Abstract
Copy number variants (CNVs) are genomic segments which are duplicated or deleted among different individuals. CNVs have been implicated in both Mendelian and complex traits, including immune and behavioral disorders, but the study of the mechanisms by which CNVs influence gene expression and clinical phenotypes in humans is complicated by the limited access to tissues and by population heterogeneity. We now report studies of the effect of 19 CNVs on gene expression and metabolic traits in a mouse intercross between strains C57BL/6J and C3H/HeJ. We found that 83% of genes predicted to occur within CNVs were differentially expressed. The expression of most CNV genes was correlated with copy number, but we also observed evidence that gene expression was altered in genes flanking CNVs, suggesting that CNVs may contain regulatory elements for these genes. Several CNVs mapped to hotspots, genomic regions influencing expression of tens or hundreds of genes. Several metabolic traits including cholesterol, triglycerides, glucose and body weight mapped to three CNVs in the genome, in mouse chromosomes 1, 4 and 17. Predicted CNV genes, such as Itlna, Defcr-1, Trim12 and Trim34 were highly correlated with these traits. Our results suggest that CNVs have a significant impact on gene expression and that CNVs may be playing a role in the mechanisms underlying metabolic traits in mice.
INTRODUCTION
Copy number variants (CNVs) are DNA segments with a variable number of repeats among individuals, ranging from kilobases to several megabases in length. CNVs are an important source of genetic variation in diverse human populations (1,2), as well as in primates (3,4) and rodents (5,6). CNVs can influence gene expression (7), presumably by altering gene dosage, through disruption or duplication of CNV regions containing genes. In humans, they are associated with a number of Mendelian and complex genetic disorders, including autoimmune disease (8), HIV infection (9,10) and autism (11). The mechanisms by which CNVs contribute to disease in humans have been difficult to study, in part due the difficulty in obtaining tissue samples and population heterogeneity. However, the presence of CNVs in mouse inbred strains, as well as the ability to manipulate the mouse genome to map gene expression and clinical traits using crosses, makes the mouse an ideal model to dissect the biological significance of CNVs. Analyses of CNVs in mouse genomes have demonstrated significant variation among mouse inbred strains (5,12) as well as non-random recurrent CNVs among members of the same inbred strain (13). Moreover, CNVs identified between mice of different inbred strains are of similar size and magnitude as those identified among different human populations (1,5,12), suggesting that the mouse could serve as a model to study the biological significance of CNVs.
In order to establish whether the mouse can be used as a model to study the impact of copy number variation, we investigated the effect of CNVs on gene expression phenotypes using a panel of CNVs previously identified in 20 mouse inbred strains (5). We asked whether CNVs influenced gene expression or clinical traits by using this set of CNVs, in conjunction with genome-wide gene expression and metabolic trait data from two independent mouse crosses between strains C57BL/6J and C3H/HeJ. We and others have shown that the genetics of gene expression can be used as a link between DNA variation and phenotypic traits to prioritize candidate genes and to identify causal relationships between chromosomal regions and clinical phenotypes (14–16). Here we show that mouse CNVs resulted in altered gene expression in the genes mapping to CNVs, which was highly correlated with copy number. We also observed an effect in genes flanking CNVs, suggesting that CNVs can influence gene expression through disruption of regulatory sequences. Our results also show that expression QTL (eQTL) hotspots mapped to CNVs, suggesting that regulatory elements present in CNVs and/or eQTL mapping to CNVs may be influencing the expression of hundreds of genes in trans. Furthermore, we found that a variety of metabolic traits, including body weight, cholesterol and glucose levels, mapped to a subset of the CNVs. Our results indicate that mouse inbred strains can be used to examine the mechanisms by which CNVs influence complex traits.
RESULTS
To assess the impact of CNVs on gene expression, we examined 19 CNVs variable between the mouse strains C57BL/6J (B6) and C3H/HeJ (C3H). These CNVs are distributed among 11 chromosomes and range in size from 47 kb to 1.9 Mb in length, with a median size of 195 kb, and contain a total 54 genes. Sixteen of 19 CNVs contain at least one gene, and 14 CNVs contain more than one gene. CNV genomic positions and array comparative genome hybridization (aCGH) ratios can be found in Supplementary Material, Table S1. These 19 CNVs represent the entire set of CNVs variable between B6 and C3H identified by Graubert et al. (5). The range in size of the CNVs in part reflects a limitation of the aCGH platform and of the CNV calling algorithm employed. The aCGH platform contained 385 000 probes spaced ∼5 kb apart, and probe density in different genomic locations also contributed to the resolution of the platform. In addition, the CNV detection algorithm employed by Graubert et al. (5) required a change in intensity in at least five probes in a segment, resulting in an increased ability to detect larger CNVs.
To ensure that the CNVs reported by Graubert et al. were also present in the parental mice used in our crosses, we validated three CNVs by qPCR on the genomic DNA from parental mice of cross 2, as well as in B6 and C3H mice recently obtained from the Jackson Laboratories (Supplementary Material, Table S2). We observed a high correlation between the published log2(C3H/B6) ratios and our qPCR validation data, with a Pearson's correlation coefficient of 0.98 and 0.99 to the samples from cross 2 and Jackson Laboratories mice, respectively. This correlation confirms that the same CNVs found in the published data are present in the F2 population used in the current study. In addition, since both the mice employed in the CGH arrays and in our mouse crosses were obtained from the Jackson Laboratories, we expect different mice of the same strain to be genetically identical.
We used microarray gene expression levels in F2 mice from an intercross between B6 and C3H strains (17) and eQTL mapping to determine if the expression levels in the genome were controlled by CNVs. We observed that a large number of eQTL overlapped CNV regions in adipose (Fig. 1A), brain, liver and muscle tissues (Supplementary Material, Fig. S2), suggesting that regulatory elements or genes mapping to CNVs may be controlling the expression of hundreds of genes in cis or trans. However, because the resolution provided by the intercross yields QTL regions which are several megabases in size, causal relationships between CNVs and trans eQTL are difficult to establish. For this reason, we focused on genes that mapped within CNVs.
Figure 1.
Mouse CNVs influence gene expression levels. (A) The number of eQTL with LOD score >4.3 in adipose tissue and the position of CNVs in the mouse genome. The genomic locations of CNVs are marked by red diamonds and red vertical bars and eQTL counts are plotted as the average count in a 2 Mb moving window. (B) Microarray expression levels are variable in genes found within or near CNVs in a segment of chromosome 7 carrying two CNVs. The absolute fold change in gene expression between B6 and C3H groups is shown for 150 genes in a 6 Mb window. (C) The concordance between gene expression log2(C3H/B6) ratios and CGH log2(C3H/B6) ratios. The CGH ratios are shown as an average of a five probe moving window.
To determine if CNVs influenced gene expression, we measured RNA expression levels for each gene and the genotype at the nearest SNP to the gene. We used the SNP genotypes to determine the parental origin of each genomic segment in order to separate F2 mice into three groups: homozygous for B6, homozygous for C3H or heterozygous, and to compare gene expression levels among the groups. Using this approach, we determined that 45 of 54 (83%) CNV genes were differentially expressed between mouse strains B6 and C3H (Fig. 1B, Table 1 and Supplementary Material, Table S3). Each of the genes identified was differentially expressed in at least one of the four tissues analyzed, and several of these genes were differentially expressed in multiple tissues. We hypothesize that the level of expression of these genes varies in response to the change in copy number in the DNA segment in which they are found.
Table 1.
Genes in CNVs vary in gene expression
| Gene name | P-value | Tissue | Sex | Cross |
|---|---|---|---|---|
| 9230105E10Rik | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| 9430025M13Rik | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Cd244 | 1.00E−16 | A, B, L | All, F, M | Cross 1 and cross 2 |
| Chi3l3 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| D14Ertd449e | 1.00E−16 | A, B, L M | All, F, M | Cross 1 and cross 2 |
| Defcr3 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Entpd4 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Glo1 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Gvin1 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Itlna | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Klra18 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Klra19 | 1.00E−16 | L | All, F, M | Cross 1 and cross 2 |
| Klra21 | 1.00E−16 | A, L, M | All, F, M | Cross 1 and cross 2 |
| Klra8 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| MMT00030144 | 1.00E−16 | L | All, F, M | Cross 1 and cross 2 |
| MMT00046617 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| MMT00078062 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Trim12 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Trim34 | 1.00E−16 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Klrk1 | 1.78E−15 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Klra9 | 4.55E−13 | L, M | All, F, M | Cross 1 |
| Anxa11 | 1.26E−07 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| Loxl2 | 2.72E−06 | A, B, L, M | All, F, M | Cross 1 and cross 2 |
| A530023O14Rik | 6.71E−04 | L | All, F, M | Cross 1 |
| MMT00007806 | 1.14E−03 | A, M | All, F | Cross 2 |
| Cfh | 1.47E−03 | A, B | All, F, M | Cross 1 and cross 2 |
| Csf2ra | 1.52E−03 | A, B, L M | All, F, M | Cross 1 and cross 2 |
| V1rd14 | 2.65E−03 | A, B, M | All, F, M | Cross 1 and cross 2 |
| Mrpl17 | 2.83E−03 | A, B, M | All, F, M | Cross 1 and cross 2 |
| Dnahc8 | 3.32E−03 | L, M | All, M | Cross 1 |
| 3000002G13Rik | 4.76E−03 | L | All, F | Cross 1 |
| Slc25a37 | 5.53E−03 | B, L, M | All, F, M | Cross 1 and cross 2 |
| Dchs1 | 6.72E−03 | A, M | All, F, M | Cross 1 and cross 2 |
| MMT00060041 | 7.96E−03 | B, L | All, M | Cross 2 |
| MMT00068533 | 8.71E−03 | L | M | Cross 2 |
| MMT00071942 | 0.013 | A, L | All, F, M | Cross 1 and cross 2 |
| MMT00068366 | 0.013 | A, B, M | All, F | Cross 1 |
| 2010016B13Rik | 0.018 | A, B, M | All, F, M | Cross 2 |
| MMT00013484 | 0.019 | L | M | Cross 2 |
| Nalp1 | 0.020 | B, L, M | All, F, M | Cross 1 and cross 2 |
| Glp1r | 0.022 | A | F | Cross 2 |
| MMT00013481 | 0.033 | A, L, M | F, M | Cross 1 |
| MMT00061970 | 0.042 | A | M | Cross 2 |
| MMT00046650 | 0.048 | L | All | Cross 1 |
| Chia | 0.049 | L | All | Cross 2 |
Tissues: A, Adipose; B, Brain; L, Liver; M, Muscle. Sex: F, Females and M, Males. P-value corresponds to the ANOVA for the group in bold.
To determine whether CNV mapping genes were indeed regulated in cis, we performed a classic cis–trans test in the CNV mapping genes Klrk1, CD244 and Trim12 (Fig. 2). We carried out the cis–trans test using semi-quantitative sequence analysis to examine the allele ratios in the genes in adipose cDNA from B6, C3H and B6×C3H F1 mice from cross 1. Each of these genes maps to CNVs with higher copy in B6, and consistent with the copy number change, we find that transcript levels in these genes show a greater B6 allele fraction in the F1 mice. These results provide further evidence that CNV mapping genes are indeed regulated in cis.
Figure 2.
CNV mapping genes are regulated in cis. Allelic imbalance shows that CNV genes (A) Klrk1, (B) CD244 and (C) Trim12 are regulated in cis. We used semi-quantitative sequence analysis in adipose cDNA from B6, C3H and B6×C3H F1 in cross 1, where we determined the ratio of the B6 and C3H alleles in the overall transcript levels of the F1 mice.
To rule out the possibility of a recombination event between each CNV mapping gene and the nearest SNP to the gene, we examined the haplotype structure of the cross and the genotype profiles surrounding each CNV. The distribution of the distances between each CNV gene and the nearest SNP is shown in Supplementary Material, Fig. S1A, where we observed a median and mean distance of 880 kb and 1.27 Mb, respectively. We expect the degree of correlation between nearby SNPs to be very high in an F1 intercross, so that the likelihood of observing a recombination event within this range is very low. Indeed, there are blocks of highly correlated SNPs that span ∼20 Mb in chromosome 1 (Supplementary Material, Fig. S1B), with similar results observed in other chromosomes. Furthermore, we observed an average of one recombination event per chromosome in each F2, as illustrated for chromosome 1 in Supplementary Material, Fig. S1C. No recombination break points were observed at the location of genes mapping to CNVs (Supplementary Material, Fig. S1D). One possible exception to this is for CNV 19 at the proximal end of chromosome 7, where the nearest SNP is found 14 Mb from the CNV. Although we cannot rule out the possibility of a recombination event between CNV 19 and the nearest SNP, the high correlation observed between nearby SNPs and the number of recombination evens per chromosome suggests that this is unlikely.
We next asked whether the increase or decrease in gene expression was concordant with the direction of the change in copy number. We observed that gene expression was concordant with CNVs in 84% of the CNV genes in adipose, with similar results observed in other tissues (Fig. 1C and Table 2). We used a binomial random model to determine the significance of these observations. The results of this analysis, shown in Table 2, indicated that gene expression differences were concordant with copy number in adipose (P = 1.14E-08), brain (P = 0.02), liver (P = 8.03E-05) and muscle (P = 0.01) tissues. In order to test the overall effect of CNVs on gene expression, we also considered all genes differentially expressed. We observed that CNVs had an effect on gene expression in adipose (P = 5.97E-07), brain (P = 0.01), liver (P = 3.22E-07) and muscle (P = 1.52E-03) tissues (Supplementary Material, Table S4). For CNVs carrying more than one gene, we asked how the overall expression levels were affected by each CNV using the regularized gamma function. Our results indicated that 11 out of 14 (79%) CNVs which contain more than one gene had a significant impact on gene expression levels in adipose, brain, liver and muscle tissues (Supplementary Material, Table S5). Similar results were observed when we examined the effect of CNVs on gene expression using non-parametric analysis.
Table 2.
Concordance between CNV and gene expression in cross 1
| Tissue | Sex | Binomial | Permutation | Concordance |
|---|---|---|---|---|
| P-value | False discovery | |||
| Adipose | All | 1.14E−08 | <0.1% | 84% |
| Adipose | Female | 2.60E−08 | <0.1% | 81% |
| Adipose | Male | 1.53E−07 | <0.1% | 89% |
| Brain | All | 0.02 | 2.9% | 80% |
| Brain | Female | 0.11 | 12.6% | 69% |
| Brain | Male | 0.01 | 2.9% | 85% |
| Liver | All | 8.03E−05 | <0.1% | 75% |
| Liver | Female | 9.67E−03 | 1.2% | 72% |
| Liver | Male | 9.84E−07 | <0.1% | 77% |
| Muscle | All | 0.01 | 2.4% | 81% |
| Muscle | Female | 0.05 | 4.3% | 81% |
| Muscle | Male | 5.47E−03 | 0.7% | 80% |
To estimate the false discovery rate in these observations, we repeated our analysis 1,000 times using a randomly permuted sample in which the mouse genotypes were randomly reassigned in each sample (Table 2, Supplementary Material, Tables S4, S5 and Fig. S3). We observed that the P-values from permuted samples followed the expected uniform distribution. These results support our hypothesis that CNVs have a significant impact on gene expression levels in mice.
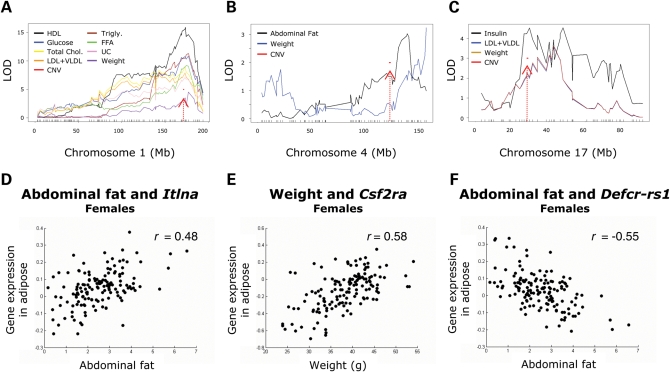
We next asked whether CNVs played a role in the etiology of metabolic traits using two different approaches: (i) by looking for clinical quantitative trait loci (cQTL) near CNVs and (ii) by looking for correlations between gene expression levels and the metabolic traits. In the QTL analysis, we observed that three CNVs overlapped several cQTL for weight, cholesterol, triglycerides, glucose and insulin levels on chromosomes 1, 4 and 17 (Fig. 3A–C). Furthermore, we observed that several of the genes mapping within CNVs were significantly correlated with metabolic traits (Fig. 3D–F) and Supplementary Material, Table S5). For example, Itlna was correlated with abdominal fat weight (r = 0.48), Csf2ra was correlated with body weight (r = 0.58) and Defcr-rs1 was correlated with abdominal fat (r = −0.55) in females. We used the false discovery rate (18) to account for multiple testing and selected all genes correlated with a trait at an FDR cutoff of 1% (Supplementary Material, Table S6).
Figure 3.
Impact of CNVs on metabolic traits. (A–C) Clinical QTL mapping reveals that CNVs overlap multiple cQTL in chromosomes 1, 4 and 17. (D–F) The expression levels of CNV genes are correlated with metabolic traits in B6×C3H F2 mice. The expression levels correspond to adipose tissue and F2 female mice. r Values correspond to the Pearson's correlation coefficient.
In order to test a causal relationship between genetic variation, differences in gene expression and clinical traits, we used the Network Edge Orienting algorithm (NEO) (19). In essence, NEO uses structural equation modeling to test the superiority of the model where genetic variation (M) influences gene expression (A), which in turn influences a trait (B), so that M → A → B. We applied NEO to each gene expression-clinical trait pair (A and B) and the nearest SNP to the gene (M) in cross 1. The two main outputs of NEO are Local Edge Orienting (LEO) scores and Root Mean Square Error of Approximation (RMSEA). RMSEA provides a measure of the goodness-of-fit for the model under investigation and LEO provides a measure of how superior the model under investigation is, relative to other models. In general, LEO scores greater than 1 and RMSEA scores less than 0.05 suggest a causal relationship between transcript levels and a clinical trait (A → B). The results of the NEO causality test for genes that met these criteria are shown in Table 3, where we provide statistical evidence for causal relationships between CNV mapping genes and clinical traits. We found that Trim12 and Trim34 in a CNV are causal for plasma cholesterol, Itlna and Gvin1 are causal for insulin levels; ltlna and Gvin1 transcript levels are causal for plasma insulin levels and Trim12 is causal for plasma glucose levels.
Table 3.
NEO causality test
| Marker: M | Chr | Transcript: A | Reactive trait: B | LEO A to B | RMSEA A to B |
|---|---|---|---|---|---|
| rs3717027 | 7 | Trim34 | Total cholesterol | 3.23 | 0 |
| rs3717027 | 7 | Trim34 | LDL plus VLDL | 3.18 | 0 |
| rs3717027 | 7 | 9230105E10Rik | Unesterified cholesterol | 2.32 | 0 |
| rs3654285 | 1 | Itlna | Log plasma insulin | 2.29 | 0 |
| rs3717027 | 7 | Trim12 | Unesterified cholesterol | 2.21 | 0 |
| rs3682831 | 1 | LOC381294 | Total cholesterol | 1.95 | 0.04 |
| rs3682831 | 1 | LOC381294 | LDL plus VLDL | 1.86 | 0.03 |
| rs3682831 | 1 | LOC381294 | Log plasma insulin | 1.78 | 0 |
| rs3717027 | 7 | Trim12 | Total cholesterol | 1.77 | 0 |
| rs3717027 | 7 | 9230105E10Rik | Glucose | 1.63 | 0 |
| rs3717027 | 7 | Trim12 | LDL plus VLDL | 1.6 | 0 |
| rs3654285 | 1 | Itlna | Plasma insulin | 1.49 | 0 |
| rs3654285 | 1 | Itlna | Plasma insulin | 1.49 | 0 |
| rs3696743 | 6 | Klra9 | Abdominal fat | 1.47 | 0 |
| rs3696743 | 6 | Klra19 | Abdominal fat | 1.45 | 0 |
| rs3711721 | 7 | Gvin1 | Glucose | 1.44 | 0 |
| rs3711721 | 7 | Gvin1 | Unesterified cholesterol | 1.42 | 0.03 |
| rs3717027 | 7 | Trim12 | Glucose | 1.32 | 0 |
| rs3671079 | 7 | 9430025M13Rik | Glucose | 1.21 | 0 |
| rs3654285 | 1 | Itlna | Plasma insulin | 1.17 | 0 |
| rs3711721 | 7 | Gvin1 | Free fatty acids | 1.14 | 0 |
| rs3682831 | 1 | LOC381294 | Unesterified cholesterol | 1.06 | 0.04 |
LEO, Local Edge Orienting score; RMSEA, Root Mean Square Error of Approximation.
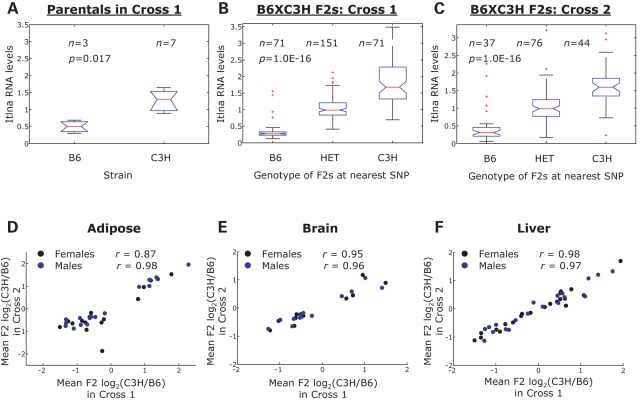
To determine the reproducibility of our results, we studied the effect of CNVs on gene expression in an independent intercross between B6 and C3H (20). We determined the overlap between the genes identified in each of the two crosses using the hypergeometric distribution (Table 4) and observed a significant overlap in adipose (P = 2.21E−09), brain (P = 4.60E−08) and muscle tissues (P = 2.29E−08) tissues. We also observed a significant overlap in the liver when females (P = 2.29E−08) and males (P = 2.94E−08) were analyzed separately. Gene expression and CGH log2(C3H/B6) ratios for Itlna, a gene mapping within a CNV in mouse chromosome 1, are shown in B6 and C3H parental strains, as well as in B6×C3H F2 mice from the first and second crosses (Fig. 4A–C). We observed consistent differences in gene expression that were concordant with CNV in both crosses (Table 2 and Supplementary Material, Table S4). Furthermore, we determined that gene expression levels were highly correlated between the two crosses in adipose (r = 0.87), brain (r = 0.95), liver (r = 0.97) and muscle (r = 0.98) tissues in females, with similar results observed in males (Fig. 4D–F, Table 4 and Supplementary Material, Fig. S4). Overall, the results obtained in the first and second crosses supported the notion that CNV has a significant effect on gene expression and metabolic traits.
Table 4.
Replication in two independent crosses
| Tissue | Sex | Total no. of genes in CNV | Differentially expressed genes |
Hypergeometric P-value | Correlation in gene expression: R | ||
|---|---|---|---|---|---|---|---|
| in Cross 1 | in Cross 2 | Overlap | |||||
| Adipose | All | 54 | 21 | 20 | 17 | 2.21E−09 | 0.92 |
| Adipose | Females | 54 | 22 | 21 | 16 | 1.89E−06 | 0.87 |
| Adipose | Males | 54 | 22 | 16 | 14 | 2.62E−07 | 0.98 |
| Brain | All | 54 | 16 | 15 | 12 | 4.60E−08 | 0.95 |
| Brain | Females | 54 | 14 | 13 | 9 | 9.18E−06 | 0.95 |
| Brain | Males | 54 | 15 | 13 | 11 | 1.61E−08 | 0.96 |
| Liver | All | 54 | 28 | 3 | 1 | 0.53 | 0.44 |
| Liver | Females | 54 | 25 | 17 | 16 | 2.29E−08 | 0.97 |
| Liver | Males | 54 | 25 | 17 | 14 | 2.94E−05 | 0.97 |
| Muscle | All | 54 | 22 | 15 | 12 | 2.97E−05 | 0.92 |
| Muscle | Females | 54 | 16 | 14 | 9 | 1.94E−04 | 0.98 |
| Muscle | Males | 54 | 19 | 16 | 10 | 1.30E−03 | 0.94 |
Figure 4.
Effect of CNV on gene expression replicates in two independent crosses. (A) Itlna microarray expression levels in the parental inbred strains B6 and C3H from the first cross; the P-value corresponds to a two-sided t-test. (B) Itlna expression levels in B6×C3H F2 mice in first cross and (C) second cross. The P-values correspond to a one-way ANOVA test among the B6, HET and C3H F2 groups. Gene expression levels are highly correlated between the first and second crosses in adipose (D), brain (E) and liver (F) tissues. Each dot plotted represents a differentially expressed gene found in a CNV. The expression values shown correspond to the mean log2(C3H/B6) ratio of the C3H to B6 F2 groups in each cross. r Values correspond to the Pearson's correlation coefficient.
We observed that a total of 1253 eQTL mapped to CNV genomic locations, suggesting that eQTL or regulatory elements mapping to CNVs may be influencing gene expression in trans (Fig. 5, Supplementary Material, Table S7). We illustrate this observation for a CNV in the proximal end mouse chromosome 3, which is found within the 95% confidence interval of 27 eQTL. All of these eQTL showed a LOD score greater than 4.3, and 22 of the 27 eQTL showed peak marker positions at or very near the CNV. Five of eQTL appear to be mapping in cis, whereas the remaining genes are mapping in trans (Fig. 5A). Since no genes mapped within this CNV (Fig. 5B), we examined the degree of conservation in the genomic sequence of this CNV using the VISTA genome browser (Fig. 5C). Previous reports have shown that highly conserved non-coding sequences represent putative regulatory elements (21), involved in both local and distant gene regulation. Interestingly, we found such highly conserved non-coding sequences within the CNV, suggesting a potential role for CNVs and/or regulatory elements within CNVs in the context of eQTL hotspots.
Figure 5.
Mouse eQTL hotspot coincides with highly conserved non-coding sequences present in mouse CNVs. (A) A CNV in chromosome 3 coincides with the 95% confidence interval of 27 eQTL. Each curve represents an eQTL with LOD score >4.3 from female liver in cross 1. The red bar marks the position of the CNV. (B) The location of the CNV and neighboring genes in a 2 Mb window from the UCSC genome browser in Build37. Cross-species conservation for the CNV sequences is shown in (C), where the base mouse genome sequence (Build 37) is compared with the rat, human, rhesus macaque, dog, horse and chicken sequences. The degree of conservation is plotted on the y-axis as % identity over a 100 bp window. Highly conserved regions are highlighted under the curve, with a red color specifying non-coding sequences.
DISCUSSION
In this study, we have employed a combined genomics and genetics approach to ask whether CNVs in the mouse genome have a significant impact on gene expression levels and metabolic phenotypes. To this end, we have used microarray gene expression as well as metabolic traits measured in an F2 population obtained from an intercross between mouse inbred strains C57BL/6J and C3H/HeJ. We have found that CNVs play a significant role in gene expression and clinical traits in mice. We observed that 83% genes (45/54) found in CNV regions between B6 and C3H were differentially expressed (Table 1). These genes include a number of genes known to play a role in disease susceptibility in mouse and humans (22–27). For example, Glo1 is associated with autism susceptibility in humans (24), Alzheimer's disease (27) and anxiety (25) in mice. Furthermore, Cfh is associated with age-related macular degeneration in humans (23) and reduced visual perception (26) in Cfh−/− mice. Interestingly, C3H mice, which have both lower Cfh expression and copy number, become blind as they age despite the addition of the gene Pde6b traditionally believe to cause blindness in C3H mice (28).
We observed that gene expression was generally increased in genes found in higher copy CNVs and decreased in genes found in lower copy CNVs (Table 2 and Supplementary Material, Table S4). However, this was not always the case, since ∼20% of genes were expressed in the opposite direction as the copy number change, and roughly 15% of genes found in CNVs were not differentially expressed. There are several reasons that could explain these observations. First, gene expression may be influenced by additional regulators or transcription factors controlling gene expression in trans. Similarly, we cannot exclude the possibility that gene expression may be disrupted if regulatory regions are affected by the CNV or the possibility of transcriptional silencing due to DNA methylation. Another possibility involves the reliability in CNV detection, particularly in defining CNV boundaries with confidence when aCGH data are used (2). Finally, our analysis suggests tissue specific expression. We observed a larger number of genes whose expression was concordant with copy number in tissues which are largely homogenous, such as adipose tissue, but a lower degree of concordance in tissues with highly specialized sections, such as the brain (Table 2 and Supplementary Material, Table S4).
Our results also suggest that CNVs can influence complex phenotypes. We observed that several genes present in CNVs were highly correlated to metabolic traits such as body weight and adiposity (Fig. 3 and Supplementary Material, Table S6) and that cQTL for these traits map to CNVs. In addition, Klra8 was highly correlated with coronary artery calcification and Csf2ra was correlated with body weight, fat mass and insulin levels (Supplementary Material, Table S6). Both Klra8 and Cs2ra have been shown to play a role in immune-related functions such as in cytomegalovirus resistance (29) and immune cell differentiation, respectively. Interestingly, we find evidence that CD244 was correlated with femoral bone mineral density (r = −0.38, Supplementary Material, Table S6), a gene recently linked to rheumatoid arthritis (30), an autoimmune condition associated with bone loss in arthritis patients (31).
cQTL mapping to CNVs suggests that CNV genes, or regulatory elements within the CNVs, are contributing to trait development. The CNV genes Itlna, Trim12 and Trim34 were correlated to weight, triglycerides, adiposity, glucose and insulin levels (Fig. 3 and Supplementary Material, Table S6), and several metabolic traits show peak QTL mapping at or very near the location of these genes (Fig. 3). In addition, our causality test also suggests that transcript levels in the genes Trim12 and Trim34 are causal for plasma cholesterol and that Itlna and Gvin1 transcript levels are causal for plasma insulin levels, among others (Table 3). However, although this genetic and statistical evidence allows us to generate strong hypotheses for the involvement of CNV genes in clinical traits, the use of mouse knock-outs and transgenics is still necessary to validate these causal relationships.
Recent studies have shown that both inherited and de novo CNVs are associated with autism (11,32,33) and schizophrenia (34,35) in humans, suggesting that CNVs may play a role in the etiology of complex traits. However, a deeper understanding of complex traits in humans necessitates the use of model organisms that can permit investigation of the molecular mechanisms underlying these traits. The vast number genetic tools available to the scientific community, as well as the availability of genome-wide gene expression profiles and extensive behavioral phenotyping databases (36,37), make the mouse an ideal choice for the dissection of CNVs. But can the mouse be used as a model to study CNVs? Our results suggest that it can. Even in the relatively small set of 19 CNVs variable between strains B6 and C3H, we observed a significant impact on gene expression levels in cis (Table 1) and possibly a much larger effect on genes regulated in trans (Fig. 1A, Fig. 5 and Supplementary Material, Fig. S2). Furthermore, the presence of highly conserved non-coding sequences in mouse CNVs (Fig. 5) suggests that these CNVs carry regulatory elements that influence the expression levels of tens or hundreds of eQTL in trans. We believe that our current work can serve as a starting point for the use of the mouse as a model to dissect the contribution of CNVs in complex traits.
MATERIALS AND METHODS
Copy number variation dataset
A set of CNVs in 20 mouse inbred strains were previously identified by Graubert et al. (5) using aCGH. A set of 19 CNVs were identified between strains B6 and C3H. The putative genomic start and end positions of each CNV were obtained from the published data (5) and aCGH intensity data were obtained from the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) accession number GSE5805.
Mouse crosses
We analyzed two independent mouse intercrosses between strains C57BL/6J (B6) and C3H/HeJ (C3H) previously generated in the Lusis laboratory by crossing the parental strains to generate F1 mice, and then further intercrossing F1's to generate F2 mice. The first cross was performed between strains B6 and C3H on an ApoE−/− background. Three hundred and forty-four F2 mice, 166 females and 168 males, were generated and fed a chow diet (Ralston-Purina Co, St Louis, MO, USA) until 8 weeks of age, then fed a high fat ‘western’ diet (Teklad 88137, Harlan Teklad, Madison WI, USA) for 16 weeks until euthanasia at 24 weeks of age. The second cross was performed between the same strains B6 and C3H on a wild-type background to generate 309 F2 mice, 145 females and 164 males. Mice were fed a chow diet until 8 weeks of age, and then fed a high fat ‘western diet’ for 12 weeks until euthanasia at 20 weeks of age. Since the second cross consisted of both B6×C3H and C3H×B6 F2 mice, we restricted our analysis to the B6×C3H mice, consistent with the direction of the first cross. A detailed description of each cross is found in the articles published by Wang et al. (17) and Farber et al. (20). We referred to these crosses as cross 1 and cross 2, respectively. All mice were housed under specific pathogen-free conditions and according to NIH guidelines.
Gene expression
RNA expression levels were measured by microarray using total RNA. Microarray analysis was carried out in adipose, brain, liver and muscle tissues from parental strains or F2 mice as described (17,20). Gene expression data are available on GEO for adipose (GSE3086), brain (GSE3087), liver (GSE2814) and muscle (GSE3088) tissues in the first cross, and in the second cross for adipose (GSE11065), brain (GSE12798), liver (GSE11338) and muscle (GSE12795) tissues.
Cis–trans test
We used adipose RNA from B6, C3H and B6×C3H F1 mice from cross 1 to generate cDNA (ABI 4367381). We used PCR amplification on the cDNA using the following primers for the genes Krlk1(5′caa cct gga tca gtt tct gaag3′ and 5′agg agc cat ctt ccc actg3′), CD244 (5′ttc tgc tgt gtc ctg ctg ac3′ and 5′gcc ttc agg tta ggg gtc tc3′) and Trim12 (5′tgg aaa gaa act cca gct cttc3′ and 5′gag cct ctg tga cct ctt gc3′). We then cleaned the PCR products using ExoSAP-IT (USB 78200), followed by sequencing of the PCR product at the UCLA genotyping and sequencing core (www.genoseq.ucla.edu). We used semi-quantitative sequence analysis to quantitative the peak heights for the B6 and C3H alleles rs30851140 (in Klrk1), rs31537914 (in CD244) and rs31924865 (in Trim12) using Chromas version 2.13 as described (38).
CNV validation
We extracted genomic DNA from the liver tissue of B6 and C3H parental mice from cross 2, and from ear tissue of B6 and C3H mice obtained from the Jackson Laboratories (Bar Harbor, Maine). We used qPCR for three CNV using primer sequences published by Graubert et al. (5): CNV1 (5′cag aat atg taa atg tta gtc ccc aaag3′ and 5′gct tca acc acc tgg aag agat3′), CNV6 (5′ggc ata ggt act atc caa gta caa ggt3′ and 5′cct ccc cat cct cag tta tct ct3′) and CNV14 (5′cca gtg ctt gag gca aat ca3′ and 5′tgg gag cat gcg ctt taa cc3′). We used the single copy gene β-Actin to normalize each sample (5′agc cat gta cgt agc cat cc3′ and 5′ctc tca gct gtg gtg gtg aa3′).
QTL mapping
Expression and clinical QTL mapping was performed using the Rqtl package in R (www.rqtl.org). We used marker regression without imputation, and 95% confidence intervals were determined using a 1.5 LOD drop.
NEO causality test
We used the NEO package in R to test causality using the simple model M → A → B, where A is transcript levels, B is clinical traits and M is the genotype of the nearest SNP to the gene. We tested each CNV gene–trait pair using expression and clinical trait data from cross 1, and selected causal gene–trait pairs where LEO scores>1 and RMSEA scores <0.05, as described (19,20).
Conservation
We used the VISTA genome browser (http://pipeline.lbl.gov/) to determine the degree of conservation between mouse and human sequences using the genomic location of mouse CNVs. We used mouse Build 34 as the reference genome and determined conservation to human sequences as described (39).
Statistical analysis
Gene expression
For each gene, we used SNP genotyping to determine the parental origin of a chosen genomic segment in the F2 mice, which allowed us to separate mice into three distinct groups, those carrying genomic segments homozygous for B6, homozygous for C3H or heterozygous. To determine if genes were differentially expressed, we used one-way ANOVA to compare microarray gene expression levels between B6 homozygous, C3H homozygous and heterozygous groups. To determine whether B6 homozygotes or C3H homozygotes were higher in gene expression levels, one-tailed right and left-handed t-tests were used to compare the means between the two groups.
Binomial test
To test the effect of CNVs on gene expression levels, we used the binomial probability of observing both a CNV and a change in gene expression levels for genes overlapping CNVs, given the genome-wide likelihood of observing gene expression differences. Three inputs were used for the binomial test: (i) the probability of a given gene being differentially expressed, P=number of genes where the ANOVA test gave P-value less than 0.05, divided by the total number of genes in microarray, (ii) the total number of genes overlapping CNV regions, n, and (iii) the total number of ‘successes’, genes which overlap CNVs and were also differentially expressed, x. We took as P-value 1 minus the binomial cumulative distribution function, with parameters p, n and x.
We also tested whether copy number was concordant with gene expression; for example, so that if a CNV was higher in B6, the level of gene expression was also higher in B6. We again used the binomial test, where number of successes x was defined as the number of genes where the level of expression was both higher in B6 relative to C3H and the CNV change was also higher in B6, plus the number of genes where the level of expression was both higher in C3H and the CNV change was also higher in C3H. Each binomial test was performed using the binocdf function in MATLAB.
Gamma test
For CNVs that overlapped more than one gene, we tested the effect of the CNV on gene expression as follows: if a given CNV overlaps x ≥ 2 genes, and the probability that each ith gene is differentially expressed is pi, then in the random model that the p are uniform on (0,1), a=p1xp2···px has a cumulative distribution function gammainc(−log(a), x, ‘upper’), in MATLAB notation. A regularized incomplete gamma function (40) serves as a suitable P-value for the probability that the at least one of the genes is differentially expressed.
Correlation
We correlated gene expression levels of genes overlapping CNVs to metabolic quantitative traits measured in each F2 mouse. Metabolic traits were measured as described (17,20). For each gene–trait pairing, Pearson's correlation coefficient r was calculated for the vector of gene expression values and corresponding vector of trait values across F2 mice. For each gene–trait correlation, we computed r using only F2 mice were both gene expression and trait data were available for the given gene–trait pair.
Overlap between replication crosses
We used a hypergeometric test to determine the overlap in the set of genes hypothesized to be influenced by CNVs in the two mouse crosses. We employed the hypergeometric test to obtain P-values for the intersection of the sets (i) the CNV genes differentially expressed by ANOVA in cross 1 and (ii) the CNV genes differentially expressed by ANOVA in cross 2, being unusually large in the universe of all CNV genes.
Furthermore, we tested whether the directionality of gene expression differences was consistent between the two crosses using Pearson's correlation. The overlap in the genes identified in each cross was calculated in adipose, brain, liver and muscle tissues separately.
Multiple comparisons
To determine false discovery (e.g. due to multiple testing), we permuted mouse and genotype relationships and repeated ANOVA, t-test, binomial test and gamma test panels 1000 times. We estimated the rate of false discovery by comparing the P-values obtained from the unpermuted (‘true’) data to their relative rank in the distribution of P-values from the permuted datasets. If the P-value was smaller than all permuted P-values, we assigned a false discovery of <0.1% due to the limited resolution inherent in using 1000 permutations. To assign a false discovery rate for the gene–trait correlations, we used the modified Benjamini and Hochberg FDR approach described by Storey (18) and selected a 1% FDR cutoff based on the correlation P-value distributions in each tissue.
All statistical analyses were performed using MATLAB.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the USPHS National Research Service Award GM07104; National Institutes of Health training grant 5T32HD07228 and program project grant NIH/NHLBI HL28481 and HL30568.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eric Schadt for his help in eQTL mapping, Rosetta Inpharmatics for funding of expression microarrays and Hannah Qi, Xuping Wang and Judy Wu for their help in tissue collection and trait measurements.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 2.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babcock M., Yatsenko S., Hopkins J., Brenton M., Cao Q., de Jong P., Stankiewicz P., Lupski J.R., Sikela J.M., Morrow B.E. Hominoid lineage specific amplification of low-copy repeats on 22q11.2 (LCR22s) associated with velo-cardio-facial/digeorge syndrome. Hum. Mol. Genet. 2007;16:2560–2571. doi: 10.1093/hmg/ddm197. [DOI] [PubMed] [Google Scholar]
- 4.Lee A.S., Gutierrez-Arcelus M., Perry G.H., Vallender E.J., Johnson W.E., Miller G.M., Korbel J.O., Lee C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum. Mol. Genet. 2008;17:1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 5.Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guryev V., Saar K., Adamovic T., Verheul M., van Heesch S.A., Cook S., Pravenec M., Aitman T., Jacob H., Shull J.D., et al. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 7.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaschl H., Aitman T.J., Vyse T.J. Copy number variation in the human genome and its implication in autoimmunity. Clin. Exp. Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milanese M., Segat L., Arraes L.C., Garzino-Demo A., Crovella S. Copy number variation of defensin genes and HIV infection in Brazilian children. J. Acquir. Immune Defic. Syndr. 2009;50:331–333. doi: 10.1097/QAI.0b013e3181945f39. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima T., Ohtani H., Naruse T., Shibata H., Mimaya J.I., Terunuma H., Kimura A. Copy number variations of CCL3L1 and long-term prognosis of HIV-1 infection in asymptomatic HIV-infected Japanese with hemophilia. Immunogenetics. 2007;59:793–798. doi: 10.1007/s00251-007-0252-4. [DOI] [PubMed] [Google Scholar]
- 11.Szatmari P., Paterson A.D., Zwaigenbaum L., Roberts W., Brian J., Liu X.Q., Vincent J.B., Skaug J.L., Thompson A.P., Senman L., et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.She X., Cheng Z., Zollner S., Church D.M., Eichler E.E. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan C.M., Sridhar S., Wigler M., Hall I.M. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 2007;39:1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- 14.Brem R.B., Yvert G., Clinton R., Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 15.Meng H., Vera I., Che N., Wang X., Wang S.S., Ingram-Drake L., Schadt E.E., Drake T.A., Lusis A.J. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl Acad. Sci. USA. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 17.Wang S.S., Schadt E.E., Wang H., Wang X., Ingram-Drake L., Shi W., Drake T.A., Lusis A.J. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ. Res. 2007;101:e11–e30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- 18.Storey J.D. A direct approach to false discovery rates. Journal of the Royal Statistical Society. Series B. 2002;64:479–498. [Google Scholar]
- 19.Aten J.E., Fuller T.F., Lusis A.J., Horvath S. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst. Biol. 2008;2:34. doi: 10.1186/1752-0509-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farber C.R., van Nas A., Ghazalpour A., Aten J.E., Doss S., Sos B., Schadt E.E., Ingram-Drake L., Davis R.C., Horvath S., et al. An integrative genetics approach to identify candidate genes regulating bone density: combining linkage, gene expression and association. J. Bone Miner. Res. 2009;24:104–116. doi: 10.1359/JBMR.080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahituv N., Prabhakar S., Poulin F., Rubin E.M., Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum. Mol. Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- 22.Vilarinho S., Ogasawara K., Nishimura S., Lanier L.L., Baron J.L. Blockade of NKG2D on NKT cells prevents hepatitis and the acute immune response to hepatitis B virus. Proc. Natl Acad. Sci. USA. 2007;104:18187–18192. doi: 10.1073/pnas.0708968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junaid M.A., Kowal D., Barua M., Pullarkat P.S., Sklower Brooks S., Pullarkat R.K. Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor. Am. J. Med. Genet. A. 2004;131:11–17. doi: 10.1002/ajmg.a.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hovatta I., Tennant R.S., Helton R., Marr R.A., Singer O., Redwine J.M., Ellison J.A., Schadt E.E., Verma I.M., Lockhart D.J., et al. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- 26.Coffey P.J., Gias C., McDermott C.J., Lundh P., Pickering M.C., Sethi C., Bird A., Fitzke F.W., Maass A., Chen L.L., et al. Complement factor H deficiency in aged mice causes retinal abnormalities and visual dysfunction. Proc. Natl Acad. Sci. USA. 2007;104:16651–16656. doi: 10.1073/pnas.0705079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F., Wollmer M.A., Hoerndli F., Munch G., Kuhla B., Rogaev E.I., Tsolaki M., Papassotiropoulos A., Gotz J. Role for glyoxalase I in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2004;101:7687–7692. doi: 10.1073/pnas.0402338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoelter S.M., Dalke C., Kallnik M., Becker L., Horsch M., Schrewe A., Favor J., Klopstock T., Beckers J., Ivandic B., et al. ‘Sighted C3H’ mice—a tool for analysing the influence of vision on mouse behaviour? Front. Biosci. 2008;13:5810–5823. doi: 10.2741/3118. [DOI] [PubMed] [Google Scholar]
- 29.Scalzo A.A., Fitzgerald N.A., Simmons A., La Vista A.B., Shellam G.R. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J. Exp. Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki A., Yamada R., Kochi Y., Sawada T., Okada Y., Matsuda K., Kamatani Y., Mori M., Shimane K., Hirabayashi Y., et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat. Genet. 2008;40:1224–1229. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 31.Berglin E., Lorentzon R., Nordmark L., Nilsson-Sojka B., Rantapaa Dahlqvist S. Predictors of radiological progression and changes in hand bone density in early rheumatoid arthritis. Rheumatology (Oxford) 2003;42:268–275. doi: 10.1093/rheumatology/keg077. [DOI] [PubMed] [Google Scholar]
- 32.Christian S.L., Brune C.W., Sudi J., Kumar R.A., Liu S., Karamohamed S., Badner J.A., Matsui S., Conroy J., McQuaid D., et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol. Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 34.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A., et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 35.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 36.Grubb S.C., Maddatu T.P., Bult C.J., Bogue M.A. Mouse phenome database. Nucleic Acids Res. 2008;37:D720–D730. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Williams R.W., Manly K.F. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 38.Doss S., Schadt E.E., Drake T.A., Lusis A.J. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abramowitz M., Stegun I.A. Handbook of Mathematical Functions with Formulas, Graphs, and Mathematical Tables. Washington: US Government Printing Office; 1965. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.