Abstract
The identification of complex disease susceptibility loci has been accelerated considerably by advances in high-throughput genotyping technologies, improved insight into correlation patterns of common variants and the availability of large-scale sample sets. Linkage scans and small-scale candidate gene studies have now given way to genome-wide association scans. In this review, we summarize insights gained from the past, highlight practical issues relating to the design and analysis of current state-of-the-art GWA studies and look into future trends in the field of human complex trait genetics.
Keywords: association study, complex disease, single nucleotide polymorphism, genome-wide association scan, meta-analysis, sequencing
INTRODUCTION
Common complex diseases have traditionally been ascribed to complicated networks of genetic and environmental factors. The search for genetic susceptibility loci has been much more straightforward for Mendelian disorders than for multifactorial traits, where numerous variants of modest or small effect sizes contribute to the genetic background of disease. The common disease–common variant and multiple rare variant hypotheses had been proposed as distinct scenarios and polarized the field of complex disease genetics for some time. However, emerging evidence indicates that the genetic aetiology of complex traits is likely to be based on a combination of multiple rare and common susceptibility loci.
The field of human complex trait genetics has undergone major transformation over the past decade. Researchers have gradually moved from family-based approaches for investigating linkage to association studies offering (and, lately, delivering) the promise of complex disease locus robust identification. The journey has witnessed study design trends come and go, with valuable lessons learnt from each such era. Rapid technological developments, coupled with the availability of larger sample sizes and a better understanding of human genome sequence variation, continue to facilitate progress in the field. In this review, we aim to distil lessons from the past few years in the field of complex disease genetics, describe the present state-of-the-art for finding common susceptibility loci and look into emerging themes for the near future.
PAST
Genetic association studies have, over the last decade, evolved from genome-wide linkage scans to candidate gene approaches, to gene-centric designs aiming to capture the majority of common variation and, ultimately, to genome-wide association (GWA) scans. Several factors have influenced this trajectory, including our understanding of human genome sequence variation, and ongoing development of genotyping technologies (moving from low- to medium- to high-throughput approaches).
Family-based linkage studies prevailed in the literature for several years as they constituted the only means of targeting variation genome-wide at the time. Linkage studies tended to lead to the identification of numerous peaks that were rarely reproduced in independent studies. For example, in type 2 diabetes (T2D), although more than 40 linkage scans have been performed, the overall picture has been one of multiple modest signals, few of which show evidence of replication [1, 2]. Linkage signals typically encompass several megabases of sequence and the resulting localization resolution is low [although this improved marginally when single nucleotide polymorphism (SNP)-based linkage scans were introduced] [3, 4]. Consortia formed for the meta-analysis of linkage scans of particular phenotypes served to distil the number of statistically believable linkage peaks [2] and promising signals were traditionally followed up by fine-mapping experiments [5]. Very few such endeavours have led to the identification of causal disease susceptibility variants [6, 7]. This is perhaps not surprising, as linkage disequilibrium (LD) mapping efforts under linkage peaks tended to make use of SNPs with common minor allele frequencies (MAFs), whereas linkage signals were more likely to reflect more penetrant effects of rare variants. Moreover, because of the relatively small number of families and microsatellite markers used, most of these studies may have been underpowered to detect many of the effects that association approaches have thus far discovered.
The field shifted towards association studies, exemplified over the last decade by the candidate gene study. Candidate gene studies focused on a few, if not just a single, variant(s) within a biologically plausible candidate gene. They were typically carried out in a few hundreds of disease cases and controls, or in a few hundreds of nuclear families, consisting of affected offsprings and unaffected parents. The latter approach (transmission disequilibrium test) [8] reached high popularity levels in the nineties due to its property of being robust to population stratification. Although several notable exceptions exist (for example [9, 10] from the field of T2D), candidate gene studies on the whole did not deliver many robustly replicating disease susceptibility loci. This irreproducibility of results could be ascribed to a combination of several contributing factors: low power (as a result of small sample sizes) to detect what we now recognize as modest or small effects; limited understanding of disease aetiopathogenesis leading to inappropriate selection of candidate loci; low thresholds for declaring significance and over-interpretation of results; and inadequate capture of variation across the genes of interest.
The International HapMap Project [11] greatly increased our understanding of correlation patterns (LD) between common variants across the genome. This enabled the selection of maximally informative, non-redundant sets of markers across genes or regions of interest. A wide variety of haplotype-based and pairwise tagging methods were developed [12–15]. Tag SNP studies continue to be carried out; they employ information from relevant HapMap populations to select SNPs capturing the majority of common variation across targeted loci. These markers are then genotyped and analysed in the datasets of interest, and inferences about their proxy variants are made on the basis of the association patterns observed.
Advances in high-throughput, high-accuracy genotyping platforms marked a new era for association studies, enabling the concurrent examination of hundreds of thousands of SNPs. Sufficient power in GWA studies was facilitated by the availability of large-scale sample collections. Over the last few years, GWA scans have succeeded in detecting and establishing complex trait associations, and have started to provide valuable insights into disease aetiopathogenesis.
PRESENT
GWA studies undoubtedly constitute the present state-of-the-art in efforts to elucidate the genetic aetiology of complex phenotypes. Several commercial products offering the potential to simultaneously assay hundreds of thousands of SNPs genome-wide are available from companies such as Affymetrix and Illumina. These have varying SNP content and density, and have been designed using diverse marker selection strategies (Table 1). For example, arrays with an exon-centric SNP content, such as the Illumina Human-1, reflect strategies focusing on potentially functional variants. LD-based platforms contain tag sets of SNPs selected to maximize the amount of common variation captured on the basis of HapMap data. Affymetrix platforms comprise quasi-randomly distributed SNPs or a combination of random and tag SNPs. In recognition of their potential role in complex disease susceptibility, copy number variants (CNVs) are also increasingly featured.
Table 1:
Overview of marker content and array design across commercially available platforms and coverage of common variation (MAF > 0.05) based on HapMap phase II data
| Platform | Number of markers | Array design | Coverage in CEUa (%) | Coverage in JPTb + CHBc (%) | Coverage in YRId (%) | Source |
|---|---|---|---|---|---|---|
| Illumina Human-1 | More than 109 000 | Gene | 26 | 28 | 12 | [16] |
| Illumina HumanHap300 | 317 511 | Tag | 75 | 63 | 28 | [16] |
| Affymetrix SNP Array 5.0 | 500 568 | Random | 65 | 66 | 41 | [16] |
| Illumina HumanHap550 | 555 352 | Tag | 87 | 83 | 50 | [17] |
| Illumina Human610 | 620 901 | Tag, CNVe | 89 | 86 | 58 | [18] |
| Illumina HumanHap650Y | 660 917 | Tag | 87 | 84 | 60 | [17] |
| Affymetrix SNP Array 6.0 | More than 1 800 000 | Random + Tag, CNVe | 83 | 84 | 62 | [17] |
| Illumina Human1M | 1 199 187 | Tag, CNVe | 93 | 92 | 68 | [17] |
aUtah residents with ancestry from northern and western Europe.
bJapanese from Tokyo, Japan.
cHan Chinese from Beijing, China.
dYoruba from Ibadan, Nigeria.
eCopy number variation.
Table 1 summarizes the extent to which different platforms capture common (MAF > 0.05) variation based on published evaluations in the three different HapMap phase II populations [11]. Coverage in European- and East Asian-descent populations is very high and has substantially improved with next generation chips. Information capture in African-descent populations is lower, reflecting higher recombination rates and lower levels of inter-marker correlation. However, it has been shown theoretically that coverage of all common variation based on HapMap has been overestimated and that larger sample sizes and denser marker sets are required for more accurate estimation of tagging SNP efficacy [19, 20]. Overestimation of previously reported coverage estimates has also been empirically confirmed by the analysis of sequence-derived variation data from 76 genes in HapMap samples [21]. Although variation capture is an important consideration in GWA study design, it is not the sole determinant of power.
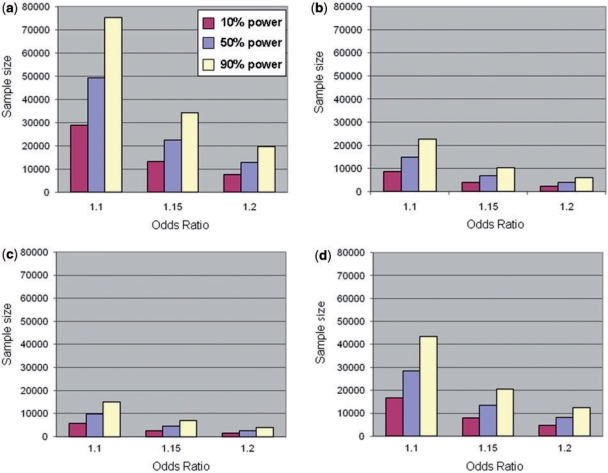
The statistical power of a GWA study to detect variants associated with disease is a function of sample size, the susceptibility locus effect magnitude, risk allele frequency of the queried SNP and its correlation with the causal variant. Although the allelic architecture of complex traits has not been fully characterized yet, recent GWA scans and follow-up studies have highlighted that common susceptibility loci are likely to have modest or small effect sizes [allelic odds ratios (ORs) between 1.1 and 1.5]. In a genome-wide setting, the large number of tests performed requires stringent thresholds for declaring statistical genome-wide significance (P = 5 × 10−8) [22, 23], necessitating large-scale sample sizes. For example, in order to achieve 90% power to detect a risk allele with 0.20 frequency and an allelic OR of 1.2 (at the genome-wide significance level), more than 6000 affected individuals and twice as many controls would be required (Figure 1). To achieve the same power to detect similar effects at lower frequency variants (frequency of 0.05 or less), a GWA study would need upwards of 20 000 cases (Figure 1).
Figure 1:
Number of affected individuals required (given a case/control ratio of 1:2) in order to achieve 10, 50 and 90% power to detect an effect at α = 5 × 10−8 for variants with modest to low effect sizes (allelic odds ratios 1.10, 1.15 and 1.20) and varying risk allele frequencies: (a) 0.05, (b) 0.20, (c) 0.50 and (d) 0.90. Calculations assume complete LD between the causal and genotyped variant.
Along with sample size considerations, GWA studies have also given rise to several logistical challenges: for example, issues relating to automated but accurate genotype calling, programmatic data handling and parsing, genotype quality control (QC) standards and analytical considerations that did not previously apply to smaller scale studies.
Genotype calling is the process by which hybridization intensities on genome-wide chips are translated into genotypes. Typically, intensities are normalized and transformed into coordinates which yield distinct genotype clouds. As high call rate and accuracy of genotype calling are important factors in safe-guarding QC standards in GWA scans, a variety of genotype calling algorithms have been developed and continue to evolve [24–27]. The possible adverse effects of inaccurate genotype calling in downstream analyses have been recognized for a while [28]. Therefore, inspection of intensity plots for interesting association signals is an essential aspect of genotype QC.
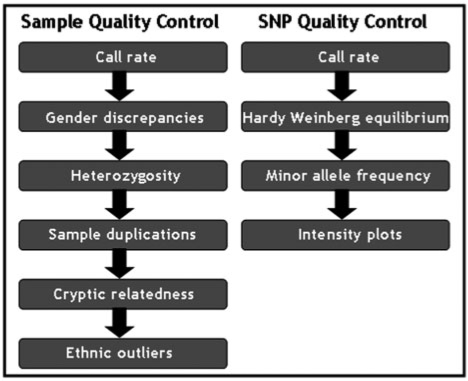
Genotype QC is an extremely important step in GWA studies, as it can dramatically reduce the number of false positive associations. The field has converged to an essential set of QC checks; Figure 2 summarizes the sample- and SNP-based QC steps that are typically employed.
Figure 2:
Flowchart of the main quality control steps in a GWA study.
SNP call rate is a good indicator of genotype probe performance. Removing SNPs with a greater proportion of missing genotypes is essential to control for false positives, as spurious associations can arise due to non-random missingness. Checking for gross departure from Hardy–Weinberg equilibrium (HWE) could help in identifying SNPs with genotyping errors (e.g. excess of heterozygotes). As clustering algorithms tend to perform less well for SNPs with low-frequency alleles, it is current practice in GWA studies to exclude rare SNPs from single point analyses (these are underpowered to detect effects anyway). Genotype calling algorithms have the potential to make incorrect calls. Therefore, inspecting intensity plots, though not feasible on a genome-wide scale, is necessary for SNPs with interesting association signals.
Sample call rate is a good indicator of hybridization performance; high rates of missingness usually indicate low DNA quality or problematic arrays. Discrepancies in gender assignment (SNP data versus phenotype data) can help identify sample mix-ups. Excess genome-wide heterozygosity may indicate possible contamination leading to a larger proportion of heterozygous genotypes. Accidentally duplicated and related individuals in large-scale studies can be identified through identity-by-descent estimation given identity-by-state information in a relatively large homogeneous sample [29]. Typically, the sample with the lowest call rate from each pair of related individuals is removed. Finally, ethnic outliers can be detected and either removed or accounted for in downstream analyses.
Population stratification can be a major confounding factor in GWA studies, both for case/control designs and population-based quantitative analyses. If undetected, it can lead to false positive associations due to differences in allele frequency between the different populations [30]. To guard against it, most GWA scans attempt to match cases and controls for broad ethnic background from the outset and then rely on statistical approaches to detect population substructure and correct for it [29, 31, 32]. Genomic control (λ) is an estimate of the degree of inflation of the test statistics genome-wide and can serve as a crude correction factor [31]. Principal component analysis [32] and multidimensional scaling [29] are methods employed to identify individuals of different ethnic origin visualized onto a two-dimensional projection on axes of genetic variation. Inferred principal components can be included as covariates in association analyses.
Directly typed SNPs in GWA studies are typically analysed by single-point methods, most frequently under the additive or multiplicative model. General models are less frequently tested as they increase dimensionality; dominant and recessive models are equally parsimonious but generally less powerful than the additive model. Multimarker tests (such as sliding haplotype window analyses) are less feasible at the genome-wide scale. However, imputation approaches have recently been developed to take into account information from multiple surrounding markers in order to infer genotypes at untyped loci [33]. Imputation therefore currently allows testing for association at >2.5 million markers genome-wide, thus maximizing information output from GWA studies, and additionally serves as an ideal tool for the combination of data from GWA scans that have been carried out on different platforms. The analysis of imputed data necessitates taking into account uncertainty by analysing the full genotype probability distribution appropriately.
The sheer number of SNPs tested for association with disease raises important statistical considerations about type I error and statistical significance levels. To account for the inflation in false positives, a variety of approaches, such as the conservative Bonferroni correction and the less stringent control of the false discovery rate [34], have been proposed. Obtaining empirical P-values after hundreds of thousands or millions of permutations are an alternative but prohibitively computer-intensive way to assess statistical significance. To overcome the multiple testing problem, stringent genome-wide significance thresholds have been proposed: adjustment for 1–2 million independent tests at common variants genome-wide has resulted in the aforementioned generally accepted significance threshold of P = 5 × 10−8 [22, 23]. In practise, most GWA studies prioritize signals for follow-up on the basis of their relative statistical strength for association and on evidence accrued from bioinformatics approaches. Replication in independent datasets (of the same variant, in the same direction, under the same model) constitutes the gold standard in genetic association studies of any scale.
T2D serves as a prime example of the success of the GWA scan approach. Over the past 2 years, multiple GWA scans have been published, greatly accelerating progress in identifying novel susceptibility variants for the disease [24, 35–42]. This first wave of studies collectively raised the number of established T2D loci to 11.
Approaches aiming to identify complex trait susceptibility loci have recently also extended to the meta-analysis of diverse scans carried out for the same phenotype. This move in the field has been brought about by the realization that effect sizes for common variants are becoming increasingly low. As Figure 1 attests, sample size is one of the most important factors in boosting power for an association study. Synergy across research groups, leading to the synthesis of GWA scan results, can greatly increase sample size and, hence, power to detect small individual effects. Several design and analytical challenges are associated with GWA scan meta-analysis (reviewed in [43]). These collaborative efforts have recently started to successfully extend the list of robustly replicating associations with complex traits [44–48]. For example, the Diabetes Genetics Initiative, Finland–United States Investigation of NIDDM and Wellcome Trust Case Control Consortium T2D scans undertook a three-way meta-analysis, which led to the identification of 6 novel susceptibility loci [44].
FUTURE
The first wave of GWA studies and meta-analyses conducted indicate that only a small amount of the genetic variance underlying the heritable component of common complex traits has been identified. For example, in the case of T2D, the so far identified loci account for <4% of the estimated heritability (reviewed in [49]). This reflects the fact that current studies involving thousands of individuals are still underpowered to discover most of the common genetic variants with the very modest to low effect sizes that are likely to exist. It is anticipated that sample sizes of many tens of thousands or even hundreds of thousands will be required to fulfil this purpose. The identification of further common variants with small effect sizes may not have immediate consequences in disease prediction and prognosis, but will hopefully continue to provide novel insights into implicated biological pathways, pointing to new targets for therapy. Therefore, the future is poised to continue in the same trend of large-scale consortia being formed to facilitate the accumulation of data and the combination of expertise, in order to make the next generation of GWA scan meta-analyses possible. These will in turn start to enable the investigation of gene–gene and gene–environment interactions, currently hindered by low power.
The associated SNPs uncovered by GWA scans are unlikely to be the functional polymorphisms. One of the major challenges that the field of complex disease genetics faces over the next few years is how best to explore information in association regions delineated by recombination hotspots, typically spanning several kilobases, in order to identify the truly causal variants. Deep resequencing in samples of interest and subsequent large-scale follow-up of interesting markers through fine-mapping is an emerging study design paradigm, enabled by next generation sequencing technologies. However, several study design issues remain unclear, including the choice of resequencing and fine-mapping samples and their ethnicity, sample size, spectrum of typed marker allele frequency and analytical approach. It is generally recognized that the benefits of fine-mapping will be finite, particularly in regions of very strong LD, and that functional studies will be necessary in order to pinpoint the truly causal variant. The availability of global gene expression profiles coupled with genotype data from the same samples can also serve as a valuable resource, as associated variants might display strong cis associations with expression of a nearby gene whose expression levels are causally linked with the underlying phenotype or disease trait [50].
The future of genetic association studies is poised to have an increasing focus on CNVs; this will be facilitated by ongoing efforts to provide a catalogue of structural variants (e.g. the CNV project [51]). Along with rare variants, CNVs could account for some of the missing complex trait heritability. For example, schizophrenia studies have uncovered CNV associations [52, 53] in a disease where GWA studies have not returned significant evidence for robust common SNP associations (reviewed in [54]).
Current studies are focused on common variants, which invariably have small effects. However, the field is now starting to recognize the role of rare variants, which can have larger effect sizes, in complex disease susceptibility. The analysis of lower frequency polymorphisms necessitates larger sample sizes and tailored analytical approaches in order to increase power [55]. The 1000 genomes project [56] will improve our understanding of variation at the lower end of the frequency spectrum and is expected to enhance information capture and interpretation in genetic association studies.
There is little doubt that large-scale sequencing studies will constitute the way forward for characterizing the allelic architecture of complex disease. Several challenges with respect to the design, analysis and interpretation of such studies continue to emerge and will undoubtedly keep researchers busy for the foreseeable future. The landscape of human complex disease genetics has witnessed major changes over the past 10 years, and is poised to change even more dramatically in the near future.
Key Points.
The genetic aetiology of complex traits is likely to be based on a combination of multiple common and rare susceptibility loci.
Genome-wide linkage scans and small-scale candidate gene studies had not previously met with widespread success.
GWA studies follow a hypothesis-free approach and interrogate the majority of common SNPs across the human genome.
Sufficiently large sample sizes, stringent genotype QC, use of appropriate significance thresholds and replication of findings in independent datasets have been crucial determinants of GWA study success.
Further advances in genotyping and next generation sequencing technologies, facilitating the study of rare and structural variation, hold the promise of an improved understanding of the allelic architecture of complex disease.
FUNDING
Wellcome Trust (WT088885/Z/09/Z).
Biographies
Kalliope Panoutsopoulou is a postdoctoral research fellow at the Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK, working towards the identification of genetic variants conferring susceptibility to osteoarthritis.
Eleftheria Zeggini is an investigator in Human Genetics at the Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK, where she leads the Applied Statistical Genetics team. Her research focus is on design, analysis and interpretation issues in large-scale complex disease association and resequencing studies.
References
- McCarthy MI. Growing evidence for diabetes susceptibility genes from genome scan data. Curr Diab Rep. 2003;3:159–67. doi: 10.1007/s11892-003-0040-y. [DOI] [PubMed] [Google Scholar]
- Guan W, Pluzhnikov A, Cox NJ, et al. Meta-analysis of 23 type 2 diabetes linkage studies from the International Type 2 Diabetes Linkage Analysis Consortium. Hum Hered. 2008;66:35–49. doi: 10.1159/000114164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Shephard N, Liu G, et al. Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet. 2004;75:54–64. doi: 10.1086/422195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet. 2004;75:687–92. doi: 10.1086/424696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Morris AP, McCarthy MI, et al. How useful is the fine-scale mapping of complex trait linkage peaks? Evaluating the impact of additional microsatellite genotyping on the posterior probability of linkage. Genet Epidemiol. 2005;28:1–10. doi: 10.1002/gepi.20023. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen D, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Ann Hum Genet. 1995;59:323–6. doi: 10.1111/j.1469-1809.1995.tb00751.x. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–72. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, et al. International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–7. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Miretti MM, Broxholme J, et al. A comparison of tagging methods and their tagging space. Hum Mol Genet. 2005;14:2757–67. doi: 10.1093/hmg/ddi309. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–62. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635–43. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- Whole-genome genotyping &. CNV analysis: human610-quad beadchip. [(9 March 2009, date last accessed)]. http://www.illumina.com/pages.ilmn?ID=248.
- Weale ME, Depondt C, Macdonald SJ, et al. Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet. 2003;73:551–65. doi: 10.1086/378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles MM. Quantification and correction of bias in tagging SNPs caused by insufficient sample size and marker density by means of haplotype-dropping. Genet Epidemiol. 2008;32:20–8. doi: 10.1002/gepi.20258. [DOI] [PubMed] [Google Scholar]
- Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nat Genet. 2008;40:841–3. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Cooper JD, Todd JA, et al. A method to address differential bias in genotyping in large-scale association studies. PLoS Genet. 2007;3:e74. doi: 10.1371/journal.pgen.0030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Inouye M, Small KS, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–6. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DG, Walker NM, Smyth DJ, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, et al. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–17. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–5. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Salonen JT, Uimari P, Aalto JM, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81:338–45. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinsthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–7. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The copy number variation project. [(21 May 2008, date last accessed)]. http://www.sanger.ac.uk/humgen/cnv.
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:178–9. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Craddock N, Daly M, et al. Psychiatric GWAS Consortium Coordinating Committee, Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–56. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 genomes project. [(9 March 2009, date last accessed)]. http://www.1000genomes.org.