Abstract
Aggregation and cleavage are two hallmarks of Tau pathology in Alzheimer disease (AD), and abnormal fragmentation of Tau is thought to contribute to the nucleation of Tau paired helical filaments. Clearance of the abnormally modified protein could occur by the ubiquitin–proteasome and autophagy–lysosomal pathways, the two major routes for protein degradation in cells. There is a debate on which of these pathways contributes to clearance of Tau protein and of the abnormal Tau aggregates formed in AD. Here, we demonstrate in an inducible neuronal cell model of tauopathy that the autophagy–lysosomal system contributes to both Tau fragmentation into pro-aggregating forms and to clearance of Tau aggregates. Inhibition of macroautophagy enhances Tau aggregation and cytotoxicity. The Tau repeat domain can be cleaved near the N terminus by a cytosolic protease to generate the fragment F1. Additional cleavage near the C terminus by the lysosomal protease cathepsin L is required to generate Tau fragments F2 and F3 that are highly amyloidogenic and capable of seeding the aggregation of Tau. We identify in this work that components of a selective form of autophagy, chaperone-mediated autophagy, are involved in the delivery of cytosolic Tau to lysosomes for this limited cleavage. However, F1 does not fully enter the lysosome but remains associated with the lysosomal membrane. Inefficient translocation of the Tau fragments across the lysosomal membrane seems to promote formation of Tau oligomers at the surface of these organelles which may act as precursors of aggregation and interfere with lysosomal functioning.
INTRODUCTION
Tau is a neuronal cytosolic protein that stabilizes microtubules and thus enables them to carry out their role as cytoskeletal elements and tracks for axonal transport. Tau is a highly soluble, natively unfolded protein which contains very little secondary structure. Its domains can be broadly subdivided into the C-terminal half, which contains three or four imperfectly repeated motifs involved in binding to microtubules, and the N-terminal half that projects away from the microtubule surface. Abnormal aggregates of Tau [neurofibrillary tangles, consisting of paired helical filaments (PHFs)] are hallmarks of several neurodegenerative diseases including Alzheimer disease (AD) and other tauopathies (1,2). The aggregation of Tau in the brain is based on the repeat domain and its tendency to convert to cross-β-structure (3). Elevation of fragmented Tau in the cerebrospinal fluid is an early marker of AD (4,5). Limited proteolysis of other pathogenic proteins, such as polyglutamine proteins and α-synuclein, plays a critical role in the pathogenesis of neurodegenerative diseases (6,7). Likewise, Tau aggregation can be accelerated by proteolytic cleavage which generates amyloidogenic fragments (8–11). Consequently, cellular conditions that increase Tau levels and fragmentation could favor formation of aggregates.
Intracellular levels of proteins are regulated by a balanced equilibrium between their synthesis and degradation. Two major proteolytic systems contribute to protein degradation inside cells, the ubiquitin–proteasome system and the autophagy–lysosomal system. The identification of ubiquitin in PHFs in AD brain has led to the speculation that the ubiquitin–proteasome system may have an important role in Tau degradation (12). Since then, a number of studies have investigated the effect of proteasomal inhibition on Tau metabolism. Some groups reported that proteasomal inhibition caused elevation of Tau (13,14), whereas other studies suggested that Tau is not a proteasome substrate (15–17). Recently, several groups showed that Tau hyperphosphorylated at Ser-Pro or Thr-Pro motifs flanking the repeat domain can be ubiquitinated by the CHIP–hsc70 complex and degraded through the proteasome system (18,19). However, Tau phosphorylated at the KXGS (K, lysine; X, any amino acid; G, Glycine; S, Serine) motifs in the repeat domain is not degraded by this pathway (20). Therefore, until now, it remains unclear whether Tau is normally degraded by the proteasome system or not. The other major route for the degradation of proteins in eukaryotic cells is through the autophagy–lysosomal system, which includes three main pathways for the delivery of cargo to lysosomes: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (21). The alteration of the lysosomal system in AD has already been extensively demonstrated (22–24). There is evidence showing that cathepsin B closely associates with intracellular neurofibrillary tangles in AD brains (25). Tau can be degraded by lysosomal proteases such as cathepsin D in vitro and in the cytosol (26,27), and inhibition of lysosomes can increase Tau levels (28). In addition, Tau was found in lysosomes of skeletal muscles in chloroquine myopathy (29) and in neurons in both AD brain and control brain (30). Intriguingly, wild-type Tau protein has been proposed to be necessary for proper macroautophagy function, at least in a disease context. Nevertheless, the involvement of different autophagy pathways in Tau clearance remains poorly understood. Recently, induction of macroautophagy by rapamycin has been shown to promote degradation of detergent-insoluble mutant hTau40/P301L (31). Although in this report basal macroautophagy did not contribute to Tau clearance, studies in another cell model support clearance of Tau by basal macroautophagy (32). Thus, more research is required to clarify this controversy and also to elucidate the possible contribution of other autophagic pathways in Tau clearance. Among them, CMA is a type of autophagy that degrades soluble cytosolic proteins containing a lysosomal targeting motif (biochemically related to the pentapeptide KFERQ) (33). The motif in the substrate proteins is recognized by a cytosolic chaperone, the heat shock cognate of 70 kDa (hsc70), which transfers the substrate to lysosomes where it binds to the lysosome-associated membrane protein type 2A (LAMP-2A). After crossing the lysosomal membrane, substrate proteins are degraded in the lysosomal lumen (34,35). In AD brain, hsc70 is associated with neurofibrillary tangles (36) and has been described to interact with Tau (19). Tau contains two CMA-targeting motifs (336QVEVK340 and 347KDRVQ351) in its C-terminal region. However, up-to-date, the role of CMA in the Tau degradation has not been investigated yet.
In this study, we use an inducible mouse neuroblastoma N2a cell model of tauopathy expressing the repeat domain of Tau with a FTDP-17 mutation (TauRDΔK280) (37), to test whether the autophagy–lysosomal system plays a role in the degradation of the protein and its aggregates. We found that macroautophagy can efficiently degrade both soluble mutant Tau and its aggregates, whereas proteasomal degradation plays only a minor role in this system. Inhibition of macroautophagy leads to enhanced aggregation and cytotoxicity of the mutant Tau. Using an inducible N2a cell line expressing the full-length wild-type isoform hTau40, we further demonstrate that macroautophagy can also degrade full-length Tau. Phosphorylation in the repeat domain, which regulates the affinity of Tau for microtubules, does not alter its sensitivity to autophagy–lysosomal degradation. Surprisingly, we also found that the lysosomal protease cathepsin L was involved in the cleavage of TauRDΔK280, generating a fragment which strongly promotes aggregation to PHFs. To determine the mechanism involved in the targeting of Tau to lysosomes for this partial cleavage we tested whether full-length Tau or Tau fragments were substrates for CMA. We found that TauRDΔK280 and its F1 fragment interact with the cytosolic hsc70 chaperone and this facilitates its binding to lysosomes in a manner depending on the CMA receptor LAMP-2A. However, contrary to typical CMA substrate proteins, these forms of Tau are not translocated into lysosomes by CMA. Instead, association of Tau to lysosomes through the CMA machinery seems to contribute to the cleavage of this mutant form of Tau and the organization of the protein into oligomeric complexes that may interfere with normal lysosomal function. We propose that changes in lysosomal degradation of Tau as well as direct effects of Tau on lysosomes can contribute to pathogenesis in AD and other tauopathies.
RESULTS
Effects of perturbation of the autophagy–lysosomal pathway on the cleavage and aggregation of TauRDΔK in N2a cells
We developed a cell model of tauopathy by expressing the repeat domain of Tau with the ΔK280 mutation (denoted as TauRDΔK) in N2a cells in an inducible fashion (37). In this cell model, part of the Tau protein undergoes stepwise proteolytic processing to generate three fragments, first F1, then F2 and F3 (Fig. 1A). The fragments have a high propensity for β-structure so that they function as seeds of aggregation of TauRDΔK or full-length Tau (hTau40ΔK) (9). Nevertheless, switching off the mutant Tau expression leads to the disappearance of the aggregates, showing that the cells retain the machinery to degrade such aggregates. We therefore analyzed the pathways involved in Tau degradation in this cell model and found that proteasome inhibitors such as MG132 and epoxomicin showed no effect on the clearance of Tau aggregates detected in the pellet of sarkosyl extraction in the cells (Fig. 1B). Thus, we focused on the autophagy–lysosomal pathway as the alternative route for the degradation of aggregates (38). Activation of macroautophagy is mediated by a protein complex containing phosphatidyl-inositol-kinase (PI3K). Consequently, inhibitors of PI3K-type III, such as 3-methyladenine (3-MA) can be used to block macroautophagy without affecting other autophagic pathways (39). In contrast, blockage of the activity of lysosomal proteases, for example neutralizing the acid lysosomal pH by treatment with NH4Cl, inhibits all types of lysosomal proteolysis (i.e. macroautophagy, microautophagy or CMA). After treatment of cells with the two types of inhibitors followed by Sarkosyl extraction to separate soluble and insoluble Tau, we found that NH4Cl treatment dramatically reduced the generation of fragments F2 and F3 and thus inhibited Tau aggregation (as nearly no F2 and F3 were found in the sarkosyl pellets; Fig. 1C, lanes 5–8). This result indicated that lysosomal proteases were involved in the generation of F2 and F3. Previously, we showed that F1 was generated by a thrombin-like activity (37). Consistent with this observation, NH4Cl treatment did not influence the generation of F1, which is a prerequisite for further cleavage to F2 and F3. Interestingly, treatment with 3-MA did not inhibit the generation of F2 and F3, on the contrary, it increased the level of all the Tau fragments (80 and 60% increase for F2 and F3, respectively), including aggregates (Fig. 1C, lane 4). This result suggested that macroautophagy was involved in the degradation of both the soluble Tau constructs and aggregates, but surprisingly that the lysosomal cleavage of Tau did not result from macroautophagy delivery to the lysosomal compartment. The presence of Tau in purified fractions of autophagic vacuoles (AVs) (Supplementary Material, Fig. S1A) and the partial co-localization of Tau with microtubule-associated light chain 3 (LC3), an autophagosome marker (Supplementary Material, Fig. S1B) confirmed that macroautophagy plays a role in Tau degradation in this cell model, as previously reported elsewhere (32). The identity and purity of the AV-enriched fractions were confirmed by immunoblotting using antibodies to LC3 (Supplementary Material, Fig. S1A).
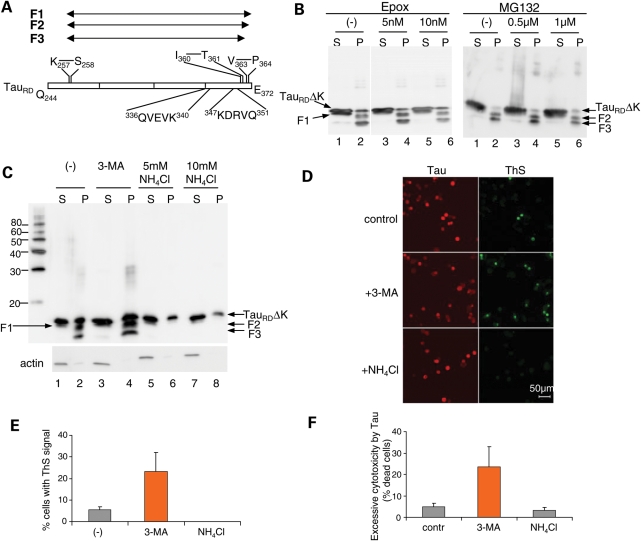
Figure 1.
Effects of NH4Cl and 3-MA on the cleavage, aggregation and cytotoxicity of TauRDΔK in N2a cells. (A) Diagram of TauRD showing cleavage sites and CMA recognition motifs. TauRD is first cleaved after K257 to generate F1, then after V363 to generate F2, and further after I360 to produce F3. The two CMA recognition motifs are 336QVEVK340 and 347KDRVQ351. (B and C) Blot analysis of effects of proteasomal inhibitors (epoxomicin or MG132) (B) or autophagy–lysosomal inhibitors (3-MA or NH4Cl) (C) on cleavage and oligomerization of TauRDΔK. N2a cells overexpressing TauRDΔK were treated with these inhibitors for 5 days. Lanes labeled P (=pellet) denote sarkosyl-insoluble Tau species, S (=supernatant) indicates soluble proteins. Note that NH4Cl but not 3-MA inhibits the cleavage and aggregation of TauRDΔK. (D) ThS staining showing the influence of 3-MA and NH4Cl on Tau aggregation. (E) Quantification of ThS positive cells, which increase ∼5-fold with 3-MA but not with NH4Cl. (F) Effects of NH4Cl and 3-MA on the cytotoxicity of TauRDΔK in N2a cells. The excessive cytotoxicity by Tau was obtained by subtracting cytotoxicity without Tau from cytotoxicity with Tau expression. Note that 3-MA but not NH4Cl aggravates TauRDΔK cytotoxicity.
Next, we determined the effects of 3-MA or NH4Cl on Tau aggregation by immunofluorescence using ThS to label Tau aggregates in N2a cells. Upon treatment with 3-MA, the fraction of ThS positive cells increased almost 5-fold, confirming that macroautophagy plays a major role in preventing accumulation of Tau aggregates (Fig. 1D and E). On the other hand, NH4Cl treatment dramatically inhibited Tau aggregation and ThS staining, consistent with the view that lysosomes are responsible for the generation of the Tau fragments F2 and F3 which in turn are necessary as seeds for PHF aggregation.
We further verified the different effects of 3-MA and NH4Cl on Tau aggregation by examining their influence on cytotoxicity, because we had previously shown that in this cell model the cytotoxicity was correlated with Tau aggregation (37). If 3-MA treatment increases the amount of Tau aggregates due to the inhibition of their degradation through macroautophagy, then higher cytotoxicity should be observed. Accordingly, lower cytotoxicity should be obtained for NH4Cl treatment, assuming that this inhibits the generation of fragments F2 and F3 and thereby prevents Tau aggregation (Fig. 1D, bottom). Expression of TauRDΔK caused a small but significant increase of LDH release after 2 days of expression (Fig. 1F, column 1). NH4Cl treatment slightly decreased cytotoxicity, whereas 3-MA dramatically enhanced the toxicity (Fig. 1F, columns 2 and 3). This result confirms that macroautophagy plays a role in the degradation of the toxic protein aggregates, while inhibition of lysosomal proteases results in the inhibition of Tau aggregation, likely by preventing its limited cleavage into pro-aggregating fragments.
Localization of Tau fragments in N2a cells
To further verify that F2 and F3 are produced by lysosomal proteases, we determined the distribution of these fragments in different subcellular fractions of the N2a cell model under normal conditions or when lysosomal proteolysis is inhibited (treatment with NH4Cl). The enrichment of organelles in these fractions has been previously well characterized (40). The expressed protein TauRDΔK was visible in most fractions except mitochondria (Fig. 2A). However, the combination of TauRDΔK and all fragments F1, F2 and F3 was unique to the lysosomal fraction (lane 5), whereas F1 occurred also in the cytosol and ER fractions (Fig. 2C). The NH4Cl treatment dramatically inhibited the production of F2 and F3 which were no longer detectable in the lysosomal fraction (Fig. 2A, lane 11).These data argue that F2 and F3 can be generated by lysosomal proteases, whereas F1 is generated in the cytosol (see below, Fig. 3).
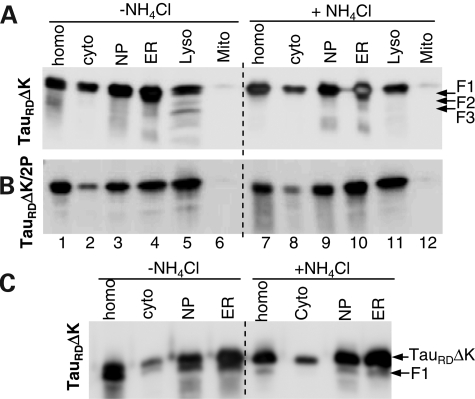
Figure 2.
Distribution of Tau constructs in subcellular fractions of N2a cells. N2a cells induced to express TauRDΔK (A and C) or TauRDΔK/2P (B) were treated without or with NH4Cl for 5 days. The cell homogenates (homo) were fractionated into nuclear pellet (NP), containing nuclei and unbroken cells, cytosol (cyto), ER, mitochondria (Mito) and lysosomes (Lyso). To clearly distinguish the F1 band from the intact TauRDΔK band, the same samples but only half of the amount used in (A) were loaded in (C). Note that fragments F2 and F3 were only detected in the lysosomal fraction from N2a cells expressing TauRDΔK, whereas F1 can be detected in the cytosol or ER fractions.
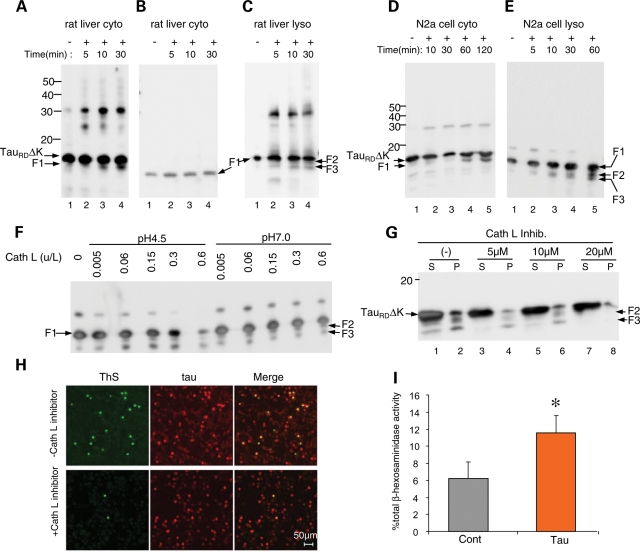
Figure 3.
Cleavage of Tau constructs by cytosol, lysosomal matrix or cathepsin L and release of lysosomal hydrolase β-hexosaminidase into the cytosol of N2a cells caused by expression of TauRDΔK. (A) TauRDΔK incubated with cytosol fraction from rat liver. Note the appearance of fragment F1 (but not F2 or F3) and oligomeric aggregates (mostly dimers). (B) F1 incubated with cytosol fraction from rat liver. Note that there is no cleavage of F1. (C) F1 incubated with lysosomal matrix fraction from rat liver. Note cleavage of F1 to F2 and F3, as well as generation of oligomers (dimers and trimers). (D) TauRDΔK incubated with cytosol fraction from N2a cells. Note appearance of fragment F1 and oligomers. (E) F1 incubated with lysosomal fraction from N2a cells. Note fragmentation to F2 and F3. (F) Digestion of F1 by cathepsin L. Recombinant F1 (0.1 µg/µl) was incubated with 0.005, 0.06, 0.15, 0.3 or 0.6 U/l cathepsin L in 100 mm Na acetate pH 4.5 or 100 mm Tris–HCl pH 7.0 for 60 min. Note that fragments F2 and F3 (arrows) are generated by cathepsin L at both pH 7.0 and pH 4.5. (G) Cathepsin L inhibitor reduces generation of F2 and F3. N2a cells expressing TauRDΔK were treated with cathepsin L inhibitor for 3 days at concentrations up to 20 µm. Lanes labeled P (=pellet) denote sarkosyl-insoluble Tau species, S (=supernatant) indicates soluble proteins. (H) Cathepsin L inhibition also inhibits aggregation of Tau. N2a cells expressing TauRDΔK were treated without or with cathepsin L inhibitor for 3 days. ThS staining reveals that inhibition of cathepsin L reduces Tau aggregation as well. (I) Expression of TauRDΔK causes the release of lysosomal hydrolase β-hexosaminidase. N2a cells expressing TauRDΔK were induced to express Tau for 2 days. The cytosolic distribution of β-hexosaminidase is expressed as the percentage of total β-hexosaminidase activity and is shown as mean ± SEM of four different experiments (*P < 0.05).
As a control, we analyzed an inducible N2a cell line expressing TauRDΔK/2P. This ‘anti-aggregation’ mutant cannot aggregate because each of the two hexapeptide motifs in the repeats R2 and R3 contain an Ile to Pro mutation which prevents β-structure (37). This mutant Tau and its F1 fragment were again found in most fractions including lysosomes (Fig. 2B), indicating that the delivery of Tau to lysosomes does not depend on aggregation. Notably, the anti-aggregation mutant did not generate the F2 and F3 fragments, consistent with the observation that a high β-propensity is required for the cleavage (9).
Cleavage of Tau constructs by cytosol fraction, lysosomal matrix fraction or lysosomal proteases
To further clarify the origin of the proteolytic processing of TauRDΔK, we incubated the protein separately with a cytosol fraction or with a lysosomal matrix fraction (containing the proteases inside lysosomes, but without the lysosomal membrane) prepared from rat liver (41) (Fig. 3A–C) or from our N2a cell model (Fig. 3D and E). We generated recombinant TauRDΔK or F1 fragment, incubated the proteins with the indicated fractions and monitored the time course of cleavage of TauRDΔK to F1 by blotting. Figure 3A and D demonstrates that a protease in the cytosol fractions cleaves TauRDΔK to F1, whereas the F1 fragment undergoes no further cleavage. An analogous time course experiment was performed with lysosomal fractions from liver or the N2a cells, monitoring the fragmentation of F1 to F2 and F3 (Fig. 3C and E). These experiments confirmed that F1 is generated by the cytosolic fraction, whereas F2 and F3 are generated by lysosomal proteases (Fig. 3A and D).
In an attempt to identify the lysosomal proteases responsible for the cleavage of F1, we examined whether recombinant F1 can be cleaved by cathepsin D, B or L. The assays were done both at pH 4.5 (the pH of the lysosomal lumen) and pH 7.0 (pH of cytosol) in order to allow for the possibility that lysosomal proteases might leak into the cytosol (as discussed below). Although cathepsin D has been shown to be able to degrade full-length Tau (26,27) (confirmed here as well), it does not degrade the Tau construct used here at the concentrations employed (data not shown). Cathepsin B also did not digest F1 to F2 or F3 at either pH (data not shown). In contrast, cathepsin L degraded F1 completely and rapidly at acid pH and was effective even at neutral pH, generating fragments F2 and F3 (Fig. 3F). To validate that F1 is indeed cleaved by cathepsin L in N2a cells, we treated N2a cells expressing TauRDΔK with the specific Cathepsin L Inhibitor IV (Sigma) and checked its influence on the generation of F2 and F3. Treatment of the N2a cells with this selective inhibitor also strongly reduced the amount of F2 and F3 in a concentration-dependent fashion (Fig. 3G), arguing that cathepsin L is indeed the protease responsible for the generation of F2 and F3. Consistent with this, the inhibitor also decreased the fraction of ThS positive cells (Fig. 3H), as expected from the decrease of the F2 and F3 fragments that nucleate the aggregation process.
These results confirm thus that the generation of F2 and F3 occurs at the lysosomal compartment and is mainly a result of specific cleavage of Tau by cathepsin L. These observations prompt the question why the Tau constructs are not completely degraded in the lysosomes. An alternative possibility is that fragmentation also occurs by proteases that have leaked out of the lysosomes into the cytosol, considering that cathepsin L can cleave F1 even at neutral pH (Fig. 3F). Such a leakage of lysosomal proteases has been suggested for several cellular conditions before (26,42,43). We therefore asked whether lysosomal leakage was promoted by the expression of TauRDΔK. This was measured by the activity of β-hexosaminidase (a lysosomal hydrolase) in the cytosolic fraction of N2a cells expressing TauRDΔK and of control N2a cells. Indeed, TauRDΔK nearly doubled the amount of β-hexosaminidase activity in the cytosolic fraction, confirming increased lysosomal leakage (Fig. 3I). These results support that part of the F1 cleavage could occur in the cytosol in close proximity to the leaky lysosomes. However, the pronounced inhibitory effect of NH4Cl on the generation of the fragments, still argues that most of the cleavage occurs in the lysosomal lumen as it can be dramatically decreased when the acid lysosomal pH is neutralized (Fig. 1C).
Clearance of Tau by the autophagy–lysosomal system
The results described so far show that inhibition of macroautophagy promotes Tau aggregation (Fig. 1C–E). However, in these experiments, when macroautophagy is inhibited, the cells continue to express Tau. It is therefore not clear, whether Tau aggregation is enhanced because the degradation of aggregates by macroautophagy is suppressed or because newly synthesized Tau is not degraded properly and therefore keeps accumulating and aggregating. To solve this problem, we took advantage of the Tet-On inducible cell model that allows switching off the Tau expression without interfering with protein synthesis [this point is important because inhibitors of protein synthesis such as cycloheximide are known to interfere with autophagy-mediated protein clearance (44,45)].
We first induced the expression of TauRDΔK for 6 days to generate aggregates (Fig. 4A, lanes 1, 2). Then the protein expression was switched off, and cells were maintained untreated or treated for 2 days with 3-MA, or NH4Cl, as labeled. Without inhibitor treatment, the majority of both soluble and aggregated TauRDΔK were removed after 2 days of switching off (Fig. 4A, lanes 3 and 4). Treatment with 3-MA (lanes 5 and 6) or NH4Cl (lanes 7 and 8) dramatically inhibited the degradation of the aggregates so that the level of insoluble Tau protein was raised ∼3-fold over the switch-off control (Fig. 4B). This result indicated that lysosomes contribute to the degradation of Tau aggregates mostly through macroautophagy. 3-MA and NH4Cl treatment also retarded the degradation of soluble Tau constructs, but this effect was not as strong as that on the aggregates. The observed effect of 3-MA on the levels of F2 and F3 supports that once in the aggregate fraction these products can be removed by macroautophagy and provides an explanation to the increased levels of F2 and F3 observed in Figure 1C. Thus, the higher levels of F2 and F3 did not resulted from enhanced processing but from decreased clearance by macroautophagy.
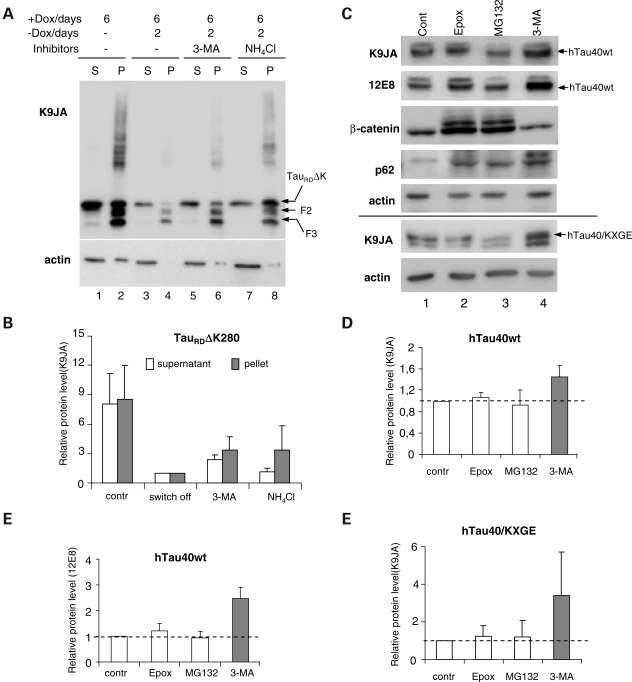
Figure 4.
Degradation of Tau by macroautophagy. (A) Blot analysis of effects of inhibiting macroautophagy or lysosomes on Tau clearance. N2a cells expressed TauRDΔK for 6 days, then expression was switched off and cells were treated with or without inhibitors for 2 days. Equal amounts of protein were loaded and checked against actin (A, lower panel). Lanes 1 and 2 show that expression of TauRDΔK generates aggregates in the pellet, fragmentation and a higher molecular weight smear. Stopping expression allows cells to clear most soluble TauRDΔK and aggregates (lane 4), but clearance is impaired if autophagy (3-MA, lane 6) or lysosomal proteases (NH4Cl, lane 8) are inhibited. (B) Quantification of effect of 3-MA and NH4Cl on TauRDΔK280 clearance. (C) Blot analysis of effects of inhibiting proteasome, macroautophagy or lysosomes on hTau40wt or hTau40/KXGE clearance. N2a cells expressed stably hTau40wt or transiently hTau40/KXGE were induced to express Tau for 2 days, then expression was switched off and cells were treated for 1 day with proteasome inhibitors (MG132 or epoxomicin) or 3-MA, or without inhibitor (control). Total Tau and Tau phosphorylated in the repeat domain(KXGS motifs) were determined with antibodies K9JA and 12E8. The level of β-catenin or p62 was measured to check the extent of the inhibition of the proteasomal or macroautophagy system. (D–F) Quantification of the effect of inhibitors on the clearance of hTau40wt (45% increase with 3-MA) (D), phospho-Tau (140% increase) (E) or hTau40/KXGE (240% increase) (F). Values are expressed as relative protein amount compared with the control (mean ± SE of three experiments). Inhibiting the proteasome has a minor effect, whereas inhibiting macroautophagy or lysosomes causes build-up of Tau. Note that Tau phosphorylated at KXGS motifs is a preferred target for macroautophagy (E, column 4).
Next, we examined whether full-length soluble wild-type Tau can also be degraded by macroautophagy. Since there is a debate on the role of proteasome in the degradation of Tau (13,15,16,18), we initially tested the effects of proteasome inhibitors such as MG132 and epoxomycin, both of which had only a small effect on the level of Tau (Fig. 4C, top, lanes 2 and 3). In contrast, autophagy inhibitor 3-MA increased the level of hTau40wt (45% increase, Fig. 4C, top, lanes 4 and D, column 4), demonstrating that the degradation of full-length Tau also occurred by the lysosomal system. β-Catenin, a well-characterized proteasome substrate, is shown as positive control in Figure 4C. Degradation of full-length wild-type Tau via macroautophagy was considerably slower than of p62, a protein known to undergo degradation both by the proteasome and macroautophagy (Fig. 4C). Since recent reports demonstrated that Tau phosphorylated at the KXGS motifs of the repeat domain is less susceptible to proteasomal degradation mediated by Hsp90 (20,46), we suspected that this kind of phosphorylated Tau may be preferentially degraded via the lysosome. In N2a cells, part of the expressed Tau is phosphorylated at the KXGS motifs of the repeats [presumably by the kinase MARK, 37)]. Thus we checked the effects of inhibitors on the degradation of KXGS-phosphorylated full-length Tau, using an antibody that selectively recognizes phosphorylated KXGS motifs in Tau (12E8). Again, proteasomal inhibitors had little effect on the level of KXGS-phosphorylated protein (Fig. 4C, second panel, lanes 2–3 and E, columns 2–3). In contrast, 3-MA increased the level of KXGS-phosphorylated Tau even to a higher extent than for unmodified Tau (150% increase, Fig. 4C, second panel, lane 4 and E, column 4), indicating that macroautophagy also plays a major role in the degradation of the KXGS-phosphorylated protein. Furthermore, we made a Tau construct hTau40/KXGE in which all four serines of the KXGS motifs in the repeat domain were mutated to glutamic acid (E) in order to mimic Tau phosphorylation at these sites. We checked the influence of both the proteasomal and the lysosomal system on the degradation of this pseudophosphorylated Tau and found that only macroautophagy inhibition caused a dramatic elevation of the protein level (240%, Fig. 4C, panel 8, lane 4 and F, column 4). This result strengthens our observation that Tau phosphorylated at the repeat domains is degraded by macroautophagy.
Mutant Tau uses CMA in an unconventional way
As discussed above, the general inhibition of lysosomes by NH4Cl disrupted the generation of fragments F2 and F3 and thus inhibited aggregation of TauRDΔK, whereas inhibition of macroautophagy (by 3-MA) increased F2 and F3 as well as soluble and aggregated TauRDΔK by inhibiting degradation. These data suggested that F1 can reach the lysosomal compartment by a pathway different from macroautophagy in order to undergo cleavage. We asked whether another form of autophagy, CMA, was the mechanism for delivery of Tau into the lysosomal lumen responsible for the generation of the pathogenic fragments. CMA has been shown to contribute to the degradation of α-synuclein, the protein that accumulates in Lewy bodies in brains of Parkinson disease patients (47,48). We noted that the Tau sequence contains two putative CMA-targeting motifs (33). These are 336QVEVK340 and 347KDRVQ351, both in repeat R4 just upstream of the C-terminal end of F2 or F3 (Fig. 1A). To determine whether Tau could be translocated into the lysosomal lumen via CMA, we used a previously optimized in vitro assay with intact isolated lysosomes that allows measuring binding, uptake and degradation via CMA (41,49,50). An essential requirement to assess direct translocation is that the integrity of the lysosomal membrane is preserved during the incubation, as leakage of lysosomal enzymes will lead to degradation of the substrates outside lysosomes. This was a major concern in these studies in light of the increased levels of lysosomal enzymes observed in cells expressing the pathogenic forms of Tau (Fig. 3I). We first examined the effects of Tau constructs on the stability of the lysosomal membranes by monitoring the release of β-hexosaminidase in isolated lysosomes incubated directly with the different Tau proteins (51). As illustrated in Supplementary Material, Figure S2, under the incubation conditions of the assay, none of the Tau constructs caused an increase of enzyme activity outside lysosomes above that of the control, indicating that lysosomal membranes are not directly perturbed by the Tau constructs. These results are clearly in contrast to the increased leakage observed in the culture cells expressing TauRDΔK, thus suggesting that the disruptive effect of the pathogenic products on the lysosomal membrane may happen when they are present inside the lysosomal lumen but not when presented to the cytosolic side of the membrane.
Because the integrity of the lysosomal membrane was comparable to lysosomes incubated alone, we were able to use these assays to test the binding and uptake of Tau by CMA into lysosomes. We found that all Tau constructs associated with lysosomes and that their association was enhanced when lysosomal proteolysis was blocked (Fig. 5A). Lysosomal association of full-length wild-type and mutant Tau was comparable to that observed with other CMA substrates (ribonuclease A is shown here), whereas truncated TauRDΔK and its F1 fragment displayed significantly higher association (Fig. 5B). Interestingly, incubation of these two forms of Tau with lysosomes lead to the formation of oligomers of the protein (well-resolved dimers and trimers when lysosomal proteases were inhibited and additional intermediate oligomeric forms in the absence of protease inhibitors) (Fig. 5A, panels 3 and 4), indicating that the interaction of these truncated forms of Tau with the lysosomal membrane promotes their oligomerization. This is consistent with the well-known tendency of Tau to self-associate on negatively charged surfaces [e.g. arachidonic acid micelles, carboxylated beads and even microtubules (52–55)].
Figure 5.
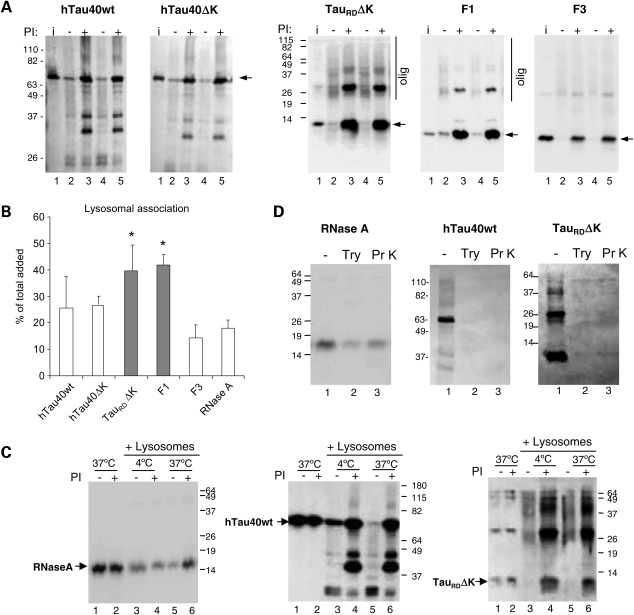
Association of different forms of Tau with isolated lysosomes. (A) Association of the indicated forms of Tau (2 µg) with isolated lysosomes untreated or pre-treated with protease inhibitors (PI). Lanes 2–3, 4–5 show two independent incubations. Input (lane 1): 1/10 of total protein added to lysosomes. Arrows denote intact Tau constructs. Higher molecular weight forms of Tau (olig) can be detected for TauRDΔK and to a lesser extent for F1 (panels 3 and 4,). For hTau40wt and hTau40ΔK, with protease inhibitors, more intact Tau was detected (panels 1 and 2, lanes 3, 5). (B) Quantification of the amount of the indicated forms of Tau associated to lysosomes when incubated under the same conditions. Values are expressed as percentage of the total protein added to the reaction and are mean ± SEM of six different experiments (*P < 0.05). Binding of ribonuclease A (RNase A), under the same conditions is shown as reference. (C) Tau fails to translocate across the lysosomal membrane. Intact rat liver lysosomes treated or not with protease inhibitors were incubated with RNase A and the indicated forms of Tau at 4°C (lanes 3, 4) or 37°C (lanes 5, 6). Increase association at 37°C in the presence of protease inhibitors (indicative of lysosomal uptake) only occurred for RNase A. The similarity of Tau patterns at both temperatures shows that there is no uptake of Tau by CMA. Lanes 1 and 2 show the proteins incubated alone (1/10 of the total protein was added into the gel). (D) Lysosomal Tau is associated to the cytosolic side of the lysosomal membrane as determined by protease protection assay. Lysosomes previously treated with lysosomal protease inhibitors and incubated with the indicated proteins were recovered by centrifugation and resuspended in medium alone (−) or supplemented with trypsin (Try) or proteinase K (Pr K). Note that Tau is on the cytosolic side of the lysosomes because it is completely digested after protease treatment, whereas the fraction of RNase A translocated into lysosomes becomes protected from the exogenous proteases.
According to the usual interpretation of in vitro uptake assay, the fact that inhibition of lysosomal proteolysis increased the amount of Tau associated to lysosomes suggested that part of these Tau proteins was translocated into lysosomes. However, as an independent test for Tau uptake across the lysosomal membrane, we compared the association of Tau constructs to lysosomes at 4 and 37°C. This assay is based on the fact that the binding of protein to lysosomal membranes takes place even at low temperatures, whereas the translocation of protein across the membrane is temperature-dependent and it will not take place below 20°C (56). As shown in Figure 5C, in contrast to the well-characterized CMA substrate RNase A for which inhibition of lysosomal proteases only increased the amount associated to lysosomes (uptake) at 37°C (Fig. 5C left panel compare lanes 4 and 6), the pattern of the association of all Tau constructs to lysosomes (full size and TauRDΔK shown here) was very similar at both temperatures, supporting the view that they were not being translocated into the lysosomal lumen, but that they bind with very high affinity to the membrane. Oligomeric forms of TauRDΔK were found associated with lysosomes even at low temperatures, further supporting their formation at the lysosomal membrane.
Finally, we confirmed that the association of Tau occurred only on the cytosolic side of the lysosomal membrane. This was done by analyzing the resistance of Tau associated with lysosomes to exogenously added proteases, reasoning that if Tau were bound to the external surface it would be accessible for digestion. As shown in Figure 5D, proteinase K or trypsin rapidly degraded Tau associated with lysosomes, showing that Tau proteins were not protected by the lysosomal membrane, whereas the same treatment in lysosomes incubated with RNase A revealed the presence of a portion of the protein protected from the exogenous protease and hence located in the lysosomal lumen (Fig. 5D, left panel). Taken together, these results demonstrate that although Tau binds to lysosomes, there is no detectable uptake of any of the Tau proteins analyzed in this study across the lysosomal membrane.
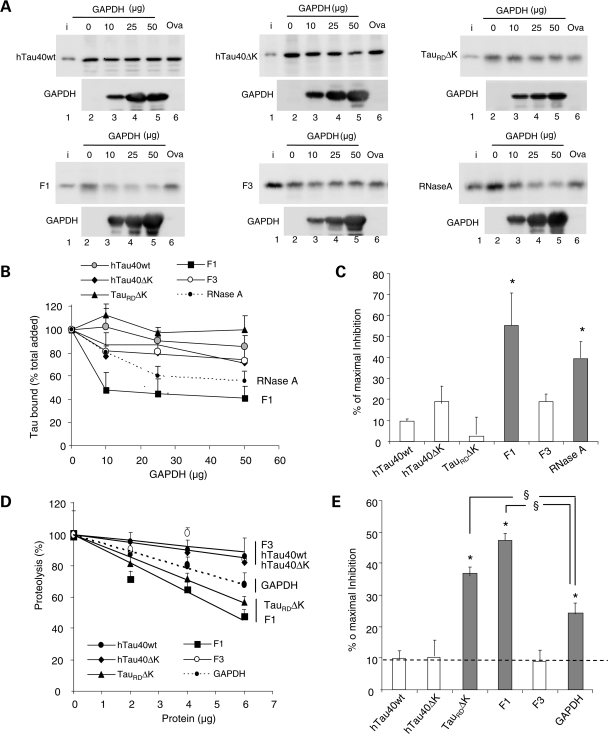
To gain further insight into the nature of the association of the different forms of Tau with the lysosomal membrane and whether binding was taking place through components shared with CMA, we performed a competition assay with a well-characterized CMA substrate [glyceraldehyde-3-phosphate-dehydrogenase (GAPDH)] and a non-CMA substrate (ovalbumin). Neither GAPDH nor ovalbumin made a significant difference to the level of the different forms of Tau associated with lysosomes, except for the F1 fragment, which could be efficiently competed by the CMA substrate but not by ovalbumin (Fig. 6A–C). The lack of competition by GAPDH implies that full-length Tau does not use CMA components to bind to lysosomes or that the affinity of Tau for the CMA components was highly superior to that of the standard CMA substrates. To discriminate between these two possibilities, we checked whether the different forms of Tau that could not be competed with CMA substrates, had an effect on uptake of other substrates by CMA. As shown in Figure 6D and E, whereas full-length Tau proteins and the F3 fragment did not have a significant effect on the degradation of a pool of CMA substrates by lysosomes, both TauRDΔK and its F1 fragment inhibited the degradation of CMA substrates, and they did so to an extent significantly higher than other well-characterized CMA substrate proteins (GAPDH shown here; Fig. 6D and E). These results are in agreement with the higher binding to lysosomes observed for TauRDΔK and F1 (Fig. 5B). On the basis of the lack of inhibitory effect of full-size Tau proteins on uptake of CMA substrates and the inability of CMA substrates to compete with Tau binding, we conclude that association of full-size Tau with lysosomes, at least in this system, is independent of CMA. In contrast, mutant TauRDΔK and its F1 fragment exert an inhibitory effect on CMA as a result of their high affinity for the CMA machinery. Further studies are required to clarify why—despite the fact that CMA substrates can efficiently compete lysosomal binding of F1 (Fig. 6A, bottom left panel)—this fragment exerts such pronounced inhibitory effect on CMA (Fig. 6D and E). In this respect, we observed that whereas CMA substrates such as GAPDH can still compete lysosomal binding of F1 in its monomeric state, the effect of GAPDH on the oligomers of F1 bound to the lysosomal membrane was negligible (Supplementary Material, Fig. S3). This result suggests that the oligomers of the truncated F1 Tau protein detected at the lysosomal membrane are likely the main cause for the inhibitory effect of the Tau fragments on CMA.
Figure 6.
Competition of the lysosomal association of different forms of Tau by substrates of CMA. (A) Effects of CMA substrate (GAPDH) and CMA non-substrate (ovalbumin) on binding and uptake of Tau by lysosomes. The different forms of Tau proteins (2 µg) and the indicated increasing concentrations of GAPDH or ovalbumin (Oval) were incubated with intact isolated lysosomes previously treated with protease inhibitors. Lane 1 shows 1/10 of the input (i) and insets show the amount of GAPDH associated with the lysosomal membranes. Only association of the F1 fragment and of RNase A (shown here as a positive control) could be competed by increasing concentrations of GAPDH, whereas ovalbumin did not modify lysosomal binding. (B) The percentage of the different Tau proteins and of RNase A bound to the lysosomal membrane in the presence of increasing concentrations of GAPDH was quantified in four experiments similar to the ones shown in (A). Values are mean ± SEM. (C) The maximal inhibition on Tau binding attained by GAPDH was calculated in four experiments similar to the ones shown in (A). Values are mean ± SEM (*P < 0.05). (D and E) Effect of different Tau proteins or GAPDH on the degradation of a pool of CMA substrates by intact lysosomes. Values expressed as % of proteolysis in the absence of Tau (D) or % of maximal inhibition of the degradation of the pool of proteins (E). Values are mean ± SEM of two experiments with triplicate samples. (*P < 0.05 compared with wild-type full-size Tau and §P < 0.05 compared with the inhibitory effect of GAPDH).
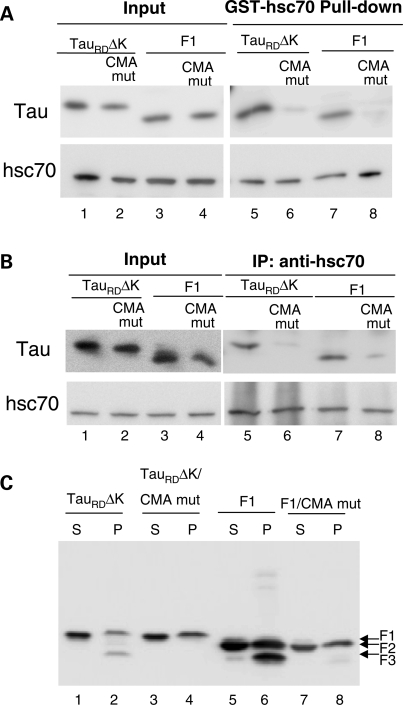
To demonstrate that association of TauRDΔK and F1 with lysosomes depends on components of the CMA machinery, we examined whether these Tau constructs were able to interact with hsc70, the chaperone reported to mediate delivery of substrates to the CMA receptor. When TauRDΔK or F1 were incubated with GST–hsc70, these Tau constructs were pulled down by GSH-beads (Fig. 7A, lanes 5 and 7), consistent with the report that hsc70 interacts with full-length Tau (57). To determine whether the hsc70/Tau interaction occurred at the CMA-targeting motifs identified in the sequence of Tau, we then mutated these motifs 336QVEVK340 and 347KDRVQ351 by replacing 336QV337 or 350VQ351 with AA residues. Mutation of these residues almost completely abolished the interaction of TauRDΔK and F1 with hsc70 (Fig. 7A, lanes 6 and 8), suggesting that these CMA motifs are important for the interaction with hsc70. We also confirmed binding of hsc70 to Tau through those domains in intact cells. The interaction of TauRDΔK and F1 constructs with endogenous hsc70 analyzed in N2a cells by immunoprecipitation, also revealed a marked reduction in Tau binding with hsc70 when the CMA motifs were no longer present (Fig. 7B). We further analyzed the direct influence of the mutation of CMA motifs on the fragmentation and degradation of these Tau constructs in N2a cells and found that the mutations of CMA motifs in TauRDΔK strongly inhibited the generation of F2 and F3, as nearly no F2 and F3 were detected in the sarkosyl pellet (Fig. 7C, lane 4). This result indicates that binding to hsc70 is essential for TauRDΔK to be targeted to lysosomes and consequently undergo cleavage. Interestingly, in contrast to the complete blockage of cleavage in mutant TauRDΔK, the mutations of CMA motifs in F1 block the generation of F3 but not F2 (Fig. 7C, lane 8), suggesting that binding to hsc70 is not necessary for F1 to be targeted to lysosomes for fragmentation. The failure of generation of F3 may be a result of the conformational changes caused by these CMA motif mutations, as we know that the cleavage of F2 to F3 is dependent on the β-structure and the disruption of the β-structure with Proline mutations could block this cleavage (9). However, it is also possible that although not necessary for targeting, the CMA motifs could be needed for interaction with the hsc70 present in lysosomes (lys-hsc70) and that this binding is required for the cleavage of F2 to F3.
Figure 7.
Effects of mutation of CMA motifs on the interaction of Tau constructs with hsc70 and Tau degradation. (A) Interaction of recombinant TauRDΔK, TauRDΔK CMA mut (CMA recognition motifs mutated), F1 or F1 CMA mut with GST-hsc70 in vitro. Note that there is an interaction between hsc70 and TauRDΔK or F1 in vitro (lanes 5, 7) and the CMA motif mutations reduced this interaction (lanes 6, 8). (B) Interaction of TauRDΔK, TauRDΔK CMA mut, F1 or F1 CMA mut with endogenous hsc70 in N2a cells. There is a strong interaction between hsc70 and TauRDΔK as well as F1 in cells (lanes 5, 7), but reduced interaction between hsc70 and the Tau mutants (lanes 6, 8). (C) Mutation of the CMA motifs blocks TauRDΔK280 cleavage and the generation of F3 from F1. S and P denote supernatant and pellet of sarkosyl extraction, respectively. Note the absence of F3 in the pellet in lanes 4 and 8.
To directly address the effect of the CMA mutations on the interaction of Tau with lysosomes, we incubated the modified proteins with intact lysosomes previously treated or not with protease inhibitors. Mutation of the CMA motifs in TauRDΔK drastically reduced the amount of protein associated to lysosomes both in the presence and in the absence of protease inhibitors (Fig. 8A and B). Levels of Tau oligomers at the lysosomal membrane were also reduced (Fig. 8A, left compare lanes 1–6 and 8–13), supporting that their formation was dependent on their delivery to lysosomes through CMA components. In contrast, mutation of the CMA motif did not have a significant effect on the association of F1 to the lysosomal membrane or the formation of F1 oligomers in lysosomes with preserved proteolytic activity (Fig. 8A, right compare lanes 1–3 and 8–10 and B, left), but it did markedly reduce the amount of protein associated to the membrane when lysosomal proteolysis was inhibited (Fig. 8A and B right). This result indicates that binding of F1 to the lysosomal membrane is independent of the interaction of hsc70 with the CMA targeting motif in this protein, but binding to hsc70 may be required to retain the protein at lysosomes until cleavage occurs.
Figure 8.
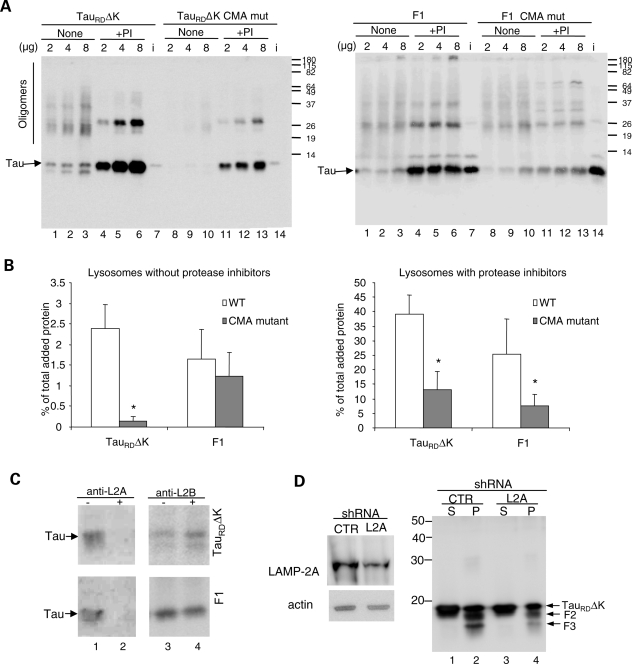
Effect of inhibition of CMA components on CMA uptake of Tau proteins. (A) Lysosomal association of TauRDΔK, F1 and its CMA mutants. Increasing concentrations of Tau proteins (as indicated) were incubated with freshly isolated intact lysosomes treated (+) or not (none) with protease inhibitors (PI). i=input (0.2 µg). (B) Quantification of lysosomal binding of the Tau proteins to untreated lysosomes (left) or lysosomes treated with protease inhibitors (right) calculated from three experiments as the ones shown in (A). Values are expressed as percentage of the total protein added and are given as mean ± SEM (*P < 0.05). (C) Effect of antibody blockage of lysosomal CMA components on the association of Tau proteins to lysosomes. The indicated Tau proteins were incubated with freshly isolated intact lysosomes pre-incubated or not with antibodies against LAMP-2A (anti-L2A) and LAMP-2B (anti-L2B). (D) Effects of knockdown of LAMP-2A on TauRDΔK280 fragmentation and aggregation. N2a cells infected with LAMP-2A (L2A) shRNA or control shRNA (CTR) lentivirus for 20 days were induced to express TauRDΔK280 for 3 days. Then sarkosyl extraction was used to separate soluble and insoluble Tau. Right panel, S and P denote sarkosyl supernatant and pellet, respectively. Left panel shows efficiency of knock-down for LAMP-2A. Actin was used as a loading control. Note: LAMP-2A shRNA knocks down the expression of LAMP-2A and reduces the generation of F2 and F3.
We then analyzed the dependence of lysosomal binding of TauRDΔK and F1 on the other main component of CMA at the lysosomal membrane, the receptor LAMP-2A. To this end, we blocked the cytosolic tail of LAMP-2A, where CMA substrates have been shown to bind, with an antibody against this region, and compared the binding levels for each of the Tau variants. As shown in Figure 8C lysosomal binding of TauRDΔK and F1 mainly takes place through LAMP-2A as it can be drastically reduced by antibodies against this protein (Fig. 8C, compare samples in lanes 1 and 2), whereas blockage of the cytosolic tail of a different LAMP-2 variant, LAMP-2B, did not have any effect (Fig. 8C, compare samples in lanes 3 and 4). In summary, our results support that TauRDΔK is targeted to lysosomes via CMA (as it interacts with both hsc70 and LAMP-2A), whereas the F1 fragment does not require the interaction with cytosolic hsc70 for lysosomal targeting, but it still binds to the CMA receptor at the lysosomal membrane.
To demonstrate the dependence on CMA of the lysosomal delivery of Tau that results in its limited cleavage, we blocked CMA in cultured N2a cells using siRNA against LAMP-2A (Fig. 8D, left panel) as described before (58). As shown in Figure 8D (right panel), inhibition of CMA resulted in a marked decrease in the generation of F2 and F3 fragments (compare lanes 2 and 4). These results confirm the requirement for binding of Tau to LAMP-2A in order to undergo lysosomal cleavage.
Taken together, our results demonstrated that mutant Tau is delivered and binds to lysosomes via CMA components, but, in contrast to conventional CMA substrates, there is no detectable uptake of Tau proteins across the lysosomal membrane. The fact that even the cleaved products remain associated to the membrane suggests that cleavage occurs in proteins tightly bound to the membrane, more likely through LAMP-2A or partially translocated. Because our data support that F2 and F3 cleavage is mediated by cathepsin L, a protease located in the lumen (Fig. 3F), and that this cleavage is still susceptible to changes in the lysosomal pH (Fig. 1C), we propose that Tau inserts into the lysosomal membrane with its C terminus first, likely through the CMA translocation complex. Once the C terminus reaches the lumen, cathepsin L cleaves F1 to form the F2 and F3 fragments that initially remain associated to the lysosomal membrane (see model in Fig. 9 and details in Supplementary Material, Fig. S4). The presence of these partially translocated products at the lysosomal membrane can thus explain their inhibitory effect on the binding/translocation of other CMA substrates. This two-step translocation pathway is reminiscent of other CMA substrates such as ribonuclease A (59).
Figure 9.
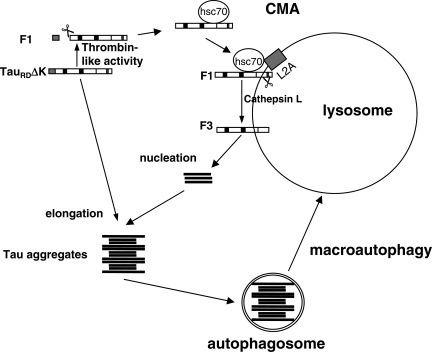
Model of fragmentation and degradation of TauRDΔK by autophagy–lysosomal system. When TauRDΔK is expressed in N2a cells, it becomes partially cleaved in the cytosol by a thrombin-like activity to generate F1. A fraction of F1 is delivered to lysosomes via CMA components (targeting through hsc70 and binding to the lysosomal membrane via LAMP-2A) where the C-terminal end is cleaved by cathepsin L to generate F2 and F3. These fragments are highly amyloidogenic, nucleate aggregation and induce the co-aggregation of intact TauRDΔK (or full-length Tau) in the cytosol. The aggregates can then be degraded through the macroautophagy pathway.
DISCUSSION
The ubiquitin–proteasome system and the autophagy–lysosome system are the two major cellular protein degradation pathways. Up to now, the degradation of Tau by the ubiquitin–proteasome system has been investigated by several authors (13,14,16,17,20), whereas the involvement of the autophagy–lysosome system has received less attention (23,31,38). Here, we demonstrated that both soluble Tau and Tau aggregates can be degraded by the autophagy–lysosomal system in a cell model of tauopathy expressing the pro-aggregation mutant TauRDΔK280. Inhibition of macroautophagy led to enhanced Tau aggregation and cytotoxicity, as seen by a strong increase of thioflavin S-stained cells, implying that macroautophagy plays an important role in preventing Tau aggregation. In agreement with this scenario, it has been reported that disruption of autophagy led to the accumulation of abnormal proteins in neurons (60,61). In AD brain, the autophagy–lysosomal system is disturbed, which may promote the accumulation of β-amyloid (24,62). Altogether, the data indicate that the autophagy–lysosomal pathway is involved in the pathogenesis of the two hallmarks of AD, amyloid plaques and neurofibrillary tangles. In addition, the presence in the sequence of Tau of two targeting motifs for CMA (336QVEVK340 and 347KDRVQ351), a form of autophagy selective for soluble cytosolic proteins, and the recently described association of Tau with hsc70 (19), the cytosolic chaperone responsible for targeting of the CMA substrates to lysosomes, prompted us to investigate its possible contribution to Tau clearance. Although full-size wild-type and mutant were able to associate to lysosomes, at least in our experimental model, association was independent of CMA, since binding of these proteins to isolated lysosomes did not modify or was modified by binding of CMA substrates (Fig. 6). In addition, binding of full-size Tau did not result in its translocation into the lysosomal lumen (Fig. 5C and D), thus supporting that full-size Tau proteins are not normally amenable to degradation via CMA. In contrast, the truncated mutant protein and its F1 fragment behaved in the binding and competition assays as bonafide CMA substrates (Fig. 6). To our surprise, our in vitro data revealed that there was no complete translocation of truncated mutant Tau into the lysosomal lumen for degradation, even though the targeting to the lysosomal membrane and association to the CMA translocating machinery (LAMP-2A and lys-hsc70) occurs. Instead, the inserted protein seems to undergo regulated cleavage by cathepsin L, a lumenal resident protein that results in the generation of the pathogenic F2 and F3 fragments on the surface of the lysosomes. The partial translocation of a protein via CMA observed here is reminiscent of the blockage in lysosomal translocation for other pathogenic proteins, e.g. mutant α-synucleins. In that case, the mutant proteins are also delivered to lysosomes via CMA but they do not translocate into the lysosomal lumen resulting in CMA blockage, even though they bind with high affinity to the lysosomal membrane (47,48). In the case of Tau, the additional feature is that the lysosome-mediated cleavage favors formation of oligomeric structures directly at the lysosomal surface (Figs 5 and 8).
Using a cell model overexpressing the wild-type full-length four-repeat Tau, we further showed that even the full-length Tau can be degraded by the autophagy–lysosomal system. In support of this, it was recently reported that the activation of macroautophagy with rapamycin led to the degradation of SDS-insoluble Tau formed by full-length Tau in COS-7 cells, whereas basal macroautophagy (without rapamycin) could not degrade Tau (31). This may be due to the difference of cell lines used in the two studies. Indeed, to highlight the influence of different cell lines on Tau aggregation, we observed that there was no fragmentation and hence no aggregation of TauRDΔK280 in CHO cells (data not shown). Consistent with our data supporting macroautophagic degradation of Tau, Hamano et al. (32) recently reported that macroautophagy is involved in the clearance of Tau in another cell model.
Several groups recently reported that Tau hyperphosphorylated at the SP/TP motifs in the flanking domains of the repeats can be ubiquitinated and then targeted for proteasomal degradation by the CHIP/hsc70 complex (18,19). However, Tau phosphorylated at the KXGS motifs of the repeats does not undergo this pathway, and thus it remained unclear how degradation of this protein takes place. Interestingly, we found here that macroautophagy seems to preferentially degrade Tau phosphorylated at the KXGS motifs, as the increase in the amount of hTau40wt phosphorylated at the KXGS sites (150%, Fig. 4E) or the phosphomimetic mutant (hTau/KXGE, 240%, Fig. 4F) is much higher than that of the total hTau40wt level (45%, checked with pan-Tau antibody K9JA, Fig. 4D) after blockage of macroautophagy. Thus, depending on the state of phosphorylation, Tau can be degraded by both proteasomal and autophagy–lysosomal systems. Similarly, α-synuclein was recently reported to be degraded by both proteasomal and autophagy–lysosomal pathways too (63,64). This is actually a common characteristic to a large pool of intracellular proteins that could undergo degradation by different pathways simultaneously depending on the cell type or even in the same cell depending on the cellular conditions. Additionally, this selective degradation could also be one of the reasons why the inhibition of degradation of p62 by 3-MA is more pronounced than that for hTau40wt, because p62 is selectively removed through its interaction with LC3, whereas hTau40wt may be part of an ‘in bulk’ macroautophagic degradation. Besides this, the increase of p62 level might also be due to the fact that synthesis of p62 is continued during 3-MA treatment, whereas the Tau expression was switched off.
The interaction of Tau with cytosolic hsc70 is complex and seems to occur at different sites and, likely for different purposes. In this work, we have found that interaction of Tau with hsc70 through the CMA targeting motif is required for lysosomal targeting and cleavage of TauRDΔK280 (Figs 7C and 8A). In contrast, although F1 also binds to hsc70 through that region both in vitro and in intact cells (Fig. 7), targeting of F1 to lysosomes occurs even when this fragment cannot interact with hsc70 [as reflected by the fact that the F1 CMA-mutant still undergoes cleavage into F2 (Fig. 7C lane 8) and can bind to isolated lysosomes (Fig. 7A, right panel)]. It is possible that binding of hsc70 at the CMA-targeting motif to TauRDΔK280 facilitates its cleavage by the cytosolic protease to F1 and the subsequent delivery to the lysosomal membrane. Interestingly, even though F1 does not depend on hsc70 binding for lysosomal delivery, its binding to the lysosomal membrane is still in part selective toward CMA components, since blockage of the lysosomal CMA receptor LAMP-2A diminished F1 binding (Fig. 8C, lower panel, compare lanes 1 with the LAMP-2A blocked lane 2). Although further studies are required to elucidate the exact role of hsc70 in cleavage of F1 in lysosomes, the fact that lower amounts of F1 lacking the hsc70 binding motif remain bound to the membrane when its lysosomal cleavage is prevented (by adding lysosome protease inhibitors; Fig. 8A, right panel), supports a role for hsc70 binding in retaining the protein bound to the membrane until cleavage takes place. The aggregation of TauRDΔK280 in our N2a cell model is nucleated by small fragments F2 or F3 which are derived from F1. In generating F1 from TauRDΔK280 a protease with thrombin-like characteristics is involved which cleaves TauRDΔK280 behind K257. However, the protease(s) responsible for the generation of F2 and F3 around I360 were still unknown (10). Now, we show here that a lysosomal protease, cathepsin L, is involved in the fragmentation of F1 to generate F2 and F3. Treatment with NH4Cl, which induces a sustained rise in the lysosomal pH but not the cytosolic pH (65), dramatically inhibits the generation of F2 and F3 and thus suggests that the fragmentation takes place in lysosomes. However, we also observed lysosomal leakage in our cell model. Therefore, although F2 and F3 are produced in the lysosomes, in the long run they might induce leakage and thus get access to the cytosol where they induce the aggregation of TauRDΔK280. In line with our observation, it has been reported that lysosomal proteases are involved in the generation of N-terminal huntingtin fragments which can seed huntingtin aggregation in the cytosol (66,67) and that lysosomal proteases cause the truncation of the scrapie-associated form of PrP which may accumulate in lysosomes (68). Thus the disturbance of the autophagy–lysosomal system, which occurs in AD brain (24), may not only inhibit the degradation of Tau aggregates, but also accelerate Tau aggregation through fragmentation of Tau.
Figure 9 depicts a model of how the autophagy–lysosomal system might generate aggregates in our cell model (also see a detailed model in Supplementary Material, Fig. S4). When TauRDΔK is expressed, it is first cleaved by a thrombin-like activity in the cytosol to generate fragment F1, and this process seems facilitated by the interaction of TauRDΔK280 with cytosolic hsc70. This cleaved product is delivered to lysosomes via CMA, where its partial translocation allows cathepsin L-mediated cleavage of the C terminus of F1 to generate F2 and F3. Part of the truncated products remain initially associated to the lysosomal membrane favoring oligomerization at or near the surface of the organelle and eventually resulting in membrane disruption, lysosomal leakage and release of other F2 and F3 fragments into the cytosol where they induce TauRDΔK280 aggregation. The aggregates are then degraded through the macroautophagy pathway. Thus, the lysosome serves two seemingly contradictory functions. It promotes aggregation by generating amyloidogenic fragments through CMA, and it disposes of aggregates via macroautophagy.
MATERIALS AND METHODS
Cell culture
Inducible Tet-On, G418-resistant N2a cell lines were generated as described (37). They expressed either the repeat domain construct TauRDΔK280 (high aggregation propensity, ‘pro-aggregation’ mutant) or TauRD with mutations ΔK280, I277P and I308P (TauRDΔK280/2P) (low aggregation propensity, ‘anti-aggregation’ mutant). Transient transfection of N2a Tet-On cells with hTau40/KXGE was done with Effectene transfection reagent (Qiagen, Germany). Tet-On inducible cells were cultured in Eagle's MEM with 10% fetal calf serum (2 mm glutamine, 0.1% non-essential amino acids and 600 µg/ml G418). Expression of Tau constructs was induced by 1 µg/ml doxycycline. For all inhibitor treatment experiments, the medium was changed daily.
Proteins and biochemical assays
Cloning and expression of all Tau constructs was done as described previously (10). The mutagenesis of Tau constructs with mutated CMA motifs was performed according to standard protocols. For solubility assays, cells were collected by centrifugation at 1000 g for 5 min. The levels and solubility of Tau protein was determined by sarkosyl extraction (69) (for details see Supplementary Material). The sarkosyl-insoluble pellets were resuspended in 50 mm Tris–HCl (pH 7.4), 0.5 ml/1 g of starting material. Supernatant and sarkosyl-insoluble pellet samples were analyzed by western blotting. The amount of material loaded for supernatant and sarkosyl-insoluble pellet represented ∼0.5 and 15% of the total material present in the supernatant and pellet, respectively (the ratio between supernatant and sarkosyl-insoluble pellet applied to the gels was always 1:30). For quantification of Tau levels, the western blots were probed with pan-Tau antibody K9JA (DAKO, Glostrup, Denmark) or phosphorylation-dependent antibody 12E8 (gift from P. Seubert, Elan Pharma) which recognizes Tau phosphorylated at Ser 262 and Ser 356 in the repeat domain, and analyzed by densitometry.
Immunofluorescence
Mouse neuroblastoma N2a cells on the coverslips were fixed with 4% paraformaldehyde in PBS for 15 min, then permeabilized with 80% MeOH for 6 min at −20°C. Thioflavin S (ThS) staining was done as described before (37).To determine the colocalization of Tau with LC3, samples were incubated with antibody anti-LC3(B) and KBTR4 in 5% goat serum (PBS) at 4°C overnight. The secondary antibody was also diluted with 5% goat serum in PBS and incubated for 45 min. The cells were washed twice with PBS, once with water and mounted. Confocal microscopy was done with an LSM510 microscope (Zeiss, Oberkochen, Germany) using an ×63 objective.
Cytotoxicity assay
Cytotoxicity was assessed by a lactate dehydrogenase (LDH) assay kit (Roche Applied Science, Indianapolis, IN, USA). LDH activity was measured spectrophotometrically at 492 nm. Cell death was calculated as percent of LDH released into medium, compared with total LDH obtained after total cell lysis. After 1 day of doxycycline-induced protein expression, the medium with 10% serum was exchanged for medium with 1% serum (after washing with PBS), and after 1 additional day the medium was collected for LDH determination.
Isolation of lysosomes and autophagy-related cellular compartments
Different subcellular compartments were isolated from cultured cells as described (62) (for details see Supplementary Material). A fraction enriched in lysosomes and mitochondria (L/M fraction) was further separated by centrifugation in two successive discontinuous gradients of metrizamide/percoll as described (51). Lysosomes, collected from the 5ndash;17% metrizamide interface, were washed in 0.25 m sucrose. A cytosol fraction was obtained by centrifuging the supernatant from the L/M fraction at 100 000g for 1 h at 4°C. The pellet from this centrifugation yielded an enriched microsome fraction that is predominantly ER, but also contained small proportions of endosomes and Golgi. Rat liver lysosomes were isolated from a light mitochondrial–lysosomal fraction in a discontinuous metrizamide density gradient (70) by the modified protocol to enrich in CMA-active lysosomes as described by Cuervo et al. (50). Preparations with more than 10% broken lysosomes, measured as β-hexosaminidase latency (51), were discarded. Lysosomal matrices and membranes were isolated after hypotonic shock (71).
Autophagosomes and autophagolysosomes were isolated from cultured cells following a protocol modified from Marzella et al. (72) . From the AV/L/M fraction, lysosomes were separated from the two AV fractions using a discontinuous (50, 26, 24, 20 and 15%) metrizamide gradient where fractions migrated as follows: lysosomes (24–26% interface), autophagolysosomes (AV20; 20–24% interface) and autophagosomes (AV15, 15–20% interface). These fractions were washed by centrifugation in 0.25 m sucrose.
Lysosomal integrity measurements
The integrity of the lysosomal membrane in cultured cells was monitored by analyzing the presence of lysosomal enzymes, namely β-hexosaminidase, in the cytosol. To measure the cytosolic β-hexosaminidase activity, N2a cells were rinsed extensively with ice-cold PBS and then removed by a cell scraper in the homogenization buffer. The cytosolic fraction was isolated as described above. The β-hexosaminidase activity was then determined by the addition of 100 µl of assay buffer (0.1 m Na-acetate, pH 4.5; 1% Triton X-100, 1 mm 4-methylumbelliferyl-N-acetyl-B-d-glucopyranoside) to 5 µg cell lysates. The reaction mixture was incubated at 37°C for 1 h and stopped by the addition of 75 µl stop solution (0.5 m glycine, 0.5 m Na2CO3).The absorbance was measured at 348 nm. The cytosolic β-hexosaminidase activity was expressed as the percentage of total activity obtained from total cell homogenates.
Uptake and degradation of proteins by isolated lysosomes
The different forms of recombinant Tau were incubated in 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (10 mm MOPS pH 7.3 and 0.3 m sucrose) with untreated or protease inhibitor-treated lysosomes as described (50). After incubation for 20 min at 37°C, lysosomes were collected by centrifugation and samples were subjected to SDS–PAGE and immunoblotted with an antibody against Tau. Transport was measured and uptake was calculated as the difference between the amount of substrate associated to lysosomes (protease inhibitor-treated lysosomes) and the amount of substrate bound to their membrane (untreated lysosomes) (49). In other experiments, after the incubation with the recombinant forms of Tau, proteinase K or trypsin were added to the isolated lysosomes to remove the substrate bound to the cytosolic side of the lysosomal membrane. Uptake was measured as the amount of substrate resistant to the protease associated to lysosomes in which degradation was inhibited. Where indicated, lysosomes were previously incubated with an antibody against the cytosolic tail of LAMP-2A or against hsc70 for 10 min at room temperature before the substrates were added to the reaction.
Degradation of a pool of radiolabeled cytosolic proteins (500 dpm/μg) by intact lysosomes was measured as described (50). Proteolysis was expressed as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at the end of the incubation.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Deutsche Forschungsgemeinschaft, EU-FP7 (Memosad) and by a Breuer Foundation award (to E.M.M.), and by NIH grants AG031782, NS038370 and a Glenn Award (to A.M.C.). M.M.V. is a Fellow of the American Liver foundation and EW of the Hereditary Disease Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Hübschmann and I. Lindner (Hamburg) for preparing the recombinant Tau proteins. We thank Dr P. Seubert (Elan Pharma, South San Francisco, CA) for antibody 12E8.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 2.Garcia M.L., Cleveland D.W. Going new places using an old MAP: tau, microtubules and human neurodegenerative disease. Curr. Opin. Cell Biol. 2001;13:41–48. doi: 10.1016/s0955-0674(00)00172-1. [DOI] [PubMed] [Google Scholar]
- 3.Mandelkow E., von Bergen M., Biernat J., Mandelkow E.M. Structural principles of tau and the paired helical filaments of Alzheimer's disease. Brain Pathol. 2007;17:83–90. doi: 10.1111/j.1750-3639.2007.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson G.V., Seubert P., Cox T.M., Motter R., Brown J.P., Galasko D. The tau protein in human cerebrospinal fluid in Alzheimer's disease consists of proteolytically derived fragments. J. Neurochem. 1997;68:430–433. doi: 10.1046/j.1471-4159.1997.68010430.x. [DOI] [PubMed] [Google Scholar]
- 5.Portelius E., Hansson S.F., Tran A.J., Zetterberg H., Grognet P., Vanmechelen E., Hoglund K., Brinkmalm G., Westman-Brinkmalm A., Nordhoff E., et al. Characterization of tau in cerebrospinal fluid using mass spectrometry. J. Proteome Res. 2008;7:2114–2120. doi: 10.1021/pr7008669. [DOI] [PubMed] [Google Scholar]
- 6.Shao J., Diamond M.I. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 2007;16(Spec no. 2):R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 7.Dufty B.M., Warner L.R., Hou S.T., Jiang S.X., Gomez-Isla T., Leenhouts K.M., Oxford J.T., Feany M.B., Masliah E., Rohn T.T. Calpain-cleavage of alpha-synuclein: connecting proteolytic processing to disease-linked aggregation. Am. J. Pathol. 2007;170:1725–1738. doi: 10.2353/ajpath.2007.061232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamblin T.C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A.L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., et al. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc. Natl Acad. Sci. US.A. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y.P., Biernat J., Pickhardt M., Mandelkow E., Mandelkow E.M. Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proc. Natl Acad. Sci. USA. 2007;104:10252–10257. doi: 10.1073/pnas.0703676104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rissman R.A., Poon W.W., Blurton-Jones M., Oddo S., Torp R., Vitek M.P., LaFerla F.M., Rohn T.T., Cotman C.W. Caspase-cleavage of tau is an early event in Alzheimer disease tangle pathology. J. Clin. Invest. 2004;114:121–130. doi: 10.1172/JCI20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wille H., Drewes G., Biernat J., Mandelkow E.M., Mandelkow E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J. Cell Biol. 1992;118:573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori H., Kondo J., Ihara Y. Ubiquitin is a component of paired helical filaments in Alzheimer's disease. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- 13.David D.C., Layfield R., Serpell L., Narain Y., Goedert M., Spillantini M.G. Proteasomal degradation of tau protein. J. Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J.Y., Liu S.J., Li H.L., Wang J.Z. Microtubule-associated protein tau is a substrate of ATP/Mg(2+)-dependent proteasome protease system. J. Neural Transm. 2005;112:547–555. doi: 10.1007/s00702-004-0196-x. [DOI] [PubMed] [Google Scholar]
- 15.Feuillette S., Blard O., Lecourtois M., Frebourg T., Campion D., Dumanchin C. Tau is not normally degraded by the proteasome. J. Neurosci. Res. 2005;80:400–405. doi: 10.1002/jnr.20414. [DOI] [PubMed] [Google Scholar]
- 16.Delobel P., Leroy O., Hamdane M., Sambo A.V., Delacourte A., Buee L. Proteasome inhibition and Tau proteolysis: an unexpected regulation. FEBS Lett. 2005;579:1–5. doi: 10.1016/S0014-5793(05)01501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown M.R., Bondada V., Keller J.N., Thorpe J., Geddes J.W. Proteasome or calpain inhibition does not alter cellular tau levels in neuroblastoma cells or primary neurons. J. Alzheimers Dis. 2005;7:15–24. doi: 10.3233/jad-2005-7103. [DOI] [PubMed] [Google Scholar]
- 18.Shimura H., Schwartz D., Gygi S.P., Kosik K.S. CHIP–Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 19.Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 20.Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C.B., et al. The high-affinity HSP90–CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler D., Nixon R.A., Bahr B.A. Potential compensatory responses through autophagic/lysosomal pathways in neurodegenerative diseases. Autophagy. 2006;2:234–237. doi: 10.4161/auto.2729. [DOI] [PubMed] [Google Scholar]
- 23.Nixon R.A., Cataldo A.M., Mathews P.M. The endosomal-lysosomal system of neurons in Alzheimer's disease pathogenesis: a review. Neurochem. Res. 2000;25:1161–1172. doi: 10.1023/a:1007675508413. [DOI] [PubMed] [Google Scholar]
- 24.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 25.Ii K., Ito H., Kominami E., Hirano A. Abnormal distribution of cathepsin proteinases and endogenous inhibitors (cystatins) in the hippocampus of patients with Alzheimer's disease, parkinsonism-dementia complex on Guam, and senile dementia and in the aged. Virchows Arch. A Pathol. Anat. Histopathol. 1993;423:185–194. doi: 10.1007/BF01614769. [DOI] [PubMed] [Google Scholar]
- 26.Kenessey A., Nacharaju P., Ko L.W., Yen S.H. Degradation of tau by lysosomal enzyme cathepsin D: implication for Alzheimer neurofibrillary degeneration. J. Neurochem. 1997;69:2026–2038. doi: 10.1046/j.1471-4159.1997.69052026.x. [DOI] [PubMed] [Google Scholar]
- 27.Bednarski E., Lynch G. Cytosolic proteolysis of tau by cathepsin D in hippocampus following suppression of cathepsins B and L. J. Neurochem. 1996;67:1846–1855. doi: 10.1046/j.1471-4159.1996.67051846.x. [DOI] [PubMed] [Google Scholar]
- 28.Bendiske J., Bahr B.A. Lysosomal activation is a compensatory response against protein accumulation and associated synaptopathogenesis—an approach for slowing Alzheimer disease? J. Neuropathol. Exp. Neurol. 2003;62:451–463. doi: 10.1093/jnen/62.5.451. [DOI] [PubMed] [Google Scholar]
- 29.Oyama F., Murakami N., Ihara Y. Chloroquine myopathy suggests that tau is degraded in lysosomes: implication for the formation of paired helical filaments in Alzheimer's disease. Neurosci. Res. 1998;31:1–8. doi: 10.1016/s0168-0102(98)00020-0. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda K., Akiyama H., Arai T., Kondo H., Haga C., Iritani S., Tsuchiya K. Alz-50/Gallyas-positive lysosome-like intraneuronal granules in Alzheimer's disease and control brains. Neurosci. Lett. 1998;258:113–116. doi: 10.1016/s0304-3940(98)00867-2. [DOI] [PubMed] [Google Scholar]
- 31.Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 32.Hamano T., Gendron T.F., Causevic E., Yen S.H., Lin W.L., Isidoro C., Deture M., Ko L.W. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur. J. Neurosci. 2008;27:1119–1130. doi: 10.1111/j.1460-9568.2008.06084.x. [DOI] [PubMed] [Google Scholar]
- 33.Dice J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends. Biochem. Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 34.Dice J.F. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 35.Massey A.C., Zhang C., Cuervo A.M. Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 36.Hamos J.E., Oblas B., Pulaski-Salo D., Welch W.J., Bole D.G., Drachman D.A. Expression of heat shock proteins in Alzheimer's disease. Neurology. 1991;41:345–350. doi: 10.1212/wnl.41.3.345. [DOI] [PubMed] [Google Scholar]
- 37.Khlistunova I., Biernat J., Wang Y., Pickhardt M., von Bergen M., Gazova Z., Mandelkow E., Mandelkow E.M. Inducible expression of Tau repeat domain in cell models of tauopathy: aggregation is toxic to cells but can be reversed by inhibitor drugs. J. Biol. Chem. 2006;281:1205–1214. doi: 10.1074/jbc.M507753200. [DOI] [PubMed] [Google Scholar]
- 38.Nixon R.A. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Seglen P.O., Gordon P.B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuervo A.M., Hu W., Lim B., Dice J.F. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuervo A.M., Dice J.F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 42.Boland B., Campbell V. Abeta-mediated activation of the apoptotic cascade in cultured cortical neurones: a role for cathepsin-L. Neurobiol. Aging. 2004;25:83–91. doi: 10.1016/s0197-4580(03)00034-4. [DOI] [PubMed] [Google Scholar]
- 43.Cataldo A.M., Paskevich P.A., Kominami E., Nixon R.A. Lysosomal hydrolases of different classes are abnormally distributed in brains of patients with Alzheimer disease. Proc. Natl Acad. Sci. USA. 1991;88:10998–11002. doi: 10.1073/pnas.88.24.10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence B.P., Brown W.J. Inhibition of protein synthesis separates autophagic sequestration from the delivery of lysosomal enzymes. J. Cell Sci. 1993;105(Pt 2):473–480. doi: 10.1242/jcs.105.2.473. [DOI] [PubMed] [Google Scholar]
- 45.Abeliovich H., Dunn W.A., Jr, Kim J., Klionsky D.J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo W., Dou F., Rodina A., Chip S., Kim J., Zhao Q., Moulick K., Aguirre J., Wu N., Greengard P., et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl Acad. Sci. USA. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Vicente M., Talloczy Z., Kaushik S., Massey A.C., Mazzulli J., Mosharov E.V., Hodara R., Fredenburg R., Wu D.C., Follenzi A., et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J. Clin. Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 49.Salvador N., Aguado C., Horst M., Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J. Biol. Chem. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 50.Cuervo A.M., Dice J.F., Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 51.Storrie B., Madden E.A. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 52.Wilson D.M., Binder L.I. Polymerization of microtubule-associated protein tau under near-physiological conditions. J. Biol. Chem. 1995;270:24306–24314. doi: 10.1074/jbc.270.41.24306. [DOI] [PubMed] [Google Scholar]
- 53.Ackmann M., Wiech H., Mandelkow E. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J. Biol. Chem. 2000;275:30335–30343. doi: 10.1074/jbc.M002590200. [DOI] [PubMed] [Google Scholar]
- 54.Barghorn S., Mandelkow E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry. 2002;41:14885–14896. doi: 10.1021/bi026469j. [DOI] [PubMed] [Google Scholar]
- 55.Chirita C.N., Necula M., Kuret J. Anionic micelles and vesicles induce tau fibrillization in vitro. J. Biol. Chem. 2003;278:25644–25650. doi: 10.1074/jbc.M301663200. [DOI] [PubMed] [Google Scholar]
- 56.Aniento F., Roche E., Cuervo A.M., Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- 57.Sarkar M., Kuret J., Lee G. Two motifs within the tau microtubule-binding domain mediate its association with the hsc70 molecular chaperone. J. Neurosci. Res. 2008;86:2763–2773. doi: 10.1002/jnr.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cuervo A.M., Terlecky S.R., Dice J.F., Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J. Biol. Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 60.Komatsu M., Chiba T., Mizushima N. [Physiological role in constitutive autophagy] Tanpakushitsu Kakusan Koso. 2006;51:1515–1518. [PubMed] [Google Scholar]
- 61.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 62.Yu W.H., Cuervo A.M., Kumar A., Peterhoff C.M., Schmidt S.D., Lee J.H., Mohan P.S., Mercken M., Farmery M.R., Tjernberg L.O., et al. Macroautophagy–a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb J.L., Ravikumar B., Atkins J., Skepper J.N., Rubinsztein D.C. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 64.Rideout H.J., Lang-Rollin I., Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int. J. Biochem. Cell Biol. 2004;36:2551–2562. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Ling H., Vamvakas S., Gekle M., Schaefer L., Teschner M., Schaefer R.M., Heidland A. Role of lysosomal cathepsin activities in cell hypertrophy induced by NH4Cl in cultured renal proximal tubule cells. J. Am. Soc. Nephrol. 1996;7:73–80. doi: 10.1681/ASN.V7173. [DOI] [PubMed] [Google Scholar]
- 66.Lunkes A., Lindenberg K.S., Ben-Haiem L., Weber C., Devys D., Landwehrmeyer G.B., Mandel J.L., Trottier Y. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol. Cell. 2002;10:259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y.J., Sapp E., Cuiffo B.G., Sobin L., Yoder J., Kegel K.B., Qin Z.H., Detloff P., Aronin N., DiFiglia M. Lysosomal proteases are involved in generation of N-terminal huntingtin fragments. Neurobiol. Dis. 2006;22:346–356. doi: 10.1016/j.nbd.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Caughey B., Raymond G.J., Ernst D., Race R.E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenberg S.G., Davies P. A preparation of Alzheimer paired helical filaments that displays distinct tau proteins by polyacrylamide gel electrophoresis. Proc. Natl Acad. Sci. USA. 1990;87:5827–5831. doi: 10.1073/pnas.87.15.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wattiaux R., Wattiaux-De Coninck S., Ronveaux-dupal M.F., Dubois F. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J. Cell Biol. 1978;78:349–368. doi: 10.1083/jcb.78.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohsumi Y., Ishikawa T., Kato K. A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J. Biochem. 1983;93:547–556. [PubMed] [Google Scholar]
- 72.Marzella L., Ahlberg J., Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.