Abstract
Tcap/telethonin encodes a Z-disc protein that plays important roles in sarcomere assembly, sarcomere-membrane interaction and stretch sensing. It remains unclear why mutations in Tcap lead to limb-girdle muscular dystrophy 2G (LGMD2G) in human patients. Here, we cloned tcap in zebrafish and conducted genetic studies. We show that tcap is functionally conserved, as the Tcap protein appears in the sarcomeric Z-disc and reduction of Tcap resulted in muscular dystrophy-like phenotypes including deformed muscle structure and impaired swimming ability. However, the observations that Tcap integrates into the sarcomere at a stage after the Z-disc becomes periodic, and that the sarcomere remains intact in tcap morphants, suggest that defective sarcomere assembly does not contribute to this particular type of muscular dystrophy. Instead, a defective interaction between the sarcomere and plasma membrane was detected, which was further underscored by the disrupted development of the T-tubule system. Pertinent to a potential function in stretch sensor signaling, zebrafish tcap exhibits a variable expression pattern during somitogenesis. The variable expression is inducible by stretch force, and the expression level of Tcap is negatively regulated by integrin-link kinase (ILK), a protein kinase that is involved in stretch sensing signaling. Together, our genetic studies of tcap in zebrafish suggested that pathogenesis in LGMD2G is due to a disruption of sarcomere–T-tubular interaction, but not of sarcomere assembly per se. In addition, our data prompted a novel hypothesis that predicts that the transcription level of Tcap can be regulated by the stretch force to ensure proper sarcomere–membrane interaction in striated muscles.
INTRODUCTION
Muscular dystrophies are inherited diseases of skeletal muscles characterized by loss of muscle strength and integrity. These are a heterogeneous group of genetic disorders; to date, 31 distinct muscular dystrophies have been described and 25 causative genes have been identified (1). These genes encode proteins located in different cellular compartments and execute distinct functions (2). Dystrophin (3) and the dystrophin-associated protein complex (DAPC) (4), including dystroglycans and sarcoglycans, which locate on the sarcolemma, are critical for the structural integrity of membranes (5). Dysferlin, which is also on the sarcolemma, is responsible for membrane repair (6). POMT1 in the sarcoplasmic reticulum (7) and fukutin in the Golgi apparatus (8) may mediate post-translational modifications. Laminin in the extracellular matrix, lamin A and C in the nucleus, and actin, myosin, titin and Tcap in the sarcomere are all structural proteins, but the pathogenesis may actually involve signaling (2).
Tcap (titin-cap), also known as telethonin, is the causative gene for autosomal recessive limb-girdle muscular dystrophy type 2G (LGMD2G) (9,10), a group of muscular dystrophies characterized by predominant weakness and wasting of muscles of the pelvic and shoulder girdle (11). Tcap encodes a 19 kDa sarcomeric protein that is located in the periphery of Z-discs, a sarcomeric structure that defines the border of the sarcomere and serves as both a structural anchor and a signaling center (12–14). Structural studies suggested that Tcap glues two parallel titin molecules within the same sarcomere by directly binding to titin's N-terminal Z1Z2 domains in a palindromic arrangement, which dramatically increases the mechanical resistance ability of titin (15). Tcap has been previously proposed as a downstream target of titin kinase and regulates sarcomere assembly (16). However, this statement was not supported by later genetic studies of titin C-terminal truncation mutants in mice (17). In the heart, Tcap has been suggested to interact with titin and muscle LIM protein (MLP) and form a stretch sensor complex in the Z-disc (18). It has also been found that Tcap interacts with MinK, the regulatory β-subunit of the delayed rectifier potassium current channel that is located on T-tubular membranes surrounding the Z-discs, suggesting a function of Tcap in sarcomere–membrane interactions and the mechano-electrical feedback system (19). In addition to structural functions, the signaling functions of Tcap have been recently suggested due to the discovery of its physical interactions with myostatin (20), calsarcin (21), Ankrd2 (22), PKD (23) and BMP10 (24). In contrast to extensive structural and biochemical studies, genetic studies of tcap have not been conducted in any animal model, which severely hindered our understanding of its in vivo function in muscular dystrophy.
The mouse has been the predominant animal model for understanding the pathological mechanism involved in muscular dystrophy. For example, the mdx mouse, a dystrophin-null model, has been investigated to study Duchenne Muscular Dystrophy (DMD) (25). The zebrafish has recently emerged as a promising model organism for the study of muscular dystrophies because of its unique embryology and powerful genetic tools (26,27). For example, studies of the zebrafish sapje mutant, which bears a nonsense mutation in dystrophin, suggested that the progressive muscle degeneration phenotype is caused by muscle attachment failure (28). Studies of the candyfloss mutant implicate an extracellular matrix adhesion failure in laminin α2-deficient congenital muscular dystrophy type 1A (MDC1A) (29).
In the present study, we aim to understand the pathogenesis of LGMD2G by exploiting zebrafish genetics. We cloned tcap, a zebrafish Tcap homologue, which exhibits an interesting variable expression pattern in developing skeletal muscles. Depletion of tcap by an antisense morpholino resulted in muscular dystrophy-like phenotypes. Detailed analysis uncovered a disrupted sarcomere–membrane interaction but normal sarcomere assembly. Further studies suggested that the transcription of Tcap is regulated by stretch force and integrin-linked kinase (ILK), a protein kinase involved in stretch sensing signaling. These results provide novel mechanistic insights on how disruption of tcap may lead to LGMD2G.
RESULTS
tcap exhibits a variable expression pattern in developing somites
In GenBank, a zebrafish tcap gene has been reported on chromosome 3 (GenBank access number XM_679011). The 1.2 kb transcript derived from its two exons encodes 179 amino acids. The identity/similarity of the zebrafish Tcap protein to its orthologues in fugu, mouse and human are 65/76%, 39/57% and 38/54%, respectively (Supplementary Material, Fig. S1). Zebrafish Tcap is expressed in somites from the 21-somite stage as revealed by whole mount in situ hybridization (ISH) and real-time reverse transcriptase-polymerase chain reaction (RT–PCR) (Supplementary Material, Table S1). The expression gradually increases throughout the development and peaks at 48 h post-fertilization (hpf), before dropping at 72 hpf. After 96 hpf, the expression of Tcap continues to increase and reach much higher levels in the adult (Supplementary Material, Table S1). In contrast to strong expression pattern in both the heart and somites in adults, strong expression of Tcap in somites can be detected during embryogenesis by using whole mount ISH, while only low-level expression can be detected in the heart. This finding was supported by a stable transgenic line encompassing a 2.5 kb upstream sequence of tcap. Strong GFP reporter expression driven by this promoter can be detected in somites during embryogenesis but not in the heart (Supplementary Material, Fig. S2).
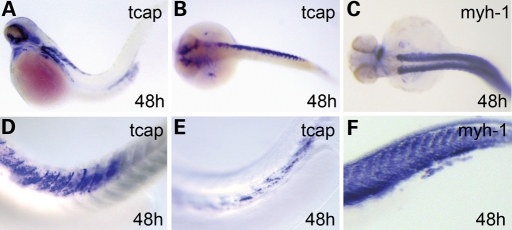
Interestingly, we observed a unique expression pattern of Tcap at 48 hpf that is different in each individual embryos. Unlike most sarcomeric genes, such as myosin heavy chain 1 (myh1) that is ubiquitously expressed in all myoblast cells (Fig. 1C and F), tcap mRNA was detected in a subpopulation of myoblast cells. In some embryos, tcap transcripts were detected anteriorly only (Fig. 1A), posteriorly only (not shown), dorsally only (Fig. 1D) or ventrally only (Fig. 1E). In other embryos, tcap transcripts were restricted to one side of the embryo instead of the typical bilateral expression pattern (Fig. 1B). This non-uniform expression pattern was confirmed by cross-section after ISH staining (not shown). The variable expression of Tcap correlates with the onset of body twitch, suggesting that the transcription of tcap might respond to stretch force, a hypothesis that will be tested later.
Figure 1.
tcap expression pattern revealed by whole mount in situ hybridization. (A–F) Tcap had a variable expression pattern in the somite at 48 hpf, different from the bilateral and ubiquitous expression of sarcomeric gene like myh1 (C and F). Tcap transcripts can be enriched in the anterior (A), posterior (not shown), dorsal (D) or ventral (E) part of the body, and even the unilateral (B). (A and D–F) Lateral view, anterior to the left. (B and C) Dorsal view, anterior to the left.
Tcap integrates into the Z-disc after α-actinin becomes periodic
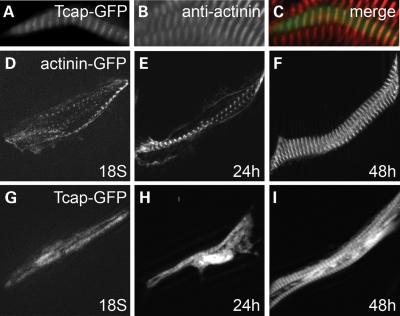
To confirm that zebrafish tcap is functionally conserved with its mammalian orthologues, we generated a construct encoding a Tcap-GFP fusion protein to examine its sub-cellular localization. Indeed, a striated GFP expression pattern can be detected in muscle cells at 48 hpf (Fig. 2A), overlapping with anti-actinin antibody staining that labels sarcomeric Z-discs (Fig. 2B and C). To compare the timing of Z-disc assembly, we also generated another construct encoding an α-actinin-GFP fusion protein. Injection of this construct resulted in mosaic expression in individual myoblast cells, which can be used to reveal the process of Z-disc assembly in a single myoblast. We detected several myofibrils with striated α-actinin pattern at 18S, which later fuse laterally to form one to two major myofibrils in each cell at 24 hpf. The Z-disc undergo lateral growth, as reflected by the broader Z-discs at 48 hpf (Fig. 2D–F). In contrast to α-actinin, Tcap-GFP appears to express throughout the cytoplasm at 18S (Fig. 2G). The striated pattern cannot be detected until 24 hpf. This late incorporation of Tcap protein into the Z-disc suggests that Tcap is not involved in the early stages of sarcomere assembly.
Figure 2.
Tcap is a Z-disc protein but assembles after α-actinin becomes periodic. (A–C) Tcap-GFP fusion protein showed a striated pattern (A, green) and co-localized with the Z-disc protein α-actinin (B, anti-actinin, red; C, overlap). (D–I) α-actinin2-GFP revealed the processes of sarcomere assembly (D, 18 somite; E, 24 hpf) including lateral alignment (F, 48 hpf). Tcap-GFP still disperses in the cytoplasm when α-actinin2 already restricts to periodic Z-lines (G). Shown in (D–F) are different cells from (G–I). Tcap-GFP was restricted to mature Z-disc after 24 hpf (H and I).
Reduction of Tcap leads to muscular dystrophy-like phenotypes without affecting sarcomere assembly
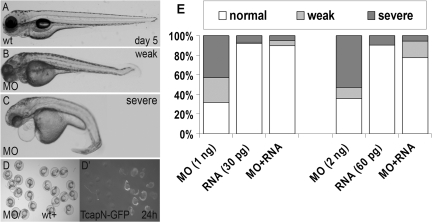
To study the functions of Tcap, we knocked down its expression by injecting MO-Tcap, an ATG morpholino targeting the translation start site of tcap. Injection of 1 ng MO-Tcap resulted in 25.6% embryos (n = 113) exhibiting weak phenotypes, including reduced body length, slight body curvature, reduced touch sensitivity and abnormal swimming behavior (Fig. 3B and Supplementary Material, Movie S1) and 42.5% embryos exhibiting severe phenotypes, including more reduced muscle mass, more severe body curvature, much reduced touch sensitivity or the complete loss of swimming ability (Fig. 3C). Injection of 2 ng MO-Tcap resulted in even more severe phenotypes with 11.3% embryos (n = 97) exhibiting weak phenotypes and 52.6% embryos exhibiting severe phenotypes (Fig. 3E). The effectiveness of the MO-Tcap morpholino is demonstrated by the much reduced GFP fluorescence after co-injection of this morpholino with mRNA encoding a Tcap-GFP fusion protein (Fig. 3D). The specificity of this ATG morpholino was confirmed by the rescue experiment (Fig. 3E and Supplementary Material, Table S2). Co-injection of 30 pg of tcap mRNA with 1 ng MO reduced the percentage of embryos with severe phenotypes from 42.5 to 5%, while co-injection of 60 pg mRNA with 2 ng MO reduced the percentage of embryos with severe phenotypes from 52.6 to 5.7%.
Figure 3.
Tcap MO result in specific phenotypes. (A–C) Representative pictures of day 5 wild-type fish (A), weak and severe phenotypes of tcap morphants. A weak phenotype included shorter body length and a slight body curvature (B), less sensitive to touch and abnormal swimming pattern. A severe phenotype included severe body curvature (C), muscular disarray, much slower response to touch, and even lost of swimming ability. Both groups showed eye and jaw hypoplasia, and pericardiac edema can be occasionally detected in severe group. (D) Co-injection of tcap ATG morpholino can abolish the expression of N-terminal-Tcap-GFP chimeric mRNA, indicating a high knockdown efficacy. (D), bright field. (D′) Green fluorescence. The GFP fluorescence level in the fish group injected with both MO and RNA (left in D and D′) is much weaker than the fish group injected with RNA only (right in D and D′). (E) Phenotypes in tcap morphants can be rescued by co-injection of tcap RNA. The graph shows a dosage-dependent effect of MO-Tcap, and co-injection of tcap RNA can reduce the morphant percentage at both concentrations of MO-Tcap.
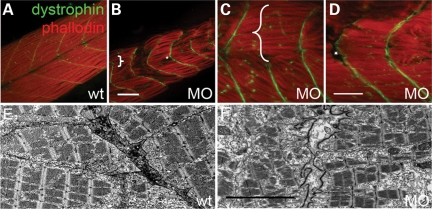
Detailed analysis of the morphant phenotypes further confirmed defective muscle development. The integrity of myoseptum is affected, as revealed by immunostaining using an anti-dystrophin antibody. In contrast to a V-shaped myoseptum in wild-type fish (Fig. 4A), a U-shaped myoseptum is detected in tcap morphants. The angle of the myoseptum increases from 95° in wild-type fish to 132° in morphants. Degenerating myoblasts can be detected in some somite segments, as revealed by phalloidin staining. In certain regions, the myosepta are discontinued and the myofibrils extend beyond the somite boundary, spanning the length of two somites (bracket in Fig. 4B and C). In other regions, myofibrils from neighboring somite segments are detached from the myoseptum and separated from each other (asterisk in Fig. 4B and D). This defective cell–cell interaction is supported by images from electron microscopy (EM) (Fig. 4E and F), which further suggested an abnormal extracellular matrix structure. The plasma membrane appears intact, as suggested by the existence of dystrophin staining at both ends of the gap. Indeed, injection of Evans blue dye into the circulation did not result in the leakage of fluorescence in myocytes (data not shown) (28).
Figure 4.
Tcap knockdown leads to muscular dystrophy-like phenotypes. (A–D) Representative pictures of anti-dystrophin antibody stains myoseptum in green and phalloidin stains F-actin in red. Unlike wild-type fish which have a V-shaped myoseptum and well-organized myofibers (A), tcap morphants usually exhibit a U-shaped myoseptum, which is discontinuous or missed in some regions (brace in B and C) and the myofibers grow through adjacent somites. Disrupted myofiber organization with uneven lengths as well as detachment from the myoseptum (asterisk in B and D) are often observed. Scale bar, 50 µm. (E and F) Representative electron microscopy pictures showing a disrupted extracellular matrix structure in a tcap morphant (F) in comparison to wild-type fish (E). Scale bar, 5 µm.
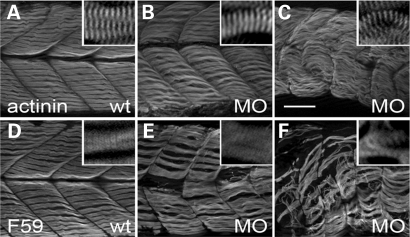
Surprisingly, our EM studies suggested that sarcomere assembly is not affected in the tcap morphants. Indeed, well-assembled sarcomeres can be detected even in the most severe morphant, as revealed by immunostaining using either the anti-α-actinin antibody that labels Z-discs or F59 antibody that labels thick filaments (Fig. 5). Instead, what has been affected is the organization of the myofibrils, which become wavy and twisted, especially in the tail region. Within each somite segment, the length of each myofibril became uneven and shorter, and the gaps between the neighboring myofibrils increased (Fig. 5). The well-developed sarcomere structure also suggested that the differentiation of the myoblast is not affected in tcap morphants. This statement was supported by the undisturbed myoD staining pattern during embryogenesis, as revealed by ISH (Supplementary Material, Fig. S3).
Figure 5.
Tcap knockdown did not affect sarcomere assembly. (A–C) Anti-α-actinin staining of wild-type fish (A) and tcap morphants with either a weak (B) or severe (C) phenotype. Although the myofibrils in tcap morphants appeared wavy, disoriented and uneven in length, they still had a well-organized striated pattern (inset), indicating undisrupted Z-disc assembly. (D–F) F59 staining of wild-type fish (D) and tcap morphants with either a weak (E) or severe (F) phenotype, which suggested that thick filament assembly was not disrupted either. Scale bar, 50 µm.
Tcap knockdown affects T-tubule development
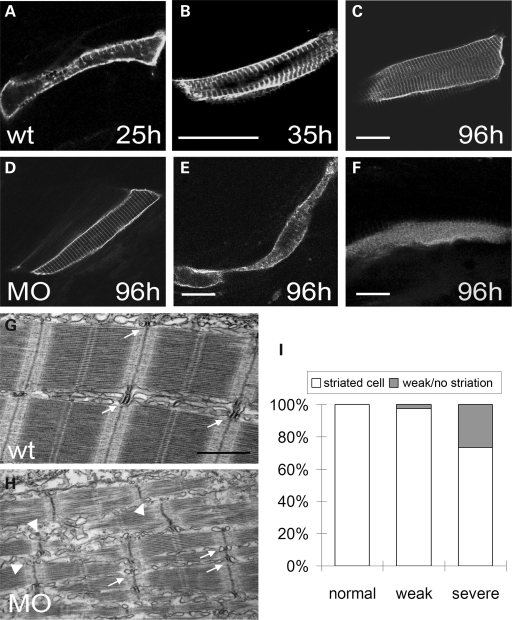
Tcap has previously been shown to interact with MinK on T-tubules in the rat ventricle, which prompted us to examine the development of the T-tubule system in zebrafish tcap morphants. For this purpose, we generated a membrane-tagged GFP construct (30) driven by a β-actin enhancer (31). Injection of this construct resulted in mosaic green fluorescence in the sarcolemma of individual myoblasts, which later invaginated to interact with Z-discs and form the T-tubule system. The invagination initiated at around 25 hpf (Fig. 6A), continued at 35 hpf and beyond (Fig. 6B) and finally formed the highly organized striated pattern at 96 hpf (Fig. 6C). This T-tubule development process is defective in tcap morphants. The striated T-tubule pattern was either weaker (Fig. 6E) or disappeared (Fig. 6F) in 26% of myoblasts in tcap morphants with severe-phenotype class (Fig. 6I and Supplementary Material, Table S3). The disruption of a proper interaction between the T-tubule system and the sarcomeric Z-disc was also confirmed by EM studies. In contrast to a perfect registration between the T-tubule system and the edge of each Z-disc in wild-type embryos (arrows in Fig. 6G and H), the T-tubule become detached from the edge of Z-discs in tcap morphants. The alignment of the Z-disc between adjacent myofibrils also appears irregular in some regions, especially near the tips of myofibrils (arrowhead in Fig. 6H).
Figure 6.
Tcap knockdown affects T-tubule development. (A–C) Time course of T-tubule development in wild-type fish. Membrane-targeted GFP under the control of β-actin enhancer was injected to fluorescently label the sarcolemma that invaginated and formed the striated T-tubule system. The invagination initiated at around 25 hpf sporadically (A), continued through all direction (B) and finally formed an organized striated pattern at 96 hpf (C). Scale bar, 20 µm. (D–F) Representative pictures of a cell with normal striations (D), and cells with either weak striation (E) or no striation (F) in tcap morphants that have been co-injected with a membrane-targeted GFP construct. Scale bar, 20 µm. (G–H) Representative pictures of electron microscopy showing T-tubule and Z-disc alignment in wild-type fish (G) or tcap morphants (H). Unlike wild-type fish with well-aligned T-tubules and Z-discs (arrow), the T-tubules in some regions of morphants were misaligned or missing (arrowhead). Scale bar, 1 µm. (I) Graphic summary of the percentage of striated cell, cell with weak striation or no striation in different classes of tcap morphants.
The transcription of tcap can be regulated by stretch force and ILK
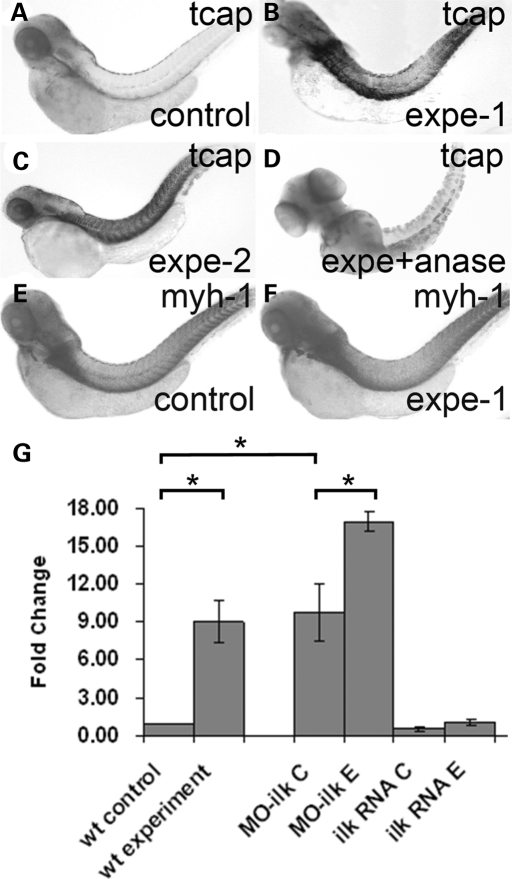
The timing of the development of the T-tubule system correlates with a twitching movement of the embryos, which prompted us to propose a hypothesis predicting that the body movement facilitates the invagination of the sarcolemma to form a T-tubule system that registers with Z-discs. Consistent with this hypothesis, we found that inhibition of body movement by anesthesia delayed the development of the T-tubule system (Supplementary Material, Table S4). Our observation that tcap exhibits a variable expression pattern at 48 hpf and that the T-tubule system is disrupted in tcap morphants further prompted us to propose that tcap regulates T-tubule development via responding to the stretching force during the twitch movement. To test this hypothesis, we increased the stretching force by hardening the chorion surrounding the embryos. This design was based on the fact that, in wild-type fish, larva break out of the chorion on day 2 and thus relieve themselves from the stretching force generated from twitching within a limited space. Treatment of the embryos with bleach hardens the chorion and prolongs the stretch force. We found that if the hardened chorion is removed manually after day 3, the larva bends to one side and lays down, but will resume its swimming ability later. The transcriptional expression of tcap in these bleach-treated embryos is significantly increased, as shown by whole mount ISH (Fig. 7A and B), and confirmed by RT–PCR (Fig. 7G). The induction of tcap transcription depends upon body movement, as evidenced by the observation that the upregulation of tcap is abolished by preventing body movement using anesthesia (Fig. 7D). To exclude the possibility that the induction of tcap transcription is due to side effects associated with chemical treatment, we physically restricted the movement space of the larva by embedding the embryos within their chorion in 1.5% agarose gel. A similar increase of tcap expression levels was observed (Fig. 7C). In contrast to tcap, the expression of other sarcomeric genes, including myosin heavy chain 1, titin N2B isoform and MLP, was not affected by these treatments (Fig. 7E and F and data not shown). In summary, these data suggested that tcap is a unique sarcomeric gene whose transcription can be regulated by stretch-related force.
Figure 7.
tcap expression level can be altered by stretch movement and modulated by ILK. (A–D) tcap expression levels can be altered by two treatments mimicking increased stretch force. Shown are whole mount in situ hybridization at 4 dpf. Under two different experimental treatments that restrict the surrounding space of fish embryos in order to increase stretch force (for detail refer to methods), the tcap expression level was increased on day 4 (B and C) compared with that in control fish (A). Co-treatment with anesthesia ablated the upregulation of Tcap expression (D). This upregulation was not observed in other sarcomeric genes such as myh-1 (E and F). (G) ILK negatively regulated Tcap level. Shown is a graphic summary of the result of quantitative RT–PCR. The expression of Tcap in treated embryos is significantly increased compared with that in wild-type fish (P = 0.02). Reduction of ilk by injection of morpholino significantly increased Tcap expression (P = 0.03) while overexpression of ilk by injection of mRNA decreased Tcap expression (P = 0.09). The inducibility of Tcap expression is still maintained in either manipulation of ILK. *P < 0.05.
Tcap has been shown to interact with titin and MLP and form a Z-disc stretch sensor complex (18). Recently, MLP in this complex has been found to interact physically with ILK, a protein kinase that interacts with the integrin complex on the sarcolemma and transmits stretch sensing signaling (32,33). This report suggested a cross-talk between the two stretch sensor complexes located in the membrane and sarcomeric Z-disc, respectively. To further test this hypothesis, we genetically manipulated ILK during zebrafish embryogenesis. Reduction of ilk expression by injection of a morpholino significantly increased tcap expression by 9.7-fold (P = 0.03), while overexpression of ilk by injection of an mRNA did not significantly affect, despite marginally reduced, tcap expression (P = 0.09) (Fig. 7G). The inducibility of tcap expression by bleach treatment remains unchanged in either gain- or loss-of-function of ILK. In summary, these data supported the statement that the two stretch sensor complexes cross-talk and suggested that the transcript level of tcap is negatively controlled by ILK.
DISCUSSION
Functions of Tcap in sarcomere assembly
In this study, we cloned zebrafish tcap and conducted the first loss-of-function analysis of Tcap in a vertebrate animal model to understand its functions in normal muscle development and in disease progression. Zebrafish tcap is expressed in striated muscles and exhibits conserved subcellular expression in Z-discs. Compared with α-actinin, Tcap appears in the Z-disc at a later stage after the sarcomeres become striated. This finding is consistent with a previous report that Tcap-GFP fusion protein can only be seen in mature Z-lines but not in pre-myofibril Z-bodies in developing skeletal muscle cells (34). The late appearance of Tcap in the Z-disc suggests that this gene is not required for sarcomere assembly, a statement that was confirmed by our observation of myofibrils with normal sarcomere structures even in tcap morphants with the most severe phenotypes. This statement may appear at odds with the conclusion based on the overexpression data, which demonstrated that forced expression of either Tcap or the titin Z1Z2 domain disrupted sarcomere assembly in cultured myocytes (13). However, our statement is consistent with a report on a human LGMD2G patient, who has a homozygous mutation in TCAP that led to a truncated protein but exhibited a normal sarcomere structure (9). Therefore, our current data strongly suggest that disrupted sarcomere structure is not the major pathogenesis event in Tcap-mediated muscular dystrophy type 2G. It is noteworthy to point out that our conclusion regarding functions of Tcap in sarcomere assembly needs to be taken with caution and shall be further confirmed by generating stable mutants in both fish and other model organisms. First, the morpholino approach used here might not be able to completely eliminate tcap expression. Second, it remains unclear whether there is another zebrafish Tcap homologue that compensates for loss of function of the present tcap gene. We have searched the zebrafish genome database and identified another potential zebrafish tcap homologue with low homology. However, preliminary studies revealed a strong expression of this gene in the heart but not somites (data not shown). Thus, it is likely that this potential homologue will not impact our conclusions here on functions of tcap in somites.
Functions of Tcap in T-tubule development
Even without an obvious sarcomere defect, Zebrafish morphants with reduced expression of tcap did exhibit muscular dystrophy-like phenotypes, including reduced muscle mass (Figs 3C and 4B) and impaired swimming ability (see Supplementary Material, movie). Severe sarcomere–membrane interaction defects were apparent, including the change in the myoseptum from being V-shaped to U-shaped, a discontinued myoseptum and detachment of the neighboring myofibrils in the same somite segment. The myofiber detachment phenotype in tcap morphants has also been reported in other zebrafish models of muscular dystrophy, such as the dystrophin mutant and laminin mutant (28,29). In tcap morphants, the sarcolemma appears intact without apparent leakage, which is more similar to the laminin mutant than the dystrophin mutant. The phenotypes of defected myosepta and elongated myofibrils extending beyond somite boundary resemble those previously noted in zebrafish embryos depleted of obscurin, a giant sarcomere-associated protein interacting with titin and small ankyrin-1, and linking sarcomere and sarcoplasmic reticulum (35). Depletion of obscurin also caused disruption of organized arrangement of T-tubules and junctional sarcoplasmic reticulum around Z-discs. Our detailed analysis in zebrafish further suggested that a disrupted sarcomere–membrane interaction, such as a defective T-tubule system, is an important pathogenesis event in Tcap-based LGMD 2G. T-tubules are critical for regulating muscle contractions and force generation, especially for excitation–contraction coupling (36). Disruption of the T-tubule system has been shown to lead to diminished force production and muscle weakness. For example, a recent study reported that the loss of myotubularin function resulted in T-tubule disorganization in zebrafish and human myotubular myopathy; the morphant fish shared a similar muscle weakness phenotype with tcap morphants (37). BIN1/amphiphysin2 and caveolin-3 are both required for T-tubule formation and maintenance, and their mutations have been also associated with muscular dystrophy (38–41). It is noteworthy to point out that, in mammals, T-tubules are primarily located over the A–I junction, at some distance from the Z-disc. Thus, it is a possibility that disruption of T-tubule is a secondary defect that is more directly related to changes in cytoskeletal architecture in LGMD2G patients.
The correlation between the timing of the expression of Tcap, the development of the T-tubule system and the development of body twitching movements prompted us to propose a hypothesis that the stretching force during the early body movement might play an important role in inducing Tcap expression, which regulates the invagination of the sarcolemma to develop the T-tubule system. This model is supported by our genetic data, which show that enhanced stretching force induced tcap gene transcription, and that blocking either body movement or tcap expression disrupted T-tubule development. As suggested by a previous study in rat ventricles, it is possible that Tcap, which is located in Z-discs, exerts this function by physically interacting with a T-tubule protein such as MinK (19). The upregulation of tcap transcription and, subsequently, the accumulation of Tcap protein facilitate the interaction between the sarcomeric Z-disc and T-tubule. The Tcap interaction partner in skeletal muscles that may mediate its function in T-tubule development is yet to be identified.
Functions of Tcap in stretch sensing signaling
The Z-disc used to be considered as a structural component of the sarcomere that is important only for the cross-linking of thin filaments and transmission of force generated by the myofilaments. Recently, many new proteins have been found to be located within the Z-disc and are linked to cardiomyopathy and/or muscular dystrophy (14). Various signaling molecules interact with these Z-disc proteins, several of which shuttle between the Z-disc and nucleus. Therefore, the Z-disc is now regarded as an important signaling center for the reception, transduction and transmission of mechanical and biochemical signals (42,43). Studies of the MLP−/− mouse suggested that Tcap, titin and MLP form such a ‘stretch sensor’ complex in the Z-disc region (18). Sarcomeres of MLP−/− mice exhibit wider Z-discs, misalignment of Z-disc components and altered Tcap localization from myofilaments to the cytosolic compartment. Here, we reported that zebrafish Tcap exhibits a variable expression pattern during somite development, which is inducible by stretch force. Consistent with this discovery, it has been previously reported that the transcription of Tcap is under developmental and functional regulation. Down-regulation of tcap has been reported in denervated myoblasts (44) or in neurogenic atrophy (45). It is of great interest to further investigate whether tcap transcription is regulated in adult hearts/somites by mechanical stress, and how this change of tcap transcription contributes to the pathogenesis of either cardiomyopathy or muscular dystrophy.
Currently, there are two major paradigms for the molecular identities of mechanosensors in striated muscles (46). The centralized models propose that a stretch signal is generated in close proximity to the membrane, while the decentralized models propose that mechanical stress applied at the cell surface is transmitted throughout the cell via the cytoskeleton. While integrins and related ILK signaling are major components of the former models (33), MLP, Tcap and titin are key components of the latter models. The physical interaction between ILK and MLP (32), and the genetic relationship between ILK and Tcap, as shown here, strongly support the statement that the two models are not mutually exclusive. Our observation that Tcap is required for the development of T-tubule system during early embryogenesis underscored an important function of Tcap in the communication between the Z-disc and sarcolemma. Therefore, our data suggested that the elaborate regulation of Tcap at the transcription level is an important signaling event during mechanotransduction. Further investigations are warranted to investigate the relationship among Tcap, ILK and MLP during mechanotransduction signaling.
MATERIALS AND METHODS
Zebrafish husbandry
Wild-type zebrafish (Danio rerio) were raised in the laboratory and handled according to standard procedures outlined in a protocol approved by the Mayo Institutional Animal Care and Use Committee.
Bioinformatics
ClastW alignments and phylogenetic tree analyses of zebrafish Tcap (GenBank access number XM_679011), fugu Tcap (modified from ENSTRUP00000041638 by including 39 bp upstream encoding sequences to the first exon ENSTRUE00000280130), human Tcap (GenBank access number NM_003673) and mouse Tcap (GenBank access number NM_011540) were done using DSGene software (Accelrys Inc.).
Molecular cloning
Full-length tcap (forward primer: 5′-ATATGCAGGTTTGCTCAGTCCTGG-3′, reverse primer: 5′-TTCCTGCTTTTCAGCCTCTC-3′) was cloned into the pEGFP-C1 vector using XhoI/ EcoRI sites. Full-length α-actinin 2 (GenBank access number NM_001037573, forward primer: 5′-AACCGCGAAAATGATGAATCAGAT-3′, reverse primer: 5′-GAAGGTCACTTTCTCCATATAGGG-3′) was cloned into the pEGFP-N1 vector using XhoI/ EcoRI sites. Membrane-targeted GFP was generated as previously described (30) by cloning the first 20 amino acids of zebrafish GAP43 (GenBank access number NM_131341, 5′-CACGAAACCATGCTGTGC TGCTCAGAAGAACTAAACCGGTTGAGAAGAATGAAGAGTCCGATCAGCTG-3′) into the pEGFP-N1 vector using BamhI/NcoI sites.
Tcap was subcloned into the pCS2+ vector for sense-capped RNA synthesis using BamhI/EcoRI sites. Full-length ilk (GenBank access number NM_200571, forward primer: 5′-GCCGAAATGGATGACATCTTC-3′, reverse primer: 5′-TGGTTATTT GTCTTGCATCTTC-3′) was cloned into the pCS2+ vector using BamhI/EcoRI sites. A chimera construct encoding Tcap-GFP fusion protein was generated by linking the N-terminal of tcap (5′-ATGCAGGTTTGCTCAGTCCTGGAGAA-3′) to the AgeI site of the GFP fragment (digested from pEGFP-C1 vector using AgeI/ XhoI sites) and then subcloned into the pCS2+ vector using EcoRI/XhoI sites.
Morpholino oligonucleotides and sense-capped RNA
The morpholino-modified oligonucleotides were purchased from Gene Tools, LLC (Corvallis, OR, USA). Morpholinos were prepared and injected as previously described (47). The sequence for the tcap MO targeting the ATG was 5′-TCTCCAGGACTGA GCAAACCTGCAT. The sequence for the ilk MO targeting the ATG was 5′-GGCACTGAGTGAAGATGTCATCCAT (33).
Sense-capped RNA was synthesized using the mMESSAGE mMASCHINE system (Ambion) for tcap, tcapN-GFP and ilk.
Whole-mount in situ hybridization
Whole-mount in situ hybridization (ISH) was performed as previously described (48). A tcap riboprobe (forward primer: 5′-ACACTCAACCAGGACAAGAGAC-3′, reverse primer: 5′-CCTAATACGACTCACTATAGGCCCAGCCTTTATGAACATTTCATC-3′), a myh1 (GenBank access number NM_001077464) riboprobe (forward primer: 5′- TTATCCAACTCCAAAAGAAACTC-3′, reverse primer: 5′-CCTAA TACGACTCACTATAGGGCAGTGTTAGATCTCTCCACATC-3′) and a MyoD riboprobe (forward primer: 5′-GAGGACCTGCGAATTTTAGACTGTA-3′, reverse primer: 5′-CCTAATACGACTCACTATAGGCAAACTCGACACCACT GAGTTTTTA-3′) were generated using AmpliScribe T7-flash transcription kit (epicenter biotechnologies).
Immunostaining
Immunostaining was performed as previously described (49). The following antibodies were used at the indicated dilutions: anti-sarcomeric α-actinin (clone EA53; Sigma) at 1:1000, F59 (Developmental Studies Hybridoma Bank, DSHB) at 1:10 and dystrophin (sigma) at 1:100. Phalloidin was used at 1:10 (Invitrogen). Alexa-conjugated secondary antibodies were used (Invitrogen). Following staining, embryos were imaged using Zeiss Axioplan 2 microscope equipped with an ApoTome (Carl Zeiss).
Transmission electron microscopy
Fish embryos fixed in Trump's solution overnight at 4°C were processed by the Mayo Clinic's Electron Microscopy Core Facility and imaged using a Philips CM10 Transmission Electron Microscope.
Real-time PCR
Twenty fish embryos were frozen in liquid nitrogen and homogenized with a pestle. RNA was extracted using RNeasy Mini Kit (Qiagen). One microgram total RNA was then reverse transcribed by SuperScript III First-Strand Synthesis System for RT–PCR (Invitrogen). To analyze the expression level of tcap, real-time PCR was performed in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) using iQ SYBR Green supermix (Bio-Rad). We used 18S as an internal control to normalize tcap levels. The following primers were used: tcap (forward primer: 5′-GGGAGGAAA ACCCAAATAAGAG-3′, reverse primer: 5′-TGGAGAGGACGCACCTGCCA-3′), and 18S (forward primer: 5′-CACTTGTCCCTCTAAGAAGTTGCA-3′, reverse primer: 5′-GGTTGATTCCGA TAACGAACGA-3′).
tcap induction assay
Experiment 1: the embryos with chorion were treated with bleach (sunbrite, 75 µl in 20 ml E3 water) for 90 s on day 1 and were rinsed with E3 water for three times and raised normally or in 1X tricaine solution (western chemical, 16 mg tricaine, 2.1 mL 1 m Tris pH9 and 97.9 ml E3 water) to anesthetize the embryos. Experiment 2: the embryos with chorion were embedded in 1.5% agarose (Fisher Biotech) in 60 mm dish on day 1 and raised normally. The embryos were manually freed from the chorion by forceps at desired stages, usually day 3 or day 4, and used for ISH or RNA extraction followed by real-time PCR.
Transgenic fish line
The tcap upstream 2.5 kb sequence (−2457∼+47, 0 was set as the transcription initiation site, forward primer: 5′-TCATTGGAGAAACAACTGTCAC-3′, reverse primer: 5′-TCTCTTGTCCTGGTTGAGTG-3′) was cloned into a Tol2 vector (50) using XhoI/SmaI sites. The construct was co-injected with tol2 transponase into one-cell stage embryos. Founders were raised and identified according to an established protocol as previously described (50).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (HL81753 to X.X.); and the Muscular Dystrophy Association (MDA#80 to X.X.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Beninio Jomok for taking care of the zebrafish facility.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dalkilic I., Kunkel L.M. Muscular dystrophies: genes to pathogenesis. Curr. Opin. Genet. Dev. 2003;13:231–238. doi: 10.1016/s0959-437x(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 2.Davies K.E., Nowak K.J. Molecular mechanisms of muscular dystrophies: old and new players. Nat. Rev. Mol. Cell. Biol. 2006;7:762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M., Monaco A.P., Kunkel L.M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 5.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl Acad. Sci. USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lennon N.J., Kho A., Bacskai B.J., Perlmutter S.L., Hyman B.T., Brown R.H., Jr Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 2003;278:50466–50473. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 7.van Reeuwijk J., Maugenre S., van den Elzen C., Verrips A., Bertini E., Muntoni F., Merlini L., Scheffer H., Brunner H.G., Guicheney P., et al. The expanding phenotype of POMT1 mutations: from Walker–Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum. Mutat. 2006;27:453–459. doi: 10.1002/humu.20313. [DOI] [PubMed] [Google Scholar]
- 8.Ishii H., Hayashi Y.K., Nonaka I., Arahata K. Electron microscopic examination of basal lamina in Fukuyama congenital muscular dystrophy. Neuromuscul. Disord. 1997;7:191–197. doi: 10.1016/s0960-8966(97)00462-8. [DOI] [PubMed] [Google Scholar]
- 9.Moreira E.S., Wiltshire T.J., Faulkner G., Nilforoushan A., Vainzof M., Suzuki O.T., Valle G., Reeves R., Zatz M., Passos-Bueno M.R., et al. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat. Genet. 2000;24:163–166. doi: 10.1038/72822. [DOI] [PubMed] [Google Scholar]
- 10.Olive M., Shatunov A., Gonzalez L., Carmona O., Moreno D., Quereda L.G., Martinez-Matos J.A., Goldfarb L.G., Ferrer I. Transcription-terminating mutation in telethonin causing autosomal recessive muscular dystrophy type 2G in a European patient. Neuromuscul. Disord. 2008;18:929–933. doi: 10.1016/j.nmd.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guglieri M., Straub V., Bushby K., Lochmuller H. Limb-girdle muscular dystrophies. Curr. Opin. Neurol. 2008;21:576–584. doi: 10.1097/WCO.0b013e32830efdc2. [DOI] [PubMed] [Google Scholar]
- 12.Valle G., Faulkner G., De Antoni A., Pacchioni B., Pallavicini A., Pandolfo D., Tiso N., Toppo S., Trevisan S., Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- 13.Mues A., van der Ven P.F., Young P., Furst D.O., Gautel M. Two immunoglobulin-like domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett. 1998;428:111–114. doi: 10.1016/s0014-5793(98)00501-8. [DOI] [PubMed] [Google Scholar]
- 14.Faulkner G., Lanfranchi G., Valle G. Telethonin and other new proteins of the Z-disc of skeletal muscle. IUBMB Life. 2001;51:275–282. doi: 10.1080/152165401317190761. [DOI] [PubMed] [Google Scholar]
- 15.Zou P., Pinotsis N., Lange S., Song Y.H., Popov A., Mavridis I., Mayans O.M., Gautel M., Wilmanns M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
- 16.Mayans O., van der Ven P.F., Wilm M., Mues A., Young P., Furst D.O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 17.Gotthardt M., Hammer R.E., Hubner N., Monti J., Witt C.C., McNabb M., Richardson J.A., Granzier H., Labeit S., Herz J. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J. Biol. Chem. 2003;278:6059–6065. doi: 10.1074/jbc.M211723200. [DOI] [PubMed] [Google Scholar]
- 18.Knoll R., Hoshijima M., Hoffman H.M., Person V., Lorenzen-Schmidt I., Bang M.L., Hayashi T., Shiga N., Yasukawa H., Schaper W., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa T., Ono Y., Tsuchiya H., Katayama Y., Bang M.L., Labeit D., Labeit S., Inagaki N., Gregorio C.C. Specific interaction of the potassium channel beta-subunit minK with the sarcomeric protein T-cap suggests a T-tubule-myofibril linking system. J. Mol. Biol. 2001;313:775–784. doi: 10.1006/jmbi.2001.5053. [DOI] [PubMed] [Google Scholar]
- 20.Nicholas G., Thomas M., Langley B., Somers W., Patel K., Kemp C.F., Sharma M., Kambadur R. Titin-cap associates with, and regulates secretion of, Myostatin. J. Cell. Physiol. 2002;193:120–131. doi: 10.1002/jcp.10158. [DOI] [PubMed] [Google Scholar]
- 21.Frey N., Olson E.N. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 22.Kojic S., Medeot E., Guccione E., Krmac H., Zara I., Martinelli V., Valle G., Faulkner G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J. Mol. Biol. 2004;339:313–325. doi: 10.1016/j.jmb.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 23.Haworth R.S., Cuello F., Herron T.J., Franzen G., Kentish J.C., Gautel M., Avkiran M. Protein kinase D is a novel mediator of cardiac troponin I phosphorylation and regulates myofilament function. Circ. Res. 2004;95:1091–1099. doi: 10.1161/01.RES.0000149299.34793.3c. [DOI] [PubMed] [Google Scholar]
- 24.Nakano N., Hori H., Abe M., Shibata H., Arimura T., Sasaoka T., Sawabe M., Chida K., Arai T., Nakahara K., et al. Interaction of BMP10 with Tcap may modulate the course of hypertensive cardiac hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H3396–H3403. doi: 10.1152/ajpheart.00311.2007. [DOI] [PubMed] [Google Scholar]
- 25.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 26.Bassett D.I., Currie P.D. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum. Mol. Genet. 2003;12(Spec no. 2):R265–R270. doi: 10.1093/hmg/ddg279. [DOI] [PubMed] [Google Scholar]
- 27.Guyon J.R., Steffen L.S., Howell M.H., Pusack T.J., Lawrence C., Kunkel L.M. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta. 2007;1772:205–215. doi: 10.1016/j.bbadis.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Bassett D.I., Bryson-Richardson R.J., Daggett D.F., Gautier P., Keenan D.G., Currie P.D. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- 29.Hall T.E., Bryson-Richardson R.J., Berger S., Jacoby A.S., Cole N.J., Hollway G.E., Berger J., Currie P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl Acad. Sci. USA. 2007;104:7092–7097. doi: 10.1073/pnas.0700942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao T., Roeser T., Staub W., Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–2967. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
- 31.Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 32.Postel R., Vakeel P., Topczewski J., Knoll R., Bakkers J. Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev. Biol. 2008;318:92–101. doi: 10.1016/j.ydbio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 33.Bendig G., Grimmler M., Huttner I.G., Wessels G., Dahme T., Just S., Trano N., Katus H.A., Fishman M.C., Rottbauer W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J.M., Sanger J.W. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskeleton. 2005;61:34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raeker M.O., Su F., Geisler S.B., Borisov A.B., Kontrogianni-Konstantopoulos A., Lyons S.E., Russell M.W. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev. Dyn. 2006;235:2018–2029. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- 36.Zhang P., Chen X., Fan M. Signaling mechanisms involved in disuse muscle atrophy. Med. Hypotheses. 2007;69:310–321. doi: 10.1016/j.mehy.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 37.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O.A., Ochoa G.C., Farsad K., Wenk M.R., De Camilli P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 39.Razzaq A., Robinson I.M., McMahon H.T., Skepper J.N., Su Y., Zelhof A.C., Jackson A.P., Gay N.J., O'Kane C.J. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galbiati F., Engelman J.A., Volonte D., Zhang X.L., Minetti C., Li M., Hou H., Jr, Kneitz B., Edelmann W., Lisanti M.P. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J. Biol. Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- 41.Nixon S.J., Wegner J., Ferguson C., Mery P.F., Hancock J.F., Currie P.D., Key B., Westerfield M., Parton R.G. Zebrafish as a model for caveolin-associated muscle disease; caveolin-3 is required for myofibril organization and muscle cell patterning. Hum. Mol. Genet. 2005;14:1727–1743. doi: 10.1093/hmg/ddi179. [DOI] [PubMed] [Google Scholar]
- 42.Frank D., Kuhn C., Katus H.A., Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 2006;84:446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 43.Pyle W.G., Solaro R.J. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 2004;94:296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- 44.Mason P., Bayol S., Loughna P.T. The novel sarcomeric protein telethonin exhibits developmental and functional regulation. Biochem. Biophys. Res. Commun. 1999;257:699–703. doi: 10.1006/bbrc.1999.0531. [DOI] [PubMed] [Google Scholar]
- 45.Schroder R., Reimann J., Iakovenko A., Mues A., Bonnemann C.G., Matten J., Gautel M. Early and selective disappearance of telethonin protein from the sarcomere in neurogenic atrophy. J. Muscle Res. Cell Motil. 2001;22:259–264. doi: 10.1023/a:1012242011109. [DOI] [PubMed] [Google Scholar]
- 46.Knoll R., Hoshijima M., Chien K. Cardiac mechanotransduction and implications for heart disease. J. Mol. Med. 2003;81:750–756. doi: 10.1007/s00109-003-0488-x. [DOI] [PubMed] [Google Scholar]
- 47.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 48.Sun X., Zhang R., Lin X., Xu X. Wnt3a regulates the development of cardiac neural crest cells by modulating expression of cysteine-rich intestinal protein 2 in rhombomere 6. Circ. Res. 2008;102:831–839. doi: 10.1161/CIRCRESAHA.107.166488. [DOI] [PubMed] [Google Scholar]
- 49.Seeley M., Huang W., Chen Z., Wolff W.O., Lin X., Xu X. Depletion of zebrafish titin reduces cardiac contractility by disrupting the assembly of Z-discs and A-bands. Circ. Res. 2007;100:238–245. doi: 10.1161/01.RES.0000255758.69821.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R., Xu X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: an inhibitory mechanism of ventricle-specific gene expression. Dev. Dyn. 2009;238:1564–1573. doi: 10.1002/dvdy.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.