp320, Thr321, Val322, Leu382, Thr385, and Ile386) play important roles in substrate–enzyme interactions based on mutagenesis and homology modeling studies (Table I). Several residues in the non-SRS regions, including Lys99, Arg137, Gln141, Phe186, Phe205, Val227, Lys453, Arg455, and Thr501, also appear to play a role in the ligand–CYP1A2 interactions. The functional effect of mutants of these residues appears to be dependent on the substrate.
Table I.
Identified Amino Acid Residues of CYP1A2 that may Play a Role in CYP1A2–Ligand Interactions by Mutagenesis and/or Molecular Modeling Studies
| Residue | Possible location | Assays | References |
|---|---|---|---|
| Thr78 | – | Homology modeling | (79) |
| Thr87 | – | Homology modeling | (79) |
| Phe88 | – | Homology modeling | (79) |
| Lys94 | Helix B | SDM | (10) |
| Lys99 | Helix B | SDM | (80) |
| Lys105 | Helix B | SDM | (10) |
| Arg108 | SRS1 | Homology modeling | (81) |
| SDM | (82) | ||
| Phe114 | SRS1 | Homology modeling | (9) |
| Thr115 | SRS1 (Helix B′) | Homology modeling | (83) |
| Ile117 | SRS1 | SDM Homology modeling | (9) |
| Asp119 | SRS1 (Helix B′–C) | Homology modeling | (83) |
| Thr124 | SRS1 (Helix B′–C) | SDM | (11) |
| Homology modeling | (83) | ||
| Ser126 | SRS1 | Random mutagenesis | (84) |
| Ser129 | SRS1 | Random mutagenesis | (84) |
| Arg135 | Helix C | SDM | (10) |
| Arg136 | Helix C | SDM | (10) |
| Arg137 | Helix C | SDM | (80) |
| Gln141 | – | Homology modeling | (81) |
| His163 | – | SDM | (85) |
| Phe181 | – | Homology modeling | |
| Thr184 | – | Homology modeling | |
| Phe186 | – | Random mutagenesis | (84) |
| Phe205 | – | Homology modeling | (9) |
| Thr223 | SRS2 (Helix F) | SDM | (11) |
| Homology modeling | (83) | ||
| Glu225 | SRS2 | Random mutagenesis | (84) |
| Phe226 | SRS2 | Homology modeling | (9) |
| SDM | (84,86) | ||
| Random mutagenesis | (84) | ||
| Val227 | – | SDM | (11) |
| Lys250 | SRS3 | SDM | (87) |
| Arg251 | SRS3 | SDM | (87) |
| Lys253 | SRS3 | SDM | (87) |
| Asn259 | SRS3 | Homology modeling | (79) |
| Phe266 | – | Homology modeling | (79) |
| Asn312 | SRS4 | SDM | (11) |
| Asp313 | SRS4 (Helix I) | Homology modeling | (81,83) |
| SDM | (82) | ||
| Gly316 | SRS4 (Helix I) | Homology modeling | (83) |
| Glu318 | SRS4 (Helix I) | SDM | (10,88–92) |
| Glu319 | SRS4 (Helix I) | SDM | (10,88–91) |
| Asp320 | SRS4 (Helix I) | Random mutagenesis | (84) |
| Thr321 | SRS4 (Helix I) | Homology modeling | (81) |
| SDM | (82) | ||
| Val322 | SRS4 | Random mutagenesis | (84) |
| Phe330 | – | Homology modeling | (79) |
| Leu382 | SRS5 | SDM | (11) |
| Homology modeling | (83) | ||
| Pro383 | SRS5 | Homology modeling | (83) |
| Thr385 | SRS5 | Homology modeling | (81) |
| SDM | (82) | ||
| Random mutagenesis | (84) | ||
| Ile386 | SRS5 | Random mutagenesis | (84) |
| Lys401 | – | SDM | (10) |
| Tyr437 | – | Homology modeling | (79) |
| Lys440 | – | SDM | (10) |
| Lys453 | – | SDM | (10,80) |
| Arg455 | – | SDM | (10,80) |
| Arg456 | Near heme | Homology modeling | (83) |
| Ile462 | Helix L | SDM and homology modeling | (9) |
| Lys463 | Helix L | SDM | (10) |
| Tyr495 | Near strand β4-1 | Homology modeling | (83) |
| Thr501 | – | Homology modeling | (9) |
| SDM | (84,86) |
SDM site-directed mutagenesis, SRS substrate recognition sequence
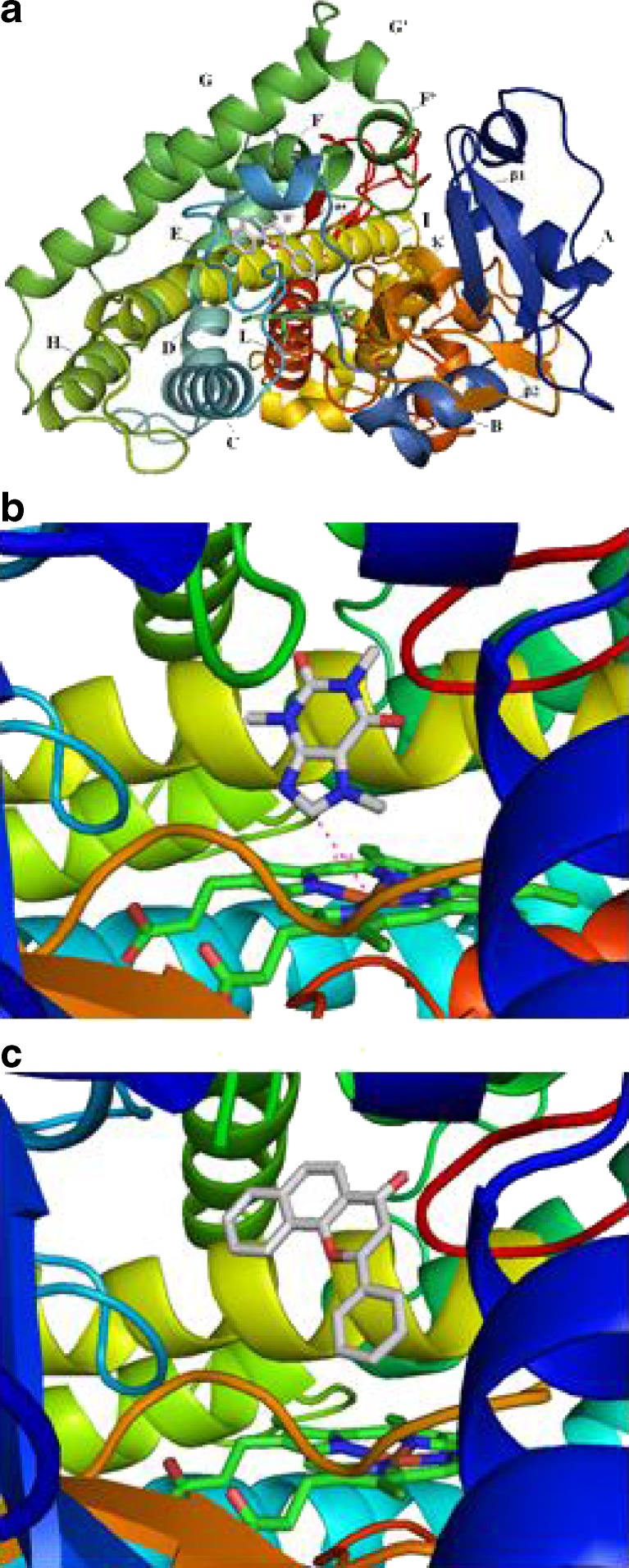
CYP1A2 substrates generally contain planar ring that can fit the narrow and planar active site of the enzyme. The planar active site architecture in CYP1A2 is well adapted for the oxidation of relatively large aromatic compounds which is conserved among all CYP1 members. Sansen et al. (12) recently reported the crystal structure of CYP1A2 in complex with α-naphthoflavone (ANF) which has been refined to 1.95 Å (PDB ID: 2HI4). In the 2HI4 structure in complex with ANF, the rather compact active site is closed without clear solvent or substrate access channels with a relatively small volume of the cavity of 375 Å3 (Fig. 1) (13) which is 44.2% larger than that of CYP2A6 (260 Å3) (12).
Fig. 1.
The structures of human CYP1A2 in complex with ANF (PDB ID = 2HI4; a and c) or caffeine b. The distance from the metabolic site of caffeine to the heme is 4.34 Å
The 2HI4 structure contains 12 α-helices designated A–L and four β-sheets designated 1–4 (12). As observed with other mammalian CYPs of known structure, the most conserved regions are the core of the protein forming the heme binding site and the proximal surface that is considered to offer binding sites for cytochrome P450 reductase and cytochrome b5. The most distinct regions between known CYP structures are the portions that constitute the distal surfaces of the substrate binding cavity, the helix B–C and F–G regions, and the C-terminal loop following helix L. In the 2HI4 structure, the α-helical hydrogen-bonding manner is lost at residues Val220 and Lys221, resulting in one helical turn in the middle of helix F to unwind (12). Two water molecules fill the space and thus form water-bridged contacts between Val220 carbonyl oxygen and Thr223 Oγ, and Lys221 carbonyl oxygen and His224 amide nitrogen, respectively.
Sansen et al. (12) have reported that the substrate binding cavity of CYP1A2 is lined by residues on helices F and I that define a relatively planar binding platform on either side. Helix I bends while it crosses the heme prosthetic group, placing its residues in one flat side of the substrate binding cavity. Coplanarity is thus formed through the Ala317 side chain, the Gly316–Ala317 peptide bond, and the Asp320–Thr321 peptide bond. On the other side of the cavity, the side chain of Phe226 of helix F forms a parallel substrate binding surface. The active site cavity of CYP1A2 is stabilized through a strong hydrogen-bonding interaction between the side chain of Thr223 on helix F and the side chain of Asp320 on helix I. Both Thr223 and Asp320 play a role in forming an extensive network of hydrogen-bonded water molecules and side chains, including Tyr189, Val220, Thr498, and Lys500.
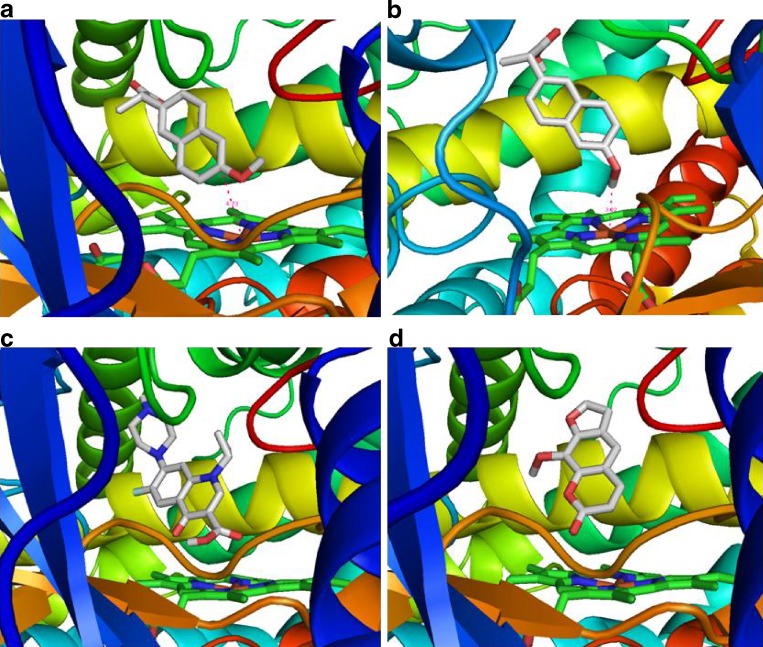
Our docking studies using the published crystal structure of CYP1A2 by Sansen et al. (12) have identified Phe226, Ala317, Gly316, Phe125, and Thr124 as the most important residues to influence the inhibitory potency of a series of natural compounds from Chinese herbal medicines [Zhou SF, et al. (2009) unpublished data]. Based on the structure of 2HI4, we have successfully docked two substrates (amitriptyline and naproxen) and two competitive inhibitors (ciprofloxacin and methoxsalen) into its active site (Fig. 2). The distance from the site of metabolism of amitriptyline and naproxen to the heme group is 4.73 and 3.22 Å, respectively. Both ciprofloxacin and methoxsalen are orientated above the heme group.
Fig. 2.
The docking of two substrates (amitriptyline and naproxen) and two competitive inhibitors (ciprofloxacin and methoxsalen) into the active site of CYP1A2 (PDB ID = 2HI4). The distance from the site of metabolism of amitriptyline and naproxen to the heme group is 4.73 and 3.22 Å, respectively
SUBSTRATE SPECIFICITY OF HUMAN CYP1A2
Phenacetin, caffeine, and theophylline have been frequently used as model substrates for evaluating the activity of CYP1A2 in vivo (14). Industry investigators have used phenacetin O-deethylation as the most common marker reaction for CYP1A2 activity in the in vitro study of 45% new drugs (87 out of 194 studies) (15). Caffeine 3-demethylation and ethoxyresorufin O-deethylation are also commonly used marker reactions for CYP1A2. To a less extent, tacrine (1,2,3,4-tetrahydro-9-aminoacridine, 1-hydroxylation), melatonin (6-hydroxylation) and tizanidine (oxidation) are also used as probes for CYP1A2 in vivo. For in vitro studies, phenacetin (O-deethylation), 7-ethoxycoumarin (O-deethylation), and 7-ethoxyresorufin (O-deethylation) are commonly used probes for determining CYP1A2 activity (16).
CYP1A2 metabolizes a variety of clinically important drugs with variable contributions to their overall elimination (Table II) (17). Drugs that are markedly (>30%) metabolized by CYP1A2 include caffeine, clozapine, ropivacaine, olanzapine, tizanidine, theophylline, tacrine, aminopyrine, zolmitriptan, melatonin, propranolol, duloxetine, leflunomide, promazine, verapamil, thioridazine, nabumetone, ropinirole, and riluzole. CYP1A2 can metabolize some drugs to a minor to moderate extent (10–30%), including mexiletine, paracetamol, flutamide, lidocaine, imipramine, paraxanthine, propafenone, terbinafine, bortezomib, aminoflavone, 5,6-dimethylxanthenone-4 acetic acid (a phase II anticancer drug), trazodone, R-warfarin, R-acenocoumarol, propofol, antipyrine, pentoxifylline, alosetron, furafylline, and tegafur. In addition, CYP1A2 plays a minor role (<10%) in the metabolism of fluvoxamine, pranidipine, zolpidem, thalidomide, cyclobenzaprine, naproxen, coumarin, ondansetron, haloperidol, guanabenz, rofecoxib, efavirenz, bufuralol, cinnarizine, flunarizine, azelastine, almotriptan, carvedilol, amitriptyline, nortriptyline, clomipramine, carbamazepine, albendazole, methadone, amiodarone, and diphenhydramine.
Table II.
Clinical Drugs as Substrates of Human CYP1A2
| Substrate | Drug class | Metabolic pathway(s) | Estimated contribution by CYP1A2 (%)a |
|---|---|---|---|
| R-Acenocoumarol | Oral anticoagulant | 6-Hydroxylation | 10–20 |
| Acetaminophen (paracetamol) | Analgesic drug | Oxidation (to form NAPQI) | 10–15 |
| Albendazole | A benzimidazole anthelmintic drug | 4-Hydroxylation | 5–10 |
| Almotriptan | Selective agonist of 5-HT1B/1D receptor (for treatment of migraine) | N-demethylation | 5–10 |
| Alosetron | Selective 5-HT3 antagonist (for irritable bowel syndrome) | N-dealkylation and hydroxylation | 10–20 |
| Amiodarone | Antiarrhythmic drug | N-deethylation | 5–10 |
| Aminoflavone (NSC686288) | Anticancer agent | 3-Hydroxylation | 10–20 |
| Aminopyrine | Analgesic drug | N-demethylation | 40–50 |
| Amitriptyline | Tricyclic antidepressant | 10-Hydroxylation | 5–10 |
| Antipyrine | Analgesic drug | N-demethylation and 3- and 4-hydroxylation | 10–25 |
| Azelastine | Antihistamine | N-demethylation | 5–10 |
| Bortezomib | Proteasome inhibitor (for treatment of multiple myeloma) | Deboronation and dehydrogenation | 10–25 |
| Bufuralol | β-Blocker | 1′-Hydroxylation | 5–10 |
| Caffeine | CNS stimulant | N-demethylation | 95 |
| Carbamazepine | Anticonvulsant | 2- and 3-Hydroxylation | 5–10 |
| Carvedilol | β-Blocker | 8-Hydroxylation and O-demethylation | 5–10 |
| Cinnarizine | Antihistamine drug | N-dealkylation | 5–10 |
| Clomipramine | Tricyclic antidepressant | N-demethylation | 5–10 |
| Clozapine | Atypical antipsychotic drug | N-demethylation and N-oxidation | 40–55 |
| Coumarin | Natural compound as anticancer agent | 7-Hydroxylation | 5–10 |
| Cyclobenzaprine | Tricyclic antidepressant | N-demethylation | 5–10 |
| 5,6-Dimethylxanthenone-4 acetic acid | Anticancer drug | 6-Methyl hydroxylation | 10–20 |
| Diphenhydramine | Antihistamine drug | N-demethylation | 5–10 |
| Duloxetine | Antidepressant | 4-, 5-, and 6-Hydroxylation | 30–40 |
| Efavirenz | Non-nucleoside reverse transcriptase inhibitor | 7- and 8-Hydroxylation | 5–10 |
| Erlotinib | Epidermal growth factor receptor tyrosine kinase inhibitor (for cancer treatment) | O-demethylation | 5–10 |
| Estradiol | Steroid in oral contraceptives | 2- and 4-hydroxylation | 5–10 |
| Flunarizine | Antihistamine | N-dealkylation | 5–10 |
| Flutamide | Non-steroidal antiandrogen for prostate cancer treatment | 2-Hydroxylation | 10–25 |
| Fluvoxamine | SSRI | O-demethylation | 5–10 |
| Furafylline | Antiasthmatic agent | 10–20 | |
| Guanabenz | α2-Adrenoreceptor agonist (antihypertensive) | N-hydroxylation | 5–10 |
| Haloperidol | Antipsychotic drug | N-dealkylation | 5–10 |
| Imatinib | Bcr-Abl tyrosine kinase inhibitor (as anticancer agent) | O-demethylation | |
| Indiplon | Hypnotic drug | N-demethylation | 5–10 |
| Imipramine | Tricyclic antidepressant | N-demethylation | 10–20 |
| KR-62980 | Selective peroxisome proliferator-activated receptor-γ modulator | Hydroxylation | 5–10 |
| KR-63198 | Selective peroxisome proliferator-activated receptor-γ modulator | Hydroxylation | 5–15 |
| Leflunomide | Disease-modifying anti-inflammatory agent | N―O bond cleavage | 40–55 |
| Lidocaine | Antiarrhythmic and local anesthetic drug | N-deethylation and 3-hydroxylation | 10–20 |
| Maprotiline | Antidepressant | 5–10 | |
| Melatonin | Pineal hormone | 6-Hydroxylation and O-demethylation | 40–60 |
| Mexiletine | Antiarrhythmic drug | Para- and 2-hydroxylation | 10–20 |
| Mianserin | Antidepressant | N-oxidation and N-demethylation | 5–10 |
| Mirtazapine | Antidepressant | N-oxidation, 8-hydroxylation, and N-demethylation | 5–15 |
| Naproxen | NSAID | O-demethylation | 5–10 |
| Nabumetone | NSAID | Aliphatic hydroxylation | 30 |
| Nicotine | Agent for treating nicotine dependence | N-hydroxylation and C-oxidation | 5–10 |
| Nortriptyline | Tricyclic antidepressant | 10-Hydroxylation | 5–10 |
| Olanzapine | Atypical antipsychotic drug | N-demethylation, and 4- and 7-hydroxylation | 30–40 |
| Ondansetron | Selective serotonin 5-HT3 receptor antagonist (antiemetic) | 7- and 8-Hydroxylation | 5–10 |
| Paraxanthine | CNS stimulant | 8-Hydroxylation | 80 |
| Pentoxifylline | A drug used to treat intermittent claudication | Xanthine 7-demethylation | 10–20 |
| Phenacetin | Analgesic drug | O-deethylation | 86 |
| Pranidipine | Calcium channel blocker | Methyl hydroxylation | 5–10 |
| Promazine | Antipsychotic drug | N-demethylation and 5-sulfoxidation | 30–45 |
| Propafenone | Antiarrhythmic drug | 5-Hydroxylation and N-desalkylation | 10–25 |
| Propofol | Sedative agent | 4-Hydroxylation | 10–25 |
| Propranolol | β-Blocker | N-desisopropylation | 30–50 |
| Riluzole | Antiglutamate agent for treatment of ALS | N-hydroxylation, and possibly 4-, 5-hydroxylation and O-dealkylation | 75–80 |
| Rofecoxib | Selective COX-2 inhibitor (withdrawn from the market) | 5-Hydroxylation | 5–10 |
| Ropinirole | Non-ergoline dopamine agonist (for treatment of Parkinson’s disease) | N-deethylation | 30–40 |
| Ropivacaine | Local anesthetic drug | 3-Hydroxylation | 50–65 |
| Selegiline | Selective irreversible MAO-B inhibitor (for treatment of Parkinson’s disease) | N-demethylation | 5–15 |
| Tacrine | Centrally acting cholinesterase inhibitor (for treatment of Alzheimer’s disease) | 1-, 2-, 4-, and 7-hydroxylation | 50–65 |
| Tegafur (tetrahydrofuranyl-5-fluorouracil) | Anticancer drug (a prodrug) | N-dealkylation | 10–20 |
| Terbinafine | Antifungal agent | N-demethylation and side chain oxidation | 10–25 |
| Thalidomide | Antiangiogenic agent (for treatment of multiple myeloma) | 5′-, 5-, and 6-hydroxylation | 5–10 |
| Theophylline | Bronchodilator | N-demethylation | 90–95 |
| Thioridazine | Antipsychotic drug | 5-Sulfoxidation and N-demethylation | 35–45 |
| Tizanidine | Muscle relaxant | 80–95 | |
| Trazodone | Antidepressant | Hydroxylation, dealkylation, and N-oxidation | 15–20 |
| Triamterene | Potassium-sparing diuretic | 4′-Hydroxylation | 5–10 |
| Verapamil | Calcium channel blocker | N-demethylation | 20–30 |
| R-Warfarin | Oral anticoagulant | 6-, 8-, and 10-Hydroxylation | 10–20 |
| WHI-P131 (JANEX-1) | Janus kinase-3 inhibitor (an anti-inflammatory agent) | 7-O-demethylation | 5–15 |
| Zileuton | Specific inhibitor of 5-lipoxygenase (for treatment of asthma) | Ring hydroxylation | 5–15 |
| Zolmitriptan | Selective agonists of 5-HT1B/1D receptor (for treatment of migraine) | N-demethylation and N-oxidation | 30–40 |
| Zolpidem | Non-benzodiazepine hypnotic drug | Ring hydroxylation | 5–10 |
| Zotepine | Atypical antipsychotic drug | 2- and 3-Hydroxylation | 5–10 |
| Zoxazolamine | Muscle relaxant | 6-Hydroxylation | 5–10 |
Abbreviations: ALS amyotrophic lateral sclerosis, CNS central nervous system, COX cyclooxygenase, MAO monoamine oxidase, NSAID non-steroid anti-inflammatory drug, SSRI selective serotonin reuptake inhibitor
aThe estimation contribution is based on inhibition studies using chemical inhibitors and/or inhibitory antibodies, and pharmacogenetic studies where poor and extensive metabolizers of CYP1A2 are compared. Data are from Rendic (17) and our own databases of CYPs
CYP1A2 is involved in the metabolism of a number of natural compounds, probably resulting in toxic metabolites (17,18). Estragole is bioactivated to reactive metabolites by CYP1A2, and CYP1A2 and 2C9 are the major enzymes responsible for conversion of methyleugenol to 1′-hydroxymethyleugenol (19). Aristolochic acids (AAs), naturally occurring nephrotoxins, and rodent carcinogens found in some herbal medicines have been associated with the development of urothelial cancer in humans. Both CYP1A1 and 1A2 contribute to their activation (20).
CYP1A2 together with CYP1A1 and 1B1 play important roles in the bioactivation of a variety of carcinogenic polycyclic aromatic hydrocarbons [PAHs, e.g., benzo[a]pyrene (B[a]P)], heterocyclic aromatic amines/amides (e.g., 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 2-amino-3,4-dimethylimidazo[4,5-f]quinoline), and mycotoxins found in some grains such as aflatoxin B1 (AFB1) and the specificity of the substrates often overlaps (21). AFB1 is a potent hepatotoxin and procarcinogen in a number of animals and is associated epidemiologically with a high incidence of primary hepatocellular carcinoma in humans. The critically reactive metabolite of AFB1 is the exo 8,9-epoxide formed by a two-electron oxidation mainly catalyzed by CYP3A4, with contribution from CYP1A2 and other CYPs (22). Oxidation of the chemicals by CYP1A1 and 1A2 serves as an initial step in the conversion of the substrates to more polar metabolites, resulting in increased excretion.
Moreover, CYP1A2 metabolizes several important endogenous substrates, such as melatonin, bilirubin, uroporphyrinogen, estrone and estradiol, and arachidonic acid (17). CYP1A1, 1A2, and 1B1 6-hydroxylate melatonin, with minor contribution from CYP2C19 (23). CYPA2 catalyzed uroporphyrinogen oxidation to form uroporphyrin (24). In addition, CYP1A2, 1A1, and 3A4 are involved in the metabolism of estradiol and estrone (25).
INDUCTION OF CYP1A2 THROUGH AROMATIC HYDROCARBON RECEPTOR (AHR)
CYP1A2 and 1A1 are highly inducible at both mRNA and protein levels by a variety of chemicals, smoking, and several dietary factors (26,27). Many potent inducers of CYP1A1 such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), 3-methylcholanthrene, and β-naphthoflavone are also potent inducers of CYP1A2. However, rifampicin, a prototypical inducer of CYP3A, is only a weak inducer of CYP1A2 (28).
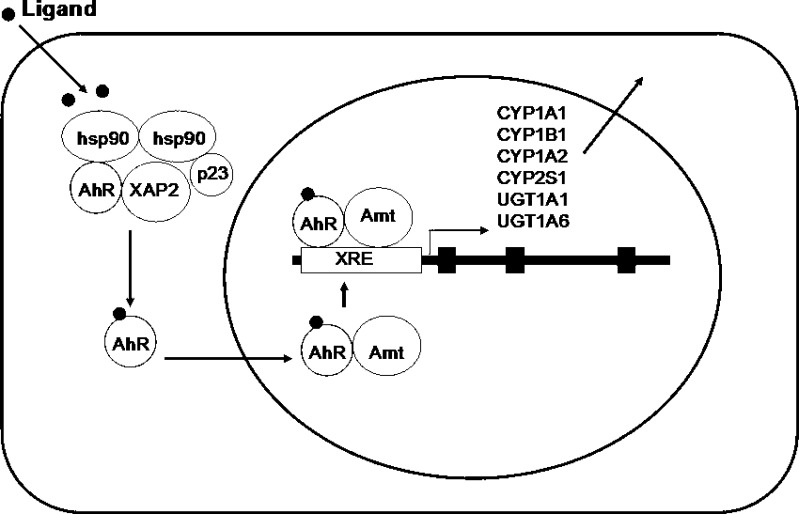
CYP1A2 is induced through AhR-mediated transactivation following ligand binding and nuclear translocation (Fig. 3), but the extent of induction is generally lower than that of CYP1A1 (29). AhR is a ligand-activated transcription factor and a basic helix-loop-helix (bHLH) protein belonging to the Per-Arnt-Sim (PAS, where Per stands for Drosophila period clock protein, Arnt refers to AhR nuclear translocator, and Sim is Drosophila single-minded protein) family of transcription factors (26,27). The bHLH motif is located in the N-terminal of the protein and is a common entity in a variety of transcription factors. Members of the bHLH superfamily have two functionally distinctive and highly conserved domains (30). The first is the basic region (b) which is involved in the binding of the transcription factor to DNA, while the second is the helix-loop-helix (HLH) region which facilitates protein–protein interactions. AhR contains two PAS domains, PAS-A and PAS-B, which are stretches of 200–350 amino acids that exhibit a high sequence homology to the protein domains that were found in the Drosophila genes period (Per) and single-minded (Sim) and in Arnt. The AhR exists as cytoplasmic aggregates bound to two 90-kDa heat-shock proteins (Hsps), the cochaperone prostaglandin E synthase 3 (p23), and a 43-kDa immunophilin-like protein hepatitis B virus X-associated protein 2 (XAP2, also called AhR interacting protein 1, AIP1; or AhR-associated protein 9, ARA9) (31). These other proteins are involved in the correct folding and stabilization of AhR. Upon binding a ligand, after the replacement of its associated molecule with Arnt to form a heterodimer with release of 90 kDa HSPs, AhR translocates into the nucleus (Fig. 3). This heterodimer interacts with a 5′-GCGTG-3′ DNA sequence, the core binding motif of the xenobiotic response element or dioxin response element of the target genes, located and present in multiple copies in the upstream region of the CYP1A1 gene promoter. The AhR-regulated genes include CYP1A1, 1A2, 1B1, 2S1, UGT1A1, 1A6, and GSTA1 (32). The regulation of expression of CYP1A1/1A2 is complex because gene transcription not only involves the AhR but also a number of transcription factors, and is potentially influenced by the actions of transcriptional coactivators and corepressors. AhR-mediated signaling pathways provide a first line of defense against potentially toxic environmental contaminants. However, induction of metabolic processes by the AhR can also produce highly carcinogenic metabolites, creating a link between AhR activation and chemical carcinogenesis.
Fig. 3.
A schematic illustration of the aromatic hydrocarbon receptor (AhR)-mediated induction of phase I and phase II drug metabolizing enzymes and drug transporters such as human CYP1A1, 1B1, 1A2, and 2S1, UGT1A1 and 1A6, and MDR1/ABCB1. The AhR exists as cytoplasmic aggregates bound to two 90-kDa heat-shock proteins (Hsps), the cochaperone p23 and a 43-kDa immunophilin-like protein hepatitis B virus X-associated protein 2 (XAP2). Upon binding a ligand, after the replacement of its associated molecule with AhR nuclear translocator (Arnt) to form a heterodimer with release of 90-kDa heat-shock proteins, AhR translocates into the nucleus. This heterodimer interacts with a 5′-GCGTG-3′ DNA sequence, the core binding motif of the xenobiotic response element (XRE) or dioxin response element of the target genes encoding CYP1A1, 1B1, 1A2, and 2S1, and uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) and UGT1A6
Induction of CYP1A1/1A2 is generally a means of maintaining the homeostasis of the chemical environment in cells by increasing the metabolic clearance of substrates. Since CYP1A1/1A2 catalyzes the metabolic activation of PAHs and heterocyclic aromatic amines/amides to ultimate carcinogens, it is expected that induction of the enzyme is detrimental in humans exposed to high levels of PAHs and heterocyclic aromatic amines/amides such as by cigarette smoking. Induction of the enzyme in humans exhibits large variations; high inducibility may impose additional risk for lung cancer to individuals who are smokers. Furthermore, CYP1A2 can metabolize a range of substrates; induction of the enzymes by one substrate may increase the metabolism of other chemicals (for instance, clinical drugs), resulting in unexpected drug–drug interactions.
INHIBITION OF CYP1A2 AND CLINICAL RELEVANCE
Several therapeutic drugs including carbamazepine, dihydralazine, furafylline, isoniazid, rofecoxib, clorgyline, thiabendazole, and zileuton are mechanism-based inhibitors of CYP1A2 (17). In addition, desethylamiodarone also inactivated CYP1A2 but amiodarone inactivated CYP3A4 only (33). The metabolite of thiabendazole, 5-hydroxythiabendazole, is also a mechanism-based inhibitor of CYP1A2 (34). Antofloxacin, an 8-NH2 derivative of levofloxacin, was found to be a mechanism-based inhibitor of rat CYP1A2 (35). Oltipraz, a chemo-protective agent, is a competitive and mechanism-based inhibitor of CYP1A2 (36). trans-Resveratrol inactivates CYP1A2, but not CYP1A1 (37).
Typical competitive CYP1A2 inhibitors are relatively small molecules often containing methyl, chloro, or fluoro substitutions such as tolfenamic acid, ciprofloxacin, furafylline, and fluvoxamine. These compounds can fit well into the active site of CYP1A2. ANF is commonly used as selective inhibitors for CYP1A2 in reaction-phenotyping studies (38,39). ANF is a potent competitive inhibitor of CYP1A2 with Ki values of 1–50 nM (38,39). Fluvoxamine, a selective serotonin reuptake inhibitor (SSRI), was a very potent and selective CYP1A2 inhibitor with Ki of 0.035–0.24 μM (40). Other SSRIs, including fluoxetine, norfluoxetine, sertraline, and paroxetine, also inhibited CYP1A2-mediated 7-ethoxyresorufin O-deethylase activity (41). The fluoroquinolone antibiotics such as levofloxacin, ciprofloxacin, moxifloxacin, gatifloxacin, caderofloxacin, and antofloxacin are weak to moderate inhibitors of CYP1A2 (42–44).
Some natural compounds can inhibit CYP1A2 and 1A1. Rutaecarpine, evodiamine, and dehydroevodiamine are quinazolinocarboline alkaloids isolated from Evodia rutaecarpa, which has been used in traditional Chinese medicine for the treatment of gastrointestinal disorder, headache, and hypertension. They are all inhibitors of CYP1A1 and 1A2, with rutaecarpine being the most potent (45). Tanshinone I, tanshinone IIA, and cryptotanshinone extracted from the commonly used Chinese herbal medicines Salvia miltiorrhiza (Danshen) are potent competitive inhibitors of CYP1A2, with Ki of 0.48, 1.0, and 0.45 μM, respectively (46). Because of the potential effects on drug disposition and inhibition of toxicological processes, natural products such as flavonoids are the focus of much current interest, from the perspectives of both nutrition and pharmacotherapy. These natural compounds may be involved in the prevention of malignant transformation by reducing the formation of carcinogens through inhibition of enzymes such as CYP1A1 and 1A2, both of which are known to be involved in carcinogen activation.
Overall, CYP1A2 is subject to reversible and/or irreversible inhibition by a number of drugs, natural substances, and other compounds. Competitive inhibitors of CYP1A2 can readily fit into the active site of CYP1A2. The inhibitory potency of compounds for CYP1A2 is determined by their physico-chemical features. It appears that planar molecules with a small volume to surface area are the most potent inhibitors of CYP1A2. Inactivation and reversible inhibition of CYP1A2 by drugs may cause important drug interactions.
PHENOTYPES OF CYP1A2 KNOCKOUT MICE
The Cyp1a2−/− mice are viable and fertile; histologic examination of 15-day embryos, newborn pups, and 3-week-old mice revealed no abnormalities (47). Cyp1a1 and Cyp1b1 mRNA levels appeared unaffected by loss of the Cyp1a2 gene and inducibility of Cyp1a1 by dioxin was not altered by the absence of Cyp1a2 (48). When the muscle relaxant zoxazolamine (a known substrate for Cyp1a2 (49)) was administered to the Cyp1a2−/− mice, they exhibited dramatically lengthened paralysis times relative to the Cyp1a2+/+ wild-type animals, and the Cyp1a2+/− heterozygous mice showed an intermediate effect. Using Cyp1a2−/− mice, it was found that 87% of caffeine clearance depended on Cyp1a2 in this species (50). Notably, Cyp1a2−/− mice exhibit increased toxicity from some compounds that are predominantly Cyp1a2 substrates. By cDNA microarray approach, it has been found that at least 15 genes are up- or down-regulated (51). The gene exhibiting the greatest down-regulation was insulin-like growth factor binding protein-1, showing only 12% expression of that in the wild-type mouse. These altered genes are associated with cell-cycle control, insulin action, lipogenesis, and fatty acid and cholesterol biosynthetic pathways. Histologically, the Cyp1a2−/− mouse exhibited an approximately 50% decrease in lipid stored in hepatocytes, and 50% increase in lipid present in interstitial fat-storing cells compared with that in the wild-type (52). These data suggest that CYP1A2 might have additional hepatic endogenous functions heretofore not revealed.
POLYMORPHISMS OF CYP1A2 AND CLINICAL IMPACT
There are wide interindividual differences (10- to 200-fold) in CYP1A2 (also called phenacetin O-deethylase) expression and activity (7). Approximately 15- and 40-fold interindividual variations in CYP1A2 mRNA and protein expression levels have been observed in human livers (2). These findings may reflect a genetically determined difference in constitutive and/or inducible CYP1A2 gene expression. Unimodal, bimodal, and trimodal distributions of CYP1A2 activity when measured by caffeine urinary metabolic ratios have been observed in different study populations (10). The frequency of poor metabolizers in non-smokers was 5% in Australians, 14% in Japanese, and 5% in Chinese (8). There is also marked racial difference in CYP1A2 activity. Swedes had a 1.54-fold higher CYP1A2 activity than Koreans (53). A lower CYP1A2 activity has been found in Asian and African populations compared to Caucasians (54). Environmental factors have been thought to influence the interindividual differences in CYP1A2 activity and expression. Cigarette smoking and intake of oral contraceptive steroids are well-established inducers of CYP1A2 activity. However, it has been suggested that approximately 35% to 75% of the interindividual variability in CYP1A2 activity is due to genetic factors (55). There is a large variability in the clearance of drugs that are mainly metabolized by CYP1A2. Identification of genetic, epigenetic, and environmental factors that regulate CYP1A2 expression and activity can assist in the optimization of therapeutic regimens of drugs that are mainly eliminated by CYP1A2.
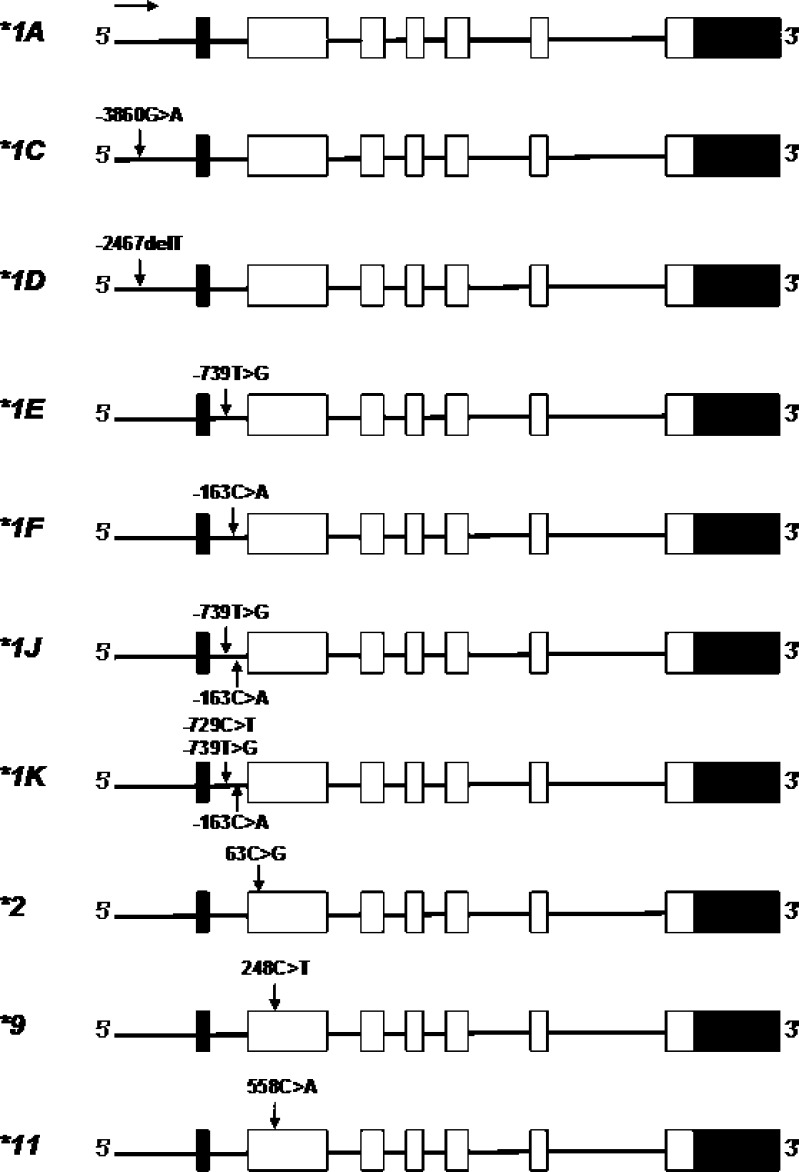
To date, more than 15 variant alleles (*1B to *16) and a series of subvariants of the CYP1A2 gene have been identified (Fig. 4; http://www.imm.ki.se/CYPalleles, access date 12 June 2009), and 177 SNPs have been found in the CYP1A2 upstream sequence, six introns and seven exons in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/, access date 12 June 2009). CYP1A2*1A is referred to as the wild type (reference). Among the SNPs located in seven exons, there are 22 non-synonymous that change amino acid sequence.
Fig. 4.
Important alleles of CYP1A2 in humans
In addition, there are several SNPs identified in the 5′-flanking region and intron 1 of CYP1A2 (56). Aitchison et al. (56) identified several SNPs in the 5′-flanking region of CYP1A2, including −3594T>G, −3598G>T, and −3605insT. The −3594T>G SNP had a 1.72% frequency in 174 Caucasians. In the 5′-flanking region, reported SNPs in Japanese include −3860G>A, −3598G>T, −3594T>G, −3113G>A, −3053A>G, −2847T>C, 2808A>C, 2667T>G, and −2467delT (57,58). In intron 1, SNPs including −739T>G, −729T>G, −733G>C, −587A>G, −367C>T, −163C>A, and −99C>T have been reported. −163C>A in intron 1 is present with considerable frequencies in most populations examined. −3860G>A, −3113A>G, −2464delT, −739T>G, and −729C>T have been found with different frequencies in various ethnic groups (57).
The most extensively studied polymorphisms are −3860G>A (CYP1A2*1C), −2467delT (*1D), −739T>G (*1E), and −163C>A located in intron 1 (*1F), which were first reported in a Japanese population. Of the polymorphic CYP1A2 alleles showing variability in the promoter region, CYP1A2*1C, *1D, *1F, and *1K have been associated with altered enzyme activity. The CYP1A2*1C allele contains −3860G>A in the 5′-flanking region of CYP1A2, which was first found in Japanese with a frequency of 23.3% (27/116) (59). This allele was reported to cause decreased enzyme activity as measured by the rate of caffeine 3-demethylation in smokers of Japanese (n = 50), probably due to decreased inducibility and expression of the enzyme (59). The CYP1A2*1F allele (i.e., −163C>A in intron 1) is common with high and comparable frequencies in various population studies (58,60–62). Sachse et al. (60) first reported this SNP in a healthy German population comprising 185 non-smokers and 51 smokers (n = 236). The frequency of this SNP in German Caucasians was 67.8%, with 45.8% being homozygotes (108/236) for −163A/A and 44.1% being heterozygotes (104/236) for −163C/A. In Japanese, the frequency of −163C>A is high (62.8%) (57). This SNP had a frequency of 55.9% in Swedes (61). The −163C>A in intron 1 caused increased enzyme activity when measured by molar ratios of 17X/137X in plasma compared with the A/C and C/C genotypes in 51 German-Caucasian smokers (10). Smokers (n = 51) with the −163C/C (n = 5) or −163C/A (n = 24) genotype had a 67.1% and 55.7% lower CYP1A2 activity, respectively, compared with those with the −163A/A genotype (n = 22) (0.82 and 0.88 vs 1.37; p = 0.008).
CYP1A2*1J (−163C>A; −739T>G) and *1K (−163C>A; −739T>G; −729C>T, all located in intron 1) have been first detected in Ethiopian non-smokers (63). The −739T>G had a frequency of 3.2% in Japanese, while the −729C>T SNP was not found in the Japanese population (57). The CYP1A2*1K haplotype was associated with 40% lower inducibility in vitro, and non-smokers heterozygous for *1K had significantly lower CYP1A2 activity compared with the wild type (63). The −729C>T SNP, which was responsible for a lower activity of *1K, abolishes a binding site for an Ets nuclear factor, resulting in highly decreased CYP1A2 expression and caffeine metabolism (63). CYP1A2*1K was found to be rare in Swedes (0.3%) and absent in Koreans (61). Thus, the effect of CYP1A2*1K could not be shown in these populations.
CYP1A2*2 carries a 63C>G mutation that causes a Phe21Leu substitution, which was first detected from one subject out of 157 Chinese subjects with an allele frequency of 0.32% (64). Its functional impact is unknown. The *2 allele was absent in British (n = 114) (65) and Italian populations (n = 500) (66). The CYP1A2*3 contains 2385G>A in exon 4 causing Asp348Asn and the synonymous 5347T>G (Asn516Asn), with a frequency of 1% in French population (67). The *4 allele contains 2499A>T in exon 5 leading to Ile386Phe. The *5 allele entails 3497G>A in exon 6 leading to Cys406Tyr. The *6 contains 5090C>T in exon 7 resulting in Arg431Trp (67). All these alleles (*3–*6) were first detected in a French population with very low frequencies (≤0.5–1.0%). CYP1A2*7 contains a 3533G>A mutation at the splice donor site of intron 6. This mutation was found in a 70-year-old patient who had very high plasma concentrations of clozapine when administered at normal dose. This SNP caused RNA splicing defect and led to loss of CYP1A2 activity.
There are significant ethnic differences in the distribution of common and rare CYP1A2 SNPs and haplotypes (Table III). The −3860G>A (*1C) SNP is less prevalent in Caucasians than in Asians (0.21–0.25) (58). The frequency of *1C was significantly lower in the Turkish population (0.04) (68) than in Japanese (0.21) (58) and Chinese (0.25), while the frequency of *1C was relatively equal in the Turkish and Egyptian populations (0.07) (69). The frequency of the −2467delT (CYP1A2*1D) allele is lower in Caucasians compared with Asians and Africans. The allelic frequency of the *1D allele is 4.1–7.9% in Caucasians (65,70), while the −2467T deletion appeared at much higher frequencies of 42.0–43.8% in Japanese (57,58) and of 40% in Egyptians (69). In Turkish, the frequency of *1D is very high (92%) (68). The −739T>G SNP (CYP1A2*1E and *1G) was frequent in Ethiopians (0.10) (63), Saudi Arabians (0.096) (63), and Japanese (0.082) (58). However, British (65), German-Caucasian (70), Spaniard (63), Turkish (68), and Egyptian (69) populations had a low frequency for this allele (0.0044, 0.016, 0.017, 0.01, and 0.03, respectively).
Table III.
Frequencies of Important SNPs in Human CYP1A2 Gene
| Ethnic group | No. of subjects | −3860G>A (*1C) | −3113A>G | −2467delT (*1D) | −739T>G (*1E, *1G, *1J, and *1K) | −729C>T (*1K) | −163C>A (*1F, *1J, and*1K) | 5347T>C (*1B, *1H, *1G, *3, *8, *15, and *16) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Caucasian | |||||||||
| British | 114 | 0.009 | 0.048 | 0.004 | 0.333 | 0.382 | (65) | ||
| German | 236 | 0.32 | (60) | ||||||
| Italian | 95 | 0.04 | 0.24 | 0.33 | (93) | ||||
| Spanish | 117 | 0.02 | 0.005 | (63) | |||||
| Swedish | 194 | 0.008 | 0.023 | 0.193 | 0.023 | 0.003 | 0.286 | (61) | |
| Swedish | 1,170 | 0.29 | (94) | ||||||
| Turkish | 110 | 0.04 | 0.92 | 0.01 | 0.27 | (68) | |||
| Asian | |||||||||
| Chinese | 27 | 0.22 | 0.09 | 0.50 | 0.09 | 0.30 | 0.20 | (95) | |
| Chinese | 168 | 0.33 | (96) | ||||||
| Japanese | 159 | 0.21 | 0.42 | 0.08 | 0.61 | (58) | |||
| Japanese | 250 | 0.24 | 0.03 | 0.44 | 0.03 | 0.32 | 0.19 | (57) | |
| Korean | 150 | 0.267 | 0.027 | 0.707 | 0.027 | 0 | 0.373 | (61) | |
| Saudi Arabian | 173 | 0.10 | 0.04 | (63) | |||||
| African | |||||||||
| Egyptian | 212 | 0.07 | 0.40 | 0.03 | 0.68 | (69) | |||
| Ethiopian | 173 | 0.10 | 0.03 | 0.40 | (63) | ||||
| Tanzanian | 71 | 0.51 | (97) | ||||||
| Tunisian | 98 | 0.07 | 0.075 | 0.13 | 0.44 | (98) | |||
| Zimbabwean | 143 | 0.43 | (97) | ||||||
A number of clinical studies have been conducted to examine the impact of CYP1A2 polymorphisms on drug clearance and drug response. For example, Obase et al. (71) examined the effect of genetic polymorphisms in the 5′-flanking region to intron 1 of the CYP1A2 gene on theophylline metabolism in Japanese asthmatic patients (n = 75) and healthy volunteers (n = 159). Among asthmatic patients, theophylline clearance was significantly lower in patients with the polymorphism at site −2964G>A whose genotype was G/A or A/A than in those whose genotype was G/G (G/A or A/A vs G/G = 0.029 vs 0.034 L/h/kg). Therapeutic drug monitoring may be needed in patients with the A allele at site −2964 in the CYP1A2 gene, because theophylline clearance is lower in these patients, particularly in young asthmatic individuals. Resistance to clozapine therapy due to low plasma drug levels has been reported in smoking schizophrenic patients harboring the −163A/A (CYP1A2*1F) genotype (72,73). Eap et al. (72) reported four smoking patients who did not respond to clozapine therapy at usual dosage and found these individuals were ultra-rapid metabolizers of CYP1A2 carrying the *1F allele. On the other hand, higher plasma concentrations of clozapine and its metabolite N-desmethylclozapine have been observed in patients carrying two CYP1A2 variants associated with reduced enzyme activity (−3860A, −2467del, −163C, −739G, and/or −729T) compared with those with one or none (74).
Both smoking and CYP2D6 genotype, but not CYP2C9 genotype, can affect thioridazine plasma levels (75). The mean steady-state plasma concentration of trazodone was significantly lower in smokers than in non-smokers (76). Subjects with −163A/A or −163C/A genotype had lower plasma melatonin concentrations after administration of 6 mg melatonin compared with −163C/C carriers (77). A recent study in patients with rheumatoid arthritis (n = 105) revealed that the CYP1A2*1F (−163C>A) allele affected the risk for leflunomide-induced organ toxicities (78). Patients with −163C/C genotype had a 9.7-fold higher risk for overall leflunomide-induced toxicity compared with patients with the CYP1A2*1F C/A or A/A genotype.
CONCLUSIONS AND FUTURE PERSPECTIVES
CYP1A2 is one of the major hepatic CYPs in human liver that metabolizes about 15% of clinical drugs such as clozapine, theophylline, tacrine, and zolmitriptan. CYP1A2 is one of the major enzymes that bioactivate a number of procarcinogens. This enzyme also metabolizes several important endogenous compounds such as steroids, retinols, melatonin, uroporphyrinogen, and arachidonic acids, suggesting its potential role in some physiologic processes in addition to xenobiotic metabolism. Because CYP1A2 has a relatively small active center due to several surrounding aromatic residues, its substrates and inhibitors are usually small, lipophilic, and planar molecules. The elucidation of the CYP1A2 crystal structure has provided deep insights into the mechanism for the interaction of ligands with CYP1A2. The structure has offered clear clues on how CYP1A2 exhibits its substrate specificity. This information may help with the design of new drugs with minimal interaction with CYP1A2.
There is an increasing number of clinical drug interactions caused by CYP1A2 induction and/or inhibition and timely identification of these drug interactions is important in clinical practice. The interaction may be substrate and inhibitor dependent and thus caution should be taken when conducting in vitro–in vivo extrapolation. Polymorphisms of CYP1A2 may alter drug clearance and drug response and thus dosage adjustment and therapeutic drug monitoring for drugs with a narrow therapeutic index may be required. There are increased pharmacogenetic studies on the impact of common CYP1A2 polymorphisms on drug clearance and response, but the evidence is still preliminary. The clearance of several drugs may be altered by the genotype of CYP1A2 and thus patients’ response to these drugs may vary considerably. More well-designed studies are warranted to explore the genotype–phenotype relationships of CYP1A2 in terms of drug clearance and response. Personalized pharmacotherapy and individualized dosing of drugs would need incorporation of both genetic and environmental factors.
The pharmaceutical industry is likely to screen drug candidates regarding their CYP1A2-binding properties early in drug development. Such candidates may be dropped if they have poor pharmacokinetic properties and where alternatives are available. This candidate selection might eventually lead to a less relevant role of this enzyme for newer drugs.
Acknowledgements
The authors appreciate the grant support of RMIT Health Innovations Research Institute, RMIT University, Bundoora, Victoria 3083, Australia and National Institute of Complementary Medicine, New South Wales, Australia. Mr. Zhi-Wei Zhou is a holder of RMIT University International Postgraduate Scholarship, Victoria, Australia. Dr. Li-Ping Yang was and Mr. Ya-He Liu is a holder of the Australian Postgraduate (PhD) Award funded by the Commonwealth Government of Australia. We also appreciate the support of Professor Hualiang Jiang, Center for Drug Discovery and Design, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China.
References
- 1.Corchero J, Pimprale S, Kimura S, Gonzalez FJ. Organization of the CYP1A cluster on human chromosome 15: implications for gene regulation. Pharmacogenetics. 2001;11:1–6. doi: 10.1097/00008571-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, Kimura S. Human CYP1A2: sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expression. Mol Endocrinol. 1989;3:1399–1408. doi: 10.1210/mend-3-9-1399. [DOI] [PubMed] [Google Scholar]
- 3.Jaiswal AK, Nebert DW, McBride OW, Gonzalez FJ. Human P(3)450: cDNA and complete protein sequence, repetitive Alu sequences in the 3′ nontranslated region, and localization of gene to chromosome 15. J Exp Pathol. 1987;3:1–17. [PubMed] [Google Scholar]
- 4.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 5.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhou SF, Di YM, Chan E, et al. Clinical pharmacogenetics and potential application in personalized medicine. Curr Drug Metab. 2008;9:738–784. doi: 10.2174/138920008786049302. [DOI] [PubMed] [Google Scholar]
- 7.Gunes A, Dahl ML. Variation in CYP1A2 activity and its clinical implications: influence of environmental factors and genetic polymorphisms. Pharmacogenomics. 2008;9:625–637. doi: 10.2217/14622416.9.5.625. [DOI] [PubMed] [Google Scholar]
- 8.Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295. [DOI] [PubMed]
- 9.Lewis DF, Lake BG, Dickins M, Ueng YF, Goldfarb PS. Homology modelling of human CYP1A2 based on the CYP2C5 crystallographic template structure. Xenobiotica. 2003;33:239–254. doi: 10.1080/0049825021000048791. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu T, Tateishi T, Hatano M, Fujii-Kuriyama Y. Probing the role of lysines and arginines in the catalytic function of cytochrome P450d by site-directed mutagenesis. Interaction with NADPH-cytochrome P450 reductase. J Biol Chem. 1991;266(6):3372–3375. [PubMed] [Google Scholar]
- 11.Liu J, Ericksen SS, Sivaneri M, Besspiata D, Fisher CW, Szklarz GD. The effect of reciprocal active site mutations in human cytochromes P450 1A1 and 1A2 on alkoxyresorufin metabolism. Arch Biochem Biophys. 2004;424:33–43. doi: 10.1016/j.abb.2003.12.040. [DOI] [PubMed] [Google Scholar]
- 12.Sansen S, Yano JK, Reynald RL, et al. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 13.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 14.Faber MS, Jetter A, Fuhr U. Assessment of CYP1A2 activity in clinical practice: why, how, and when? Basic Clin Pharmacol Toxicol. 2005;97:125–134. doi: 10.1111/j.1742-7843.2005.pto_973160.x. [DOI] [PubMed] [Google Scholar]
- 15.Yuan R, Madani S, Wei XX, Reynolds K, Huang SM. Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos. 2002;30:1311–1319. doi: 10.1124/dmd.30.12.1311. [DOI] [PubMed] [Google Scholar]
- 16.Waxman DJ, Chang TK. Use of 7-ethoxycoumarin to monitor multiple enzymes in the human CYP1, CYP2, and CYP3 families. Methods Mol Biol. 2006;320:153–156. doi: 10.1385/1-59259-998-2:153. [DOI] [PubMed] [Google Scholar]
- 17.Rendic S. Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002;34:83–448. doi: 10.1081/DMR-120001392. [DOI] [PubMed] [Google Scholar]
- 18.Zhou SF, Koh HL, Gao YH, Gong ZY, Lee EJD. Herbal bioactivation: the good, the bad and the ugly. Life Sci. 2004;74:935–968. doi: 10.1016/j.lfs.2003.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueng YF, Hsieh CH, Don MJ, Chi CW, Ho LK. Identification of the main human cytochrome P450 enzymes involved in safrole 1'-hydroxylation. Chem Res Toxicol. 2004;17:1151–1156. doi: 10.1021/tx030055p. [DOI] [PubMed] [Google Scholar]
- 20.Stiborova M, Frei E, Wiessler M, Schmeiser HH. Human enzymes involved in the metabolic activation of carcinogenic aristolochic acids: evidence for reductive activation by cytochromes P450 1A1 and 1A2. Chem Res Toxicol. 2001;14:1128–1137. doi: 10.1021/tx010059z. [DOI] [PubMed] [Google Scholar]
- 21.Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutat Res. 1998;400:201–213. doi: 10.1016/s0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 22.Crespi CL, Penman BW, Steimel DT, Smith T, Yang CS, Sutter TR. Development of a human lymphoblastoid cell line constitutively expressing human CYP1B1 cDNA: substrate specificity with model substrates and promutagens. Mutagenesis. 1997;12:83–89. doi: 10.1093/mutage/12.2.83. [DOI] [PubMed] [Google Scholar]
- 23.Skene DJ, Papagiannidou E, Hashemi E, et al. Contribution of CYP1A2 in the hepatic metabolism of melatonin: studies with isolated microsomal preparations and liver slices. J Pineal Res. 2001;31:333–342. doi: 10.1034/j.1600-079X.2001.310408.x. [DOI] [PubMed] [Google Scholar]
- 24.Lambrecht RW, Sinclair PR, Gorman N, Sinclair JF. Uroporphyrinogen oxidation catalyzed by reconstituted cytochrome P450 1A2. Arch Biochem Biophys. 1992;294:504–510. doi: 10.1016/0003-9861(92)90717-B. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki H, Shaw PM, Guengerich FP, Shimada T. Roles of cytochromes P450 1A2 and 3A4 in the oxidation of estradiol and estrone in human liver microsomes. Chem Res Toxicol. 1998;11:659–665. doi: 10.1021/tx970217f. [DOI] [PubMed] [Google Scholar]
- 26.Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: transcription, receptor regulation, and expanding biological roles. Curr Drug Metab. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- 27.Ma Q, Lu AY. CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos. 2007;35:1009–1016. doi: 10.1124/dmd.107.015826. [DOI] [PubMed] [Google Scholar]
- 28.Backman JT, Granfors MT, Neuvonen PJ. Rifampicin is only a weak inducer of CYP1A2-mediated presystemic and systemic metabolism: studies with tizanidine and caffeine. Eur J Clin Pharmacol. 2006;62:451–461. doi: 10.1007/s00228-006-0127-x. [DOI] [PubMed] [Google Scholar]
- 29.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 30.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carver LA, Bradfield CA. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J Biol Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 32.Yueh MF, Huang YH, Hiller A, Chen S, Nguyen N, Tukey RH. Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1. J Biol Chem. 2003;278:15001–15006. doi: 10.1074/jbc.M300645200. [DOI] [PubMed] [Google Scholar]
- 33.Ohyama K, Nakajima M, Suzuki M, Shimada N, Yamazaki H, Yokoi T. Inhibitory effects of amiodarone and its N-deethylated metabolite on human cytochrome P450 activities: prediction of in vivo drug interactions. Br J Clin Pharmacol. 2000;49:244–253. doi: 10.1046/j.1365-2125.2000.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thelingwani RS, Zvada SP, Hugues D, Ungell AL, Masimirembwa CM. In vitro and in silico identification and characterisation of thiabendazole as a mechanism-based inhibitor of CYP1A2 and simulation of possible pharmacokinetic drug–drug interactions. Drug Metab Dispos. 2009;37:1286–94. [DOI] [PubMed]
- 35.Zhu Q, Liao J, Xie L, Wang GJ, Liu XD. Mechanism-based inhibition of CYP1A2 by antofloxacin, an 8-NH2 derivative of levofloxacin in rats. Xenobiotica. 2009;39:293–301. doi: 10.1080/00498250802709428. [DOI] [PubMed] [Google Scholar]
- 36.Langouet S, Furge LL, Kerriguy N, Nakamura K, Guillouzo A, Guengerich FP. Inhibition of human cytochrome P450 enzymes by 1, 2-dithiole-3-thione, oltipraz and its derivatives, and sulforaphane. Chem Res Toxicol. 2000;13:245–252. doi: 10.1021/tx990189w. [DOI] [PubMed] [Google Scholar]
- 37.Chang TK, Chen J, Lee WB. Differential inhibition and inactivation of human CYP1 enzymes by trans-resveratrol: evidence for mechanism-based inactivation of CYP1A2. J Pharmacol Exp Ther. 2001;299:874–82. [PubMed]
- 38.Shimada T, Yamazaki H, Foroozesh M, Hopkins NE, Alworth WL, Guengerich FP. Selectivity of polycyclic inhibitors for human cytochrome P450s 1A1, 1A2, and 1B1. Chem Res Toxicol. 1998;11:1048–1056. doi: 10.1021/tx980090+. [DOI] [PubMed] [Google Scholar]
- 39.Cho US, Park EY, Dong MS, Park BS, Kim K, Kim KH. Tight-binding inhibition by α-naphthoflavone of human cytochrome P450 1A2. Biochim Biophys Acta. 2003;1648:195–202. doi: 10.1016/s1570-9639(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 40.Jensen KG, Poulsen HE, Doehmer J, Loft S. Kinetics and inhibition by fluvoxamine of phenacetin O-deethylation in V79 cells expressing human CYP1A2. Pharmacol Toxicol. 1995;76:286–288. doi: 10.1111/j.1600-0773.1995.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen BB, Maenpaa J, Pelkonen O, et al. Selective serotonin reuptake inhibitors and theophylline metabolism in human liver microsomes: potent inhibition by fluvoxamine. Br J Clin Pharmacol. 1995;39:151–159. doi: 10.1111/j.1365-2125.1995.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker AC, Preston T, Heaf D, Kitteringham NR, Choonara I. Inhibition of caffeine metabolism by ciprofloxacin in children with cystic fibrosis as measured by the caffeine breath test. Br J Clin Pharmacol. 1994;38:573–576. doi: 10.1111/j.1365-2125.1994.tb04399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granfors MT, Backman JT, Neuvonen M, Neuvonen PJ. Ciprofloxacin greatly increases concentrations and hypotensive effect of tizanidine by inhibiting its cytochrome P450 1A2-mediated presystemic metabolism. Clin Pharmacol Ther. 2004;76:598–606. doi: 10.1016/j.clpt.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Wei MJ, Zhao CY, Qi HM. Determination of the inhibitory potential of 6 fluoroquinolones on CYP1A2 and CYP2C9 in human liver microsomes. Acta Pharmacol Sin. 2008;29:1507–1514. doi: 10.1111/j.1745-7254.2008.00908.x. [DOI] [PubMed] [Google Scholar]
- 45.Ueng YF, Jan WC, Lin LC, Chen TL, Guengerich FP, Chen CF. The alkaloid rutaecarpine is a selective inhibitor of cytochrome P450 1A in mouse and human liver microsomes. Drug Metab Dispos. 2002;30:349–353. doi: 10.1124/dmd.30.3.349. [DOI] [PubMed] [Google Scholar]
- 46.Qiu F, Zhang R, Sun J, et al. Inhibitory effects of seven components of danshen extract on catalytic activity of cytochrome P450 enzyme in human liver microsomes. Drug Metab Dispos. 2008;36:1308–1314. doi: 10.1124/dmd.108.021030. [DOI] [PubMed] [Google Scholar]
- 47.Liang HC, Li H, McKinnon RA, et al. CYP1A2(−/−) null mutant mice develop normally but show deficient drug metabolism. Proc Natl Acad Sci U S A. 1996;93:1671–1676. doi: 10.1073/pnas.93.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang HC, McKinnon RA, Nebert DW. Sensitivity of CYP1A1 mRNA inducibility by dioxin is the same in Cyp1a2+/+ wild-type and Cyp1a2−/− null mutant mice. Biochem Pharmacol. 1997;54:1127–1131. doi: 10.1016/S0006-2952(97)00263-3. [DOI] [PubMed] [Google Scholar]
- 49.Lasker JM, Huang MT, Conney AH. In vivo activation of zoxazolamine metabolism by flavone. Science. 1982;216:1419–1421. doi: 10.1126/science.7089530. [DOI] [PubMed] [Google Scholar]
- 50.Buters JT, Tang BK, Pineau T, Gelboin HV, Kimura S, Gonzalez FJ. Role of CYP1A2 in caffeine pharmacokinetics and metabolism: studies using mice deficient in CYP1A2. Pharmacogenetics. 1996;6:291–296. doi: 10.1097/00008571-199608000-00002. [DOI] [PubMed] [Google Scholar]
- 51.Tsuneoka Y, Dalton TP, Miller ML, et al. 4-Aminobiphenyl-induced liver and urinary bladder DNA adduct formation in CYP1A2−/− and CYP1A2+/+ mice. J Natl Cancer Inst. 2003;95:1227–1237. doi: 10.1093/jnci/djg025. [DOI] [PubMed] [Google Scholar]
- 52.Smith AG, Davies R, Dalton TP, et al. Intrinsic hepatic phenotype associated with the Cyp1a2 gene as shown by cDNA expression microarray analysis of the knockout mouse. EHP Toxicogenomics. 2003;111:45–51. [PubMed] [Google Scholar]
- 53.Kall MA, Clausen J. Dietary effect on mixed function P450 1A2 activity assayed by estimation of caffeine metabolism in man. Hum Exp Toxicol. 1995;14:801–807. doi: 10.1177/096032719501401004. [DOI] [PubMed] [Google Scholar]
- 54.Relling MV, Lin JS, Ayers GD, Evans WE. Racial and gender differences in N-acetyltransferase, xanthine oxidase, and CYP1A2 activities. Clin Pharmacol Ther. 1992;52:643–658. doi: 10.1038/clpt.1992.203. [DOI] [PubMed] [Google Scholar]
- 55.Rasmussen BB, Brix TH, Kyvik KO, Brosen K. The interindividual differences in the 3-demethylation of caffeine alias CYP1A2 is determined by both genetic and environmental factors. Pharmacogenetics. 2002;12:473–478. doi: 10.1097/00008571-200208000-00008. [DOI] [PubMed] [Google Scholar]
- 56.Aitchison KJ, Gonzalez FJ, Quattrochi LC, et al. Identification of novel polymorphisms in the 5′ flanking region of CYP1A2, characterization of interethnic variability, and investigation of their functional significance. Pharmacogenetics. 2000;10:695–704. doi: 10.1097/00008571-200011000-00004. [DOI] [PubMed] [Google Scholar]
- 57.Soyama A, Saito Y, Hanioka N, et al. Single nucleotide polymorphisms and haplotypes of CYP1A2 in a Japanese population. Drug Metab Pharmacokinet. 2005;20:24–33. doi: 10.2133/dmpk.20.24. [DOI] [PubMed] [Google Scholar]
- 58.Chida M, Yokoi T, Fukui T, Kinoshita M, Yokota J, Kamataki T. Detection of three genetic polymorphisms in the 5′-flanking region and intron 1 of human CYP1A2 in the Japanese population. Jpn J Cancer Res. 1999;90:899–902. doi: 10.1111/j.1349-7006.1999.tb00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T. Genetic polymorphism in the 5'-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J Biochem. 1999;125:803–808. doi: 10.1093/oxfordjournals.jbchem.a022352. [DOI] [PubMed] [Google Scholar]
- 60.Sachse C, Brockmoller J, Bauer S, Roots I. Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br J Clin Pharmacol. 1999;47:445–449. doi: 10.1046/j.1365-2125.1999.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghotbi R, Christensen M, Roh HK, Ingelman-Sundberg M, Aklillu E, Bertilsson L. Comparisons of CYP1A2 genetic polymorphisms, enzyme activity and the genotype–phenotype relationship in Swedes and Koreans. Eur J Clin Pharmacol. 2007;63:537–546. doi: 10.1007/s00228-007-0288-2. [DOI] [PubMed] [Google Scholar]
- 62.Han XM, Ouyang DS, Chen XP, et al. Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br J Clin Pharmacol. 2002;54:540–543. doi: 10.1046/j.1365-2125.2002.01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aklillu E, Carrillo JA, Makonnen E, et al. Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol Pharmacol. 2003;64:659–669. doi: 10.1124/mol.64.3.659. [DOI] [PubMed] [Google Scholar]
- 64.Huang JD, Guo WC, Lai MD, Guo YL, Lambert GH. Detection of a novel cytochrome P450 1A2 polymorphism (F21L) in Chinese. Drug Metab Dispos. 1999;27:98–101. [PubMed] [Google Scholar]
- 65.Sachse C, Bhambra U, Smith G, et al. Polymorphisms in the cytochrome P450 CYP1A2 gene (CYP1A2) in colorectal cancer patients and controls: allele frequencies, linkage disequilibrium and influence on caffeine metabolism. Br J Clin Pharmacol. 2003;55:68–76. doi: 10.1046/j.1365-2125.2003.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pucci L, Geppetti A, Maggini V, Lucchesi D, Maria Rossi A, Longo V. CYP1A2 F21L and F186L polymorphisms in an Italian population sample. Drug Metab Pharmacokinet. 2007;22:220–222. doi: 10.2133/dmpk.22.220. [DOI] [PubMed] [Google Scholar]
- 67.Chevalier D, Cauffiez C, Allorge D, et al. Five novel natural allelic variants—951A>C, 1042G>A (D348N), 1156A>T (I386F), 1217G>A (C406Y) and 1291C>T (C431Y)—of the human CYP1A2 gene in a French Caucasian population. Hum Mutat. 2001;17:355–356. [PubMed] [Google Scholar]
- 68.Bilgen T, Tosun O, Luleci G, Keser I. Frequencies of four genetic polymorphisms in the CYP1A2 gene in Turkish population. Genetika. 2008;44:1133–1136. [PubMed] [Google Scholar]
- 69.Hamdy SI, Hiratsuka M, Narahara K, et al. Genotyping of four genetic polymorphisms in the CYP1A2 gene in the Egyptian population. Br J Clin Pharmacol. 2003;55:321–324. doi: 10.1046/j.1365-2125.2003.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skarke C, Kirchhof A, Geisslinger G, Lotsch J. Rapid genotyping for relevant CYP1A2 alleles by pyrosequencing. Eur J Clin Pharmacol. 2005;61:887–892. doi: 10.1007/s00228-005-0029-3. [DOI] [PubMed] [Google Scholar]
- 71.Obase Y, Shimoda T, Kawano T, et al. Polymorphisms in the CYP1A2 gene and theophylline metabolism in patients with asthma. Clin Pharmacol Ther. 2003;73:468–474. doi: 10.1016/S0009-9236(03)00013-4. [DOI] [PubMed] [Google Scholar]
- 72.Eap CB, Bender S, Jaquenoud Sirot E, et al. Nonresponse to clozapine and ultrarapid CYP1A2 activity: clinical data and analysis of CYP1A2 gene. J Clin Psychopharmacol. 2004;24:214–219. doi: 10.1097/01.jcp.0000116646.91923.2f. [DOI] [PubMed] [Google Scholar]
- 73.Ozdemir V, Kalow W, Okey AB, et al. Treatment-resistance to clozapine in association with ultrarapid CYP1A2 activity and the C>A polymorphism in intron 1 of the CYP1A2 gene: effect of grapefruit juice and low-dose fluvoxamine. J Clin Psychopharmacol. 2001;21:603–607. doi: 10.1097/00004714-200112000-00011. [DOI] [PubMed] [Google Scholar]
- 74.Melkersson KI, Scordo MG, Gunes A, Dahl ML. Impact of CYP1A2 and CYP2D6 polymorphisms on drug metabolism and on insulin and lipid elevations and insulin resistance in clozapine-treated patients. J Clin Psychiatry. 2007;68:697–704. doi: 10.4088/JCP.v68n0506. [DOI] [PubMed] [Google Scholar]
- 75.Berecz R, de la Rubia A, Dorado P, Fernandez-Salguero P, Dahl ML, Llerena A. Thioridazine steady-state plasma concentrations are influenced by tobacco smoking and CYP2D6, but not by the CYP2C9 genotype. Eur J Clin Pharmacol. 2003;59:45–50. doi: 10.1007/s00228-003-0576-4. [DOI] [PubMed] [Google Scholar]
- 76.Mihara K, Kondo T, Suzuki A, et al. Effects of genetic polymorphism of CYP1A2 inducibility on the steady-state plasma concentrations of trazodone and its active metabolite m-chlorophenylpiperazine in depressed Japanese patients. Pharmacol Toxicol. 2001;88:267–270. doi: 10.1034/j.1600-0773.2001.d01-115.x. [DOI] [PubMed] [Google Scholar]
- 77.Hartter S, Korhonen T, Lundgren S, et al. Effect of caffeine intake 12 or 24 hours prior to melatonin intake and CYP1A2*1F polymorphism on CYP1A2 phenotyping by melatonin. Basic Clin Pharmacol Toxicol. 2006;99:300–304. doi: 10.1111/j.1742-7843.2006.pto_491.x. [DOI] [PubMed] [Google Scholar]
- 78.Bohanec Grabar P, Rozman B, Tomsic M, Suput D, Logar D, Dolzan V. Genetic polymorphism of CYP1A2 and the toxicity of leflunomide treatment in rheumatoid arthritis patients. Eur J Clin Pharmacol. 2008;64(9):871–876. doi: 10.1007/s00228-008-0498-2. [DOI] [PubMed] [Google Scholar]
- 79.Lewis DF, Lake BG. Molecular modelling of CYP1A subfamily members based on an alignment with CYP102: rationalization of CYP1A substrate specificity in terms of active site amino acid residues. Xenobiotica. 1996;26:723–753. doi: 10.3109/00498259609046745. [DOI] [PubMed] [Google Scholar]
- 80.Mayuzumi H, Sambongi C, Hiroya K, Shimizu T, Tateishi T, Hatano M. Effect of mutations of ionic amino acids of cytochrome P450 1A2 on catalytic activities toward 7-ethoxycoumarin and methanol. Biochemistry. 1993;32:5622–5628. doi: 10.1021/bi00072a018. [DOI] [PubMed] [Google Scholar]
- 81.Dai R, Zhai S, Wei X, Pincus MR, Vestal RE, Friedman FK. Inhibition of human cytochrome P450 1A2 by flavone: a molecular modeling study. J Protein Chem. 1998;17:643–650. doi: 10.1007/BF02780965. [DOI] [PubMed] [Google Scholar]
- 82.Hadjokas NE, Dai R, Friedman FK, et al. Arginine to lysine 108 substitution in recombinant CYP1A2 abolishes methoxyresorufin metabolism in lymphoblastoid cells. Br J Pharmacol. 2002;136:347–352. doi: 10.1038/sj.bjp.0704711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lozano JJ, Pastor M, Cruciani G, et al. 3D-QSAR methods on the basis of ligand–receptor complexes. Application of COMBINE and GRID/GOLPE methodologies to a series of CYP1A2 ligands. J Comput Aided Mol Des. 2000;14:341–353. doi: 10.1023/A:1008164621650. [DOI] [PubMed] [Google Scholar]
- 84.Parikh A, Josephy PD, Guengerich FP. Selection and characterization of human cytochrome P450 1A2 mutants with altered catalytic properties. Biochemistry. 1999;38:5283–5289. doi: 10.1021/bi990142+. [DOI] [PubMed] [Google Scholar]
- 85.Mayuzumi H, Shimizu T, Sambongi C, Hiroya K, Hatano M. Essential role of His163 of cytochrome P450 1A2 in catalytic functions associated with cytochrome b5. Arch Biochem Biophys. 1994;310:367–372. doi: 10.1006/abbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 86.Cvrk T, Strobel HW. Photoaffinity labeling of cytochrome P4501A1 with azidocumene: identification of cumene hydroperoxide binding region. Arch Biochem Biophys. 1998;349:95–104. doi: 10.1006/abbi.1997.0464. [DOI] [PubMed] [Google Scholar]
- 87.Krainev AG, Shimizu T, Hiroya K, Hatano M. Effect of mutations at Lys250, Arg251, and Lys253 of cytochrome P450 1A2 on the catalytic activities and the bindings of bifunctional axial ligands. Arch Biochem Biophys. 1992;298:198–203. doi: 10.1016/0003-9861(92)90113-B. [DOI] [PubMed] [Google Scholar]
- 88.Shimizu T, Murakami Y, Hatano M. Glu318 and Thr319 mutations of cytochrome P450 1A2 remarkably enhance homolytic O-O cleavage of alkyl hydroperoxides. An optical absorption spectral study. J Biol Chem. 1994;269:13296–13304. [PubMed] [Google Scholar]
- 89.Shimizu T, Ito O, Hatano M, Fujii-Kuriyama Y. CO binding studies of engineered cytochrome P450ds: effects of mutations at putative distal sites in the presence of polycyclic hydrocarbons. Biochemistry. 1991;30:4659–4662. doi: 10.1021/bi00233a004. [DOI] [PubMed] [Google Scholar]
- 90.Krainev AG, Shimizu T, Ishigooka M, Hiroya K, Hatano M, Fujii-Kuriyama Y. Absorption spectral study of cytochrome P450d–phenyl isocyanide complexes: effects of mutations at the putative distal site on the conformational stability. Biochemistry. 1991;30:11206–11211. doi: 10.1021/bi00111a003. [DOI] [PubMed] [Google Scholar]
- 91.Hiroya K, Murakami Y, Shimizu T, Hatano M, de Montellano PR Ortiz. Differential roles of Glu318 and Thr319 in cytochrome P450 1A2 catalysis supported by NADPH-cytochrome P450 reductase and tert-butyl hydroperoxide. Arch Biochem Biophys. 1994;310:397–401. doi: 10.1006/abbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 92.Nakano R, Sato H, Watanabe A, Ito O, Shimizu T. Conserved Glu318 at the cytochrome P450 1A2 distal site is crucial in the nitric oxide complex stability. J Biol Chem. 1996;271:8570–8574. doi: 10.1074/jbc.271.15.8570. [DOI] [PubMed] [Google Scholar]
- 93.Pavanello S, Pulliero A, Lupi S, Gregorio P, Clonfero E. Influence of the genetic polymorphism in the 5'-noncoding region of the CYP1A2 gene on CYP1A2 phenotype and urinary mutagenicity in smokers. Mutat Res. 2005;587:59–66. doi: 10.1016/j.mrgentox.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 94.Nordmark A, Lundgren S, Ask B, Granath F, Rane A. The effect of the CYP1A2 *1F mutation on CYP1A2 inducibility in pregnant women. Br J Clin Pharmacol. 2002;54:504–510. doi: 10.1046/j.1365-2125.2002.01673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X, Wang L, Zhi L, et al. The G-3113A polymorphism in CYP1A2 affects the caffeine metabolic ratio in a Chinese population. Clin Pharmacol Ther. 2005;78:249–259. doi: 10.1016/j.clpt.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 96.Han XM, Ou-Yang DS, Lu PX, et al. Plasma caffeine metabolite ratio (17X/137X) in vivo associated with G-2964A and C734A polymorphisms of human CYP1A2. Pharmacogenetics. 2001;11:429–435. doi: 10.1097/00008571-200107000-00006. [DOI] [PubMed] [Google Scholar]
- 97.Dandara C, Basvi PT, Bapiro TE, Sayi J, Hasler JA. Frequency of −163 C>A and 63 C>G single nucleotide polymorphism of cytochrome P450 1A2 in two African populations. Clin Chem Lab Med. 2004;42:939–941. doi: 10.1515/CCLM.2004.152. [DOI] [PubMed] [Google Scholar]
- 98.B'Chir F, Pavanello S, Knani J, Boughattas S, Arnaud MJ, Saguem S. CYP1A2 genetic polymorphisms and adenocarcinoma lung cancer risk in the Tunisian population. Life Sci. 2009;84(21–22):779–784. doi: 10.1016/j.lfs.2009.03.008. [DOI] [PubMed] [Google Scholar]