Abstract
TATA binding protein (TBP), a universal transcription factor, is broadly required by nuclear RNA polymerases for the initiation of transcription. TBP contains a polymorphic polyglutamine tract in its N-terminal region, and expansion of this tract leads to spinocerebellar ataxia type 17 (SCA17), one of nine dominantly inherited neurodegenerative diseases caused by polyglutamine expansion in the affected proteins. The expanded polyglutamine proteins are ubiquitously expressed, but cause selective and characteristic neurodegeneration in distinct brain regions in each disease. Unlike many other polyglutamine proteins, whose functions are not yet fully understood, TBP is a well-characterized transcription factor that is restricted to the nucleus. Thus, investigating how mutant TBP mediates neuropathology should help elucidate the mechanisms by which transcriptional dysregulation contributes to neuronal dysfunction and/or neurodegeneration in polyglutamine diseases. To this end, we characterized cellular and mouse models expressing polyQ-expanded TBP. The cell model exhibits characteristic features of neuronal dysfunction, including decreased cell viability and defective neurite outgrowth. We found that the high-affinity nerve growth factor receptor, TrkA, is down-regulated by mutant TBP in cells. Down-regulation of TrkA also occurs in the cerebellum of SCA17 transgenic mice prior to Purkinje cell degeneration. Mutant TBP binds more Sp1, reduces its occupancy of the TrkA promoter and inhibits the activity of the TrkA promoter. These findings suggest that the transcriptional down-regulation of TrkA by mutant TBP contributes to SCA17 pathogenesis.
INTRODUCTION
TATA binding protein (TBP) is a universal transcription factor that is essential for the activity of all three nuclear RNA polymerases. TBP is part of multiple complexes, including the general transcription factor complex transcription factor IID (TFIID). TBP confers site-specific DNA binding activity to the TFIID complex that mediates transcriptional initiation at RNA polymerase II-transcribed genes. DNA binding by TFIID is followed by the entry of other general transcription factors and RNA polymerase through either a sequential assembly or a preassembled holoenzyme pathway (1,2). Within the TFIID complex, TBP is associated with transcription co-factors, referred to as TBP-associated factors (TAFs), that can influence promoter binding or transcription activity via interactions with upstream-binding activators and other components of the basal transcription machinery (3,4). These interactions allow TBP to retain specificity in transcriptional initiation while functioning as a general transcription factor.
An N-terminal expansion in the polyglutamine (polyQ) tract of TBP leads to the autosomal dominant disorder spinocerebellar ataxia type 17 (SCA17). Whereas the range of the normal polyQ tract is 29–42 glutamines, which is encoded by a polymorphic mixed CAG/CAA trinucleotide repeat, mutant TBP contains an expanded polyQ tract (47–55 glutamines) (5). SCA17 is characterized by late-onset neurological symptoms (5,6) that are similar to those of Huntington's disease (HD) and include psychiatric abnormalities, progressive dementia, ataxia and seizures (5,7,8). SCA17 patients typically have marked cerebellar atrophy and Purkinje cell loss, with less pronounced neurodegeneration in other brain regions (5,7,8–11).
SCA17 is one of nine inherited disorders caused by an expansion of the polyQ tract in the affected proteins. These proteins have no homology other than the polyQ domain and are ubiquitously expressed. Yet despite their widespread expression throughout the brain and body, mutant proteins with expanded polyQ domains cause selective neurodegeneration in distinct brain regions in each polyQ disease (12). Most polyQ proteins are distributed in both the cytoplasm and nucleus, whereas TBP is predominantly, if not exclusively, localized to the nucleus. Although polyQ proteins have different functions and structures, they induce common pathological changes characterized by the accumulation of mutant polyQ proteins and transcriptional dysregulation (12–14). Since the protein context can significantly modulate polyQ toxicity and lead to the selective pathology of each polyQ disease, understanding the normal function of polyQ proteins could help us understand the pathogenesis of polyQ diseases. Indeed, each polyQ disease protein is found to interact abnormally with different proteins to mediate pathological changes (12,15,16). The well-characterized function of TBP makes SCA17 an ideal polyQ disease model to study how polyQ expansion alters protein function and induces selective neurodegeneration.
We previously established transgenic SCA17 mice that show pathological and behavioral changes similar to those of other polyQ diseases and recapitulate features of the patient phenotype (17). Microarray studies, however, showed no global transcriptional changes in these SCA17 mice. This finding suggests that mutant TBP may selectively alter gene transcription important for neuronal function. Here, we show that mutant TBP inhibits neurite outgrowth in cultured PC12 cells and reduces the expression of TrkA, a receptor for nerve growth factor (NGF) that promotes neuronal differentiation and survival. We also found that mutant TBP binds more tightly to the transcription factor Sp1 and reduces the association of Sp1 with the TrkA promoter. These findings suggest that polyQ expansion alters the association of TBP with Sp1 to down-regulate the expression of TrkA, which can contribute to the selective neuropathology of SCA17.
RESULTS
Mutant TBP suppresses neurite outgrowth of PC12 cells
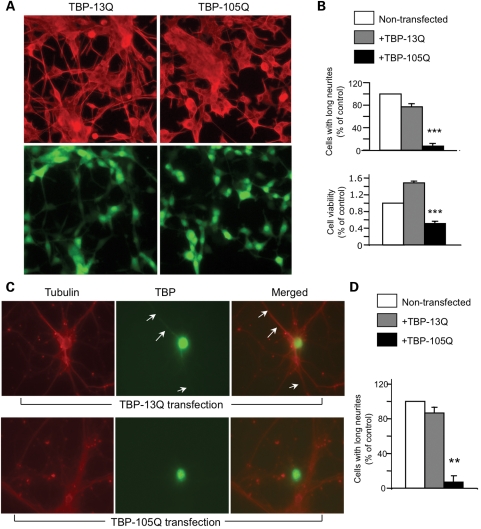
We previously established PC12 cell lines that stably expressed TBP containing 13Q (TBP-13Q) or 105Q (TBP-105Q) (17). These cells also co-express GFP, allowing for selection of TBP-expressing cells. These cells were stimulated with NGF, which can differentiate PC12 cells and promote their neurite outgrowth. Cells expressing TBP-105Q showed shorter neurite extension than those expressing TBP-13Q after NGF treatment (Fig. 1A). Quantifying the number of cells with neurites longer than two cell bodies confirmed a defect in neurite outgrowth of TBP-105Q cells compared with TBP-13Q cells (upper panel in Fig. 1B). To determine whether mutant TBP affects the viability of PC12 cells, we treated the stably transfected PC12 cells with the apoptotic stimulator staurosporine and then measured cell viability via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. TBP-105Q cells showed a greater decrease in their viability than non-transfected PC12 cells or TBP-13Q transfected cells (lower panel in Fig. 1B).
Figure 1.
Defective neurite outgrowth of neurons expressing TBP-105Q. (A) TBP stably transfected PC12 stable cell lines were treated with NGF (100 ng/ml, 48 h). Upper panels are images showing tubulin labeling. Lower panels are images showing GFP that reflects the expression of transfected TBP. Cells expressing TBP-105Q show defective neurite outgrowth. (B) Stably transfected PC12 cell lines were treated with staurosporine (50 µm, 5 h) and then assessed for their neurites (upper panel) and viability using the MTS assay (lower panel). ***P < 0.001 compared with TBP-13Q cells. (C) Cultured primary rat cortical neurons were infected with TBP-13Q or TBP-105Q adenovirus. The infected cells were stained with anti-tubulin to reveal neurites (arrows). (D) The quantification of neurite length demonstrates that TBP-105Q-containing cells show a significant decrease in neurite extension relative to non-infected neurons. **P < 0.01.
To verify that mutant TBP also affects neurite outgrowth of cultured primary neurons, we infected cultured rat brain cortical neurons with an adenoviral vector that co-expresses GFP and TBP-105Q or TBP-13Q. Staining of infected neurons with an antibody to tubulin allowed us to visualize the neurites of neurons expressing exogenous TBP and GFP. We then counted neurons with long neurites and found that neurons expressing TBP-105Q had a substantially lower tendency to display long neurites (Fig. 1C and D). Thus, our examination of stably transfected PC12 cells and cultured primary neurons revealed that mutant TBP could inhibit neurite outgrowth, a characteristic feature of neuronal cells.
Mutant TBP decreases the expression of TrkA in cultured cells
Neurite extension of PC12 cells is largely regulated by NGF, which acts on the receptors p75 and TrkA. Hence, we used RT–PCR analysis to determine whether the expression of either p75 or TrkA is altered in PC12 cells expressing mutant TBP. We also analyzed the same cDNA materials for GAPDH transcripts, which served as an internal control. This assay shows that TrkA, but not p75, transcript levels are lower in TBP-105Q PC12 cells than in TBP-13Q cells (Fig. 2A), suggesting that polyQ expansion can selectively inhibit the expression of TrkA. Quantification of the ratio of TrkA or p75 to GAPDH transcripts also confirmed a decrease in TrkA, but not p75, transcripts in TBP-105Q cells (Fig. 2B). Since neurite outgrowth depends more on the activation of TrkA than p75 (18–21), the reduced TrkA expression is consistent with the decrease in neurite outgrowth we saw in neuronal cells that express mutant TBP. To examine whether TrkA dysfunction is involved in the defective neurite outgrowth in TBP-105Q cells, we treated these cells with gambogic amide (500 nm), a drug that was recently found to selectively activate TrkA (22). By counting the percentage of cells with neurites (758–858 cells each group), we found that gambogic amide could increase the number of TBP-105Q cells with neurite outgrowth (1.3% for untreated versus 8.6% for gambogic amide treated cells, Fig. 2C). As NGF treatment also partially rescued the neuritic defects of TBP-105Q cells and led 16.7% of TBP-105Q cells to develop neurites (also see Fig. 1A), the partial rescue effects of gambogic amide and NGF are consistent with our previous finding that mutant TBP can also affect the expression of other molecules, such as Hsp27, to promote neuropathology (17).
Figure 2.
Decreased level of TrkA transcripts in stably TBP-105Q-transfected PC12 cells. (A) RT–PCR analysis of TrkA transcripts in PC12 cells stably transfected with TBP-13Q or TBP-105Q. The same RNAs were also subjected to RT–PCR with primers for p75. For the internal control, the primers for GAPDH were included in the PCR. (B) The ratio (n = 3) of TrkA or p75 transcripts to GAPDH transcript. TBP-105Q cells show a lower level of TrkA compared with TBP-13Q cells. **P < 0.01. (C) TBP-105Q cells were treated with gambogic acid (+GA, 500 nm), a TrkA agonist, for 5 days. Arrows indicate extended neurites. Control is cells treated with vehicle.
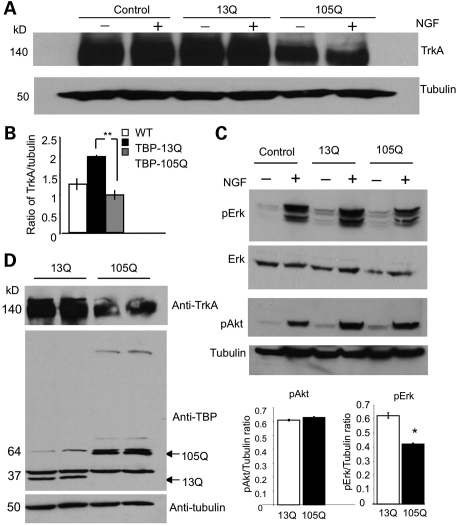
On the basis of these results, we focused on the effect of mutant TBP on TrkA. First, we wanted to test whether TrkA is also reduced at the protein level in cells that express mutant TBP. Although NGF treatment did not alter the level of TrkA, we found that TrkA is indeed decreased in TBP-105Q cells compared with non-transfected PC12 cells and TBP-13Q cells (Fig. 3A). Quantification of the ratio of TrkA to tubulin verified the decreased level of TrkA in TBP-105Q cells (Fig. 3B). Because NGF/TrkA activation increases the phosphorylation of Erk, which is more important for neuronal differentiation than Akt phosphorylation (18,19,23,24), we also examined the phosphorylation of Erk and observed that its phosphorylation is decreased in TBP-105Q cells compared with that of Akt (Fig. 3C). To rule out the possibility that decreased TrkA resulted from the selection of stably transfected cells, we infected normal PC12 cells with adenoviral vectors encoding TBP-13Q or TBP-105Q for 48 h and found that PC12 cells expressing transgenic TBP-105Q also show a decreased level of TrkA compared with cells overexpressing TBP-13Q (Fig. 3D).
Figure 3.
Decreased protein levels of TrkA in cells expressing mutant TBP. (A) Representative western blot of stably transfected PC12 cell lines treated with (+) or without (−) NGF (100 ng/ml). Cells expressing TBP-105Q show decreased levels of TrkA regardless of NGF stimulation. (B) Ratios (n = 3) of TrkA to tubulin for the results in (A) also verify the decreased level of TrkA in TBP-105Q cells. **P < 0.01. (C) Stably transfected PC12 cells were subjected to western blotting with antibodies to phosphorylated Erk (pErk), total Erk or Akt (pAkt). Note that NGF drastically increases the signal of pErk in control and TBP-13Q cells, but this increase is reduced in TBP-105Q cells. Quantitative analysis of the ratios of pErk or pAkt to tubulin confirms the decrease of pErk in TBP-105Q cells. *P<0.05 (n = 3). (D) Representative western blots of PC12 cells infected with TBP-13Q or -105Q adenovirus. Cells infected with TBP-105Q virus also show decreased levels of TrkA protein. The same samples were probed with antibodies to TrkA (upper panel), TBP (middle panel) and tubulin (lower panel).
Mutant TBP decreases the expression of TrkA in the cerebellum of SCA17 mice
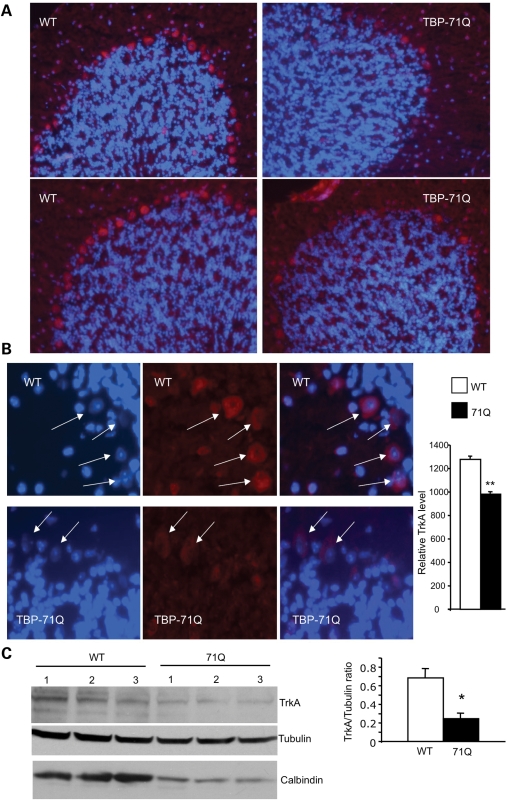
We have shown previously that Purkinje cells degenerate in SCA17 mouse brains (17), which led us to ask whether there is a decrease in TrkA in Purkinje cells. Thus we performed immunofluorescent staining of the cerebellum of SCA17 mice with the antibody to TrkA. As expected, we saw decreased staining of TrkA-positive Purkinje cells in the cerebellum (Fig. 4A). To better compare the levels of TrkA in Purkinje cells in control and SCA17 mouse brains, we performed immunofluorescent staining of these mouse brains under the same conditions, and fluorescent images were taken using the same exposure parameters. Purkinje cells from SCA17 mouse brains show cytoplasmic staining of TrkA, and their levels of TrkA fluorescent signals are also lower than those from control mouse brains (Fig. 4B), implying decreased TrkA levels in the Purkinje cells of SCA17 mouse brains. Western blot analysis of the level of TrkA also reveals that TrkA is reduced in the cerebellar tissues of SCA17 mice (Fig. 4C).
Figure 4.
Reduced TrkA in the cerebellar neurons of symptomatic SCA17 mice at 5 months of age. (A) Cerebellar sections from TBP-71Q and wild-type (WT) littermate control mice were labeled by an antibody to TrkA. Note that there is a decrease in TrkA staining in the Purkinje cell layer. Hoechst dye staining was used to reveal the nuclei of cells. (B) High-magnification (×63) micrographs showing TrkA-containing Purkinje cells (arrows). The TrkA signals relative to the nuclear Hoechst staining signals were quantified (right panel), also showing decreased TrkA in TBP-71Q cerebellum. **P < 0.01. (C) Representative western blot of cerebellar extracts of WT and TBP-71Q transgenic mice at 20 weeks of age. TrkA and calbindin levels are decreased in the TBP-71Q cerebellum compared with the WT cerebellum. The ratio of TrkA to tubulin is also presented (right panel). *P < 0.05.
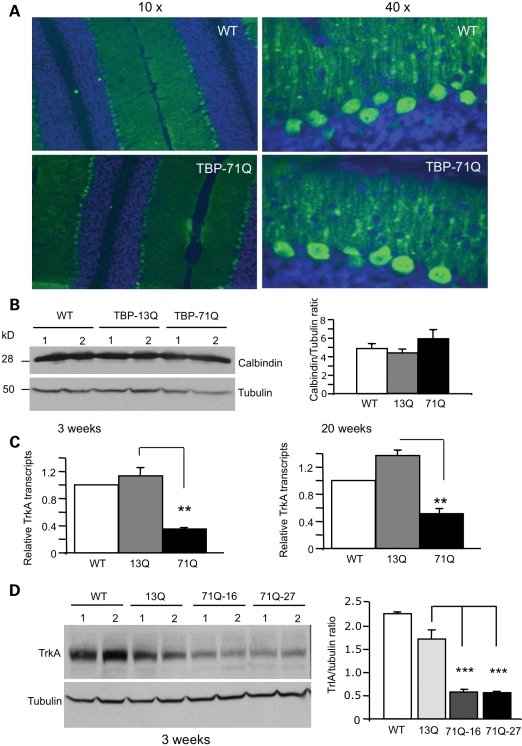
An important issue to explore is whether the reduction in TrkA levels occurs prior to neurodegeneration in SCA17 mice. We therefore examined young SCA17 mice at the age of 3 weeks. Although the timing of symptom onset in SCA17 mice varies between lines based on polyQ tract length and the level of transgene expression, obvious neurological symptoms generally are not evident before 7 weeks of age in TBP-71Q mice (17). We saw no degeneration of Purkinje cells in 3-week-old SCA17 mice, as staining of Purkinje cells with an antibody against calbindin, a cellular marker for Purkinje cells, did not reveal any significant morphological difference between wild-type control and SCA17 mouse brains (Fig. 5A). Further, western blot analysis showed no alteration of calbindin levels in the cerebellar tissues of SCA17 mice at 3 weeks of age (Fig. 5B). We then performed quantitative RT–PCR, which is more sensitive and quantitative, to detect any changes in transcripts. This assay also revealed lower levels of TrkA in SCA17 mice at 3 and 20 weeks (Fig. 5C). Further, western blot analysis shows that TrkA is significantly decreased in the cerebellar tissues from two SCA17 mouse lines (71Q-16 and 71Q-27) when compared with expression in TBP-13Q transgenic mice (Fig. 5D). Thus, these findings demonstrate that TrkA is reduced in the cerebellum of SCA17 mice before the onset of any obvious neurological symptoms or Purkinje cell degeneration.
Figure 5.
Decreased TrkA in young SCA17 mice at 3 weeks of age prior to neurodegeneration. (A) The cerebellum of TBP-71Q and TBP-13Q mice at 3 weeks of age was labeled with antibody to calbindin (green) and nuclear Hoechst staining (blue). Low (×10, left panel) and high (×40, right panel) magnification graphs show no significant difference in calbindin staining of the Purkinje cell layer. (B) Western blotting of the cerebellar tissues of wild-type (WT), TBP-13Q and TBP-71Q mice. Two mice from each group were examined for the expression of calbindin. The ratio of calbindin to tubulin is also presented (right panel). (C) Quantitative RT–PCR analysis of TrkA transcripts reveals a decreased level of TrkA in TBP-71Q cerebellum compared with WT and TBP-13Q mice. **P < 0.01. (D) Representative western blot of cerebellar extracts of 3-week-old WT, TBP-13Q and TBP-71Q (line-16 and line-27) mice. The samples were probed with antibodies to TrkA and tubulin. The ratios of TrkA to tubulin are also presented (right panel). ***P < 0.001.
Mutant TBP inhibits the activity of the TrkA promoter and its association with SP1
Since Sp1 is known to interact with TBP (25,26) and other polyQ-containing proteins (27–29) and since the promoter of the TrkA gene contains the Sp1 binding site (30), we examined whether mutant TBP binds differentially to Sp1 to affect TrkA expression. We first examined the distribution of nuclear Sp1 in the cerebellum of SCA17 mice, but did not find co-localization of Sp1 with nuclear TBP inclusions (Fig. 6A). This suggests that aggregated TBP may not be able to interact with Sp1. Previous studies (25,26) have shown that the Sp1–TBP interaction can be assessed using an in vitro binding assay. We thus followed the established assay to use GST pulldown to compare the interactions of GST–Sp1 with His-tagged TBPs (TBP-31Q and TBP-71Q). This assay revealed increased binding of Sp1 to soluble TBP-71Q (Fig. 6B). Quantitative measurement of the ratio of pulldown to input verified that mutant TBP (TBP-71Q) binds more Sp1 in vitro.
Figure 6.
Inhibitory effect of mutant TBP on the activity of the TrkA promoter. (A) Immunofluorescence staining of primary cerebellar granular neurons showing TBP-105Q aggregates (green) and Sp1 (red). No co-localization is seen between Sp1 and TBP aggregates. (B) GST–Sp1 in vitro pulldown assay shows an increased interaction of Sp1 with soluble TBP-71Q compared with TBP-31Q. The percentage of pulldown (% input) is also presented (right panel). (C) Transfected HEK293 cells (left panel) and PC12 cells (right panel) expressing the TrkA promoter reporter and TBP-13Q or TBP-71Q. HEK293 cells were also transfected with a Sp1 construct. The presence of TBP-71Q decreases TrkA promoter activity. Control: cells transfected only with TrkA promoter reporter and Sp1. (D) Stably transfected PC12 cells expressing TBP-13Q or TBP-105Q were used for ChIP assays to examine the association of Sp1 with the TrkA promoter. There is a decreased association of Sp1 with the TrkA promoter in TBP-105Q cells compared with TBP-13Q cells. The same precipitates were also used for PCR to examine the association of Sp1 with the PCNA promoter. C: no template. The ratios (n = 3) of PCR products from the immunoprecipitates (IP) to those from the inputs are also presented (lower panel). *P < 0.05.
Since the promoter of TrkA contains Sp1 binding sites (30), we asked whether mutant TBP could suppress the promoter activity via its abnormal interaction with Sp1. We generated a reporter construct that links the Sp1 binding site-containing promoter region of the TrkA gene with the red fluorescent protein DsRed, such that the expression of DsRed or the red fluorescence signal reflects the transcriptional activity of the TrkA promoter. This promoter lacks a TATA box, and overexpression of TBP can inhibit its expression, possibly via interactions with other transcription factors. Co-expression of this reporter with Sp1 and normal (TBP-13Q) or mutant TBP (TBP-71Q) in HEK293 cells revealed that reporter activity is significantly reduced by mutant TBP (left panel in Fig. 6C). Expression of the reporter in the stable PC12 cell lines confirmed decreased activity of this reporter in mutant PC12 cells that express TBP-105Q (right panel in Fig. 6C). We then used these cells to perform a chromatin immunoprecipitation (ChIP) assay in which the Sp1–DNA complex was immunoprecipitated by anti-Sp1. The precipitates were subjected to PCR with primers for the TrkA or PCNA promoter sequences, which also contain the Sp1 binding site. Although there is no significant difference in the association of the PCNA promoter with Sp1 between TBP-13Q and TBP-105Q cells, we observed a decrease in the association of Sp1 with the TrkA promoter (Fig. 6D). The ratios of PCR products from the precipitates to those from the inputs also demonstrated a specific decrease in the association of Sp1 with the TrkA promoter (Fig. 6D).
DISCUSSION
Our earlier studies have shown that mutant TBP abnormally binds TFIIB, another component of the basal transcription machinery, and reduces the expression of HSPB1 (or Hsp27), a small heat shock protein that is critical for neuronal survival and axonal integrity (17). It is also evident that polyQ expansion can alter protein–protein interactions to affect multiple targets or functions in cells. For example, mutant ataxin-1 abnormally interacts with several different proteins to cause neuropathology (31–34). Similarly, mutant huntingtin binds abnormally to Sp1, CBP, TAFII130 and other transcription factors to affect different transcriptional functions (16,35). On the basis of these previous findings, we chose to search for additional targets in SCA17 that might account for disease phenotypes, such as diminished neurite outgrowth, in order to uncover the molecular basis for the effects of mutant TBP on neuronal function.
Through our investigations of cellular and mouse models of SCA17, we were able to detect a decrease in TrkA expression in both. This decrease is specifically associated with the expression of mutant TBP as opposed to transfected or transgenic TBP containing 13Q. Thus, polyQ expansion in mutant TBP results in decreased levels of TrkA. Second, we have provided evidence for the transcriptional dysregulation of TrkA by mutant TBP, as our analysis of transcripts and promoter activity of TrkA consistently shows the inhibitory effects of mutant TBP. In addition, mutant TBP binds more Sp1 and reduces the association of Sp1 with the TrkA promoter. Combined with our earlier finding that mutant TBP does not elicit a global alteration of gene transcription (17), the current results suggest that the abnormal association of mutant TBP with transcription factors selectively alters the expression of genes that are important for neuronal function or viability, which could contribute to the selective neuropathology seen in SCA17.
The observation of decreased TrkA in mutant TBP-containing PC12 cells may largely explain the defective neurite outgrowth in these cells. It is known that neurite outgrowth of PC12 cells is triggered by NGF via its activation of membrane receptors p75 (low-affinity) and TrkA (high-affinity). Because RT–PCR analysis suggests that TrkA, but not p75, transcripts are reduced in mutant TBP-containing cells, we focused our subsequent analysis on TrkA, which is expressed in various brain regions, including the cerebellum (36–40). In addition, TrkA is critical for neuronal survival and viability, and its levels are decreased in Alzheimer's disease (41–43). We found that TrkA levels are also decreased in transgenic SCA17 mouse cerebella, and more importantly, this decrease precedes the degeneration of their Purkinje cells, suggesting that the decreased TrkA level contributes to the degeneration of Purkinje cells.
It is well known that TBP binds Sp1 (25,26,44) and that Sp1 activates the transcription of many TATA-less genes (45,46). Unlike TFIIB, which can be sequestered into nuclear TBP inclusions (17,47), Sp1 binds soluble mutant TBP and is not co-localized with nuclear TBP inclusions. Although the interaction of Sp1 with soluble, oligomeric mutant TBP cannot be excluded, the association described here is similar to the increased interaction of soluble mutant huntingtin with Sp1 (27,28). Interestingly, HD and SCA17 share similar neurological symptoms and neuropathology (6,48). Like mutant htt (49), polyQ-expanded TBP could reduce the association of Sp1 with certain promoters via its increased interaction with Sp1. In support of this idea, we found that mutant TBP decreases Sp1 occupancy at the TrkA promoter, which could account for the reduced TrkA expression. Furthermore, identification of the abnormal interaction of mutant TBP with Sp1 also suggests a potential role for this abnormal interaction in the decreased HSPB1 levels we found in our earlier study, as the human HSPB1 promoter contains conserved Sp1 binding sites (50–52).
Several important issues need to be considered regarding the implication of our findings for the pathogenesis of SCA17 and other polyQ diseases. Sp1 is ubiquitously expressed in a variety of mammalian cells and binds the GC boxes in promoter DNA to modulate expression of a large number of genes. An interesting question is why the abnormal interactions of TBP with Sp1 or other transcription factors only selectively change the expression of certain genes, which was revealed by the microarray data in our previous study (17). TBP is essential for the assembly of transcription factor complexes at different promoters. Soluble TBP containing an expanded polyQ domain is also able to stimulate promoter transcription (47,53), though it can abnormally bind other transcription factors. Thus, the effects of mutant TBP on Sp1-mediated transcriptional activity depends largely on the association of TBP with different transcription factors and the regulation by other transcriptional factors or co-factors of the interaction between Sp1 and its GC-rich binding sites (26,44,54,55). This is partially analogous to the finding that aberrant interaction between mutant huntingtin and Sp1 selectively reduces the expression of certain Sp1-dependent genes (49). The promoter of TrkA contains Sp1 binding sites but lacks a TATA box (30), and the activity of this promoter is also regulated by Sp3 (56). It is likely that the composition of promoter-bound transcriptional factor complexes, including the availability of different co-factors, plays an important role in the selective effects of mutant TBP on gene expression.
Another issue is about the relative contribution of the TrkA deficit to SCA17 pathology. Examination of the postmortem brains of a limited number of SCA17 patients has revealed neurodegeneration in different brain regions, including the cortex, striatum and cerebellum, in which pronounced Purkinje cell loss is evident (5,7,11,57,58). In transgenic SCA17 mice, there is also obvious Purkinje cell degeneration (17). Thus, the cerebellar pathology allows us to investigate the mechanism by which mutant TBP causes neurodegeneration. TrkA is expressed in Purkinje cells and is critical for neuronal function and viability (37–39). Unlike other Trk receptors, such as TrkB and TrkC, TrkA is more restricted to certain types of neurons in adult brains (37,59). Since the promoters of TrkB and TrkC do not contain consensus Sp1 binding sites (30,60), it is possible that the Sp1 binding sites in TrkA contribute to the more selective expression of TrkA in certain types of neurons. Accordingly, the decreased expression of TrkA caused by mutant TBP, with other mutant TBP-mediated effects, could more profoundly affect the viability and function of Purkinje cells in the cerebellum. Another possibility is that mutant TBP and/or a proteolytic fragment accumulates in a cell type-specific manner that also contributes to the selective neuropathology. We have found that protein context influences the interaction of Sp1 with huntingtin (61). Moreover, we have detected expanded polyQ-containing TBP fragments that lack an intact DNA-binding domain in affected brain regions of SCA17 transgenic mice (47). Thus, variable accumulation of these putative degradation products of mutant TBP, which may retain the ability to bind Sp1 and other transcription factors but are not capable of facilitating transcriptional initiation, could contribute to selective neuropathology.
The findings in our study also support the idea that the altered protein function caused by polyQ expansion can lead to neuropathology in polyQ diseases (34). Since the function of TBP has been well characterized, further exploration of the effects of polyQ expansion on the function of TBP will give us valuable information about the selective neurodegeneration in polyQ diseases.
MATERIALS AND METHODS
Plasmids and reagents
SCA17 mice were generated in our previous study (17) and maintained in the animal facility at Emory University under specific pathogen-free conditions in accordance with institutional guidelines of The Animal Care and Use Committee at Emory University. Mouse and human TBP cDNA constructs and plasmids were described in our previous studies (17,47). Stably transfected TBP PC12 cell lines were generated by transfecting the Tet-Off stable cell line (Clontech) with PBI–EGFP–TBP constructs (TBP-105Q and TBP-13Q) using Lipofectamine 2000 (17). TBP-13Q or -105Q and EGFP were independently expressed from a bidirectional CMV promoter in these PC12 cells. Constructs encoding Sp1-HA (27), GST-Sp1 (27) and His-TBPs (47) were described previously. Antibodies against TBP included N-12 (Santa Cruz) and 1TBP18 (QED). Other antibodies used for these studies were those against tubulin (Sigma), phospho-Erk and phospho-Akt (Cell Signaling), Sp1 (Upstate Biotechnologies), calbindin (Chemicon), TrkA (Upstate Biotechnologies) and 12CA5 (Roche). Adenovirus expressing GFP and TBP-13Q or -105Q was made by Welgen (Worcester, MA, USA).
Immunofluorescence staining, cell culture and cell viability assays
The cerebellum of 71Q-TBP (line-16 and line-27) transgenic and littermate control mice were rapidly isolated and sectioned (10 µm) with a cryostat at −20°C. The brain sections were placed on gelatin-precoated, precleaned glass slides. Sections were fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.4% Triton X-100 in PBS for 30 min, blocked with 5% normal goat serum in PBS for 1 h, and incubated with primary antibodies in 2% NGS and PBS overnight at 4°C in a moisture chamber. Hoechst was used to label the nuclei. Fluorescent images were acquired on a Zeiss microscope (Axiovert 200 MOT; Carl Zeiss Imaging) equipped with a digital camera (Hamamatsu Orca-100) and OpenLAB software (Improvision Inc.). We used ×10, ×40 and ×63 objectives for image acquisition.
Cerebellar granule cell culture was performed using the method described previously (17). Wild-type Tet-Off and stably transfected PC12 cell lines expressing either TBP-13Q or TBP-105Q were plated into 12-well plates at ∼40% confluency. Cells were treated with 100 ng/ml NGF for 48 h at 37°C in serum-free media then subjected to immunofluorescent staining with an antibody against alpha-tubulin (1:5000, Sigma) for neurite staining and Hoechst for visualizing nuclei. PC12 cells were also treated with 500 nm gambogic amide for 5 days to examine their neurites (22). Fluorescent images were captured with a Zeiss microscope (Axiovert 200 MOT; Carl Zeiss Imaging) and stored in a computer. Quantitative results of the neurite outgrowth were obtained by counting the percentage of cells that extended neurites twice the length of the cell body. Cortical neurons were dissected from embryonic day 18 rats and plated on poly-d-lysine-coated plastic culture plates (Corning Costar) in B27-supplemented medium (Invitrogen). At DIV2, neurons were infected with either adenoviral vectors expressing GFP–TBP-13Q or GFP–TBP-105Q (Welgen, Inc.). After 24 h, neurites were assessed as described above.
Cell viability was determined by a modified MTT assay (Cell Titer 96; Promega) as described previously (27). Wild-type and stably transfected PC12 cells expressing either TBP-13Q or -105Q in serum-free medium were treated with staurosporine at the final concentration of 50 nm for 5 h. A ratio of untreated and staurosporine-treated cells was used to determine cell viability.
Gene expression and western blot analyses
Total RNAs were purified from PC12 cells or mouse brains. The RNAs were reverse transcribed and used for RT–PCR. Primers used were: tgtggaagtgggggatgacg (sense) and gcactcagcaagaaagacct (antisense) for TrKA; ccacattccgacgactgatg (sense) and ccaagaatgagcgcactaac (antisense) for p75 and acgaccccttcattgacctc (sense) and gggggctaagcagttggtgg (antisense) for GAPDH. PCR conditions were 95°C for 45 s, 60°C for 1 min and 72°C for 2 min with 35 cycles. For quantitative RT–PCR, first-strand cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR of TrkA and GAPDH were assayed using TaqMan probes and reagents (Applied Biosystems). All quantitative PCR experiments were done on the Eppendorf RealPlex 4 machine.
For western blotting analysis, brain tissue samples and cultured cells were harvested and homogenized in immunoprecipitation (IP) assay buffer (50 mm Tris–HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, pH 8.0, 1 mm EGTA, pH 8.0, 0.1% SDS, 0.5% sodium deoxycholate and 1% Triton X-100, with protease inhibitors (Sigma) and sonicated for 10 s. After 30 min incubation on a rotating apparatus at 4°C, the samples were clarified at 734×g (change to g) for 5 min. Two hundred micrograms of the brain supernatant and 50 µg of cell supernatant were used for western blotting. Protein samples were resolved on 4–20% Tris-glycine polyacrylamide gels (Invitrogen). Immunoreactive bands were visualized by an enhanced Chemiluminescence Kit (ECL plus, GE Healthcare, Amersham).
Transcription reporter assay
The human TrkA promoter was cloned from genomic DNA using the primers sense 5′-ATCAAAGCTTCACCTCCGAGGCGTTC-3′ and antisense 5′-GAATTCTCTGTGCGCTCCCAGCTGC-3′ and inserted into the DsRed 2.1 vector (Clontech) using HindIII and EcoR1 restriction sites. HEK293T cells were co-transfected with the reporter vector, Sp1, TBP-31Q, TBP-105Q or a mock vector in 12-well plates using Lipofectamine (Invitrogen). The cells were harvested in 500 µl of cold 1× PBS after 48 h transfection, and 50 µl of resuspended cells was combined with 50 µl of 1× PBS in a 96-well black bottom plate. Samples from each co-transfection were plated in duplicate, and red fluorescence was detected with a FLUOstar Galaxy microplate reader (BMG LABTECH, Offenburg, Germany). For experiments using the stably transfected PC12 cell lines expressing TBP-13Q or -105Q, the reporter or empty vector was transfected. Transfections were done in duplicate in 12-well plates using Lipofectamine 2000. Fluorescence images were visualized on the Zeiss microscope (Axiovert 200 MOT; Carl Zeiss Imaging), and fluorescence levels were quantitated using OpenLAB Software (Improvision Inc.).
Chromatin immunoprecipitation
ChIP assays with semi-quantitative PCR were performed as described previously (17). TBP-13Q and -105Q PC12 cells were transfected with Sp1-HA and collected 48 h after transfection. For each IP, 850 µg of precleared whole cell lysate and 5 µg of anti-Sp1 (Millipore, Upstate Biotechnologies) or no antibody were used. Semi-quantitative PCR, using PCNA and TrkA primers in separate reactions, was performed on DNA recovered from IP samples and inputs (10% pre-cleared lysate). PCR without template DNA served as negative controls. Promoter sequences were acquired using the UCSC genome browser to include Sp1 binding sites. The size of the amplicons for PCNA and TrkA were 282 and 183 bp, respectively. Primer sequences and PCR cycling parameters were as follows: rat PCNA: sense 5′-TGGCTTTCATTTCCGTGGC-3′ and antisense 5′-AGTCACCTGCGCCCGCAAC-3, rat TrkA: sense 5′-ACATGTGAAGCAATCTGTGGCAG-3′ and antisense 5′-CGGGGCGGTGTTAAAGACTAGCC-3′. PCR conditions were as follows: 96° for 3 min, 96° for 45 s, 63° for 45 s and 72° for 1 min with 35–45 cycles. PCR products were electrophoresed on a 1.8% agarose gel. Densitometry was used to measure PCR product signals from the precipitates after subtracting the background signals from the precipitates in the absence of anti-Sp1 antibody. The ratios of the precipitated PCR products to input were then calculated.
Statistical analysis
All values are expressed as mean ± SE. We assessed statistical significance using Student's t-test and used ANOVA when multiple samples were compared. A P-value of less than 0.05 is considered to be significant.
FUNDING
This work was supported by grants from the National Institutes of Health to S.H.L. (NS0465016) and to X.J.L. (NS041669 and AG019206).
ACKNOWLEDGEMENTS
We thank Dr Keqiang Ye at Emory University for providing gambogic amide and Cheryl Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hochheimer A., Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 2.Roeder R.G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Lee T.I., Young R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 4.Malik S., Roeder R.G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 6.van Roon-Mom W.M., Reid S.J., Faull R.L., Snell R.G. TATA-binding protein in neurodegenerative disease. Neuroscience. 2005;133:863–872. doi: 10.1016/j.neuroscience.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 7.Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum. Mol. Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 8.Rolfs A., Koeppen A.H., Bauer I., Bauer P., Buhlmann S., Topka H., Schols L., Riess O. Clinical features and neuropathology of autosomal dominant spinocerebellar ataxia (SCA17) Ann. Neurol. 2003;54:367–375. doi: 10.1002/ana.10676. [DOI] [PubMed] [Google Scholar]
- 9.Bauer P., Laccone F., Rolfs A., Wullner U., Bosch S., Peters H., Liebscher S., Scheible M., Epplen J.T., Weber B.H., et al. Trinucleotide repeat expansion in SCA17/TBP in white patients with Huntington's disease-like phenotype. J. Med. Genet. 2004;41:230–232. doi: 10.1136/jmg.2003.015602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruni A.C., Takahashi-Fujigasaki J., Maltecca F., Foncin J.F., Servadio A., Casari G., D'Adamo P., Maletta R., Curcio S.A., De Michele G., et al. Behavioral disorder, dementia, ataxia, and rigidity in a large family with TATA box-binding protein mutation. Arch. Neurol. 2004;61:1314–1320. doi: 10.1001/archneur.61.8.1314. [DOI] [PubMed] [Google Scholar]
- 11.Toyoshima Y., Yamada M., Onodera O., Shimohata M., Inenaga C., Fujita N., Morita M., Tsuji S., Takahashi H. SCA17 homozygote showing Huntington's disease-like phenotype. Ann. Neurol. 2004;55:281–286. doi: 10.1002/ana.10824. [DOI] [PubMed] [Google Scholar]
- 12.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 13.Cha J.H. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 14.Sugars K.L., Rubinsztein D.C. Transcriptional abnormalities in Huntington disease. Trends. Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 15.Harjes P., Wanker E.E. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 16.Li S.H., Li X.J. Huntingtin–protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Friedman M.J., Shah A.G., Fang Z.H., Ward E.G., Warren S.T., Li S., Li X.J. Polyglutamine domain modulates the TBP–TFIIB interaction: implications for its normal function and neurodegeneration. Nat. Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- 18.Bai G., Kusiak J.W. Nerve growth factor up-regulates the N-methyl-D-aspartate receptor subunit 1 promoter in PC12 cells. J. Biol. Chem. 1997;272:5936–5942. doi: 10.1074/jbc.272.9.5936. [DOI] [PubMed] [Google Scholar]
- 19.Vaudry D., Stork P.J., Lazarovici P., Eiden L.E. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 20.Hisata S., Sakisaka T., Baba T., Yamada T., Aoki K., Matsuda M., Takai Y. Rap1-PDZ-GEF1 interacts with a neurotrophin receptor at late endosomes, leading to sustained activation of Rap1 and ERK and neurite outgrowth. J. Cell Biol. 2007;178:843–860. doi: 10.1083/jcb.200610073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isada T., Hirose M., Murata E., Okayama Y., Takatori M., Tani M., Fukazawa K., Hirata M., Taniguchi S., Shigemi K., Tanaka Y. Repeated exposures to hyperbaric air suppress neurite outgrowth in PC12 cells. J. Physiol. Sci. 2007;57:321–325. doi: 10.2170/physiolsci.SC009707. [DOI] [PubMed] [Google Scholar]
- 22.Jang S.W., Okada M., Sayeed I., Xiao G., Stein D., Jin P., Ye K. Gambogic amide, a selective agonist for TrkA receptor that possesses robust neurotrophic activity, prevents neuronal cell death. Proc. Natl Acad. Sci. USA. 2007;104:16329–16334. doi: 10.1073/pnas.0706662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu A., Prenger M.S., Norton D.D., Mei L., Kusiak J.W., Bai G. Nerve growth factor uses Ras/ERK and phosphatidylinositol 3-kinase cascades to up-regulate the N-methyl-D-aspartate receptor 1 promoter. J. Biol. Chem. 2001;276:45372–45379. doi: 10.1074/jbc.M105399200. [DOI] [PubMed] [Google Scholar]
- 24.Zentrich E., Han S.Y., Pessoa-Brandao L., Butterfield L., Heasley L.E. Collaboration of JNKs and ERKs in nerve growth factor regulation of the neurofilament light chain promoter in PC12 cells. J. Biol. Chem. 2002;277:4110–4118. doi: 10.1074/jbc.M107824200. [DOI] [PubMed] [Google Scholar]
- 25.Torigoe T., Izumi H., Yoshida Y., Ishiguchi H., Okamoto T., Itoh H., Kohno K. Low pH enhances Sp1 DNA binding activity and interaction with TBP. Nucleic Acids Res. 2003;31:4523–4530. doi: 10.1093/nar/gkg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emili A., Greenblatt J., Ingles C.J. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S.H., Cheng A.L., Zhou H., Lam S., Rao M., Li H., Li X.J. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell. Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 29.Goold R., Hubank M., Hunt A., Holton J., Menon R.P., Revesz T., Pandolfo M., Matilla-Duenas A. Down-regulation of the dopamine receptor D2 in mice lacking ataxin 1. Hum. Mol. Genet. 2007;16:2122–2134. doi: 10.1093/hmg/ddm162. [DOI] [PubMed] [Google Scholar]
- 30.Sacristan M.P., de Diego J.G., Bonilla M., Martin-Zanca D. Molecular cloning and characterization of the 5′ region of the mouse trkA proto-oncogene. Oncogene. 1999;18:5836–5842. doi: 10.1038/sj.onc.1202963. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda H., Jafar-Nejad H., Patel A.J., Sun Y., Chen H.K., Rose M.F., Venken K.J., Botas J., Orr H.T., Bellen H.J., Zoghbi H.Y. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Lam Y.C., Bowman A.B., Jafar-Nejad P., Lim J., Richman R., Fryer J.D., Hyun E.D., Duvick L.A., Orr H.T., Botas J., Zoghbi H.Y. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 33.Serra H.G., Duvick L., Zu T., Carlson K., Stevens S., Jorgensen N., Lysholm A., Burright E., Zoghbi H.Y., Clark H.B., Andresen J.M., Orr H.T. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A.B., Richman R., Hill D.E., Orr H.T., Zoghbi H.Y. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhai W., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 36.Sobreviela T., Clary D.O., Reichardt L.F., Brandabur M.M., Kordower J.H., Mufson E.J. TrkA-immunoreactive profiles in the central nervous system: colocalization with neurons containing p75 nerve growth factor receptor, choline acetyltransferase, and serotonin. J. Comp. Neurol. 1994;350:587–611. doi: 10.1002/cne.903500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muragaki Y., Timothy N., Leight S., Hempstead B.L., Chao M.V., Trojanowski J.Q., Lee V.M. Expression of trk receptors in the developing and adult human central and peripheral nervous system. J. Comp. Neurol. 1995;356:387–397. doi: 10.1002/cne.903560306. [DOI] [PubMed] [Google Scholar]
- 38.Torres J.M., Javier Naves F., Esteban I., Del Valle M.E., Vega J.A. Neurotrophin receptor proteins immunoreactivity in the rat cerebellar cortex as a function of age. Mech. Ageing Dev. 1995;83:1–9. doi: 10.1016/0047-6374(95)01616-8. [DOI] [PubMed] [Google Scholar]
- 39.Dohrman D.P., West J.R., Pantazis N.J. Ethanol reduces expression of the nerve growth factor receptor, but not nerve growth factor protein levels in the neonatal rat cerebellum. Alcohol. Clin. Exp. Res. 1997;21:882–893. [PubMed] [Google Scholar]
- 40.Riva-Depaty I., Dubreuil Y.L., Mariani J., Delhaye-Bouchaud N. Eradication of cerebellar granular cells alters the developmental expression of trk receptors in the rat inferior olive. Int. J. Dev. Neurosci. 1998;16:49–62. doi: 10.1016/s0736-5748(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 41.Hock C., Heese K., Muller-Spahn F., Hulette C., Rosenberg C., Otten U. Decreased trkA neurotrophin receptor expression in the parietal cortex of patients with Alzheimer's disease. Neurosci. Lett. 1998;241:151–154. doi: 10.1016/s0304-3940(98)00019-6. [DOI] [PubMed] [Google Scholar]
- 42.Dubus P., Faucheux B., Boissiere F., Groppi A., Vital C., Vital A., Agid Y., Hirsch E.C., Merlio J.P. Expression of Trk isoforms in brain regions and in the striatum of patients with Alzheimer's disease. Exp. Neurol. 2000;165:285–294. doi: 10.1006/exnr.2000.7447. [DOI] [PubMed] [Google Scholar]
- 43.Savaskan E., Muller-Spahn F., Olivieri G., Bruttel S., Otten U., Rosenberg C., Hulette C., Hock C. Alterations in trk A, trk B and trk C receptor immunoreactivities in parietal cortex and cerebellum in Alzheimer's disease. Eur. Neurol. 2000;44:172–180. doi: 10.1159/000008229. [DOI] [PubMed] [Google Scholar]
- 44.Segal J.A., Barnett J.L., Crawford D.L. Functional analyses of natural variation in Sp1 binding sites of a TATA-less promoter. J. Mol. Evol. 1999;49:736–749. doi: 10.1007/pl00006596. [DOI] [PubMed] [Google Scholar]
- 45.Emami K.H., Burke T.W., Smale S.T. Sp1 activation of a TATA-less promoter requires a species-specific interaction involving transcription factor IID. Nucleic Acids Res. 1998;26:839–846. doi: 10.1093/nar/26.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wierstra I. Sp1: emerging roles—beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 47.Friedman M.J., Wang C.E., Li X.J., Li S. Polyglutamine expansion reduces the association of TATA-binding protein with DNA and induces DNA binding-independent neurotoxicity. J. Biol. Chem. 2008;283:8283–8290. doi: 10.1074/jbc.M709674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevanin G., Brice A. Spinocerebellar ataxia 17 (SCA17) and Huntington's disease-like 4 (HDL4) Cerebellum. 2008;7:170–178. doi: 10.1007/s12311-008-0016-1. [DOI] [PubMed] [Google Scholar]
- 49.Chen-Plotkin A.S., Sadri-Vakili G., Yohrling G.J., Braveman M.W., Benn C.L., Glajch K.E., DiRocco D.P., Farrell L.A., Krainc D., Gines S., MacDonald M.E., Cha J.H. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol. Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Frohli E., Aoyama A., Klemenz R. Cloning of the mouse hsp25 gene and an extremely conserved hsp25 pseudogene. Gene. 1993;128:273–277. doi: 10.1016/0378-1119(93)90574-m. [DOI] [PubMed] [Google Scholar]
- 51.Gaestel M., Gotthardt R., Muller T. Structure and organisation of a murine gene encoding small heat-shock protein Hsp25. Gene. 1993;128:279–283. doi: 10.1016/0378-1119(93)90575-n. [DOI] [PubMed] [Google Scholar]
- 52.Oesterreich S., Hickey E., Weber L.A., Fuqua S.A. Basal regulatory promoter elements of the hsp27 gene in human breast cancer cells. Biochem. Biophys. Res. Commun. 1996;222:155–163. doi: 10.1006/bbrc.1996.0714. [DOI] [PubMed] [Google Scholar]
- 53.Reid S.J., Rees M.I., van Roon-Mom W.M., Jones A.L., MacDonald M.E., Sutherland G., During M.J., Faull R.L., Owen M.J., Dragunow M., Snell R.G. Molecular investigation of TBP allele length: a SCA17 cellular model and population study. Neurobiol. Dis. 2003;13:37–45. doi: 10.1016/s0969-9961(03)00014-7. [DOI] [PubMed] [Google Scholar]
- 54.Courey A.J., Holtzman D.A., Jackson S.P., Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 55.Pascal E., Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 56.Lambiase A., Merlo D., Mollinari C., Bonini P., Rinaldi A.M., D'Amato M., Micera A., Coassin M., Rama P., Bonini S., Garaci E. Molecular basis for keratoconus: lack of TrkA expression and its transcriptional repression by Sp3. Proc. Natl Acad. Sci. USA. 2005;102:16795–16800. doi: 10.1073/pnas.0508516102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujigasaki H., Martin J.J., De Deyn P.P., Camuzat A., Deffond D., Stevanin G., Dermaut B., Van Broeckhoven C., Durr A., Brice A. CAG repeat expansion in the TATA box-binding protein gene causes autosomal dominant cerebellar ataxia. Brain. 2001;124:1939–1947. doi: 10.1093/brain/124.10.1939. [DOI] [PubMed] [Google Scholar]
- 58.Zuhlke C., Hellenbroich Y., Dalski A., Kononowa N., Hagenah J., Vieregge P., Riess O., Klein C., Schwinger E. Different types of repeat expansion in the TATA-binding protein gene are associated with a new form of inherited ataxia. Eur. J. Hum. Genet. 2001;9:160–164. doi: 10.1038/sj.ejhg.5200617. [DOI] [PubMed] [Google Scholar]
- 59.Numakawa T., Takei N., Yamagishi S., Sakai N., Hatanaka H. Neurotrophin-elicited short-term glutamate release from cultured cerebellar granule neurons. Brain Res. 1999;842:431–438. doi: 10.1016/s0006-8993(99)01867-3. [DOI] [PubMed] [Google Scholar]
- 60.Lei L., Parada L.F. Transcriptional regulation of Trk family neurotrophin receptors. Cell. Mol. Life Sci. 2007;64:522–532. doi: 10.1007/s00018-006-6328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cornett J., Smith L., Friedman M., Shin J.Y., Li X.J., Li S.H. Context-dependent dysregulation of transcription by mutant huntingtin. J. Biol. Chem. 2006;281:36198–36204. doi: 10.1074/jbc.M607839200. [DOI] [PubMed] [Google Scholar]