Abstract
Leucine rich repeat kinase 2 (LRRK2) mutations are the most common genetic cause of Parkinson's disease (PD) although LRRK2 function remains unclear. We report a new role for LRRK2 in regulating autophagy and describe the recruitment of LRRK2 to the endosomal–autophagic pathway and specific membrane subdomains. Using a novel human genomic reporter cellular model, we found LRRK2 to locate to membrane microdomains such as the neck of caveolae, microvilli/filopodia and intraluminal vesicles of multivesicular bodies (MVBs). In human brain and in cultured human cells LRRK2 was present in cytoplasmic puncta corresponding to MVBs and autophagic vacuoles (AVs). Expression of the common R1441C mutation from a genomic DNA construct caused impaired autophagic balance evident by the accumulation of MVBs and large AVs containing incompletely degraded material and increased levels of p62. Furthermore, the R1441C mutation induced the formation of skein-like abnormal MVBs. Conversely, LRRK2 siRNA knockdown increased autophagic activity and prevented cell death caused by inhibition of autophagy in starvation conditions. The work necessitated developing a new, more efficient recombineering strategy, which we termed Sequential insertion of Target with ovErlapping Primers (STEP) to seamlessly fuse the green fluorescent protein-derivative YPet to the human LRRK2 protein in the LRRK2 genomic locus carried by a bacterial artificial chromosome. Taken together our data demonstrate the functional involvement of LRRK2 in the endosomal–autophagic pathway and the recruitment to specific membrane microdomains in a physiological human gene expression model suggesting a novel function for this important PD-related protein.
INTRODUCTION
Lewy body (LB) diseases, including Parkinson's disease (PD) and dementia with LBs, are common neurodegenerative disorders. Autosomal-dominant mutations in the leucine-rich repeat kinase 2 (LRRK2) gene were recently found to be the most important genetic cause of both familial and sporadic LB disease (1–3). In addition to LB pathology, patients with LRRK2 mutations can display tau tangles, motoneuron disease features, frontotemporal lobar degeneration or pure nigral degeneration (2,4), suggesting that LRRK2 may have a central role across the spectrum of neurodegenerative disease and may lie upstream in the disease-related chain of events, preceding the deposition of alpha-synuclein and tau (5). This hypothesis is supported by the recent discovery that LRRK2 protein accumulates in early-stage alpha-synuclein pathological lesions in LB disease (6), and multiple system atrophy (MSA) (7).
The function of LRRK2 is currently unclear, partly due to difficulties in its detection and unknown relevance of overexpression studies (8,9). LRRK2 is a G protein and a kinase activated by nucleotide-dependent dimerization (10). LRKK2 pathogenic mutations may affect dimerization (the R1441C/G mutations) or the kinase domain (G2019S or I2020T) (11). LRRK2 is expressed in most cells, reflecting involvement in basic cellular functions (2). Inside the cell LRRK2 associates with lipid rafts (12,13) but a detailed and physiologically-relevant characterization of the sub-cellular distribution of wild-type (WT) and mutant LRRK2 protein has been lacking. LRRK2 regulates neurite process morphology (14) perhaps through phosphorylation of ezrin, radixin and moesin (ERM proteins) (15), which are involved in neurite growth cone morphology, motility and process formation (16).
The endosomal–autophagic pathway is key in understanding neurodegeneration. The accumulation of autophagic vacuoles (AVs), which reflects autophagic stress, especially within dystrophic neurites, is a widespread and early pathological feature of neurodegeneration, particularly in PD (17). Interestingly, LRRK2 regulation of neurite processes needs autophagy (18). However, a role for LRRK2 in autophagy and its location in autophagic organelles has not been addressed.
To allow physiological expression and rapid detection of LRRK2 protein in human cells we developed a novel recombineering strategy, which we have termed Sequential insertion of Target with ovErlapping Primers (STEP), to fuse the bright fluorescent protein YPet to LRRK2 within a bacterial artificial chromosome (BAC) vector carrying the human genomic LRRK2 locus. Using this expression system in human cells we found that LRRK2 is specifically located to membrane microdomains and in cytoplasmic puncta which correspond to multivesicular bodies (MVBs) and AVs. The R1441C mutation induced autophagic stress characterized by the accumulation of abnormal MVBs and enlarged AVs with high levels of p62 (sequestosome-1) and by the appearance of skein-like inclusions. Finally, siRNA-mediated LRRK2 knockdown increased autophagic activity and prevented starvation-induced cell death when autophagy was inhibited. Our data support a novel role for LRRK2 and suggest a disease mechanism in PD involving dysfunction of the endosomal–autophagic pathway.
RESULTS
Construction and expression of LRRK2 genomic DNA fusion constructs
In order to express LRRK2 at more physiologically-relevant levels we used a BAC containing the human genomic LRRK2 locus. The BAC RP11-568G5 contained all LRRK2 51 exons and a ≈20 kb upstream region but lacked an alternative canonical poly(A) site, which we added using recombineering (19) to create BAC-LRRK2-WT.
To achieve LRRK2 protein detection at physiologically-relevant levels, we next inserted YPet into BAC-LRRK2-WT (BAC-YPet-LRRK2-WT), generating a fusion reporter YPet-LRRK2 protein. To create BAC-YPet-LRRK2-WT with efficiency we developed the new and broadly applicable strategy STEP for seamless insertion of reporter genes in genomic DNA vectors (Fig. 1A). Resulting bacterial colonies were screened by YPet-exon1 junction PCR and NotI digestion and correct sequence was confirmed across the modified region. We performed the insertion of the reporter gene with different lengths of homology and found that the efficiency, as measured by the percentage of correct colonies produced in the second recombination step, increased steeply in line with the total length of homology, from 1% with 276 bp homology, 10% with 299 bp and 50–96% with ≥310 bp.
Figure 1.
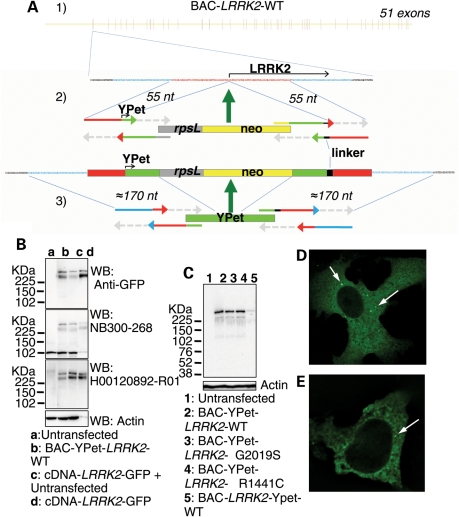
Construction and expression of fluorescently N-terminal tagged genomic fusion constructs of LRRK2 using STEP recombination. (A1) BAC-LRRK2-WT containing the LRRK2 human genomic locus was built by combining BAC RP11-115F18 and BAC RP11-568G5 inserts. (A2) In the first stage of STEP recombination a linear cassette containing the selection/counter-selection gene RpsL-neo was inserted in LRRK2 exon 1 at the starting ATG by homologous recombination. The RpsL-neo cassette was created by a single PCR reaction incorporating four 80-mer overlapping primers. The cassette contained at the 3′ and 5′ ends 55 bp homology regions which flanked the first ≈55 bp and last ≈55 bp, respectively, of Ypet and a peptide linker. (A3) The RpsL-neo gene was replaced by a marker-less linear cassette containing the full sequence of Ypet and additional homology regions created again by a single PCR reaction. Homologous recombination occurred efficiently through the extended ≈170 bp homology regions. (B) Protein lysates from cells transfected to express either cDNA-LRRK2-GFP or BAC-YPet-LRRK2-WT were used for immunoblotting using either an anti-GFP antibody (which recognizes YPet and GFP) or two anti-LRRK2 antibodies. The amount of tagged LRRK2 protein was equalized by loading 1 µg of total protein for cDNA-LRRK2-GFP and 100 µg of total protein for BAC-YPet-LRRK2-WT. A control was included containing 100 µg of protein from untransfected cells together with 1 µg of lysate containing overexpressed cDNA derived protein. (C) BAC-YPet-LRRK2-WT was engineered to carry either the G2019S or the R1441C mutations using recombineering. The N-terminally tagged BAC-YPet-LRRK2-WT, BAC-YPet-LRRK2-G2019S or BAC-YPet-LRRK2-R1441C as well as the C-terminally tagged BAC-LRRK2-Ypet-WT expressed full length LRRK2 protein as detected using anti-GFP antibodies. (D and E) Cellular expression of BAC-YPet-LRRK2-WT revealed a cytoplasmic pattern with the appearance of fine cytoplasmic puncta in Vero (D) and HEK293 (E) cells as detected by IF using anti-GFP antibodies.
BAC-YPet-LRRK2-WT expressed full-length LRRK2 protein of comparable size to a cDNA based C-terminal green fluorescent protein (GFP)-tagged construct (Fig. 1B). However, BAC-YPet-LRRK2-WT expressed 25–30 times less LRRK2 protein than cDNA-LRRK2-GFP when normalized for transfection efficiency (10–20% for the BAC and 85–95% for cDNA-LRRK2-GFP), consistent with our aim of expressing LRRK2 at physiologically-relevant levels. Expression of the LRRK2 transgene was confirmed using two anti-LRRK2 antibodies (Fig. 1B). Full-length LRRK2 protein expression was also achieved from an additional C-terminal tagged genomic construct (BAC-LRRK2-YPet-WT) (Fig. 1C).
Next, the G2019S or R1441C mutations were engineered into BAC-YPet-LRRK2-WT using recombineering. These mutations did not affect the level of expression or band pattern of LRRK2 protein (Fig. 1C).
LRRK2 is localized to fine cytoplasmic puncta in cultured cells and human brain
Using fluorescence microscopy or immunofluorescence (IF) with anti-GFP antibodies, LRRK2 appeared in a cytoplasmic pattern in two different cell lines transfected with BAC-YPet-LRRK2-WT (Vero and HEK293) (Fig. 1D, E and Supplementary Material, Figure S1) in contrast to untagged YPet which was distributed throughout the nucleus and cytoplasm (Supplementary Material, Figure S4). In addition LRRK2 localized to fine cytoplasmic puncta ≤1 µm of size (Fig. 1D and E).
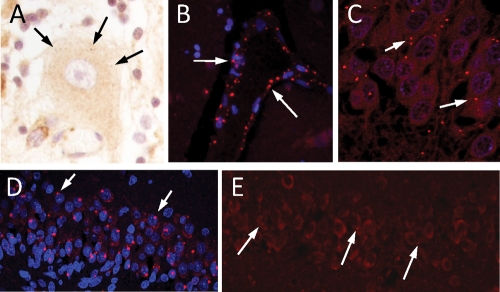
In control human brain we also found LRRK2 localizing to cytoplasmic puncta in neurons (Fig. 2A and E). However, LRRK2 puncta in neurons of the hippocampal fascia dentata were more easily identifiable in certain pathological conditions such as in a patient diagnosed with both AD and PD (Fig. 2C) and in a newborn infant who suffered cerebral hypoxia (Fig. 2D), conditions known to increase appearance of AVs. Identification of puncta in endothelial cells (Fig. 2B) may reflect LRRK2 presence in caveolae and transport vesicles involved in transcytosis (20).
Figure 2.
Appearance of LRRK2 puncta in human brain by IF. Staining of LRRK2 puncta in: (A) A Purkinje cell in a control case; (B) vascular endothelial cells in the striatum of a MSA patient; (C–E) fascia dentata of a patient diagnosed with both AD and PD (C), a 2-month-old infant (D) and an aged control (E). Anti-LRRK2 antibodies NB300-268 (A, B, C and E) or NB300-267 (D) were used.
LRRK2 localizes to specific membrane subdomains, including the neck of caveolae, microvilli and endosomal–autophagic structures
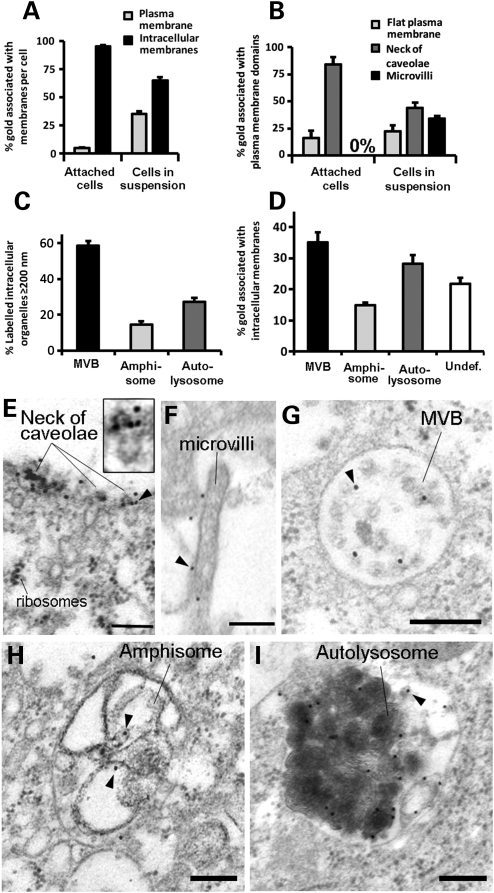
We next defined the sub-cellular localization of LRRK2 in human cells using immunoelectron microscopy (IEM) with anti-GFP antibodies (Fig. 3). Although LRRK2 was detected free in the cytosol (≈4% of total gold particles) it was mainly localized to cellular membranes (95.8 ± 0.4% of total gold particles) both at the plasma membrane (4.5 ± 0.7% of membrane gold particles) and intracellular membranes (95.4 ± 0.7% of membrane gold particles) (Fig. 3A). Interestingly, LRRK2 was concentrated on the outer portion of caveolae (84 ± 6.7% of plasma membrane gold particles), which is an important microdomain known as the neck of caveolae (Fig. 3B and E) (21). Since the neck of caveolae represents the counterpart of microvilli (21), we induced the formation of microvilli by detaching the cells from the culture substrate using trypsin. Under these conditions LRRK2 greatly redistributed to the plasma membrane (from 4.5 ± 0.7% to 35 ± 2.5% of membrane gold particles) (Fig. 3A) concentrating in the newly appeared microvilli (33.9 ± 2.6% of plasma membrane gold particles) (Fig. 3B and F) while the percentage of LRRK2 associated with the neck of caveolae decreased in the same proportion.
Figure 3.
LRRK2 localizes to the neck of caveolae, microvilli and the endosomal–autophagic pathway. (A) Quantification of the distribution of membrane-associated LRRK2 between the plasma membrane and intracellular membranous structures in attached or trypsinized cells. (B) Quantification of the distribution of LRRK2 within distinct plasma membrane domains in attached or trypsinized cells. (C) Quantification of the proportion of different organelles ≥200 nm (the resolution limit of light microscopy) labelled with LRRK2. (D) Quantification of the amount of LRRK2 within distinct intracellular membranous organelles. (E–I) IEM micrographs show LRRK2 gold labelling on the neck of caveolae (E and amplification of region identified by arrowhead in inset), microvilli (F), intraluminar vesicles of MVBs (G), amphisomes (H) and autolysosomes (I). Representative gold particles labelling LRRK2 are identified using arrowheads. HEK293 cells were transfected with BAC-YPet-LRRK2-WT and IEM was obtained with anti-GFP antibodies. Quantification of gold particles was obtained by counting eight random labelled cells. Bars represent mean + SEM. Scale bars represent 100 nm.
Within the cytoplasm of human cells LRRK2 specifically localized to endosomal–autophagic organelles including MVBs (Figs 3G, 4A3 and Supplementary Material, Figure S2D) and AVs [amphisomes (Figs 3H and 4A2) and autolysosomes (Fig. 3I and Supplementary Material, Figure S2D)] (22), all of them typically larger than 200 nm and therefore within the resolution limit of light microscopy. MVBs followed by AVs were the most abundantly labelled organelles >200 nm (Fig. 3C). Therefore most LRRK2 puncta visible by light microscopy corresponded to MVBs and to a lesser extent to AVs. However, AVs contained a higher amount of LRRK2 per organelle (9.2 ± 1.8 and 8.9 ± 1.4 gold particles per amphisome and autolysosome, respectively) than MVBs (4.7 ± 0.5 of gold particles). Within MVBs LRRK2 localized preferentially to the intraluminal vesicles (ILVs). Autophagosomes, which accounted for a small percentage of AVs (8.9 ± 0.7%), were not labelled. LRRK2 also localized to a lesser extent to morphologically undefined small vesicular structures (Fig. 3D and Supplementary Material, Figure S2E). A similar staining to that of anti-GFP antibody was obtained with an anti-LRRK2 antibody (Supplementary Material, Figure S3).
Figure 4.
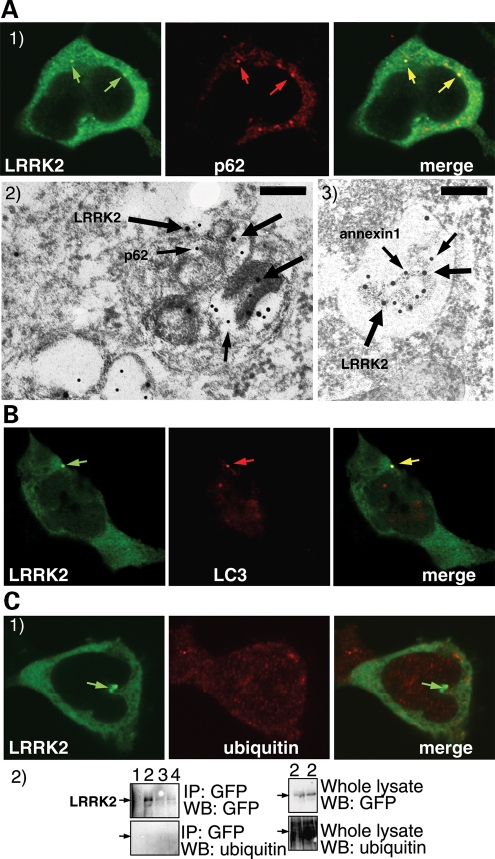
A proportion of LRRK2 puncta colocalize with the proteins p62 and LC3, but not with ubiquitin. (A) LRRK2 puncta colocalize with p62 endogenous puncta. BAC-YPet-LRRK2-WT was transfected in HEK-293 cells and double-labelling performed by IF (A1) or IEM (A2 and 3) using anti-GFP (thick arrows in A2 and 3) and either anti-p62 [A1 and 2 (thin arrows in A2)] or anti-annexin1 (A3) antibodies (thin arrows). Scale bar in image A2 represents 200 nm although in A3 represents 500 nm. (B) LRRK2 puncta colocalize with LC3 protein. BAC-YPet-LRRK2-WT was transfected in HEK-293 cells stably expressing HA-mCherry-LC3 and analysed by IF using anti-HA and anti-GFP antibodies. (C) BAC-YPet-LRRK2-WT, BAC-YPet-LRRK2-G2019S or BAC-YPet-LRRK2-R1441C were expressed in transfected cells and double labelling performed using YPet-LRRK2 fluorescence and anti-ubiquitin antibodies (BAC-YPet-LRRK2-WT in C1). Alternatively protein lysates were prepared and subjected to immunoblotting either directly or after immunoprecipitation using anti-GFP antibodies (C2). Legend of samples: 1 = Untransfected, 2 = BAC-YPet-LRRK2-WT, 3 = BAC-YPet-LRRK2-G2019S and 4 = BAC-YPet-LRRK2-R1441C. A1, B and C1 are confocal images.
The identification of LRRK2 puncta as MVBs and AVs was further supported by real-time live-cell imaging in which we found LRRK2 puncta to have a centripetal transport towards the microtubule organising centre or centrosome (23) (Supplementary Material, Figure S1B and C), which explained how LRRK2 puncta were frequently beside the centrosome (Fig. 5A2) or distributed over a radial pattern (Supplementary Material, Figure S1A).
Figure 5.
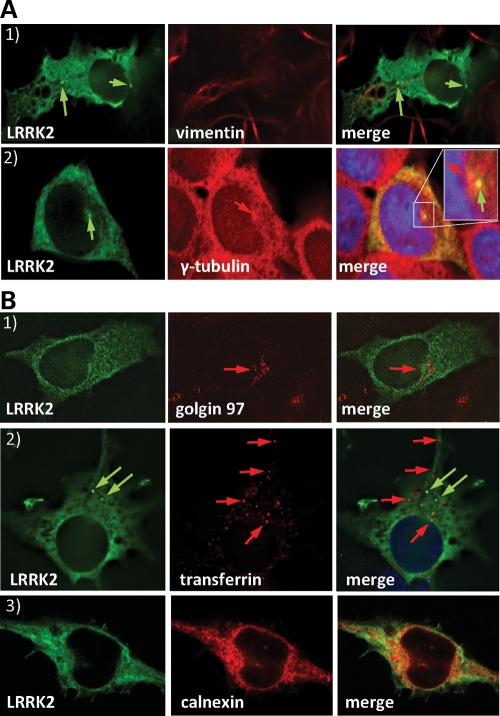
LRRK2 puncta do not colocalize with aggresome markers. LRRK2 shows a discrete colocalization with calnexin (ER) but not with the Golgi apparatus (golgin97) or recycling endosomes (transferrin). (A) BAC-YPet-LRRK2-WT was expressed in transfected cells and IF performed using anti-GFP and either anti-vimentin (A1) or anti-γ-tubulin antibodies (A2). LRRK2 puncta frequently located close to the γ-tubulin positive centrosome (A2 inset). (B) Cells transfected with BAC-YPet-LRRK2-WT were subjected to IF with anti-GFP and either anti-golgin97 (TGN) or calnexin (ER) antibodies (B1 and 3). Alternatively cells were loaded with alexa-594-transferrin (recycling endosomes) and IF performed with anti-GFP detected by alexa-488 conjugated secondary antibodies (B2).
A fraction of LRRK2 puncta colocalize with the proteins p62 and LC3, but not with aggresome markers or ubiquitin
To explore the reported relationship between LRRK2 and p62 proteins (6), we next performed IF immunolabelling in human cells using anti-GFP and anti-p62 antibodies (Fig. 4A1). Endogenous p62 appeared as fine cytoplasmic puncta in untransfected cells, excluding a transfection artefact. We found that about half of the LRRK2 puncta were positive for p62 both with BAC-YPet-LRRK2-WT and BAC-YPet-LRRK2-R1441C. Double-labelling IEM revealed amphisomes (Fig. 4A2) and autolysosomes (Supplementary Material, Figure S2D) as the organelles positive for both p62 and LRRK2. MVBs double-labelled for LRRK2 and annexin1 (Fig. 4A3), known to localize in MVBs (24), but were negative for p62 (Supplementary Material, Figure S2D). Autophagosomes were labelled with p62 only. Next we expressed BAC-YPet-LRRK2-WT and BAC-YPet-LRRK2-R1441C in a cell line stably expressing HA-mCherry-LC3, an autophagic marker (25), and found that ≈15% of LRRK2 puncta contained HA-mCherry-LC3 protein (Fig. 4B). Among AVs, LC3 mainly labels autophagosomes (22), which we found devoid of LRRK2, explaining the lower colocalization than with p62.
Overexpressed mutant LRRK2 accumulates in aggresomes (26) which contain ubiquitinated protein and p62. However, LRRK2 puncta observed upon expression of BAC-YPet-LRRK2-WT or BAC-YPet-LRRK2-R1441C showed no appreciable co-labelling for ubiquitin (Fig. 4C1) or aggresome markers (vimentin and γ-tubulin) (Fig. 5A1 and 2). Neither immunoprecipitated WT nor R1441C LRRK2 protein was detected using anti-ubiquitin antibodies (Fig. 4C2). Furthermore, no vimentin-positive basket-like structures were present (Fig. 5A1), suggesting that neither WT nor mutant LRRK2 expressed from its own genomic locus induced the formation of aggresomes.
Finally, to characterize the morphologically undefined vesicular structures labelled for LRRK2 in IEM we performed double labelling IF (Fig. 5B) with markers for the Golgi apparatus (golgin97), endoplasmic reticulum (ER) (calnexin) or clathrin coated endosomes/recycling endosomes (labelled transferrin). Discrete colocalization was observed only with calnexin (Fig. 5B3) suggesting LRRK2 partially localized to the ER.
LRRK2 knockdown increases autophagic activity and prevents bafilomycin A1-induced cell death under cell starvation stress
The localization of LRRK2 in MVBs and AVs and co-localization with p62 and LC3 suggested a function for LRRK2 in relation to the endosomal–autophagic pathway. We therefore tested if siRNA-mediated knockdown of LRRK2 modified the levels of the autophagy marker LC3-II. We used Flp-mediated site-specific chromosomal insertion of BACs to generate human cell lines stably expressing BAC-YPet-LRRK2-WT in which we could easily confirm LRRK2 siRNA knockdown. Detection of the expression of full length Ypet-LRRK2 and demonstration of siRNA-mediated knockdown of LRRK2 using two different siRNA oligonucleotides were achieved using anti-GFP antibodies (Fig. 6A).
Figure 6.
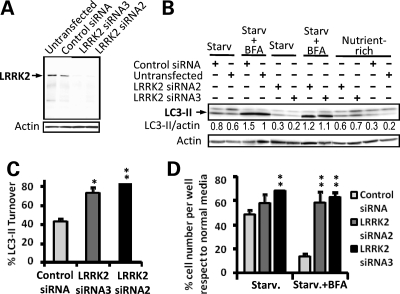
LRRK2 knockdown increases autophagic activity and prevents Bafilomycin-induced cell death under starvation. (A) Demonstration of siRNA mediated LRRK2 knockdown. siRNA mediated knockdown of wild-type Ypet-LRRK2 in clonal cell lines carrying BAC-YPet-LRRK2-WT-FRT shown by western blot using anti-GFP antibodies. Forty micrograms of protein were loaded per lane. (B) Upon LRRK2 knockdown, LC3-I and LC3-II endogenous expression was assessed by western blot under nutrient-rich or 4.5 h starvation in HBSS with or without 200 nM of the autophagic inhibitor bafilomycin A1 (BFA). Ten micrograms of protein were loaded per lane. Mean LC3-II densitometric relative values versus actin of three experiments are shown. (C) Quantification of LC3-II turnover under starvation upon LRRK2 siRNA knockdown after normalization against actin using a densitometric analysis. The amount of LC3-II under starvation was subtracted from the amount of LC3-II under starvation in the presence of the inhibitor of late stages of autophagy BFA, and the difference expressed as a percentage relative to the amount of LC3-II under starvation in the presence of BFA. (D) Quantification of the relative amount of cells per well surviving 4.5 h of starvation treatment in HBSS with or without BFA upon LRRK2 knockdown with respect to nutrient rich conditions. Values are expressed as percentages against the values obtained from cells cultured in normal media for each condition. Bars represent mean + SEM. Experiments were performed in triplicate. Statistical comparisons are made against the control using Student's t test; *P < 0.05, **P < 0.01.
We next assessed LC3-II levels after LRRK2 knockdown under nutrient-rich or starvation conditions. In nutrient-rich conditions LRRK2 siRNA knockdown produced a trend towards an increase of LC3-II levels [LC3-II band intensity 0.6 ± 0.07 arbitrary units (AU) with LRRK2 siRNA2 and 0.7 ± 0.07 AU with LRRK2 siRNA3 versus 0.37 ± 0.07 AU with control siRNA, P = 0.1 and P = 0.08, respectively] (Fig. 6B), suggesting a possible upregulation of basal autophagy upon LRRK2 knockdown. When autophagy was induced by cell starvation LC3-II levels increased as expected in control cells but significantly decreased when LRRK2 was knocked down with either LRRK2 siRNA2 (LC3-II band intensity 0.34 ± 0.04 AU, P < 0.05) or LRRK2 siRNA3 (LC3-II band intensity 0.2 ± 0.03 AU, P < 0.01) with respect to control siRNA (LC3-II band intensity 0.86 ± 0.04 AU) (Fig. 6B). Since LC3-II is itself degraded by autophagy we determined autophagic activity by comparing LC3-II levels in the presence or absence of 200 nM bafilomycin A1 (BFA), a specific inhibitor of autophagic degradation (27) under starvation conditions [LC3-II turnover (28)]. LRRK2 siRNA knockdown did not substantially affect the levels that LC3-II reached in the presence of BFA suggesting that the recruitment of LC3-II to autophagosomes was unaffected (Fig. 6B). However, LC3-II turnover increased significantly upon LRRK2 knockdown (Fig. 6C), suggesting an increased autophagic activity after LRRK2 knockdown and indicating that LRRK2 may normally act as a negative regulator of autophagy.
In support of this hypothesis, we found that LRRK2 siRNA knockdown showed a tendency to revert the negative influence of nutrient starvation on cell number relative to nutrient-rich conditions (29) (58 ± 7% with LRRK2 siRNA2, 68 ± 3% with siRNA3 and 48 ± 4% with control siRNA) (Fig. 6D). Moreover, LRRK2 siRNA knockdown prevented cell death when autophagy was inhibited under starvation conditions with BFA (58 ± 9% with siRNA2 P < 0.01 and 63 ± 4% with siRNA3 P < 0.01 with respect to 10 ± 2% with control siRNA) (Fig. 6D).
The LRRK2 R1441C pathogenic mutation induces the accumulation of MVBs, abnormal AVs and skein-like cellular lesions
Next we assessed whether LRRK2 pathogenic mutations, which have been proposed to have a gain of function mechanism, influenced the number of MVBs and AVs in which we found LRRK2 to be abundant. Using GFP IF we found that the R1441C mutation induced a significant increase in the number of LRRK2 puncta (Fig. 7A, B and E). The G2019S mutation also showed a clear trend towards the formation of LRRK2 puncta (Fig. 7A). Using IEM we confirmed that the R1441C mutation significantly induced an accumulation of MVBs and AVs per cell (Fig. 7C) (32% ± 14.3 increase of MVBs, 29.8% ± 7.8 increase of AVs) reflecting that the R1441C mutation caused an imbalance between the formation and the clearance of these organelles. This idea was further supported by finding that the R1441C mutation also increased the size of AVs (mean AV diameter per cell 394.4 ± 13.7 nm for WT and 488.1 ± 37.9 nm for R1441C P < 0.05) leading to an increase in the total area per cell occupied by AVs (Fig. 7D). Furthermore, enlarged AVs with the R1441C mutation contained a mixture of dark amorphous material, vesicles and incompletely digested material (Supplementary Material, Figure S2A), a type of AV never found with the WT construct. The latter findings suggest that the R1441C mutation caused a relative impairment in the degradation of the intraluminal material within AVs (30) further supported by the accumulation within AVs of the protein p62 (Supplementary Material, Figure S2B) (31). Moreover, LRRK2 protein itself was recruited from the small vesicle pool to the AVs where it also accumulated (Supplementary Material, Figure S2C). We propose that although cytosolic LRRK2 protein is known to be degraded by the proteasome (32), membrane-bound LRRK2 protein may normally be degraded by autophagy and that degradation of mutant R1441C protein within AVs is impaired.
Figure 7.
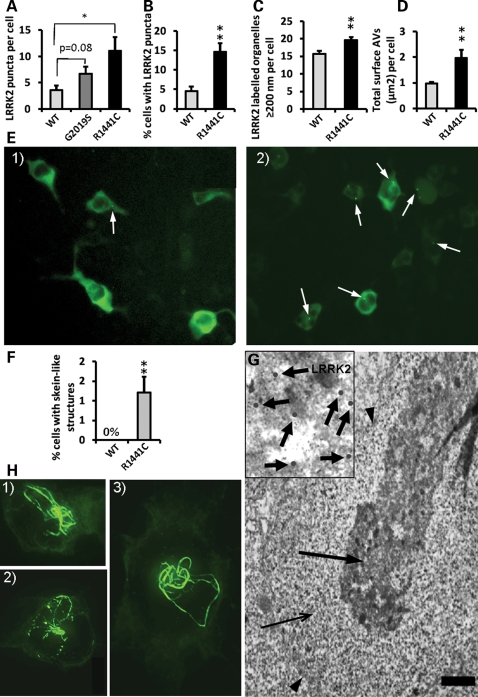
LRRK2 pathogenic mutations increase the number and size of LRRK2 puncta and induce the formation of skein-like structures when expressed from the genomic DNA constructs. (A) Expression of the R1441C LRRK2 pathogenic mutation from BAC-YPet-LRRK2-R1441C showed a ≈3-fold increase respect to WT in the number of LRRK2 puncta per cell (Vero) assessed by IF using anti-GFP antibodies (P < 0.05, Fisher's exact test). Puncta in 20 random labelled cells per genotype were counted using a 63× objective. The G2019G LRRK2 mutation showed a trend of a ≈2-fold increase which did not reach statistical significance. (B) The R1441C mutation also showed a ≈3-fold increase in the percentage of cells (HEK293) harbouring observable LRRK2 puncta assessed by IF (P = 0.0001, Fisher's exact test), illustrated in (E) [E1 (WT) and E2 (R1441C)]. (C) Quantification of the number of labelled organelles >200 nm (the resolution limit of light microscopy) assessed by IEM. (D) The R1441C induced a ≈2-fold increase in the total area per cell occupied by AVs assessed by IEM. The total cell area remained constant between genotypes. (F) The R1441C LRRK2 pathogenic mutation led to the formation of skein-like three dimensional structures in a statistically significant number of cells (P = 0.0038, Fisher's exact test) (HEK293 cells) which are illustrated in z-stack IF pictures H1 to 3 (anti-GFP antibodies). (H) The R1441C LRRK2 skein-like structures were frequently interspersed with LRRK2 puncta and pointed towards a paranuclear location. (G) The skein-like structures appeared under the electron microscope as membrane (arrowheads) contained structures heavily positive for LRRK2 (anti-GFP antibodies) and composed of a densely packed core (thick arrow and amplification in inset) and a fine granular matrix (thin arrow). Scale bar represents 2 µm. Six randomly-chosen fields of view per genotype where analysed using a 20× objective in IF quantifications except where otherwise indicated. Eight randomly-chosen labelled cells per genotype were analysed in IEM quantifications. Counting of all structures was done blind to the genotype. Bars represent mean + SEM. Statistical significances were obtained using a Student's t test except where indicated; *P < 0.05. **P < 0.01.
Expression of R1441C LRRK2 also led to the formation of skein-like three-dimensional structures (Fig. 7H1–3) in a statistically significant number of cells (1.2 ± 0.4%) (Fig. 7F). The R1441C LRRK2 skein-like structures were frequently interspersed with LRRK2 puncta and oriented towards a paranuclear location (Fig. 7H1–3). By IEM we identified the skein-like structures (Fig. 7G) as membrane-surrounded elongated structures composed of a fine granular material enclosing a densely packed core positive for LRRK2 (anti-GFP antibodies) but not for p62, suggesting they were abnormal MVBs. Since normal MVBs are necessary for autophagy (33) it is possible that the formation of abnormal MVBs contributes to the impairment in autophagic clearance observed with the R1441C mutation.
DISCUSSION
In traditional molecular biology technology genes are expressed from cDNAs driven by strong heterologous promoters. It is increasingly clear, however, that non-coding DNA, including promoters, introns, upstream and downstream sequences all contain essential information for the appropriate expression and function of genes (34). We therefore expressed WT and mutant human LRRK2 from a complete genomic DNA locus in human cells. Since LRRK2 protein has been reported to be difficult to detect in some applications (8,9), we decided to fuse a fluorescent tag to LRRK2 with a minimum alteration of potential regulatory sequences. However, this kind of seamless genetic manipulation of genomic DNA material contained in vectors such as BACs has proven to be difficult and inefficient (35). In this article, we report the development of STEP, a novel strategy based on recombineering (19) which obtains high recombination efficiency without leaving any unwanted sequences behind. Our constructs expressed full-length LRRK2 in human cells which could be easily detected in real-time fluorescence live-cell microscopy, IF, immunoblotting and IEM.
Despite the absence of a known membrane-targeting signal we have found a robust association of LRRK2 with both the plasma membrane and intracellular membranes. Interestingly, we report for the first time that LRRK2 was recruited to specific membranous microdomains such as the neck of caveolae, microvilli/filopodia and ILVs of MVBs. These three microdomains share important topological and biochemical characteristics. Topologically, they form when cellular membranes modify their curvature so that the cytosolic side protrudes on the extracellular or intravesicular side (36), a process that likely involves the rich underlying actin cytoskeleton network whose arrangement is regulated by the ERM family of proteins (37). ERM proteins are enriched precisely in the neck of caveolae, microvilli and ILVs of MVBs (21,38,39) and are regulated though phosphorylation. Since LRRK2 has been shown to phosphorylate ERM proteins (15), the specific presence of LRRK2 in the neck of caveolae, microvilli and ILVs of MVBs suggests a role for LRRK2 in regulating actin cytoskeleton reorganization and membrane curvature dynamics.
Our proposal that LRRK2 is involved in the formation of these membranous structures is further supported by two experiments. First, upon induction of formation of microvilli/filopodia LRRK2 was recruited to the plasma membrane, increasing its presence in these microdomains (Fig. 3A and B). Second, ILVs formation is the key event leading to the formation of MVBs (36) and the R1441C mutation, which seems to induce a constitutively active form of LRRK2 (11), increased the appearance of MVBs and led to the formation of skein-like structures which likely represented abnormal MVBs (Fig. 7F, H1–3 and G). Evidence of the role of such a pathway in neurodegenerative disease mechanisms is supported by mutations inducing malfunction of MVB formation that cause frontotemporal dementia and amyotrophic lateral sclerosis (40). Moreover, LRRK2 has been recently found to directly interact with the Alix-related protein His domain phosphotyrosine phosphatase/His-Domain/Type N23 protein tyrosine phosphatase (41), which is essential for endosomal cargo sorting and MVB morphogenesis (42).
In addition to localizing to the neck of caveolae, microvilli and MVBs, LRRK2 localized to AVs, where it partially colocalized with p62 and LC3. Mono-ubiquitination or Lys63-linked di-ubiquitination are the most common signals in the endocytic pathway (43) and the fact that most anti-ubiquitin sera have a bias towards K48 poly-ubiquitination (44) explains the lack of ubiquitin staining of LRRK2 puncta. LRRK2 also localized to morphologically undefined smaller vesicles. Double labelling experiments suggest that these vesicles may be part of the ER (Fig. 5B3). Unlike previous studies which mainly relied on subcellular fractionation (45), we did not find any association of LRRK2 with mitochondria (Supplementary Material, Figure S2E). Nevertheless it may be possible that LRRK2 associated with the outer membrane of engulfed mitochondria within AVs.
Our data provide for the first time direct evidence of a functional relationship between LRRK2 and autophagy. Using a physiologically-relevant human genomic DNA expression system we have shown that the R1441C LRRK2 mutation recapitulates a key and early pathological feature of neurodegenerative disorders, including PD, such as the accumulation of AVs (46–51). We found an accumulation of multivesicular structures and AVs with ‘late’ characteristics filled with incompletely degraded material (52,53) distinct from the smaller and uniformly dense mature lysosomal dense bodies (53,54). This observation suggests that the R1441C LRRK2 mutation induces an imbalance between macroautophagy induction and maturation of AVs to lysosomes, further supported by the accumulation of p62 within AVs (Supplementary Material, Figure S2B) (31,55). Interestingly, LRRK2 has been found to accumulate in neuritic varicosities and globular lesions in the brainstem of PD in association with p62 positive aggregation centres and early stage aggregation forms of alpha-synuclein (6), linking our present findings to the reported accumulation of MVBs and AVs as the main component of neuritic varicosities (54). Furthermore, tyrosine hydroxylase-positive neuritic varicosities and globular lesions appeared in R1441G LRRK2 BAC transgenic mice suffering from levodopa-responsive akinesia, similar to sporadic PD (56). Our finding that LRRK2 puncta are transported towards the cellular soma, possibly by dynein, may indicate that axonal transport deficits could further contribute to the imbalanced maturation of AVs in neurons. Conversely, LRRK2 knockdown increased the autophagic activity and counterbalanced the effect on cell death of the autophagy inhibitor BFA under starvation conditions. Therefore our study suggests that modulating LRRK2 function could help restore the altered autophagic equilibrium of many neurodegenerative disorders and lead to new treatments and neuroprotection strategies.
MATERIAL AND METHODS
Plasmid construction
BAC RP11-568G5 (Children's Hospital Oakland Research Institute in Oakland, CA, USA) contains most of the LRRK2 locus including all exons which were confirmed by exon PCR. Homologous recombination in Escherichia coli using the Red/ET recombination plasmid pSC101BADαβγtet and the RpsL-neo selection/counter selection cassette (Genebridges, Dresden) was performed according to the manufacturer's instructions to obtain the vector BAC-LRRK2-WT. Briefly a DNA fragment was amplified from BAC RP11-115F18 (Children's Hospital Oakland Research Institute in Oakland, CA, USA) using primers rpsl-neo check_S and AC107023-BACe3.6_A by PCR amplification using Bio-X-Act long (Bioline) and used to substitute the RpsL-neo cassette which was previously amplified by PCR using primers AC084290 3′-pRpsL-neo_S and pRpsl-neo_BACe3.6_A within the BAC RP11-568G5. The vector BAC-YPet-LRRK2-WT was obtained by seamless recombination of the coding sequence of YPet [obtained from the plasmid pCEP4YPet-MAMM generously provided by Patrick Daugherty though Addgene (57))] right after the starting ATG of the LRRK2 gene using the newly developed strategy STEP which is based on the Red/ET recombination system. Briefly, a PCR fragment obtained using the primers OH-GFPwtHRbis_S_S, P-N-MYPetwtHR bis_S_A, P-N-MYPetwtHR bis_A_S and OH-N-EmGFPwtHRbis_A_A was used to replace the RpsL-neo cassette previously introduced into BAC-LRRK2-WT by inclusion of homology regions by PCR using primers OH-N-YPetwtHR_S_S, P-N-YPetwtHR_A_S, OH-N-YPetwtHR_A_A and P-N-YPetwtHR_S_A. For all DNA fragments obtained by PCR using four long primers the enzyme Bio-X-Act long (Bioline) was used according to the manufacturer instructions using the external pair of primers at 0.2 µm and the internal pair of primers at 2 nm. The resulting vector also contained the peptide linker Glycine-Alanine-Glycine-Alanine-Glycine between YPet and LRRK2. A similar approach was used to obtain the C-terminally YPet tagged LRRK2 genomic construct BAC-LRRK2-Ypet-WT. Constructs were verified by sequencing across the modified region. For the introduction of the G2019S and R1441C pathogenic mutations into BAC-YPet-LRRK2-WT to produce BAC-YPet-LRRK2-G2019S and BAC-YPet-LRRK2-R1441C a two step procedure was used. In the first step a ≈500 bp genomic region around each target nucleotide was amplified by PCR using G2019S EcoRI_S and G2019S XhoI_A primers for G2019S and R1441C EcoRI_S and R1441C XhoI_A primers for R1441C. These fragments were then digested with EcoRI/XhoI and ligated into pBlueSKM (Invitrogen). Site-directed mutagenesis was then performed on these cloned fragments using Pfu Turbo Polymerase (Stratagene) as previously described (58) using primers G2019S SDM_S and G2019 SDM_A for G2019S and primers R1441C SDM_S and R1441C SDM_A for R1441C. A second step involved the introduction of mutations obtained into BAC-YPet-LRRK2-WT by using The Red/ET recombination system. Primers pairs RpSL G2019S_S/RpSL G2019S_A and RpSL R1441C_S/ RpSL R1441C_A were used for G2019S and R1441C, respectively, for the first round of recombination, whereas primer pairs G2019S_S/G2019S_A and R1441C_S/R1441C_A were used for the second round. Constructs were verified by sequencing across the modified region. Sequence of all primers are provided in Supplementary Material, Table 1.
The vector BAC-YPet-LRRK2-WT-FRT was obtained by retrofitting 1 µg of BAC-YPet-LRRK2-WT with 3 ng of pH-FRT-Hy using Cre recombinase (Novagen) according to the manufacturer instructions. The pH-FRT-Hy vector was obtained in two steps. In the first step the pcDNA5/FRT plasmid (Invitrogen) was digested with BglII and BamHI (both from New England Biolabs, NEB) and the vector backbone religated using T4 DNA ligase (NEB). Then a 2.3 kb fragment was excised with AflIII and NotI and ligated into a vector containing the R6Kγ bacterial conditional replication origin and a loxP site to allow Cre mediated recombination.
The HA-mCherry-LC3 vector was obtained by modification of EGFP-LC3 [generously provided by Karla Kirkegaard through Addgene (59)] in which the EGFP tag of EGFP-LC3 was exchanged for a red fluorescent protein tag [mCherry, a generous gift of Dr R.Y. Tsien, Department of Pharmacology, University of California at San Diego (60)] and a HA tag.
The plasmid cDNA-LRRK2-GFP was a generous gift from C. Gloeckner (Institute of Human Genetics, Munich-Neuherberg, Germany).
Cell culture
HEK293-FRT cells were obtained by transfecting HEK293 cells (ATCC #CRL-1573) with the pFRT/lacZeo plasmid (Invitrogen) using Lipofectamine (Invitrogen) according to the manufacturer's instructions and performing selection of resistant clones upon treatment with 100 µg/ml of zeocin (Invitrogen). HEK293 clonal cell lines expressing the fusion protein YPet-LRRK2 from the human LRRK2 genomic locus were obtained by cotransfecting the BAC-YPet-LRRK2-WT-FRT vector together with pOG44 (Invitrogen), which expresses Flp recombinase, into clonal HEK293-FRT cells and subsequent selection with 100 µg/ml of hygromycin (Gibco). All BAC DNA transfections were performed using Lipofectamine and Plus reagents (Invitrogen) according to the manufacturer instructions using a DNA (µg) to Lipofectamine (µl) ratio of 1:4.
Clonal HEK293 cells stably expressing HA-mCherry-LC3 were obtained by selecting with 800 µg/ml of geneticin (Gibco) after transfection with the HA-mCherry-LC3 vector using Lipofectamine.
Both HEK293 and Vero cells (a derivative of the VERO African green monkey kidney cells kindly provided by Dr Rozanne Sandri-Goldin, University of California, Irvine, CA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% foetal bovine serum (Gibco), 100 U/ml penicillin, 0.1 mg/ml streptomycin (Sigma) and 2 mm l-glutamine (Sigma).
LRRK2 knockdown and immunoblotting
LRRK2 knockdown was achieved by transfecting 3 × 105 HEK293 clonal cells stably expressing YPet-LRRK2 from the BAC-YPet-LRRK2-WT-FRT vector with LRRK2 siRNA2 (Ambion cat#AM16704 ID263837), LRRK2 siRNA3 (Ambion cat#AM16704 ID272780) or control siRNA 5′-GCGACGUUCCUGAAACCACdtdt-3′ (Ambion) using Lipofectamine 2000 (Invitrogen) as previously described (61) in six well plates. Cells were analysed 4 days after transfection. Starvation was achieved by incubating the cells in HBSS containing calcium and magnesium (Gibco) for 4.5 h as previously described (27). Bafilomycin (BFA) (Sigma) stock was dissolved in DMSO at 200 µm and used as 200 nm. The same amount of DMSO was added to control cells and cells under BFA treatment. After the treatments cells were directly scraped into the treatment/culture medium and collected by centrifugation at 900 rpm for 10′ in a Rotanta 460R centrifuge (Hettich-Zentrifugen). Cells were washed once with phosphate buffered saline (PBS) (Sigma) and the final pellet was resuspended in 50 µl Hepes-sucrose buffer [20 mm HEPES (Gibco) pH 7.2, 0.25 m sucrose (Sigma)] in the presence of a cocktail of protease inhibitors (Complete Mini, EDTA-free; Roche) and sonicated at 4°C (3 s three times) (Misonix XL-2000 sonicator). Protein samples were dissolved in Laemmli buffer (62) and run in a Criterion electrophoresis system (BioRad) using precast Criterion Tris–Hcl 4–15% acrylamide gradient gels. Transfer was performed in a Criterion blotter (BioRad) according to the manufacturer instructions using 20% methanol in the transfer buffer to PVDF type membranes (Immovilon P, Millipore). Membranes were blocked with 3% skimmed milk powder milk (Marvel) in Tris buffered saline (TBS) (Sigma) containing 0.1% Tween for 1 h at room temperature (RT). The primary antibodies, namely rabbit anti-GFP (Molecular Probes, 1/1000 dilution), rabbit anti-ubiquitin (Dako, 1/1000 dilution), mouse monoclonal anti-LC3 (Nanotools, 1/200 dilution), mouse monoclonal anti-actin (Sigma, 1/1000 dilution) mouse monoclonal anti-LRRK2 (Abnova H00120892-R01, 1/2500 dilution) and rabbit anti-LRRK2 (Novus Biologicals NB300-268, 1/2000 dilution) were incubated overnight at 4°C in TBS-Tween containing 1% milk. After washing, HRP-conjugated anti-mouse secondary antibody or HRP-conjugated protein A (Biorad) were applied followed by chemiluminescence detection using ECL-plus (Amersham). Images acquired in a Gel Doc XR System (Biorad). Western blot quantification was performed using Image J software (http://rsbweb.nih.gov/ij/). Values were always normalized against actin. Relative cell number was determined using the bicinchoninic acid (4,4′-dicarboxy-2,2′-biquinoline, Sigma) assay reagent as previously described (63).
Immunoprecipitation
For immunoprecipitation Dynabeads Protein G (Invitrogen) coated with anti-GFP antibodies were used according to the manufacturer instructions.
Immunocytochemistry and IF using human brain samples
Brains were fixed for at least 1 month by suspension in 10% buffered formalin before being sliced and multiple tissue blocks were embedded in paraffin wax), after which 6 µm sections were cut. Slides were then dewaxed at 55ºC for 10 min, treated with xylene for 5 min at RT, and hydrated in decreasing concentrations of ethanol. Endogenous peroxidase activity was blocked in 0.3% H2O2 in PBS for 1 h at RT. Antigen retrieval was performed by placing the slides in a black polyoxymethylene-poly-acetal plastic staining dish and performing intermittent microwaving of samples for a total of 10 min (Sanyo 800 W, 100% power) in 0.01 m citrate buffer pH 6 (tri-Sodium citrate, BDH) and then allowed to cool down completely. Sections were then mounted in a slide rack (Sequenza slide rack), blocked for 30 min in 10% fetal calf serum in TBS-0.1% Triton-100 and then incubated overnight at 4ºC with the primary antibody diluted in 10% FCS-TBS-T. Antibodies rabbit anti-LRRK2 (Novus Biologicals, NB300-268, 1/300 dilution) and rabbit anti-LRRK2 (Novus Biologicals, NB300-267, 1/300 dilution) were used. Detection was obtained with REAL EnVision Detection System (Peroxidase/DAB+, Rabbit/Mouse; Dako) according to the manufacturer's instructions. Sections were counterstained with haematoxylin and mounted in xylene-based permanent medium (DPX; Agar Scientific). Photomicrographs were taken using an AxioCam MRc camera and KS300 software (Zeiss). For IF sections were processed as explained and after antigen retrieval, a HRP-goat anti rabbit Tyramide Signal Amplification Kit (Invitrogen) based on Alexa Fluor 594 dye was used according to the manufacturer's instructions. Fluorescence detection was performed using the Alexa 594-tyramide solution, at a 1/75 dilution. Nuclear staining was achieved using a 1/200 dilution of TO-PRO 3 dye (Invitrogen) in the final mounting medium (Vectashield from VectorLabs). Images were captured with a Zeiss LSM510 META Confocal Imaging System.
IF on cultured cells
Cells were grown on 19 mm glass coverslips at the bottom of 12 well plates and transfected with the indicated plasmids. Four days after transfection cells were fixed in 4% paraformaldehyde in PBS for 30 min. Cells were permeabilized with IF blocking buffer (1% fish gelatine, 0.1% Triton X-100 and 10% normal goat serum in TBS) for 30 min at RT and incubated with the primary antibodies diluted in IF blocking buffer overnight at 4°C. The antibodies rabbit anti-GFP (Molecular Probes, 1/1000 dilution), rabbit anti-ubiquitin (Dako, 1/400 dilution), mouse monoclonal anti-p62 lck ligand (BD Transduction Laboratories, 1/500 dilution), mouse monoclonal anti-vimentin (Sigma, 1/500 dilution), mouse monoclonal anti-γ-tubulin (Sigma, 1/200 dilution), anti-golgin 97 (Molecular Probes, 1/500 dilution) and mouse monoclonal anti-calnexin (BD Biosciences, 1/100 dilution) were used. Transferrin receptors were labelled by incubating the cells with alexa-594-transferrin (Molecular Probes, 50 µg/ml) for 30 min at 37°C. Fluorescent secondary antibodies goat anti rabbit alexa-488, goat anti rabbit alexa-594 and goat anti mouse alexa-594 (all from Molecular Probes) were used at 1:500 dilution in IF blocking buffer. For nuclear counterstaining 4,6-diamino-2-phenylindole (Sigma) was added at 0.5–1 µg/ml. Coverslips were mounted on standard microscope glass slides using Fluoromount-G (Southern Biotech) and left at 4°C overnight before visualising. Photomicrographs were taken using an Eclipse-TE inverted microscope (Nikon) coupled to a monochrome CCD Orc camera (Hamamatsu) and a Volocity Version 4.3.2 Build 23 software (Improvision). Z-Stacks were acquired with 0.1 µm interval and images subjected to a built in deconvolution algorithm (images in Figs 5A, B1, B2, 7H and Supplementary Material, Figure S1A). Alternatively confocal images were captured with a Zeiss LSM510 META Confocal Imaging System (Figs 1D, E, 4A1, B, C1 and 5B3).
Immunoelectron microscopy
Four days post-transfection cells were washed twice with PBS and fixed with 3% paraformaldehyde and 0.05% glutaraldehyde in PBS for 30 min. Then cells were scraped and allowed to fix into a 1.5 ml microfuge tube with fresh fixative for three additional hours. To induce the formation of microvilli cells were washed twice with PBS and placed with trypsin-EDTA (Sigma) at 37°C for 5 min. Cells were then washed with normal growth medium, collected by spinning at 900 rpm for 5 min and fixed as above. The fixative was removed and substituted with PBS containing 2.3 m sucrose leaving the cells in this solution for 24 h. The cells were now slam-frozen (Reichert MM80E; Leica), freeze-substituted at −80°C in methanol for 48 h and embedded at −20°C in LR Gold acrylic resin (London Resin Company Ltd) in a Reichert freeze-substitution system (Reichert). Cells were cut into ultrathin sections of 50–80 nm using a Eichert Ultracut S ultratome and mounted onto formvar-coated nickel grids. The primary antibodies, either rabbit anti-GFP (Molecular Probes) at 1:250 dilution, goat anti-LRRK2 EB06550 (Everest) at 1:200 dilution, sheep anti-ANXA1 antibody (gift from Prof Rod Flower, William Harvey Research Institute, London, UK) (64) at 1:1000 dilution and mouse monoclonal anti-p62 lck ligand (BD Transduction Laboratories) at 1/100 dilution were incubated for 2 h. To detect the primary antibodies, cells were incubated with either Protein A-15 nm gold complex (for rabbit antibodies), donkey anti goat-15 nm gold complex, goat anti-mouse-5 nm gold complex or goat anti-sheep-5 nm gold complex, then lightly counterstained with uranyl acetate and lead citrate. All antibodies were diluted in 0.1 m phosphate buffer containing 0.1% w/v egg albumin. For control sections, the primary antibodies were omitted and replaced with 0.1 m phosphate buffer containing 0.1% w/v egg albumin; immunogold labelling was absent in these conditions. Sections were examined with a JEOL 1010 transmission electron microscope (JEOL USA, Inc.). The number of organelles and number of gold particles were counted in 8 cells. For measurement of cell area micrographs of each cell were taken at a magnification of ×4000. The cell areas were analysed from scanned micrographs using Axiovision software, version 3.4 (Zeiss). Cell area was not significantly different between WT and mutant. In all cases the analyst was blind to the sample code.
Live-cell imaging
HEK-293 cells were seeded on 25 mm coverslips and transfected as detailed above. After 4 days coverslips were placed in a LU-CPC-CEH Leiden closed perfusion chamber (Harvard Apparatus) and incubated at 37°C with normal growth medium saturated with 5% CO2 containing 25 mm Hepes. Live-cell imaging was performed using automated shutters mounted on a Nikon Eclipse TE-2000 epifluorescence microscope equipped with a YFP HYQ filter block (Nikon) in order to diminish UV light irradiation.
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported in part by grants from the National Institutes of Health (R01 NS045961-91A1), the Parkinson's Disease Society, the Alzheimer's Research Trust, the Friedreich's Ataxia Research Alliance and Ataxia UK. L.L.V. is supported by a Fundação para a Ciência e Tecnologia (Portugal) Studentship. We thank the Alzheimer's Research Trust for equipment grant support.
Supplementary Material
ACKNOWLEDGEMENTS
We thank numerous colleagues for their generous gifts of reagents as detailed in Material and Methods and to the patients and their families who support the Thomas Willis Oxford Brain Collection. We are grateful to Professor Margaret Esiri, Professor John Morris, Dr Antonio Velayos Baeza, Dr Zoe Holloway and Dr Megan Dennis for their insightful comments. We thank the Wellcome Trust Centre for Human Genetics microscopy core laboratory for assistance with confocal microscopy.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Mata I.F., Kachergus J.M., Taylor J.P., Lincoln S., Aasly J., Lynch T., Hulihan M.M., Cobb S.A., Wu R.M., Lu C.S., et al. Lrrk2 pathogenic substitutions in Parkinson's disease. Neurogenetics. 2005;6:171–177. doi: 10.1007/s10048-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 4.Gaig C., Marti M.J., Ezquerra M., Rey M.J., Cardozo A., Tolosa E. G2019S LRRK2 mutation causing Parkinson's disease without Lewy bodies. J. Neurol. Neurosurg. Psychiatry. 2007;78:626–628. doi: 10.1136/jnnp.2006.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mata I.F., Wedemeyer W.J., Farrer M.J., Taylor J.P., Gallo K.A. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends. Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Alegre-Abarrategui J., Ansorge O., Esiri M., Wade-Martins R. LRRK2 is a component of granular alpha-synuclein pathology in the brainstem of Parkinson's disease. Neuropathol. Appl. Neurobiol. 2008;34:272–283. doi: 10.1111/j.1365-2990.2007.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Song Y.J., Murphy K., Holton J.L., Lashley T., Revesz T., Gai W.P., Halliday G.M. LRRK2 and parkin immunoreactivity in multiple system atrophy inclusions. Acta. Neuropathol. 2008;116:639–646. doi: 10.1007/s00401-008-0446-3. [DOI] [PubMed] [Google Scholar]
- 8.Biskup S., West A.B. Zeroing in on LRRK2-linked pathogenic mechanisms in Parkinson's disease. Biochim. Biophys. Acta. 2009;1792:625–633. doi: 10.1016/j.bbadis.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melrose H. Update on the functional biology of Lrrk2. Future Neurol. 2008;3:669–681. doi: 10.2217/14796708.3.6.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell. Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 11.Deng J., Lewis P.A., Greggio E., Sluch E., Beilina A., Cookson M.R. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatano T., Kubo S., Imai S., Maeda M., Ishikawa K., Mizuno Y., Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum. Mol. Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 13.Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.W., Savitt J.M., Waldvogel H.J., et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 14.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., Alessi D.R. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paglini G., Kunda P., Quiroga S., Kosik K., Caceres A. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J. Cell Biol. 1998;143:443–455. doi: 10.1083/jcb.143.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu C.T. Autophagic stress in neuronal injury and disease. J. Neuropathol. Exp. Neurol. 2006;65:423–432. doi: 10.1097/01.jnen.0000229233.75253.be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plowey E.D., Cherra S.J., 3rd, Liu Y.J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Buchholz F., Muyrers J.P., Stewart A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 20.Predescu S.A., Predescu D.N., Malik A.B. Molecular determinants of endothelial transcytosis and their role in endothelial permeability. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2007;293:L823–L842. doi: 10.1152/ajplung.00436.2006. [DOI] [PubMed] [Google Scholar]
- 21.Foti M., Porcheron G.v., Fournier M., Maeder C., Carpentier L. The neck of caveolae is a distinct plasma membrane subdomain that concentrates insulin receptors in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1242–1247. doi: 10.1073/pnas.0610523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eskelinen L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 23.Hollenbeck P. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 1993;121:305–315. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White I.J., Bailey L.M., Aghakhani M.R., Moss S.E., Futter C.E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blackinton J., Thomas K.J. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell. Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 28.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 29.Boya P., Gonzalez-Polo R.A., Casares N., Perfettini J.L., Dessen P., Larochette N., Metivier D., Meley D., Souquere S., Yoshimori T., et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., Nixon R.A. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Xie C., Greggio E., Parisiadou L., Shim H., Sun L., Chandran J., Lin X., Lai C., Yang W.-J., et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J. Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 34.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 35.Tursun B., Cochella L., Carrera I., Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 37.Tsukita S., Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J. Biol. Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 38.Yonemura S., Tsukita S., Tsukita S. Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. J. Cell Biol. 1999;145:1497–1509. doi: 10.1083/jcb.145.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fevrier B., Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell. Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 41.Zheng X.Y., Yang M., Tan J.Q., Pan Q., Long Z.G., Dai H.P., Xia K., Xia J.H., Zhang Z.H. Screening of LRRK2 interactants by yeast 2-hybrid analysis. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2008;33:883–891. [PubMed] [Google Scholar]
- 42.Doyotte A., Mironov A., McKenzie E., Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piper R.C., Katzmann D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansorge O., Giunti P., Michalik A., Van Broeckhoven C., Harding B., Wood N., Scaravilli F. Ataxin-7 aggregation and ubiquitination in infantile SCA7 with 180 CAG repeats. Ann. Neurol. 2004;56:448–452. doi: 10.1002/ana.20230. [DOI] [PubMed] [Google Scholar]
- 45.West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto K., Hirai S., Iizuka T., Yanagisawa T., Watanabe M. Reexamination of granulovacuolar degeneration. Acta. Neuropathol. 1991;82:340–345. doi: 10.1007/BF00296544. [DOI] [PubMed] [Google Scholar]
- 47.Cataldo A., Hamilton D., Barnett J., Paskevich P., Nixon R. Properties of the endosomal–lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer's disease. J. Neurosci. 1996;16:186–199. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersen A., Larsen K.E., Behr G.G., Romero N., Przedborski S., Brundin P., Sulzer D. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum. Mol. Genet. 2001;10:1243–1254. doi: 10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 49.Sikorska B., Liberski P.P., Giraud P., Kopp N., Brown P. Autophagy is a part of ultrastructural synaptic pathology in Creutzfeldt-Jakob disease: a brain biopsy study. Int. J. Biochem. Cell Biol. 2004;36:2563–2573. doi: 10.1016/j.biocel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Anglade P., Vyas S., JavoyAgid F., Herrero M.T., Michel P.P., Marquez J., MouattPrigent A., Ruberg M., Hirsch E.C., Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 51.Zhu J.H., Guo F., Shelburne J., Watkins S., Chu C.T. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn W., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J. Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W.H., Cuervo A.M., Kumar A., Peterhoff C.M., Schmidt S.D., Lee J.-H., Mohan P.S., Mercken M., Farmery M.R., Tjernberg L.O., et al. Macroautophagy–a novel {beta}-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 55.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 56.Li Y., Liu W., Oo T.F., Wang L., Tang Y., Jackson-Lewis V., Zhou C., Geghman K., Bogdanov M., Przedborski S., et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen A.W., Daugherty P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 58.Wang W., Malcolm B.A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 59.Jackson W.T., Giddings T.H., Jr, Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 61.Fountaine T.M., Venda L.L., Warrick N., Christian H.C., Brundin P., Channon K.M., Wade-Martins R. The effect of alpha-synuclein knockdown on MPP+ toxicity in models of human neurons. Eur. J. Neurosci. 2008;28:2459–2473. doi: 10.1111/j.1460-9568.2008.06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 63.Tuszynski G.P., Murphy A. Spectrophotometric quantitation of anchorage-dependent cell numbers using the bicinchoninic acid protein assay reagent. Anal. Biochem. 1990;184:189–191. doi: 10.1016/0003-2697(90)90032-5. [DOI] [PubMed] [Google Scholar]
- 64.Chapman L., Nishimura A., Buckingham J.C., Morris J.F., Christian H.C. Externalization of Annexin I from a folliculo-stellate-like cell line. Endocrinology. 2002;143:4330–4338. doi: 10.1210/en.2002-220529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.