Abstract
Prenatal famine in humans has been associated with various later-life consequences, depending on the gestational timing of the insult and the sex of the exposed individual. Epigenetic mechanisms have been proposed to underlie these associations. Indeed, animal studies and our early human data on the imprinted IGF2 locus indicated a link between prenatal nutritional and DNA methylation. However, it remains unclear how common changes in DNA methylation are and whether they are sex- and timing-specific paralleling the later-life consequences of prenatal famine exposure. To this end, we investigated the methylation of 15 loci implicated in growth and metabolic disease in individuals who were prenatally exposed to a war-time famine in 1944–45. Methylation of INSIGF was lower among individuals who were periconceptionally exposed to the famine (n = 60) compared with their unexposed same-sex siblings (P = 2 × 10−5), whereas methylation of IL10, LEP, ABCA1, GNASAS and MEG3 was higher (all P < 10−3). A significant interaction with sex was observed for INSIGF, LEP and GNASAS. Next, methylation of eight representative loci was compared between 62 individuals exposed late in gestation and their unexposed siblings. Methylation was different for GNASAS (P = 1.1 × 10−7) and, in men, LEP (P = 0.017). Our data indicate that persistent changes in DNA methylation may be a common consequence of prenatal famine exposure and that these changes depend on the sex of the exposed individual and the gestational timing of the exposure.
INTRODUCTION
Adverse environmental conditions during specific windows of mammalian development can have lasting effects on metabolic pathways and physiology, thereby influencing the susceptibility to chronic diseases (1). An extensive epidemiological literature has reported associations between characteristics of early development and health outcomes later in life (2,3). Historical famines provide a quasi-experimental setting in which the long-term consequences of adverse conditions during development can be studied in humans. Studies of the Dutch Hunger Winter, a severe wartime famine at the end of WW II affecting the western part of The Netherlands, suggest that famine exposure in utero can lead to various adverse metabolic or mental phenotypes, depending on the sex of the exposed individual and the timing of the exposure during gestation (4–7).
The period around conception may be especially sensitive to famine exposure (8). Exposure to famine in this period is associated with diverse phenotypic outcomes such as an increased risk of adult schizophrenia (7) and spina bifida at birth in men (9). However, the effects of prenatal famine exposure are not limited to this developmental period. An example of a general, sex-specific late-life effect is the increase in body mass index (6,10) and various lipids in blood (4) among famine-exposed women, irrespective of the precise gestational timing of the exposure. Also, increases in cerebro-cardiovascular related deaths have been reported among individuals exposed to seasonal food shortages independent of the gestational timing in a historical cohort (11), although preliminary results from the Dutch Famine indicated that an increased risk of coronary artery disease is specific for exposure to famine early in gestation (12). Animal experiments confirm the importance of timing and sex (13,14). Thus, prenatal exposure to famine can have different long-term effects that depend on the timing of the exposure and the sex of the exposed individual.
Persistent epigenetic changes induced by environmental factors are a plausible molecular mechanism underlying the relationship between early development and later-life disease (15,16). Experiments in animal models provide strong supporting evidence. Manipulation of the maternal diet during pregnancy leads to a persistent shift in average DNA methylation levels of specific genes in offspring, resulting in permanent changes in coat color or tail shape (17,18). A proof-of-principle for complex diseases was reported by Bogdarina et al. (19), who showed in rats that a low-protein diet during pregnancy was associated with decreased DNA methylation of the Agtr1b gene promoter in offspring, explaining the increase in blood pressure among these animals. We recently showed that similar mechanisms may be operative in humans; periconceptional exposure to famine was associated with a decrease in DNA methylation of the insulin-like growth factor 2 (IGF2) differentially methylated region (DMR) (20). However, to be a valid candidate mechanism in humans, the effect of prenatal famine on DNA methylation should be widespread, mirror the epidemiological findings with sex- and timing-specific associations and affect genes in relevant pathways.
To further explore associations between prenatal famine and DNA methylation, including the role of the timing of exposure and the sex of the prenatally exposed individual, we assessed the methylation state of loci in 15 candidate genes involved in metabolic and cardiovascular disease and growth with diverse epigenetic features in our ongoing Hunger Winter Families Study (21).
RESULTS
Periconceptional exposure
We studied methylation of loci implicated in the transcriptional regulation of 15 candidate genes for metabolic and cardiovascular disease. The loci studied included imprinted loci (GNASAS, GNAS A/B, MEG3, KCNQ1OT1, INSIGF and GRB10), a putatively imprinted locus (IGF2R) and non-imprinted loci (IL10, TNF, ABCA1, APOC1, FTO, LEP, NR3C1 and CRH). The selection of the loci measured was based on a combination of factors, including binding of methylation-sensitive transcription factors, associations of DNA methylation with gene expression and the presence of DMRs for imprinted loci (a detailed overview can be found in Supplementary Material, Table S1).
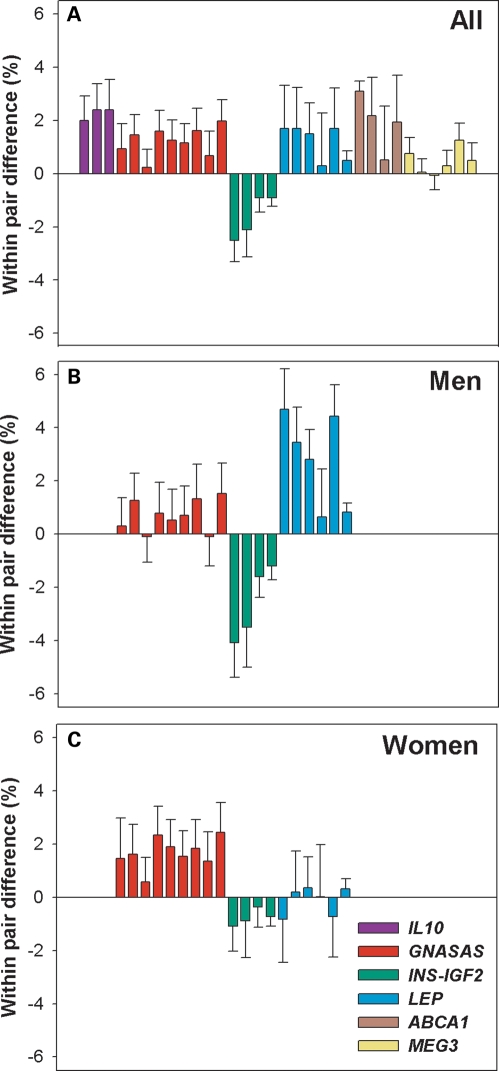
DNA methylation of these 15 loci was measured in 60 individuals conceived during the Dutch Famine (i.e. exposed periconceptionally) and compared with their unexposed same-sex sibling to minimize the possible confounding effects of familial environment and genetic background. For 6 of the 15 loci, significant differences in DNA methylation were observed (Table 1). DNA methylation was increased among famine-exposed individuals for the imprinted genes GNASAS (P = 3.1 × 10−6) and MEG3 (P = 8.0 × 10−3) and the non-imprinted IL10 (P = 1.8 × 10−6), ABCA1 (P = 8.2 × 10−4) and LEP (P = 2.9 × 10−3) proximal promoters. DNA methylation was decreased for the imprinted INSIGF promoter (P = 2.3 × 10−5), which is part of the proximal promoter of INS (22). For the remaining nine loci, there was no association with periconceptional exposure to famine. All associations remained statistically significant after Bonferroni correction for multiple testing (15 loci) with the exception of MEG3 (PBonferroni = 0.12). When analysed separately, individual CpG dinucleotides showed similar associations with famine exposure as the complete loci (Fig. 1A). The loci affected did not share obvious features with respect to sequence, epigenetic features or biological function.
Table 1.
DNA methylation and periconceptional exposure to famine
| Gene locusa | Average methylation, % (SD) | Within-pair differenceb, Δ% | Effect sizec, SD units | P-valued | Bonferroni-corrected P-valuee |
|---|---|---|---|---|---|
| IL10 | 20.8 (6.5) | 2.4 | 0.37 | 1.8 × 10−6 | 2.7 × 10−5 |
| GNASAS | 48.8 (4.7) | 1.1 | 0.24 | 3.1 × 10−6 | 4.7 × 10−5 |
| INSIGF | 84.8 (2.6) | −1.6 | −0.61 | 2.3 × 10−5 | 3.5 × 10−4 |
| LEP | 28.6 (4.9) | 1.2 | 0.24 | 2.9 × 10−3 | 4.4 × 10−2 |
| MEG3 | 54.0 (2.4) | 0.5 | 0.21 | 8.0 × 10−3 | 0.12 |
| ABCA1 | 19.9 (4.2) | 0.7 | 0.17 | 0.017 | 0.26 |
| ABCA1 methf | 36.9 (8.2) | 1.7 | 0.21 | 8.2 × 10−4 | 0.012 |
| KCNQ1OT1 | 30.1 (1.5) | −0.2 | −0.16 | 0.053 | NS |
| GRB10 | 47.2 (4.6) | 0.4 | 0.08 | 0.091 | NS |
| GNASAB | 40.3 (5.0) | 0.6 | 0.11 | 0.092 | NS |
| APOC1 | 16.7 (3.1) | −0.5 | −0.17 | 0.13 | NS |
| IGF2R | 84.1 (6.9) | −0.7 | −0.10 | 0.29 | NS |
| FTO | 97.3 (0.8) | −0.5 | −0.61 | 0.28 | NS |
| CRH | 58.9 (6.0) | 0.4 | 0.07 | 0.61 | NS |
| TNF | 9.6 (1.7) | 0.1 | 0.06 | 0.63 | NS |
| NR3C1 | 4.8 (1.1) | 0.0 | −0.01 | 0.79 | NS |
aTable sorted on P-value.
bAverage absolute difference in DNA methylation between exposed and unexposed siblings.
cObserved within-pair difference divided by the standard deviation in the sibling controls.
dTwo-sided P-value resulting from a linear mixed model accounting for family relations, bisulphite batch and age at blood draw.
eThe Bonferroni-corrected P-values. Results that were already not significant before Bonferroni correction are shown as NS.
fSeparate analysis of methylated CpGs in ABCA1. In contrast to the 3′ CpGs, 5′ CpG dinucleotides (n = 13) were methylated. The methylation of the methylated 5′ CpGs was highly correlated (r > 0.87) among themselves, but not with the methylation of the 3′ CpG dinucleotides, which had little to no methylation (<4.5%).
Figure 1.
Difference in DNA methylation of CpG dinucleotides in siblings discordant for periconceptional exposure to famine. Bars in the figures represent the average absolute within-pair difference in DNA methylation and their standard errors for CpG dinucleotides. A positive difference indicates a higher methylation level among exposed individuals. The exact location of the CpG dinucleotides can be found using Supplementary Material, Table S1. (A) The absolute within-pair difference for CpG dinucleotides for which a significant overall difference of the locus in DNA methylation was observed. (B) As in (A), but for men. Only loci showing a significant interaction between sex and exposure are depicted. (C) As in (B), but for women only.
Late gestational exposure
Epigenetic modulation may also occur during other developmental windows (23,24). We therefore also studied 62 individuals who were exposed to famine late in gestation together with their unexposed, same-sex siblings. We measured DNA methylation at four loci that were associated with periconceptional famine exposure (IL10, GNASAS, INSIGF and LEP) and at four that were not (IGF2R, APOC1, KCNQ1OT1 and CRH). These loci include four imprinted ones and diverse epigenetic features (Supplementary Material, Table S1).
No associations were observed except for a significant reduction in methylation at the GNASAS locus (P = 1.1 × 10−7, PBonferroni = 8.8 × 10−7) (Table 2). This association was consistent for the individual CpG sites within the locus (data not shown). The direction of the association was opposite to what was observed for periconceptional exposure. We then combined all periconceptional and late pregnancy exposed individuals and their controls in a single analysis to test for a statistical interaction between the famine associations with DNA methylation and the precise gestational timing of the exposure. The DNA methylation differences found for IL10, GNASAS and INSIGF were timing-specific (Table 2), but for LEP the test for interaction was not significant.
Table 2.
DNA methylation and late gestational exposure to famine
| Gene locusa | Average methylationb, % (SD) | Within-pair differencec, Δ% | Effect sized, SD units | P-valuee | Bonferroni corrected-P-valuef | Timing specificity P-valueg |
|---|---|---|---|---|---|---|
| IL10 | 20.7 (5.0) | −0.2 | −0.04 | 0.76 | NS | 1.2 × 10−3 |
| GNASAS | 48.8 (4.2) | −1.1 | −0.26 | 1.1 × 10−7 | 8.8 × 10−7 | 3.1 × 10−12 |
| INSIGF | 84.7 (2.8) | 0.0 | 0.0 | 0.95 | NS | 3.2 × 10−4 |
| LEP | 28.7 (4.6) | 0.4 | 0.09 | 0.18 | NS | 0.13 |
| KCNQ1OT1 | 30.2 (1.7) | 0.2 | 0.12 | 0.17 | NS | 0.058 |
| APOC1 | 16.7 (3.1) | −0.6 | −0.19 | 0.22 | NS | 0.90 |
| IGF2R | 84.0 (4.4) | 0.0 | 0.0 | 0.88 | NS | 0.25 |
| CRH | 58.9 (4.8) | 0.5 | 0.10 | 0.51 | NS | 0.86 |
aData in this table have been given in the same order as in Table 1.
bThe batch-corrected average methylation for the unexposed late gestation sibling controls and the standard deviation.
cAverage absolute difference in DNA methylation between exposed and unexposed siblings.
dObserved within-pair difference divided by the standard deviation in the sibling controls.
eTwo-sided P-value resulting from a linear mixed model accounting for family relations, bisulphite batch and age at blood draw. This test was performed on the late gestational exposed sibships (n = 62).
fThe Bonferroni-corrected P-values. Results that were already not significant before Bonferroni correction are shown as NS.
gTwo-sided P-value resulting from the test for timing specificity. The timing specificity was calculated by joining the data sets for both the periconceptional and late gestational siblings and their unexposed same-sex siblings and introducing an interaction term for gestational timing times exposure status. This test thus includes all 122 pairs.
Sex-specific associations
Previous work found basal DNA methylation differences in humans (25). We therefore first tested for sex differences in DNA methylation for the measured loci combining the two sibling control groups (men n = 56, women n = 66). DNA methylation was higher in men for IGF2R (2.6%, P = 0.019) and lower in men for LEP (−2.6%, P = 3.0 × 10−3), IL10 (−2.9%, P = 0.015) and APOC1 (−1.5, P = 0.015) compared with women. No significant differences were found for the other four loci (GNASAS, INSIGF, KCNQ1OT1 and CRH).
Next, we tested whether the observed significant associations with prenatal famine were sex-specific. The interaction between sex and periconceptional famine exposure was significant for LEP (P = 2.3 × 10−4), INSIGF (P = 8.5 × 10−3) and GNASAS (P = 0.027) (Table 3). For LEP and INSIGF, the association of famine exposure with DNA methylation was restricted to men (PLEP,♂= 3.6 × 10−7, PINSIGF,♂ = 6.5 × 10−6) (Fig. 1B and C). For GNASAS, the association was significant in both sexes but most pronounced in women (Pmen = 0.013; Pwomen = 1.1 × 10−5). The association between late gestational famine exposure and GNASAS methylation was independent of sex.
Table 3.
Sex-specific associations of DNA methylation with periconceptional exposure to famine
| Gene locus | Psex interactiona | Sex | Within-pair differenceb, Δ% | Effect sizec, SD units | P-valued |
|---|---|---|---|---|---|
| GNASAS | 0.027 | ♂ | 0.7 | 0.15 | 0.013 |
| ♀ | 1.5 | 0.33 | 1.1×10−5 | ||
| INSIGF | 8.5 × 10−3 | ♂ | −2.6 | −0.99 | 6.5 × 10−6 |
| ♀ | −0.8 | −0.29 | 0.11 | ||
| LEP | 2.3 × 10−4 | ♂ | 2.8 | 0.57 | 3.6 × 10−7 |
| ♀ | −0.2 | −0.04 | 0.70 |
aThe two-sided P-value from the test for sex specificity of the observed periconceptional effect of prenatal famine. This was tested by entering an interaction term of sex times the exposure status in the linear mixed model.
bAverage absolute difference in DNA methylation between exposed and unexposed siblings.
cObserved within-pair difference divided by the standard deviation in the sibling controls.
dTwo-sided P-value resulting from a linear mixed model accounting for family relations, bisulphite batch and age at blood draw.
Timing independent association
For LEP, there was no indication for a significant interaction between the famine association with DNA methylation and the gestational timing of the exposure, even though the methylation difference was significant only following periconceptional famine exposure. Since, this association was later found to be male-specific, we tested for an interaction with sex in the late exposure group. Indeed, a significantly higher LEP methylation was found for men exposed late in gestation (P = 0.017). Analysis of the whole cohort (n = 244) including both exposure groups revealed a significant association between prenatal exposure to famine irrespective of the precise gestational timing (P = 0.003). Further analysis suggested that this association was male-specific [Pinteraction = 1.3 × 10−6; men: 2.2% (0.83 SD), P = 7.5 × 10−8; women: P = 0.47].
DISCUSSION
We studied the DNA methylation levels of 15 loci for their association with prenatal exposure to the Dutch Famine at the end of WW II. For six of the loci studied, we observed significant differences in DNA methylation after famine exposure during periconception (INSIGF, GNASAS, MEG3, IL10, LEP and ABCA1). This association differed by sex for three loci (INSIGF, GNASAS and LEP). Of the eight loci tested, exposure to famine late in gestation was associated with methylation for GNASAS and LEP, which was specific for men. Of interest, the differences in DNA methylation included both increases and decreases, in one case even at the same locus after exposure during different gestational periods. Together with our previous finding that the IGF2 DMR is associated with periconceptional exposure (20), our data indicate that an adverse prenatal environment may trigger widespread and persistent changes in DNA methylation.
Our current and previous (20) observations suggest that the periconceptional period may be an especially sensitive exposure period in humans. This might be inherent to mammalian development (26,27) and this hypothesis is also supported by detailed animal studies (28,29). The association of GNASAS and LEP with late gestational exposure, however, suggests that environmentally induced DNA methylation changes may not be limited to the periconceptional period. This is in line with findings that methylation of the glucocorticoid receptor promoter depends on postnatal circumstances (e.g. maternal care in rats (24) and child abuse in humans (23)). In addition to timing-specific associations, we observed sex-specific associations for three of the six loci for which DNA methylation was significantly associated with prenatal exposure to famine. In men, LEP methylation was associated with prenatal famine irrespective of the timing of exposure. Our observation that the methylation changes in relation to the prenatal environment may be sex-specific is in agreement with the sex-specific methylation changes found in offspring of sheep that were folate- and vitamin B12-restricted during periconception (29). How such sex-specific associations can arise is currently unknown, but interactions between sex hormones and the expression of DNA methyltransferases may be a factor (30).
The differences in DNA methylation observed here are comparable, although slightly smaller than we previously found for IGF2 [absolute difference of 2.7% (5.2% relative to the mean DNA methylation level in the population)] (20). The smaller average differences may be related to the inherent stochastic nature of epigenetic processes (31) leading to a large variability in responses. The stochastic nature is strikingly illustrated by the large variation observed in the response of agouti gene methylation on maternal methyl donor supplementation even though the mice are inbred and the environmental conditions are highly controlled (17). In human studies, genetic and environmental heterogeneity may further obscure the full impact of prenatal famine. In our study, we tried to minimize the heterogeneity caused by these factors by comparing exposed individuals with their unexposed, same-sex siblings. Another potential source of heterogeneity is the cellular diversity of whole blood, the tissue we studied. This heterogeneity is less likely to play a role for the affected imprinted loci IGF2 (20), GNASAS and MEG3, since methylation is generally cell-type independent for imprinted loci (32) and the observed differences for non-imprinted loci between exposed individuals and controls were similar. With respect to our finding that GNASAS methylation was associated with exposure to famine late in gestation, it should be mentioned that a compromised late gestational development was hypothesized to lead to an immature immune system. The absolute numbers of lymphocytes were reported to be lower in children with a shorter gestation in a large population-based study (33). However, since the proportions of the different cell types were not affected, it is unlikely that this has contributed to our findings.
The changes observed are comparable with those found in the liver of rats prenatally exposed to a protein-deficient diet, where promoter methylation of the Pparα promoter was decreased from 6.1 to 4.5%, with individual CpG dinucleotides affected up to 5% and explaining up to 43% of the variance in gene expression of Pparα (34). This small absolute decrease of Pparα DNA methylation corresponded to a large ∼26% relative change. It may be hypothesized that modest absolute changes in DNA methylation may lead to significant changes in gene expression for loci with a relatively low methylation level so that relative changes are substantial. The larger relative changes of LEP in men (2.8/27.1 = 10.3%) and IL10 (2.4/20.8 = 11.5%) may be more likely to have functional consequences than those of INSIGF in men (2.6/84.8 = 3.1%).
Similar to animal studies of methionine restriction (29), we observed both increases and decreases in DNA methylation, depending on the locus studied. Our results cannot be readily explained by damage because of a deficiency in dietary methyl donors due to the famine and may thus be part of an adaptive response. To definitely prove or disprove the existence of an adaptive response, it will be necessary to characterize epigenetic responses of entire relevant pathways. Epigenetic mechanisms may contribute to the development of a thrifty phenotype (1,15), and sex differences in this respect are increasingly well described (13). It has been hypothesized that a thrifty phenotype may result from the combined effects of smaller epigenetic changes across the genome-shifting metabolic networks (35). INS (36) and LEP (35), both affected by prenatal famine exposure, were suggested to be particularly relevant in this respect. Our observations provide empirical evidence for these hypotheses in humans.
Our finding that the association of DNA methylation with prenatal conditions may depend on timing, and sex matches the specificity in phenotypic outcomes observed for prenatal famine exposure, including neonatal outcomes (9) and psychiatric (7) and metabolic traits (4,6,10). It remains to be determined whether the observed differences in DNA methylation in blood mark functional differences in relevant tissues for these traits. Previous work on IGF2 DMR has shown that methylation in blood can mark the methylation level in a relevant tissue (37). Also, the CpG sites studied in the LEP promoter were previously reported to show similar methylation in peripheral blood and adipocytes in vivo (38) and to influence LEP expression in vitro (39). Changes in DNA methylation induced early in development, for example by prenatal famine, may be propagated soma-wide, contributing to the observed correlation in DNA methylation across tissues (17,28). A preliminary analysis of our data did not reveal significant associations between DNA methylation and plasma lipids and BMI that we previously reported to depend on prenatal exposure to famine in women in this cohort (4,6). But DNA methylation at most genes relevant to these particular complex phenotypes was found to be affected in men only, but not in women. Similar to genetic studies, epigenetic studies linking variation in DNA methylation and complex disease will most likely require large study samples. Such studies may be particularly promising for IL10, which we found to be sensitive to periconceptional famine exposure. Genetic variation influencing IL10 expression at this locus is associated with schizophrenia (40,41), a phenotype also particular to the periconceptional period (7).
In summary, our study shows that exposure to famine in pregnancy may cause persistent changes in DNA methylation levels of multiple imprinted and non-imprinted genes with diverse biological functions. Our data support the hypothesis that associations between early developmental conditions and health outcomes later in life may be mediated by changes in the epigenetic information layer. Understanding how disturbances early in human development are linked to later-life disease may suggest new ways to prevent disease.
MATERIALS AND METHODS
Study population
Study participants are a subset of the population from our ongoing Hunger Winter Families Study, whose design and recruitment were described previously (21). In short, this study includes individuals exposed to famine during different periods in pregnancy and time controls born before or after the famine. Study subjects were selected from births between 1943 and 1947 at three institutions in famine-exposed cities (the midwifery training schools in Amsterdam and Rotterdam and the Leiden University Medical Center). The daily rations distributed by the authorities during the famine period from 28 November 1944 to 15 May 1945 period contained an average energy equivalent of 667 kcal (SD 151). During the famine, there was little variation in the percentage of calories from proteins (12%), fat (19%) and carbohydrates (69%) (42). In addition to time controls from these three institutions, we recruited whenever possible a same-sex unexposed sibling of these individuals to serve as sibling control. Clinical examinations, including blood collections, were completed for 311 births in these institutions and 311 sibling controls. Ethical approval for the study was obtained from the participating institutions, and all participants provided written informed consent.
The subset selected for the present study includes two exposure groups with their unexposed sibling controls. The first exposure group comprises births conceived during the height of the famine, with periconceptional and early pregnancy exposure. The second exposure group comprises births during the height of the famine, with late pregnancy exposure. Periconceptionally exposed individuals were defined as births with a mother's last menstrual period between 28 November 1944 and 15 May 1945. This group includes 60 individuals (age 58.1 years, SD 0.35), of whom 28 were male. Individuals exposed in late pregnancy were defined as births between 28 January and 30 May 1945. This group includes 62 individuals (age 58.8 years, SD 0.40), of whom 28 were male. As controls for each exposure group, we used a matched same-sex sibling (age 57.1 years, SD 5.50). The total study population therefore includes 244 individuals.
DNA methylation measurements
Genomic DNA was isolated from whole blood using the salting out method. One microgram of genomic DNA was bisulphite-treated using the EZ 96-DNA methylation kit (Zymo Research). All samples were bisulphite-treated on a total of three 96-well plates. Sibling pairs were on the same plate and periconceptionally and last trimester-exposed pairs were equally distributed over the plates. DNA methylation of CpG dinucleotides were measured by a mass spectrometry-based method (Epityper, Sequenom). The method determines the amount of DNA methylation by interrogating thousands of DNA copies assuming that 1 ng of genomic DNA equals approximately 300 copies. The quantitative nature, accuracy and reproducibility of this method have been shown extensively (20,43,44). All biochemical steps inherent to the methodology were performed according to the manufacturers' protocol. Bisulphite-converted DNA-specific PCR primers used to amplify the 15 investigated regions are summarized in the Supplementary Material, Table S1. This table includes the precise genomic location of the amplified regions and an overview of the CpG sites quantized in each amplicon. All individuals exposed early in gestation and their sibling controls were measured in triplicate on a single 384-well plate for each locus and the same was true for the late gestational exposed individuals and their sibling controls. Data quality control and filtering were done as described previously (45). Data filtering consisted of the removal of CpG dinucleotides of which the measurement could be confounded by single-nucleotide polymorphisms and of CpG dinucleotides of which the measurement success rate was <80%. Common causes of a lower success rate include fragments bordering on the upper and lower limits of the mass range that can be detected and cases of fragments of which the base of the peak signal in the mass spectrum overlapped another fragment. The success rate for the CpG-containing fragments that could be measured within the limits of the methodology was 93.3%.
Statistical analysis
The analyses were performed within sibships to minimize the possible confounding effects of differences in familial environment and genetics. We applied linear mixed models on the raw data without imputation of missing values to calculate exposure-specific differences between sibling pairs. All the analyses account for family relations, age at examination, bisulphite plate and the correlation between CpG dinucleotides. Sibship was entered as a random effect. Age at examination, bisulphite plate, exposure status and CpG dinucleotide and, where appropriate, sex were entered as fixed effects. The test for the timing specificity of the association between exposure and DNA methylation levels was done by adding a variable indicating the timing of the exposure and merging the periconceptional and last trimester data sets: timing specificity was tested by adding an interaction term of exposure status times the timing of the exposure to the linear mixed model. Testing the sex specificity of the associations between famine exposure and DNA methylation levels was done by adding a term for sex times the exposure status of the individuals in the linear mixed model.
The linear mixed model was chosen over a standard paired t-test because it allows for the analysis of multiple individual CpG dinucleotides in one test, accounts for the correlation between adjacent CpG dinucleotides, includes relevant adjustments within the model on the raw data and uses all available data. The linear mixed model reduces to a test with identical outcome to a paired t-test if the within-family difference is assessed for a single CpG dinucleotide, if no adjustments are performed and if there are no incomplete data for the sib pairs.
The basal difference in DNA methylation between men and women in the controls was calculated using ANOVA. All P-values reported are two-sided. All analyses were performed using SPSS 16.0.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Netherlands Heart Foundation (2006B083 to B.T.H.); the US National Institutes of Health (R01-HL067914 to L.H.L); The Netherlands Organization for Scientific Research NWO (911-03-016 to P.E.S.) and the European Union-funded Network of Excellence LifeSpan (FP6 036894). L.H.L. was supported by a 2008–2009 Lorentz fellowship from the Royal Netherlands Academy of Sciences.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participants of the Hunger Winter Families Study, TNO Quality of Life for contact tracing, the staff of the Gerontology and Geriatrics Study Center at the Leiden University Medical Center for performing the clinical examinations and the Central Clinical Chemical Laboratory for extracting genomic DNA.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hales C.N., Barker D.J. The thrifty phenotype hypothesis. Br. Med. Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Barker D.J. The origins of the developmental origins theory. J. Intern. Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 3.Whincup P.H., Kaye S.J., Owen C.G., Huxley R., Cook D.G., Anazawa S., Barrett-Connor E., Bhargava S.K., Birgisdottir B.E., Carlsson S., et al. Birth weight and risk of type 2 diabetes: a systematic review. JAMA. 2008;300:2886–2897. doi: 10.1001/jama.2008.886. [DOI] [PubMed] [Google Scholar]
- 4.Lumey L.H., Stein A.D., Kahn H.S., Romijn J.A. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am. J. Clin. Nutr. 2009;89:1737–1743. doi: 10.3945/ajcn.2008.27038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roseboom T., de Rooij S., Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum. Dev. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Stein A.D., Kahn H.S., Rundle A., Zybert P.A., van der Pal-de Bruin K., Lumey L.H. Anthropometric measures in middle age after exposure to famine during gestation: evidence from the Dutch famine. Am. J. Clin. Nutr. 2007;85:869–876. doi: 10.1093/ajcn/85.3.869. [DOI] [PubMed] [Google Scholar]
- 7.Susser E., Neugebauer R., Hoek H.W., Brown A.S., Lin S., Labovitz D., Gorman J.M. Schizophrenia after prenatal famine. Further evidence. Arch. Gen. Psychiatry. 1996;53:25–31. doi: 10.1001/archpsyc.1996.01830010027005. [DOI] [PubMed] [Google Scholar]
- 8.Kyle U.G., Pichard C. The Dutch famine of 1944–1945: a pathophysiological model of long-term consequences of wasting disease. Curr. Opin. Clin. Nutr. Metab. Care. 2006;9:388–394. doi: 10.1097/01.mco.0000232898.74415.42. [DOI] [PubMed] [Google Scholar]
- 9.Brown A.S., Susser E.S. Sex differences in prevalence of congenital neural defects after periconceptional famine exposure. Epidemiology. 1997;8:55–58. doi: 10.1097/00001648-199701000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Ravelli A.C., van Der Meulen J.H., Osmond C., Barker D.J., Bleker O.P. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am. J. Clin. Nutr. 1999;70:811–816. doi: 10.1093/ajcn/70.5.811. [DOI] [PubMed] [Google Scholar]
- 11.Bygren L.O., Edvinsson S., Brostrom G. Change in food availability during pregnancy: is it related to adult sudden death from cerebro- and cardiovascular disease in offspring? Am. J. Hum. Biol. 2000;12:447–453. doi: 10.1002/1520-6300(200007/08)12:4<447::AID-AJHB3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Painter R.C., de Rooij S., Bossuyt P.M., Simmers T.A., Osmond C., Barker D.J., Bleker O.P., Roseboom T.J. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am. J. Clin. Nutr. 2006;84:322–327. doi: 10.1093/ajcn/84.1.322. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert J.S., Nijland M.J. Sex differences in the developmental origins of hypertension and cardiorenal disease. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;295:R1941–R1952. doi: 10.1152/ajpregu.90724.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathanielsz P.W. Animal models that elucidate basic principles of the developmental origins of adult diseases. ILAR J. 2006;47:73–82. doi: 10.1093/ilar.47.1.73. [DOI] [PubMed] [Google Scholar]
- 15.Gluckman P.D., Hanson M.A., Cooper C., Thornburg K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterland R.A., Michels K.B. Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 17.Waterland R.A., Jirtle R.L. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterland R.A., Dolinoy D.C., Lin J.R., Smith C.A., Shi X., Tahiliani K.G. Maternal methyl supplements increase offspring DNA methylation at Axin fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 19.Bogdarina I., Welham S., King P.J., Burns S.P., Clark A.J. Epigenetic modification of the renin–angiotensin system in the fetal programming of hypertension. Circ. Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lumey L.H., Stein A.D., Kahn H.S., van der Pal-de Bruin K.M., Blauw G.J., Zybert P.A., Susser E.S. Cohort profile: the Dutch Hunger Winter families study. Int. J. Epidemiol. 2007;36:1196–1204. doi: 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- 22.Monk D., Sanches R., Arnaud P., Apostolidou S., Hills F.A., bu-Amero S., Murrell A., Friess H., Reik W., Stanier P., et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 23.McGowan P.O., Sasaki A., D'Alessio A.C., Dymov S., Labonte B., Szyf M., Turecki G., Meaney M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver I.C., Cervoni N., Champagne F.A., D'Alessio A.C., Sharma S., Seckl J.R., Dymov S., Szyf M., Meaney M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 25.El-Maarri O., Becker T., Junen J., Manzoor S.S., az-Lacava A., Schwaab R., Wienker T., Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 26.Morgan H.D., Santos F., Green K., Dean W., Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec no. 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 27.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 28.Morgan H.D., Jin X.L., Li A., Whitelaw E., O'Neill C. The culture of zygotes to the blastocyst stage changes the postnatal expression of an epigentically labile allele, agouti viable yellow, in mice. Biol. Reprod. 2008;79:618–623. doi: 10.1095/biolreprod.108.068213. [DOI] [PubMed] [Google Scholar]
- 29.Sinclair K.D., Allegrucci C., Singh R., Gardner D.S., Sebastian S., Bispham J., Thurston A., Huntley J.F., Rees W.D., Maloney C.A., et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl Acad. Sci. USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagata Y., Asada H., Tamura I., Lee L., Maekawa R., Taniguchi K., Taketani T., Matsuoka A., Tamura H., Sugino N. DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum. Reprod. 2009;24:1126–1132. doi: 10.1093/humrep/dep015. [DOI] [PubMed] [Google Scholar]
- 31.Bjornsson H.T., Fallin M.D., Feinberg A.P. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 33.Duijts L., Bakker-Jonges L.E., Labout J.A., Jaddoe V.W., Hofman A., Steegers E.A., van Dongen J.J., Hooijkaas H., Moll H.A. Fetal growth influences lymphocyte subset counts at birth: the Generation R Study. Neonatology. 2009;95:149–156. doi: 10.1159/000153099. [DOI] [PubMed] [Google Scholar]
- 34.Lillycrop K.A., Phillips E.S., Torrens C., Hanson M.A., Jackson A.A., Burdge G.C. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br. J. Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoger R. The thrifty epigenotype: an acquired and heritable predisposition for obesity and diabetes? Bioessays. 2008;30:156–166. doi: 10.1002/bies.20700. [DOI] [PubMed] [Google Scholar]
- 36.Prentice A.M., Rayco-Solon P., Moore S.E. Insights from the developing world: thrifty genotypes and thrifty phenotypes. Proc. Nutr. Soc. 2005;64:153–161. doi: 10.1079/pns2005421. [DOI] [PubMed] [Google Scholar]
- 37.Cui H., Cruz-Correa M., Giardiello F.M., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 38.Stoger R. In vivo methylation patterns of the leptin promoter in human and mouse. Epigenetics. 2006;1:155–162. doi: 10.4161/epi.1.4.3400. [DOI] [PubMed] [Google Scholar]
- 39.Melzner I., Scott V., Dorsch K., Fischer P., Wabitsch M., Bruderlein S., Hasel C., Moller P. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J. Biol. Chem. 2002;277:45420–45427. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 40.Chiavetto L.B., Boin F., Zanardini R., Popoli M., Michelato A., Bignotti S., Tura G.B., Gennarelli M. Association between promoter polymorphic haplotypes of interleukin-10 gene and schizophrenia. Biol. Psychiatry. 2002;51:480–484. doi: 10.1016/s0006-3223(01)01324-5. [DOI] [PubMed] [Google Scholar]
- 41.Ozbey U., Tug E., Namli M. Interleukin-10 gene promoter polymorphism in patients with schizophrenia in a region of East Turkey. World J. Biol. Psychiatry. 2009;1:1–8. doi: 10.1080/15622970802626580. [DOI] [PubMed] [Google Scholar]
- 42.Burger G.C.E., Drummond J.C., Sandstead H.R. Malnutrition and Starvation in Western Netherlands, September 1944–July 1945. Part I. The Hague, The Netherlands: General State Printing Office; 1948. [Google Scholar]
- 43.Coolen M.W., Statham A.L., Gardiner-Garden M., Clark S.J. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den B.D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heijmans B.T., Kremer D., Tobi E.W., Boomsma D.I., Slagboom P.E. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.