Abstract
15N-1H spin relaxation is a powerful method for deriving information on protein dynamics. The traditional method of data analysis is model-free (MF), where the global and local N-H motions are independent and the local geometry is simplified. The common MF analysis consists of fitting single-field data. The results are typically field-dependent, and multi-field data cannot be fit with standard fitting schemes. Cases where known functional dynamics has not been detected by MF were identified by us and others. Recently we applied to spin relaxation in proteins the Slowly Relaxing Local Structure (SRLS) approach which accounts rigorously for mode-mixing and general features of local geometry. SRLS was shown to yield MF in appropriate asymptotic limits. We found that the experimental spectral density corresponds quite well to the SRLS spectral density. The MF formulae are often used outside of their validity ranges, allowing small data sets to be force-fitted with good statistics but inaccurate best-fit parameters. This paper focuses on the mechanism of force-fitting and its implications. It is shown that MF force-fits the experimental data because mode-mixing, the rhombic symmetry of the local ordering and general features of local geometry are not accounted for. Combined multi-field multi-temperature data analyzed by MF may lead to the detection of incorrect phenomena, while conformational entropy derived from MF order parameters may be highly inaccurate. On the other hand, fitting to more appropriate models can yield consistent physically insightful information. This requires that the complexity of the theoretical spectral densities matches the integrity of the experimental data. As shown herein, the SRLS densities comply with this requirement.
Keywords: Protein dynamics by NMR, slowly relaxing local structure, model-free
I. Introduction
NMR is currently the most powerful method for studying protein dynamics at the residue level.1-3 The commonly used dynamic probe is the 15N-1H bond. The relaxation parameters 15N T1, T2 and 15N-1{H} NOE are measured experimentally at one or several magnetic fields. Their expressions are given by the spectral densities, J(ω), and the relevant magnetic interactions (15N-1H dipolar and the 15N CSA).4,5 The functions J(ω) are determined by the dynamic model used, and the local geometry at the N-H site.
The traditional method of data analysis is the model-free (MF) approach.6-8 MF assumes that the global motion of the protein (RC = 1/6τm) and local motion of the N-H bond (RL = 1/6τL) are ‘independent’ or ‘decoupled’, by virtue of the former being much slower than the latter (τm ≫ τL). The local ordering is measured by a squared generalized order parameter, S2, and the rate of local motion is evaluated by an effective correlation time, τe. Both parameters represent mathematical properties of the spectral density. The local geometry is simplified, with the ordering and magnetic tensor frames axial and collinear.
Three point (15N T1, T2 and 15N-{1H} NOE) data acquired at a single magnetic field pertaining to structured regions of the protein backbone can be usually analyzed with optimization (data fitting) methods using the original MF formula.6,7 Flexible residues residing in loops and mobile domains required the development of the extended MF formula.8 The latter features a fast effective local motion, τf, associated with a generalized squared order parameter, Sf2, and a slow effective local motion, τs, associated with a generalized squared order parameter, Ss2. All the modes are assumed to be independent, i.e., τm ≫ τf, τm ≫ τs, and τs ≫ τf.
The MF order parameters including the global motion correlation time, τm, are typically found to be field-dependent. This means that combined multi-field data sets cannot be fit with standard fitting schemes unless some data are excluded.9 Small anisotropies in the global diffusion tensor were found to have a very large effect on the analysis.10 Non-normal t-distribution of NOE's was detected.11 The temperature-dependence of MF order parameters was found to be unduly small.12,13 The local motion was found to be practically independent of temperature and/or experimentally measured viscosity,14-16 contrary to expectations based on typical activation energies for motions in flexible molecules. In some cases experimental relaxation parameters exceeded the extreme theoretical values.13,17 Combined analysis of N-H bond dynamics and C′-Cα bond dynamics yielded inconsistent results.13,18 We found that known functional dynamics in adenylate kinase from E. Coli is not detected with MF analysis.19,20 Similar observations were made by other workers in the field.21
These shortcomings are usually rationalized by invoking data imperfection. Alternatively the simplicity of the MF analysis may be the main underlying reason. This option can be tested by analyzing the same data with an improved version of the theory, where the simplifying MF assumptions related to liquid dynamics and local geometry are no longer invoked. This was accomplished by applying to spin relaxation in proteins22 the Slowly Relaxing Local Structure (SRLS) approach of Freed et al.23-25 which can be considered a generalized version of MF. Rather than assuming mode-independence SRLS accounts rigorously for mode-mixing through a local potential. The latter represents the spatial restrictions on N-H motion which in MF are expressed by a squared generalized order parameter. Genuine axial and rhombic order parameters are defined in SRLS in terms of the local potential. Unlike MF, SRLS allows for a full range of time scale separation between the local and global motions (e.g., they can be comparable). The magnitude, symmetry and orientation of the ordering, diffusion and magnetic tensors are all allowed to vary. In general SRLS features pure and mixed local and global dynamic modes. In the appropriate asymptotic limit it yields the pure-mode (or mode-independent) MF formulae.
Experimental 15N relaxation data were subjected in parallel to SRLS (exact solution) and MF (asymptotic solution) analyses.19,20,22,26,27 Significant improvement on many of the issues mentioned above was obtained with SRLS analysis. The goodness of fit was similar to, but the best-fit parameters significantly different from, the MF counterparts. Given that the more general SRLS contains MF as a special case, this indicates that the experimental data correspond to the general SRLS solution rather than the asymptotic MF solution. It also indicates that it is the simplicity of MF, rather than experimental imperfections, that underlie the inconsistencies mentioned above. That a similar quality of fit was obtained is related to the fitting process involving specific values of J(ω) which enter the expressions for T1, T2 and the NOE.4,5 The process whereby an oversimplified spectral density yields inaccurate best-fit parameters with good statistics is called ‘force-fitting’.
Let us point out the asymptotic nature of the MF approach. The original MF formula represents the SRLS solution in the Born-Oppenheimer (BO) asymptotic limit defined by RL ≫ RC, where the local motion, characterized by the rate RL, can be treated for frozen global motion, measured by the rate RC.27,28 In this limit the total time correlation function, C(t), may be expressed within a good approximation as the product of the time correlation function for global motion, CC(t), and the time correlation function for local motion, CL(t). When CC(t) = exp(t/τm) and the local ordering is high then the S2 from MF is a good approximation to the squared axial SRLS order parameter (S20)2, and the effective local motion correlation time, τe, is given by the ‘renormalized’ local motion correlation time, τren.27 The concept of renormalization was used in early work29 to characterize significant reduction in τL = 1/6RL by strong local potentials. It was shown29 that τren ≅ 2 τL/c20, where c20 evaluates the strength of the local axial potential. Clearly τren ⪡ τL when c20 is large. Typical values are c20 = 10 – 40 (which is to be multiplied by kBT) for squared order parameters of 0.8 – 0.95. Equation A4 of reference 6, based on the wobble-in-a-cone model, is also appropriate for relating τe to τL and S2 provided the ordering is high.27
We determined quantitatively over what range the conditions RL ≫ RC and (S20)2 ∼ 1 apply by comparing SRLS and MF results.27 The original MF formula often yields best-fit parameters which do not fulfill these requirements. These are cases where the experimental spectral density comprises mixed modes, which are incompatible with the simplified MF formula. S2 and τe can no longer be associated with the relevant physical quantities. Instead they just become fitting parameters, which have absorbed the discrepancies between the experimental and oversimplified theoretical spectral densities. Moreover, we found that often the symmetry of the local ordering at the N-H site is rhombic.26 In these cases the original MF formula is not a good approximation to the experimental spectral density even when S2 is high and RC/RL is small, because a single order parameter no longer suffices.
The extended MF formula was obtained in early work as a perturbational expansion of the SRLS solution in rhombic local ordering in the RL ≫ RC limit.30 For a 90° tilt between the (axial) magnetic frame and the main local ordering/local diffusion frame (M),27 this means that the N-H bond experiences fast diffusive local motion in the presence of very small ordering exerted by the immediate protein environment around, say, the Cαi-1-Cαi axis, or the Ni-Cαi bond. The components of the diffusion tensor are RL∥ = 1/6τL∥ and RL⊥ = 1/6τL⊥ and of the ordering tensor, S20 and S22. The protein surroundings reorient at a rate RC = 1/6τm much slower than RL∥ and RL⊥. The extended MF formula8 was offered to represent the mathematical scenario where the N-H bond experiences both fast and slow isotropic local motions with eigenvalues and squared order parameters 1/τf + 1/τm and Sf2, and 1/τs + 1/τm and Ss2, respectively. These motions are assumed to be decoupled from one another, and from the global motion, implying the conditions τf ⪡ τs ⪡ τm. In practice the extended MF formula is used when τs ∼ τm. The coefficients of the local and global motion terms in the extended MF formula are formally expressible in terms of (S20)2 and (S22)2. However, the MF parameters are totally different in implication from the SRLS parameters.
Typical best-fit values obtained with MF fitting of flexible protein residues are Ss2 ∼ 0.55, Sf2 ∼ 0.8, τs ∼ τm and τf ⪡ τm, which are just fitting parameters. This is implied by the presence of mixed modes which dominate the spectral density when τs is on the order of τm (which is typically the case for flexible residues in proteins). It should be pointed out that even if the perturbational conditions would prevail at the N-H site the MF physical picture would be puzzling, requiring two independent isotropic but restricted local motions associated with different ordering scenarios (Ss2 and Sf2) imposed by the very same protein environment reorienting with correlation time τm ∼ τs while being at the same time decoupled from τs. On the other hand, an N-H bond may reorient almost independently around Cαi-1-Cαi (i.e., mixed modes can be ignored) in the limit where RL ≫ RC when the restricting local potential is very small. In this case the physical properties of axial local diffusion and rhombic local ordering are properly described by the simplified spectral density given by eq 19 below.
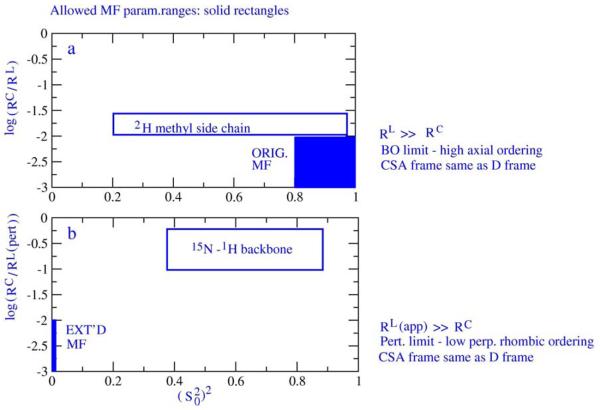
The validity ranges of the MF formulae are illustrated in Figs. 1a and 1b. The ordinates represent the logarithm of the time scale separation between the global and local motions and the abscissas represent squared order parameters. The original MF formula is applicable to a good approximation within the solid box on the right-hand side of Fig. 1a. This range is often exceeded in MF studies. We found that typical usage of the original MF formula involves discrepancies on the order of 7-8% between the squared SRLS order parameter, (S20)2, and squared generalized MF order parameter, S2, implied by limited mode-mixing effects, and by the simplified MF assumption that the 15N-1H dipolar and 15N CSA magnetic frames are collinear.27 If the effective correlation time for local motion, τe, is taken to represent the bare correlation time for local motion, τL, the latter will be underestimated five-, to twenty-fold.27 The 2H spin relaxation of side chain methyl groups is analyzed mainly with the original MF formula (e.g., refs. 31 and 32). The parameter range covered by typical best-fit parameters is shown in Fig. 1a by the rectangle labeled ‘2H methyl side chain’, which clearly digresses from the solid box in this Figure. Thus, in this application the original MF formula is mostly used outside of its validity range. The solid box in Fig. 1b shows the parameter range in which the extended MF formula is valid. In this case the abscissa represents both the axial, (S20)2, and the rhombic, (S22)2, squared order parameters which are very small. The rectangle labeled ‘15N-1H backbone’ shows the parameter range in which the extended MF formula is applied in N-H bond dynamics studies. Here the abscissa represents both Ss2 and Sf2. Clearly in this application the extended MF formula is used outside of its validity range. We found that the MF parameters τs and Ss2 exceed their formal SRLS analogues, τL⊥ and (S20)2, up to four-fold and twelve-fold, respectively.19,20,22 Significantly larger disagreements between SRLS and MF are expected when the SRLS analyses are carried out allowing for rhombic potentials. Illustrative calculations based on a recently developed fitting scheme featuring rhombic potentials are provided below.
Figure 1.
Schematic illustration of the range of validity of the original (Fig. 1a) and the extended (Fig. 1b) model-free formulae. RC (RL) represents the diffusion rate for isotropic global (local) motion. The solid rectangles delineate the valid ranges. The empty rectangles delineate the parameter ranges where these formulae are typically applied in protein dynamics research. The conditions under which the MF formulae are valid are specified on the right-hand side of Figs. 1a and 1b.
Force-fitting will also occur with SRLS versions which are over-simplified as compared to the experimental spectral densities. Therefore investigating the mechanism of force fitting with the goal of elucidating the SRLS version, which satisfactorily matches the experimental data as implied by their integrity is important. This is the subject of the present study. It is shown that model-free force-fits the experimental data because mode-mixing, rhombic potentials and general features of local geometry are not accounted for. When possible the various effects mentioned above are estimated quantitatively. Our general conclusions imply that the dynamic picture yielded by MF analysis is often distorted. We show cases where qualitatively erroneous conclusions were drawn, fictitious phenomena were detected, and known functional dynamics was missed. Conformational entropy and other thermodynamic quantities derived from MF order parameters33-36 may be inaccurate. Reliable fitting occurs with SRLS when the rhombicity of the local potential is accounted for and the local diffusion is allowed to be axial without limitations on the ratio N = RL∥/RL⊥. At this level of complexity the SRLS spectral density matches the integrity of currently available experimental data.
II. Theoretical background
1. The Slowly Relaxing Local Structure (SRLS) model
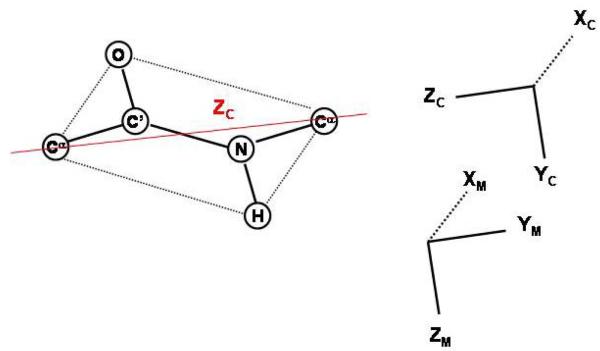
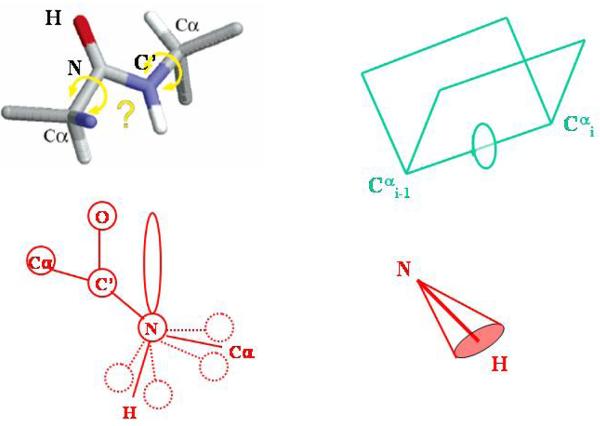
The fundamentals of the stochastic coupled rotator slowly relaxing local structure (SRLS) theory as applied to biomolecular dynamics including protein NMR were outlined recently.22,25,37 We summarize below key aspects. The various reference frames, which define the SRLS model, and their relation to N-H sites in proteins, are shown in Fig. 2. A segment of the protein backbone comprising the atoms Cαi, Ni, HNi, C′i-1, Oi-1 and Cαi-1, the equilibrium positions of which are traditionally taken to lie within the peptide plane defined by Ni, HNi, C′i-1 and Oi-1, is illustrated in Fig. 2b. The orientation of the N-H bond with respect to the magnetic field is modulated by its local motions and by the global motion of the protein. Thus, in the SRLS model we are dealing with at least two dynamic modes which we can represent by two bodies (N-H bond and protein) whose motions are coupled or mixed.23,24 For each motion two frames need to be introduced. The first is the local ordering/local diffusion frame, M, which is fixed in body 1 (in this case the N-H bond) and is usually determined by its geometric shape in the context of its motionally restricting environment. The second is the director frame, C′, whose axes represent the preferred orientation of the N-H bond (Fig. 2b) and which is fixed within the protein framework. The motion of body 1 is coupled to, or mixed with, the motion of body 2 (in this case the protein) by a local coupling or orienting potential which seeks to bring the N-H bond into alignment with the director frame. There are no limitations on the relative rates of motion of the two bodies, or the symmetry and strength of the coupling potential.
Figure 2.
(a) Various reference frames which define the Slowly Relaxing Local Structure (SRLS) model: L – laboratory frame, C – global diffusion frame associated with protein shape, C′ – local director frame associated with the stereochemistry of the local protein structure at the N-H site, M – local ordering/local diffusion frame fixed at the N-H bond, D – magnetic 15N-1H dipolar frame, CSA - magnetic 15N chemical shift anisotropy frame. (b) ZD, XD, ZM and YM reside within the peptide plane, ZD lies along the N-H bond and YD is perpendicular to the peptide plane.44 The uniaxial local director (C, assuming isotropic global diffusion) is taken to lie along the equilibrium Cαi−1−Cαi axis. The main ordering axis is taken along Cαi−1−Cαi. This implies perpendicular YM ordering with βMD = 101.3°. ‘Nearly planar YM-XM ordering’, i.e., positive ordering along XM and almost no ordering along ZM (for brevity we will denote this ordering symmetry below as ‘nearly planar YM-XM ordering/symmetry’), implies βMD = 101.3° and γMD = 90°. For high ordering the YM axis is aligned preferentially along C. The axes XCSA, YCSA and ZCSA (not shown) are defined to be aligned with the most shielded (σ11), intermediate (σ22) and least shielded (σ33) components of the 15N shielding tensor, respectively44 (information on chemical shielding and local geometry for the C′-Cα bond appears in ref. 45). The polar angle between ZD and ZCSA was set equal to 17° in our study.3 YCSA is perpendicular to the peptide plane (i.e., parallel to YD).44
The reorientation of the Ni-Hi bond is restricted due to limited bond oscillations, conformational reorientations about the adjacent dihedral angles (Φi, Ψi-1), the crankshaft motion (anti-correlated rotations about Φi and Ψi-1),38 nitrogen pyramidalization,39 peptide-plane motion around C αi-1-Cαi, etc., and any interactions with the local environment. In general, these processes imply effective Euler angles ΩMD = (αMD, βMD, γMD) which define the relative orientation of the local ordering/local diffusion frame, M, and the magnetic 15N-1H dipolar frame, D (which lies along the N-H bond). In particular, taking C αi-1-Cαi as local director, C′, and as main local diffusion axis, one has YM along the instantaneous orientation of the C αi-1-Cαi axis (i.e., ‘YM ordering’), and C′ along the equilibrium orientation of the C αi-1-Cαi axis. In this case ΩMD = (0°, 101.3°, 90°). This geometry is implicit in the 3D Gaussian Axial Fluctuations (GAF) model40 (the difference in the γMD values − 180° in 3D GAF and 90° in SRLS – is implied by the different definitions of the XM and YM axes). Similar values of ΩMD are obtained by replacing the Cαi-1-Cαi axis with the Ni-Cαi bond. The N-H bond experiences an increasing orienting potential when YM deviates from C′. The global motion of the protein (body 2) is frequently approximated as that of a cylinder, with its long axis taken to be the z-axis of the global diffusion (C) frame. For spherical (or globular) proteins the C and C′ frames are the same.
1a Geometry and observables
The various frames of the SRLS model, as applied to amide 15N spin relaxation in proteins, are shown in Fig. 2a. A formal definition, as compared to the physically descriptive presentation given above, of the various frames follows. The laboratory L frame is space-fixed, both C and C′ are protein-fixed, and the M, D (15N-1H dipolar) and CSA (15N CSA) frames are fixed with respect to the N-H bond. The L frame is considered an inertial frame with respect to which all moving frames are defined. The M frame represents both the local ordering and the local diffusion frame, which for convenience are taken to be the same. The Euler angles ΩLM are modulated by the local motion and the global motion, whereas the Euler angles ΩLC are only affected by the latter. These angles are referred to the fixed lab frame to properly describe the diffusion. The local ordering frame M tends to align with respect to a local director C′. The relative orientations of M with respect to C′ and C are defined by ΩCM and ΩC′M, respectively. The local director C′ is tilted at Euler angles ΩCC′ with respect to the cage (i.e., protein) frame C (tilted with respect to the laboratory frame at Euler angles ΩLC). The Euler angles ΩCC′ are time-independent. It is reasonable to assume that only the polar angle βC′C is important. Note that ΩLM involves the sum of rotations ΩLC + ΩCC′ + ΩC′M [here and in the following we shall employ a shorthand notation for indicating sequences of rotations; namely, for a generic rotation Ω12 = Ω2′ + Ω1, resulting from first applying Ω1 and then Ω2′ we can write the explicit relation among Wigner rotation matrices as Djmk(Ω12) = Σm′ Djmm′(Ω1) Djm′k(Ω2)]. The time dependence of the Euler angles ΩC′M is governed by the local orienting potential, which couples the two modes of motion. Through the time dependence of ΩC′M the locally reorienting N-H bond follows the slower motion of the protein.
The magnetic15 N-1 H dipolar tensor frame, D, and the magnetic 15N CSA tensor frame CSA, are also shown in Fig. 2a. The Euler angles specifying the rotation from M to D are ΩMD, and the rotation from D to CSA is given by ΩCSA. The Euler angles ΩMD and ΩD–CSA are time independent. The D frame is axially symmetric. If the M frame is also axially symmetric, then ΩMD = (0, βMD, 0), where βMD is known as ‘diffusion tilt’.
The diffusion tensor RL describing the rotational diffusion properties of the probe (N-H bond in this case) is diagonal in M, while the diffusion tensor Rc describing the rotational diffusion properties of the cage is diagonal in C. We start by assuming Smoluchowski dynamics for the coupled set of orientational coordinates ΩLM and ΩLC, according to the slowly relaxing local structure or SRLS approach. Namely, the system consists of two Brownian rotators (or ‘bodies’) -the amide group and the rest of protein - linked by an interaction potential which depends on their relative orientation. Their motions are characterized by slow diffusive changes, controlled by suitable rotational diffusion parameters. Formally the diffusion equation for the coupled system is given by:
| (1) |
where X is a set of coordinates completely describing the system:
| (2) |
where Ĵ(ΩLM) and Ĵ(ΩLC) are the angular momentum operators for the probe and the cage, respectively.
Changing to different coordinates is straighforward.28 We select the set defined by ΩC′M and ΩLC′. The Euler angles ΩC′M describe the N-H-bond--fixed M frame orientation relative to the protein-fixed C′ frame, and the Euler angles ΩLC′ the C′ frame orientation with respect to the lab frame. In the new coordinate frame one has:
| (3) |
The Boltzmann distribution Peq = exp[−U(ΩLM)/kBT]/〈exp[−U(ΩLM)/kBT]〉 is defined with respect to the probe-cage interaction potential given by:
| (4) |
This represents the expansion in the full basis set of Wigner rotation matrix elements, DLKM(ΩMC), with only lowest order, i.e. L = 2, terms being preserved. The coefficient c20 is a measure of the orientational ordering of the N-H bond with respect to the local director whereas c22 measures the asymmetry of the ordering around the director. Here we follow historical convention by using L = 2 terms as the leading terms, rather than L = 1. This is sufficient for many purposes, as we have previously shown, because NMR involves second-rank (i.e., L = 2) magnetic tensors. But the SRLS theory can readily be modified to include L = 1 terms. The current approach is in the spirit of keeping the number of parameters to a minimum.
The SRLS equation can be solved in terms of the time dependent distribution P(ΩC′M,ΩLC′,t), which describes the evolution in time and orientational space of the system. Alternatively, it is convenient to directly calculate time correlation functions , which for proper values of the coefficients J,M,K,K′ determine the experimental NMR relaxation rates. Actually, the Fourier-Laplace transforms of are needed, and they are obtained as the spectral densities at a given frequency ω:
| (5) |
As stated here, the model has eleven parameters: c0, c2 (potential parameters), (probe diffusion i =1,2,3 principal values), (global diffusion i =1,2,3 principal values) and ΩCC′ = (αCC′,βCC′,γCC′ (global diffusion tilt angles). For the sake of simplicity, we shall limit our analysis to axial probe , axial cage , and αCC′ = γCC′ = 0. The orientation of the magnetic tensors is specified by ΩMD and Ω (defined in Fig. 2a). In the past work19,20,22,26,27 we made use of eq 2 involving ΩLM and ΩLC. In the present study we have used eq 3 involving ΩLC′ and ΩC′M. The primary reason is that the use of the relative orientation of the N-H bond in the protein specified by the ΩC′M is the more natural one in terms of conventional intuition. One can simply think of the Euler angles ΩC′M as just being modulated by the local motion, whereas ΩLC′ is just modulated by the overall tumbling of the protein. Also, as we have already noted, the ΩC′M are the natural coordinates for expressing the potential energy given by eq 4 (This does, however, render the Γ̂ operator somewhat more complicated). Of course, the two forms are mathematically equivalent. The global diffusion tensor assumes the form in the C′ frame:
| (6) |
The probe diffusion tensor, defined in the M frame, is diagonal. Note that for βCC′ = 0 or Rc⊥ = Rc∥ the global diffusion tensor is diagonal and invariant in both the C and C′ frames.
1b Numerically exact treatment
We address here the problem of devising an efficient procedure for evaluating numerically accurate spectral densities. We adopt a variational scheme, based on a matrix vector-representation of eq 5, followed by an application of the Lanczos algorithm in its standard form developed for Hermitian matrices. It is convenient to express the generic correlation function as the linear combination of normalized auto-correlation functions. Defining , the spectral densities of the normalized auto-correlation functions of interest are:
| (7) |
and the generic spectral densities are:
| (7a) |
where J = 2 and M = 0 in our case. We use the shorthand notation [J ] = 2J +1. A numerical calculation is then performed by choosing a basis set of functions, representing in matrix form the transformed operator , and evaluating eq 7 directly by employing a standard Lanczos approach. The latter is reviewed here for completeness in accordance with the standard technique of Moro and Freed.41,42 Let us suppose that we are interested in calculating the Fourier-Laplace transform of the normalized auto-correlation function of an observable f (q) for a diffusive symmetrized (i.e. Hermitian) operator Γ̃ acting on coordinate q, in the form , where δf = f − 〈fpeq〉 is the observable redefined to yield an average value of zero. In the present case we consider only rotational motion in an isotropic fluid, so the relevant 〈fpeq〉 = 0. The Lanczos algorithm is a recursive procedure to generate orthonormal functions which allow a tridiagonal matrix representation of Γ̃ in terms of the coefficients αn, βn, which form the main and secondary diagonal of the tridiagonal symmetric matrix T, and the spectral density can be written in the form a continued fraction41,42. The calculation of the tridiagonal matrix elements can be carried out in finite precision by working in the vector space obtained by projecting all the functions and operators on a suitable set of orthonormal functions |λ〉. One only needs to define the matrix operator, Γ, and starting vector elements, v1, given by Γλλ′ = 〈λ|Γ̃|λ′〉, respectively.
In the case under study the SRLS diffusion operator is given by eq 3 and the starting vector is given by . A natural choice for a set of orthonormal functions is the direct product of normalized Wigner matrices. What is left is the calculation of the matrix elements Γλ,λ′ and the vector elements 〈λ|1〉. The algebraic intermediate steps are relatively straightforward and based on properties of the Wigner rotation matrices, angular momentum operators and spherical tensors; we skip the technical details and list the resulting expressions.23,24,28
1c Observables
In order to interpret 15 N-1 H dipolar and 15 N CSA auto-correlated relaxation rates we only need spectral densities with J = 2 and M = 0. Dependence upon K, K′ is slightly more complex and it discussed in detail in the following section.
According to standard analysis for the motional narrowing regime43 we can define the observable spectral density for two magnetic interactions μ and ν as the real part of the Fourier-Laplace transform of the correlation function of the second rank Wigner functions in the orientation of the magnetic tensors in the laboratory frame, (here μ,ν = D or CSA and ΩD = ΩMD,ΩCSA = ΩMD + Ω, cf. Fig. 2a):
| (8) |
and relying on standard properties of the Wigner functions, in the form:
| (8a) |
Based on the symmetry relation (cf. eq 7a) we obtain:
| (9) |
where ℜ stands for the real part. Note that for axial potentials () the second term goes to zero and we are left with standard expressions. The coefficients are readily evaluated, while can be calculated in terms of ΩMD and Ω, as in the expression: .
The spectral densities for 15N-1H dipolar and 15N CSA auto-correlation are then obtained as and , respectively. The measurable 15N relaxation quantities 15N T1, T2 and 15N-{1H} NOE are calculated as functions of JDD (ωi) and JCC (ωi), with ωi = 0,ωH, ωN, ωH − ωN and ωH + ωN, using standard expressions for NMR spin relaxation.4,5 Note that due the additional symmetry jM,K,K′ = jM,−K,−K′, only the nine distinct couples (K, K′) = (−2, 2), (−1,1), (−1, 2), (0,0), (0,1), (0, 2), (1,1), (1, 2), (2, 2) need to be considered. For dipolar auto-correlation, where ΩMD = (0,βMD,0), one has the explicit expression (denoting for brevity):
| (10) |
with only six couples (K, K′) = (0,0), (1,1), (2, 2), (0, 2), (−1,1) and (−2, 2) involved.
A convenient measure of the orientational ordering of the N-H bond is provided by the order parameters, and , which are related to the orienting potential (eq 4), hence and , via the ensemble averages:
| (11) |
One may convert to Cartesian ordering tensor components according to . Note that Sxx + Syy + Szz = 0.
In case of zero potential, , and axial diffusion, the solution of the diffusion equation associated with the time evolution operator features three distinct eigenvalues:
| (12) |
where RL∥ = 1/(6τ∥) and RL⊥ = 1/(6τ⊥) = 1/(6τ0). Only diagonal jK(ω) ≡ jKK(ω) terms are non-zero and they can be calculated analytically as Lorentzian spectral densities, each defined by width 1/τK. When the ordering potential is axially symmetric, , again only diagonal terms survive, but they are written as infinite sums of Lorenzian spectral densities, which are defined in terms of eigenvalues 1/τi of the diffusion operator and weighing factors cK,i such that:
| (13) |
The eigenvalues 1/τi represent modes of motion of the system, in accordance with the parameter range considered. Note that although in principle the number of terms in eq 13 is infinite, in practice a finite number of terms is sufficient for numerical convergence of the solution. Finally when the local ordering potential is rhombic, , both diagonal jK(ω) and non-diagonal jKK′(ω) terms are different from zero and need to be evaluated explicitly according to expressions analogous to eq 13.
Details of the implementation of SRLS in a data fitting scheme featuring axial potentials and isotropic global diffusion were outlined previously.22 For practical reasons this fitting scheme is based on pre-calculated 2D grids of spectral densities, jK(ω). The coordinates of these grids are c20 and RC. The structural parameters βMD and γMD are used to assemble JDD(ω) out of jK(ω). The set of free variables includes c20, RC and βMD. The angle γMD was fixed at 90° based on stereo-chemical considerations, and RL∥≫RL∥ (in analogy with the MF requirement that τs ≫ τf) was imposed. This scheme is computationally as fast as the commonly used MF fitting schemes.46,47
We developed recently a fitting scheme where the functions jKK′(ω) are calculated on the fly. In this case the set of free variables includes c20, c22, RL∥/RL⊥, RC and βMD. Clearly the local potential is allowed to be rhombic and the local diffusion, axial. This scheme is currently rather demanding computationally and efforts to improve its efficiency are underway. A number of conditions can be employed, however, to simplify the analysis. If the local geometry is assumed to be known, as in the 3D GAF model,40 βMD can be fixed at 101.3°. If the symmetry of the local ordering is known c22/c20 can be fixed. Note that in SRLS the global diffusion rate, RC, is determined in the same fitting process as the site-specific parameters, as is appropriate when the modes RC and RL are ‘mixed’. The next stage will be to allow the global diffusion tensor to be axially symmetric. This requires a complex fitting scheme where the RC tensor is global whereas all the other parameters are local. Instead of a single variable for global motion, RC, three variables, RC(app), RC∥/RC⊥ βCC′ will be featured. Note that RC and RC(app) define the time scale separation between the global and local motions as rates are given in SRLS in units of RL⊥.
When the local potential is axially symmetric in SRLS then formal (but not necessarily physical) analogies with the MF formulae can be specified. In this case the number of formally analogous free parameters, hence the minimum number of data points required, is the same in SRLS and MF.22 Model-free data fitting was carried out in this study with the computer programs Modelfree 4.0 (ref. 46) and Dynamics (ref. 47).
Thus, the spectral densities jK(ω) (jKK′(ω) for rhombic potentials) are the building blocks for a given dynamic model relative to the local diffusion frame, and the spectral densities Jx(ω) are the building blocks for a specific geometric implementation of this dynamic model relative to the frames of the magnetic tensors. The measurable quantities are Jx(0), Jx(ωN), Jx(ωH), Jx(ωH + ωN) and Jx(ωH − ωN). Together with the magnetic interactions they determine the experimentally measured relaxation rates according to standard expressions for NMR spin relaxation.4,5 If the equilibrium orientations of N-H and Cα-C′ are assumed to reside within the peptide plane then the functions jKK′(ω) for N-H bond dynamics can also be used to treat Cα-C′ bond dynamics. Different local geometry, specific to the Cα-C′ bond, determines the Jx(ω) functions and different magnetic interactions enter the calculation of the relaxation rates measured experimentally.
2. The model-free approach
A brief summary of the model-free approach, as formulated by its developers, is outlined below.
2a. Original MF spectral density.6,7
The basic premise is that the global motion of the protein is much slower than the local motions of the N-H bond. Consequently the global and local motions are ‘independent’, and the total time correlation function, C(t), can be expressed as:
| (14) |
The global motion is assumed to be isotropic, with CC(t) = exp(−t/τm). CL(t) is given by:
| (15) |
where τe is the effective correlation time for local motion defined as the area of CL(t) divided by (1 − S2), and τe << τm. The parameter S2, which represents the plateau value of CL(t) at long times (t ≫ τe), is taken as the square of a generalized order parameter. This definition of S2 (eqs 14-16 of ref. 6) involves the spherical harmonic functions of rank 2, whereas CL(t) at shorter times is given in terms of the Legendre polynomial of rank 2 (eqs 4 and 12 of ref. 6). All of the equations cited involve the equilibrium probability distribution function, Peq(ΩCM) where C denotes the local director fixed in the protein (called ‘molecular axis’ in ref. 6), and M the local diffusion frame (taken in MF to lie along the N-H bond). Equations 4 and 12 feature Peq(0, βCM, 0) (peq(θ) in the notation of ref. 6), whereas eqs 14-16 feature Peq(0, βCM, γCM) (peq(θ,φ) in the notation of ref. 6). Thus, there is inconsistency in the symmetry of CL(t) at short and long times, implied by M considered axial in eqs 4 and 12 and rhombic in eqs 14-16. Fourier transformation of eq 14 with eq 15 inserted for CL(t) yields:
| (16) |
where 1/τe′ = 1/τe + 1/τm.
2b. The original MF formula as SRLS asymptote
It was shown in early work30 that in the limit where RL≫ RC the following equation is valid in the perturbation limit, i.e., for very small local ordering:
| (17) |
with τK given by eq 12. S2K denotes the principal values of the ordering tensor in irreducible tensor notation (where S21 = 0.) When the symmetry of the local potential/local ordering is axial ten S22 is zero. In this case j0(ω) is given by the first term of eq 17, whereas jK≠0(ω) are given by 2τK/(1 + ω2τK2).48
The function C0(t) corresponding to j0(ω) is shown by the dashed curve in Fig. 3a, with the plateau value given by (S20)2 2 and the decay to it by τ0 = 1/6RL. The final decay of C0(t) to zero is given by τm = 1/6RC. However, the local ordering at an N-H bond is never as low as required by the perturbation limit in the local ordering, but rather quite high. Using the full SRLS solution we showed in previous work26 that for high enough (S20)2 and low enough RC/RL (see ref. 26 for quantitative evaluation of validity ranges) eq 16 is valid with S2 ∼ (S20)2 and the initial decay of C0(t) given by:
| (18) |
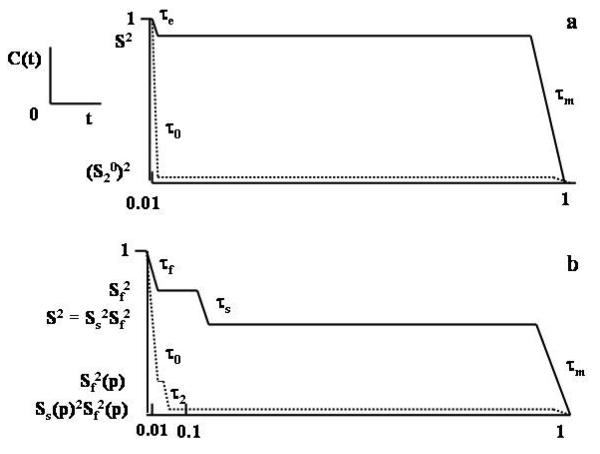
where τren is the renormalized correlation time for local motion.29 The parameter 1/6τren is the rate at which the distribution of orientations is restored to equilibrium when a spin-bearing particle reorients rapidly in the presence of a strong orienting potential.29,49 τren is reduced significantly relative to τ0, in accordance with the strength, c20, (and symmetry) of the local potential. The expression given by eq 18 is valid in SRLS in the asymptotic limit (RL ≫ RC)27 when the local potential is axial and high. In this case eq 16 is a good approximation of the SRLS solution with S2 representing (S20)2 and τe representing τren. The time correlation function corresponding to eq 16 is shown by the solid curve in Fig. 3a.
Figure 3.
(a) Time correlation function, C(t), corresponding to j0(ω) of eq 17 (and eq 16 applied in the perturbation limit) with (S20)2 → 0 and τ0/τm << 1 (dashed curve). Time correlation function, C(t), corresponding to eq 16 with (S20)2 ∼ 0.8 and τe/τm << 1 (solid curve). (b) Time correlation function, C(t), corresponding to eq 20 (and eq 19 applied in the perturbation limit) with (S20)2 → 0 and (S22)2 → 0 (Sf2 ∼ 0.25 and Ss2 ∼ 0) and τ0, τ2 << τm (τf, τs << τm) (dashed curve). Time correlation function, C(t), corresponding to eq 19 as applied to treat flexible residues in proteins with Sf2 ∼ 0.75, S2 ∼ 0.55, τs/tf ∼ 10 and τf/τm << 1 (solid curve). The index ‘p’ stands for ‘perturbational limit’. The abscissas in Figs. 3a and 3b are given in units of τL/τm. Note that τe and τf are significantly smaller than displayed (for visibility), as they represent renormalized correlation times.
The range of validity of eq 16 depends on τm and the experimental uncertainties. It can be determined by comparing results with SRLS. For example, we showed previously that for τm = 15 ns and typical experimental errors, eq 16 may be considered valid when S2 ≥ 0.8 and τ/τm ≤ 0.01 (ref. 27). When these conditions are fulfilled (see below) S2 and τe are physically meaningful. Otherwise they become parameterizing entities.
When S2 is high, the angle βCM is restricted to small values, hence the cosine squared potential of the cone model is a good approximation to U/kBT = −c20 P2cos2(βCM), where P2 denotes the Legendre polynomial of rank 2. This represents the first term of eq 4. In this case τe determined with the wobble-in-a-cone model agrees with τren, the wobbling rate, Dw, of the cone model represents RL⊥ and D∥ = RL∥ → ∞. Other models, such as the Woessner model, or jumps between symmetry-related sites,6 yield τe values which can disagree with τren (eq18). The quantity S2, taken as the square of a generalized order parameter, is in actual fact an approximation to (S20)2 when the time scale separation between the global and local motions is large enough, and the ordering high enough for the solution for the local motion to be given solely in terms of the DLMK (When (S20)2 is not very high additional local motion eigenmodes emerge. Their presence requires a more complex description of how the correlation functions of the DLMK relate to the eigenmodes of a rotor in a fairly restricted (even static) potential). Quantitative evaluations of validity ranges appear in reference 26.
The order parameter S20 is obtained in terms of U/kBT = −c20 P2cos2(βCM) based on Peq(βCM) ∝ exp(−U/kBT) (eq 4 with c22 = 0, and eq 11). Likewise conformational entropy (or any other thermodynamic quantity based on Peq) is obtained directly in SRLS. In MF the local potential has to be derived from S2 to calculate thermodynamic properties, notably residual configurational entropy. This is appropriate only when S2 is a good approximation to (S20)2, i.e., when the conditions specified in the previous paragraph are fulfilled. Since this is often not the case the MF-derived residual configurational entropy is likely to be inaccurate. The form of the potential is clearly ambiguous. As pointed out in the previous paragraph, other forms may not be compatible with the meaning of τe as given by eq 18, which can complicate their interpretation. In SRLS the potential given by eq 4 represents the leading terms in a complete expansion, and the parameters varied are the potential coefficients. The latter procedure is a general one.
For high rhombic ordering there is no analytical expression for C(t), so the ensuing spectral density, even in the RL ≫ RC (BO) limit, requires the full SRLS solution. We found that the actual local potential at N-H sites in proteins is rhombic. Note that in SRLS the conformational entropy can still be calculated based on Peq using the rhombic form of the potential (eq 4) with c20 and c22 determined with data fitting.
For rigid residues, where the fast local fluctuations at the N-H site can be considered harmonic (i.e., cos2βCM), rhombic local ordering can be treated with the 3D GAF model.40 The local geometry is pre-determined in 3D GAF by selecting Cαi−1−Cαi as the principal ordering axis (z), with x perpendicular to it within the peptide plane. Contrary to 3D GAF, the SRLS approach is applicable to arbitrary local geometry and arbitrary rates of local motion.22-25 Rhombic symmetry of the local ordering is outside the scope of MF. Taking the D and 15N CSA frames collinear in MF introduces further inaccuracies (see below).
Single-exponential approximation, τe; the effect of additional local motion eigenmodes
It was shown in early work that a single exponent, τe, is a good approximation for the multi-exponential time correlation function of the wobble-in-a-cone model.50,51 Moreover, an analytical formula which relates τe to S2 and the wobbling rate, Dw, was developed.51 This result is based on the assumption that eq 14 is valid, which implies the neglect of additional local motion eigenmodes. Table 1 shows the SRLS eigenvalues (and corresponding weights) which contribute to C(t) for a time scale separation τL/τm = 0.01 and potential strength decreasing from c20 = 20 ((S20)2 = 0.901) to c20 = ((S20)2 = 0.507). As benchmark we show the eigenvalues and associated weights when a single local motion eigenmode prevails. These include 1/τm (column 3) and (S20)2 (column 1) for the global motion, and 1/τren (column 2, calculated with eq 18) and (1 − (S20)2) (numbers in parenthesis in column 6) for the local motion. Column 7 shows the percent deviation of the correlation function for local motion from its solely DLKM-determined single-local-motion-eigenmode form (see above). Namely, for each value of c20 the numbers in column 7 (in fractional units) have to be added to wtL (numbers without parenthesis) and wtC to obtain the total weight of 1.
Table 1.
SRLS eigenvalues 1/τm (1/τL) of the global motion term (the main local motion term, i.e., the largest 1/τ(i) value of eq 13 with weight near to (1 – (S20)2), and associated weights wtC (wtL), as a function of c20 in units of kBT (and corresponding (S20)2 values) calculated for τL/τm = 0.01. The eigenvalues are given in units of RL, hence 1/τL = 6 and 1/τm = (τL/τm×6). The parameter 1/τren represents the renormalized local motion eigenvalue calculated with eq 18. The numbers in parentheses in column 6 show (1 – (S20)2). Column 7 shows the percent deviation of the correlation function for local motion from its solely DLKM-determined single-local-motion-eigenmode form (see above).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|
| c20 ((S20)2) | 1/τren | 1/τm | wtC | 1/τL | wtL | wMM, % |
| 20 (0.901) | 60 | 0.06 | 0.903 | 58.5 | 0.093(0.099) | 0.4 |
| 10 (0.803) | 30 | 0.06 | 0.800 | 27.9 | 0.172 (0.197) | 2.8 |
| 8 (0.754) | 24 | 0.06 | 0.757 | 21.6 | 0.202 (0.246) | 4.1 |
| 6 (0.671) | 18 | 0.06 | 0.676 | 15.4 | 0.239 (0.329) | 8.5 |
| 4 (0.507) | 12 | 0.06 | 0.512 | 10.0 | 0.294 (0.493) | 19.4 |
It can be seen that for 4 ≤ c20 ≤ 20 the global motion eigenvalue is given by 1/τm = 0.06 and its weight, wtC, is given within a good approximation by (S20)2. The main local motion eigenvalue, 1/τL, decreases relative to 1/τren with decreasing c20. The difference is 2.5% for (S20)2 = 0.901 (c20 = 20), 10% for (S20)2 = 0.803 (c20 = 10) and 16.7% for (S20)2 = 0.507 (c20 = 4). The deviation of the correlation function for local motion from its solely DLKM-determined single-local-motion-eigenmode form is 0.4% when (S20)2 = 0.901, 2.8% when (S20)2 = 0.803, and 19.4% when (S20)2 = 0.507. A typical (S20)2 value for rigid N-H bonds is 0.8, implying 10% error in τe calculated with the cone model and 2.8 % error in assuming that the weight of the local motion term is (1 − (S20)2). This implies 3.1% error in S2 which should be taken into consideration when the accuracy and precision of S2 are estimated in MF studies.11,13 The estimates given above are based on direct calculation. When S2 is determined with data fitting the errors can be larger.
The time scale separation between the global and local motion is evaluated in MF based on the τe/τm ratio, which is substantially smaller than the true measure, τL/τm, and which is S2-dependent. For example, for τL/τm = 0.01 the ratio τe/τm is 0.002 for (S20)2 = 0.8 (c20 = 10) and 0.003 for (S20)2 = 0.75 (c20 = 7.9). As noted above for τL/τm ≥ 0.01 mixed modes (see below) contribute significantly to the spectral density. Tables 2 and 3 show the effect of τL/τm exceeding 0.01 for (S20)2 assuming the values of 0.75 and 0.8. The numbers in parenthesis are the values corresponding to an ‘accurate’ MF formula where S2 = (S20)2 and τe = τren. It can be seen that the errors are significant for τe/τm values which might be considered in MF analyses as representing large time scale separations. For example, while the true time scale separation is τL/τm = 0.1, MF would report on τe/τm = 0.02 (0.025) for (S20)2 = 0.8 (0.75), corresponding to 5.3% (6.8%) mixed-mode contribution, implying increase in wtC and decrease in wtL, as shown in Table 2. As pointed out above, the error in the compromise value of S2 determined by data fitting may be larger than the estimates of Tables 2 and 3 which are based on direct calculations. Further inaccuracies will be implied by the MF assumption that the dipolar and 15N CSA frames are collinear, and by simultaneously (S20)2 being lower, and τL/τm being higher, than the relevant threshold values.
Table 2.
Eigenvalue (1/τm) and weight (wtC) of the global motion, eigenvalue (1/τL) and weight (wtL) of the main local motion mode, and contribution of additional local motion eigenmodes modes (wMM) to C(t) as a function of τL/τm. The last column shows τe/τm corresponding to τL/τm in column 1 (eq 18). An axial potential with coefficient c20 = 10, corresponding to (S20)2 = 0.8, was used. The terms in parenthesis represent the case in which the local motion is given by the eigenvalue 1/τren and the weight (1 – (S20)2), and the global motion by the eigenvalue 1/τm and the weight (S20)2.
| τL/τm | 1/τm | wtC | 1/τL | wtL | wMM, % | τe/τm |
|---|---|---|---|---|---|---|
| 0.01 | 0.06(0.06) | 0.805(0.803) | 27.9(30.0) | 0.172(0.2) | 2.8 | 0.002 |
| 0.030 | 0.18(0.18) | 0.813 | 28.5 | 0.167 | 3.3 | 0.006 |
| 0.050 | 0.29(0.30) | 0.819 | 29.1 | 0.161 | 3.9 | 0.01 |
| 0.100 | 0.55(0.60) | 0.833 | 30.7 | 0.147 | 5.3 | 0.02 |
| 0.200 | 1.00(1.20) | 0.858 | 33.8 | 0.119 | 8.1 | 0.04 |
Table 3.
Same as the title of Table 2 except that c20 = 7.9, corresponding to (S20)2 = 0.75, was used.
| τL/τm | 1/τm | wtC | 1/τL | wtL | wMM, % | τe/τm |
|---|---|---|---|---|---|---|
| 0.01 | 0.06(0.06) | 0.755(0.75) | 21.30(23.7) | 0.234(0.25) | 1.6 | 0.003 |
| 0.030 | 0.18(0.18) | 0.763 | 21.81 | 0.201 | 4.9 | 0.008 |
| 0.050 | 0.29(0.30) | 0.770 | 22.32 | 0.196 | 5.4 | 0.013 |
| 0.100 | 0.55(0.60) | 0.788 | 23.61 | 0.182 | 6.8 | 0.025 |
| 0.200 | 1.00(1.20) | 0.818 | 26.23 | 0.158 | 9.2 | 0.051 |
2c. The extended MF formula.8
When eq 16 cannot fit the experimental data the extended MF spectral density, given by:
| (19) |
has been used. The parameter τf is taken as the effective correlation time for the fast local motion, τs as the effective correlation time for the slow local motion, and Ss2 and Sf2 as squared generalized order parameters associated with these motions. 1/τf′ = 1/τf + 1/τm and 1/τs′ = 1/τs + 1/τm. No effort is made to define any geometric relationships between the axes of fast and slow local motions. Although eq 19 requires that τf << τs << τm, in practice this formula is used when τs is on the order of τm.
2d. The extended MF formula as a SRLS asymptote
As shown previously, an expression similar to equation 19 was obtained in early work as a perturbational expansion of the SRLS solution in rhombic local ordering in the RL >> RC limit for βMD ≠ 0° (ref. 30) Let us reiterate the basics of this derivation. JDD(ω) is given by eq 10, the functions jK(ω) are given by eq 13, and setting βMD = 90° implies A = (1.5cos2βMD − 0.5)2 = 0.25, B = 3 cos2βMD sin2βMD = 0 and C = 0.75 sin4βMD = 0.75 (A, B and C represent (d200)2, 2 (d201)2 and 2 (d202)2, respectively, where ‘d’ denotes reduced Wigner rotation matrix elements). The function JDD(ω) is then given by:
| (20) |
Assuming that J(ω) = JDD(ω) = JCC(ω) eq 20 is formally analogous to the extended MF formula8 (eq 19) with τs formally equivalent to τ0, τf to τ2, and the squared generalized order parameters, Ss2 and Sf2, related to (S20)2 and (S22)2 as:
| (21) |
and
| (22) |
The equivalence outlined above is only formal. Equation 19 is a physically vague mathematical formula whereas eq 20 is a physically precise geometric model based on SRLS. Note also that other SRLS models, such as one, which features an additional mode of internal motion, would yield the form of eq 19 in a perturbational limit.
For τf, τs << τm (representing the RL >> RC limit) and very low local ordering ((S20)2 → 0 and (S22)2 → 0 implied by the perturbation limit) one obtains Sf2 ∼ 0.25 and Ss2 ∼ 0 when eqs 21 and 22 are used. The corresponding time correlation function is shown by the dashed curve in Fig. 3b. However this C(t) function is never used to analyze N-H bond dynamics in proteins since the local ordering at the N-H site is significantly higher than Sf2 ∼ 0.25 and Ss2 → 0, and τs is on the order of τm rather than being much smaller than τm. A typical parameter set obtained with MF analysis for flexible residues in proteins is given by τf ∼ 20 ps, τs ∼ 10 ×τf, τm ≥ 10×τs, Sf2 ∼ 0.75 and Ss2 ∼ 0.55. The corresponding time correlation function C(t) is shown by the solid curve in Fig. 3b. Table V of reference 23 (where the SRLS theory has been fully developed) shows quantitatively that mode-mixing dominates the time correlation function when τm ≥ 10×τs and the ordering is (S20)2 ∼ 0.55 (corresponding to c20 ∼ 4.4). In this case the spectral densities given by eq 13 instead of spectral densities given by eq 17 are to be used. For rhombic ordering not only diagonal terms, jKK(ω), but also cross-terms, jKK′(ω), need to be considered. Jx(ω) obtained from jKK′(ω) by frame transformations (determined by the specific local geometry) are therefore complex functions. In the MF formulation J(ω) = JDD(ω) = JCC(ω) is the Fourier transform of the simple function shown by the solid line in Fig. 3b, with the plateau values determined by Sf2 and S2 = Ss2×Sf2 and the step between them monitored by τs. Therefore when force-fitting is successful, i.e., the statistical requirements are fulfilled this can be only accomplished with highly inaccurate best-fit parameters which constitute parameterizing entities. The latter are field-dependent since parameterization by force-fitting depends on which J(ω) values are to be reproduced. The fitting of larger data sets obtained by combining multi-field data is likely to fail when standard fitting schemes are used. The trends in the values of the best-fit parameters upon changing environmental conditions such as temperature and complex formation are devoid of physical meaning, and may show abrupt changes, which are not associated with genuine physical phenomena. These features are illustrated below.
Table 5.
Results of fitting with SRLS and MF the experimental data of eight VHHS residues fit with model 1 by Vugymeyster et al.11 Squared SRLS order parameters, (S20)2, obtained from the best-fit c20 values using eq 4 with c22 =0, and eq 11, and best-fit MF S2 values, are shown. The corresponding χ2 values are also given. The data in parentheses were obtained with the SRLS program where the 15N-1H dipolar and 15N CSA tensors were set deliberately collinear. %diff is the percent difference between the MF and SRLS squared order parameters divided by the SRLS value. The last two columns show the experimental NOE error and the percent difference (%Dmax) between the experimental NOE and the maximum NOE obtained for a rigid sphere.
| SRLS | MF | SRLS | MF | NOE | |||
|---|---|---|---|---|---|---|---|
| res | (S20)2 | S2 | %diff | χ2 | %err | %Dmax | |
| 45 | 0.842 | 0.804 (0.803) | −4.5 | 6.44 | 1.7 (1.7) | 2.1 | 1.4 |
| 49 | 0.853 | 0.817 (0.815) | −4.2 | 2.1 | 2.2 (2.2) | 1.9 | 0.0 |
| 57 | 0.887 | 0.847 (0.845) | −4.5 | 6.6 | 1.7 (1.8) | 2.2 | 2.1 |
| 58 | 0.908 | 0.869 (0.860) | −4.3 | 34.0 | 15.5 (20.0) | 2.2 | 7.7 |
| 59 | 0.898 | 0.855 (0.853) | −4.8 | 19.0 | 12.6 (14.2) | 2.4 | 3.5 |
| 60 | 0.849 | 0.810 (0.810) | −4.6 | 27.0 | 18.7 (19.0) | 3.3 | 16.0 |
| 69 | 0.853 | 0.815 (0.814) | −4.5 | 15.6 | 7.7 (7.9) | 2.2 | 4.5 |
| 71 | 0.841 | 0.803 (0.803) | −4.5 | 12.0 | 3.1 (3.3) | 1.9 | 3.1 |
The extended MF formula is based on the theory of moments, which is a mathematical approach that ignores physical details for convenience. The physical principles underlying NMR spin relaxation in locally orienting environments have been set forth previously.52a The important structural/electronic/charge-related information one can extract when the restrictions on the local motion are properly treated as potentials or ordering tensors have been illustrated amply in the literature (e.g., ref 52b). Within the scope of these established approaches the solution offered by the extended MF formula to N-H bond motion in proteins is physically not reasonable. The very same entity (the cylindrical N-H bond) cannot be involved in two separate motions which are isotropic (as manifested by the scalar quantities τs and τf) and at the same time restricted (as manifested by Ss2 and Sf2). The simplification to isotropic local motion, is certainly not justified for the restricted slow motion, τs, as τs ∼ τm. It is not reasonable to have no geometric relationship whatsoever between the fast and slow local motions. The very same (internal) protein environment cannot exert multiple different restrictions on the same body. The global motion (τm) cannot occur on the same time scale as the slow local motion (τs) and at the same time not lead to mixed modes. A restricted motion can be nearly ‘decoupled’ from the slowly relaxing environment, which exerts the spatial restrictions, only when these processes occur on very different time scales (RL∥/RL⊥ >> 1) and the ordering (S20 and S22) is so small that it constitutes a perturbation on the free motion, or so large that the local motion correlation times become renormalized by the strong local potential. Only in this case can mode-mixing be ignored. Accounting for the correct local geometry (βMD on the order of 90° in the present case) one may use the analytical function given by eq 20, which is assembled from the simplified functions jK(ω) given by eq 17. In this case eq 20 describes properly an axially diffusing N-H bond in the presence of a weak rhombic potential. Note that even in this limit the global and local modes are only nearly ‘independent’ since the terms (S2K)2 (τLK)/(1 + ω2(τLK2), K = 0 and K = 2, actually represents statistical interdependence.48
A numerical example, which illustrates the distorted picture obtained by using the extended MF formula outside the perturbation limit is shown in Table 4.
Table 4.
Squared axial ((S20)2) and rhombic ((S22)2) SRLS order parameters in irreducible tensor notation, corresponding order parameters (Sxx, Syy and Szz) in Cartesian tensor notation, and corresponding squared generalized MF order parameters (Sf2 and Ss2, based on using eqs 21 and 22). The coefficients c20 and c22 determine the potential U/kBT in terms of which S20 and S22 are defined (eq 11).
| c20 | c22 | (S20)2 | (S22)2 | Ss2 | Sf2 | Sxx | Syy | Szz |
|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 0.008 | 0.327 | 0.50 | 0.50 | 0.306 | −0.394 | 0.088 |
The coefficients c20 = 2 and c22 = 3 represent rhombic YM ordering with ‘nearly planar YM-XM′ symmetry, which we found previously to prevail at the N-H site.26 This symmetry is reflected clearly in the principal values, Sxx, Syy and Szz, of the Cartesian tensor. In irreducible tensor notation one has S20 = 0.089 and S22 = 0.572 (Table 4 shows the squared values of S20 and S22, which appear in eq 20). The corresponding MF parameters are Ss2 = 0.50 and Sf2 = 0.50. The physical picture of two independent isotropic local motions of the N-H bond associated with squared generalized order parameters (incidentally) equal to 0.5 is certainly different from the physical picture associated with an axial N-H bond diffusing in a well-defined rhombic local potential associated with a well-defined ordering tensor with its YM axis aligned preferentially along the Cαi-1-Cαi axis (or the Ni-Cαi bond).
In practice a reduced form of eq 19, where τf′ is set equal to zero, is used in MF studies. The reason for this simplification is that standard MF fitting schemes can typically only fit three-point single-field data sets, precluding the variation of τf as a free parameter in addition to Ss2, Sf2 and τs. Values of Sf2 are typically in the range of 0.8-0.9. The weight of the last term of eq 19 is (1 – Sf2). Hence a 20-10% contribution is being ignored when the reduced extended MF formula is used, implying further inaccuracies in the best-fit parameters. This can also be realized noticing that formally the reduced extended MF formula is given by J(ω) = Sf2×j0(ω), where j0(ω) = Ss2 τm/(1 + ω2τm2) + (1 – Ss2) τs′/(1 + ω2τs′2) has the form of the K = 0 perturbational expansion (eq 17) featuring the squared order parameter Ss2 and local motional correlation time τs′. This is analogous to JDD(ω) = A jK(0) in SRLS, with the K = 1 and K = 2 terms set equal to zero in eq 10. We call this form of JDD(ω) (and ensuing JCC(ω)) ‘combination 5’, in analogy with model 5 MF (the term ‘combination’ is used instead of ‘model’ since the hierarchy consists of different parameter combinations within the scope of the same model). Setting B j1(ω) = C j2(ω) = 0 in eq 10 was considered justified based on the relation RL∥ >> RL⊥ (implicit in the calculation) which is analogous to τs >> τf in MF.
The fact that the coefficient A is returned by the fitting scheme as 0.8 – 0.9 instead of unity means that in the presence of significant mode-mixing the K = 1 and K = 2 terms will still contribute to JDD(ω) even though RL∥ >> RL⊥. This has been verified by us with relevant calculations.19 While SRLS combination 5 is certainly a better spectral density than MF model 5, since j0(ω) SRLS accounts for mode-mixing while j0(ω) MF does not, it still misses 10-20% contributions, to be absorbed by the best-fit parameters. As shown below, a consistent physical picture is only obtained with rhombic instead of axial ordering, and arbitrary instead of very high local diffusion anisotropy, RL∥/RL⊥.
In summary, the mathematical model-free formulae were introduced as parameterizing spectral densities.6-8 Independently the stochastic SRLS model has been developed first in the perturbation limit for certain simplified geometries,48,30 in the BO limit for axial ordering and isotropic local motion,28 and in its general form.23,24 It turned out that (1) the original MF formula is given by the SRLS solution in the BO limit and the extended MF formula is given by the perturbational expansion of ref. 30, (2) N-H bond dynamics exceeds the perturbation limits and in most cases the BO limit, (3) mode-coupling and general features of local geometry, ignored in both limits, are important, and (4) MF analysis does not stop at the stage of parameterization but proceeds by interpreting the parameterizing quantities in terms of physical quantities (order parameters, correlation times) inherent to the SRLS model. This justifies the assessments associated with the implications of interpreting the MF parameters in terms of physical quantities.
2. Practical implementation of the theoretical premises of SRLS and MF
The basic idea underlying the MF approach is to reproduce the spectral density assuming statistical independence (decoupling) between the mobility of the probe and the mobility of the protein.6-8 This requires large time scale separation between these motions. Based on the theory of moments analytical expressions for the spectral density were suggested. The price paid for simplicity becomes relevant when the parameters obtained by data fitting46,47 are interpreted within the scope of specific models. The MF formulae only agree with high symmetry of the various physical quantities featured, and accommodate only simplified local geometry, besides requiring mode-decoupling. Hence the usage of MF is prone to overextension.
A different but related idea is to envision the overall system to be composed of two bodies, probe and protein, with mobilities coupled by a phenomenological potential energy function. An established set of dynamic variables is modulated according to an explicit model, typically based on stochastic operators. Contrary to MF there is no pretence for generating a universal tool. Instead there is an attempt to treat the experimentally relevant situations within the scope of rigorous formal frameworks. The computational burden is greater than that of the analytical model-free formulation.
The SRLS model features such a framework. It is based on a Smoluchowski equation representing the rotational reorientation of two interacting rotors (bodies).23,24 SRLS was applied earlier to molecular probes in complex fluids and ESR spin relaxation in bio-macromolecules.25,27 Recently we applied SRLS to NMR spin relaxation in proteins.19,20,22,26,27 In this application the two rotors are represented by the locally reorienting spin-bearing moiety (e.g., the N-H bond), and the globally reorienting protein. The global and local motions are described at the diffusive level, hence characterized by two distinct diffusion tensors. The coupling potential, which expresses the spatial restrictions imposed by the immediate protein surroundings at the site of the motion of the probe, depends upon the mutual orientation of the coupled rotors. The physical quantities may be asymmetric, and features of general local geometry are accommodated. Obviously mode-coupling is accounted for rigorously.
Results were compared with MF. SRLS is clearly the generalization of MF, yielding the latter in asymptotic limits. We found that the MF formulae are poor approximations of the experimental spectral density. On the other hand, the SRLS solution appears to match the integrity of currently available experimental data.
The practical problem with SRLS is computational efficiency, as in some cases the (numerical) calculation of the SRLS spectral densities is significantly more demanding than the instantaneous calculation of the simple analytical MF formulae. Otherwise the SRLS and MF fitting schemes are similar. In our first implementation of SRLS in a fitting scheme we pre-calculated 2D grids of spectral density values which were then used as look-up tables. This program is comparable in speed with the MF programs, and is operated in the same way. The best-fit parameters are formally (but not physically) analogous to the parameters of the extended MF formula. The deficiencies of this scheme are that (1) the global motion is isotropic and determined separately from the local motion (similar to the MF strategy), and (2) the symmetry of the local restrictions is axial, as in MF. We found that these limitations must be eliminated. To this end we developed recently a fitting program for SRLS where the generic spectral densities (eq 13) are calculated on the fly. In terms of operating it the only extra requirement on the part of the user is to determine a truncation parameter which controls the number of terms which need to be taken into account for convergence of the solution (given by eq 13 or similar equations). Several trial and error calculations carried out for typical cases suffice. Some aspects of this program are still under development. It is expected that this effort will be brought to completion shortly, at which time this general fitting scheme will be made available to the community. The 2D-grid-based fitting scheme, as well as and the 2D grids, are available upon request. The ‘Theoretical background’ section of this paper comprises all the information needed for ab initio programming.
III. Results and Discussion
1. SRLS versus MF analysis in the asymptotic limit
1a. Geometric effects: the D → CSA frame transformation
When τe is very small the second term in the MF formula (eq 16) can be ignored, yielding the so-called MF ‘model 1’. In this limit the difference between SRLS and MF consists solely of the D-CSA transformation carried out in SRLS and omitted in MF. The implications of this approximation are illustrated below using the experimental data obtained at 295 K, 11.7 T, by Vugmeyster et al.11 for eight out of the 35 residues of the Villin Headpiece Helical Subdomain (VHHS) which were analyzed with MF model 1. τm = 2.5 ns was determined based on T1/T2 ratios.53 We subjected these data to model 1 MF analysis using the program Modelfree 4.0,46 and to combination 1 SRLS analysis using our fitting scheme for axial potentials.22 From S2 MF we calculated c20 using eq 4 with c22 =0, and eq 11. In SRLS we varied c20 and calculated (S20)2 using eq 4 with c22 =0, and eq 11. The results are shown in Table 5.
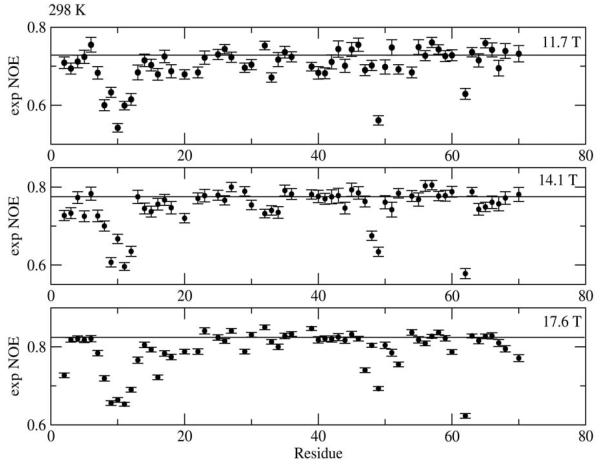
The data in parentheses, obtained by setting D and CSA deliberately collinear in the fitting program for SRLS, are practically identical with the corresponding MF data. This indicates that the two programs perform identically when the D and CSA frames coincide. Thus, the differences between corresponding data in columns 2 and 3 are due solely to the D-to-CSA frame transformation which was carried out for a tilt angle θ =17° (ref. 3) in SRLS and omitted in MF. Underestimation of (S20)2 by MF on the order of 4.5% is not negligible given that currently reported precision in S2 is, in some cases, on the order of 1% (ref. 11) and the precision in the average value of S2 on the order of 0.2% (ref. 13). The error in S2 has severe implications for conformational entropy calculations (see below). Recently θ = 21.4° was determined with an extensive Ubiquitin data set.54 The larger angle θ implies even greater inaccuracies than shown in Table 5.
MF is clearly force-fitting the experimental data yielding S2 and corresponding c20 values which are too low. Table 6 illustrates this using the experimental data of residue 49 of VHHS. Back-calculated 15N T1, T2 and 15N-{1H} NOE relaxation parameters obtained with the best-fit SRLS and MF order parameters of Table 5 are shown in Table 6a aside the experimental data. Table 6b shows the specific J(ω) values associated with the back-calculated T1, T2 and NOE data of Table 6a. It can be seen that the MF spectral density can fit the experimental data as well as the SRLS spectral density (Table 6a) by compensation of the individual J(ω) values (Table 6b). In particular, MF yields smaller JDD(ω) values and larger JCC(ω) values than their correct SRLS counterparts. Note the significantly different values of JDD(0) and JDD(ωN) versus JCC(0) and JCC(ωN) in SRLS, implied by carrying out the D-to-CSA frame transformation. Also note that (with one exception) the experimental NOE exceeds the maximum NOE as shown by %Dmax > 0 (Table 5). This feature will be discussed below in detail.
Table 6a.
Experimental and back-calculated SRLS and MF 15N T1, T2 and 15N-{1H} NOE values obtained with the best fit squared order parameters shown in Table 5 for residue 49. In this Table, and in all of the Tables and Figures below where T1, T2 and NOE were calculated, we used 15N CSA of σ∥ – σ⊥ = −170 ppm, rNH = 1.02 Ǻ (e.g., ref 11) and θ = 17° (ref. 3).
| T1 [ms] | T2 [ms] | NOE | |
|---|---|---|---|
| Exp | 381.2±6.1 | 251.5±3.1 | 0.565±0.011 |
| SRLS | 387.8 | 250.9 | 0.5533 |
| MF | 386.7 | 249.5 | 0.5758 |
Table 6b.
J(ω) values for ω = 0, ωN, ωH+ωN, ωH and ωH−ωN for dipolar auto-correlation and 15N CSA auto-correlation. The c20 values (and the corresponding (S20)2 values from Table 5) used in these calculations are given under the heading ‘Input’. τm = 2.5 ns was used. The units of J(ω) are ns/rad.
| Input | Output | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (S20)2 | c20 | JDD(0) | JDD(ωN) | JDD(ωH+ωN) | JDD(ωH) | JDD(ωH−ωN) | JCC(0) | JCC(ωN) | |
| SRLS: | 0.853 | 13.5 | 0.854 | 0.523 | 0.0168 | 0.0130 | 0.0113 | 0.651 | 0.398 |
| MF: | 0.817 | 10.8 | 0.817 | 0.500 | 0.0161 | 0.0130 | 0.0108 | 0.817 | 0.500 |
1b. Local motion effects
The 15N relaxation data of 21 VHHS residues were fit by Vugmeyster et al.11 with model 2, where the complete original MF formula (eq 16) is used. We subjected 15 out of 21 residues to SRLS analysis using combination 2 and repeated the calculations of Vugmeyster et al.11 using the same computer program (Modelfree 4.0). The average results obtained for the squared order parameters and the associated c20 values are shown in Table 7 under the heading ‘model 2’. For comparison the average results of Table 5, including c20 corresponding to the squared order parameters, are also shown under the heading ‘model 1’.
Table 7.
Average c20 and corresponding (S20)2 best-fit SRLS parameters. Average S2 and corresponding c20 best-fit MF parameters. %diff represents 100×[param(MF) − param(SRLS)]/param(SRLS).
| Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|
| SRLS | MF | %diff | SRLS | MF | %diff | |
| (S20)2 | 0.87 | 0.83 | −4.5 | 0.73 | 0.78 | +6.8 |
| c20 | 15.4 | 11.7 | −23 | 7.5 | 9.0 | +20 |
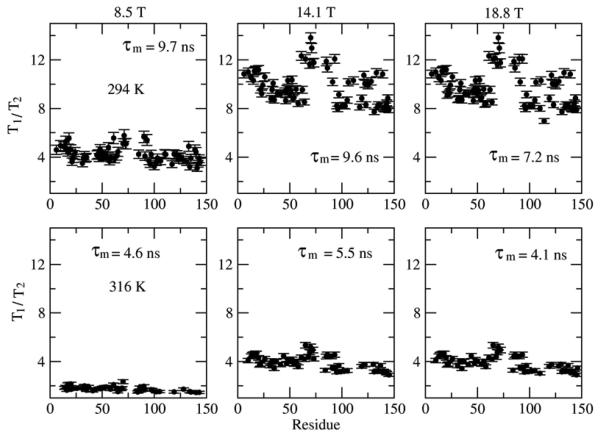
The differences featured by the ‘model 1’ SRLS and MF results stem solely from the geometric D → CSA frame transformation. The differences featured by the ‘model 2’ SRLS and MF results stem from the D → CSA frame transformation, and from the effect of additional local motion eigenmodes on the form of the local motion correlation function, accounted for in SRLS and ignored in MF. SRLS yielded <τL/τm> = 0.1 whereas MF yielded <τe/τm> = 0.02 (data not shown). If τL would be derived from τe according to eq 18 then the MF time scale separation would have been 0.09, which is close to its SRLS counterpart (differing by only 10%).
Table 7 shows that S2 over-estimates (S20)2 by nearly 7% in model 2 and under-estimates it by approximately 4.5% in model 1. As already mentioned, this has implications for the precision of S2 MF. Most importantly, it affects the accuracy of thermodynamic parameters calculated from potentials derived from S2 MF.33-36 The coefficient c20 of the general form of the potential (eq 4 with c22 = 0) is very sensitive to changes in (S20)2 when (S20)2 is high, as shown in Fig. 4 which is the graphical representation of eq 4 with c22 = 0. Because of the asymptotic form of the (S20)2 versus c20 curve as (S20)2 → 1, relatively small uncertainty in (S20)2 imply large uncertainty in c20. For example, in Table 5 the S2 errors cover the range between −4.5% and 6.8% whereas the c20 errors cover the range between −23% and 20%. Note that these large errors in the strength of the potential, hence in the probability distribution function Peq = exp(-U/BT), stem solely from the geometric effect of omitting the D-to-CSA frame transformation. Significantly larger inaccuracies are implied by also disregarding the possibility that the correlation function for local motion has a more complex form implied by the presence of additional eigenmodes, and over-simplifying the symmetry of the local ordering.
Figure 4.
Squared order parameter, (S20)2 as a function of the potential coefficient, c20 (in units of kBT) determined with eq 4 with c22 = 0, and eq 11.
As discussed above, the effective correlation time, τe, typically reported in MF studies as a ‘correlation time for local motion’, is actually a composite, approximately given by 2τL/c20 (eq 18). For S2=0.8, 0.9 and 0.95, corresponding to c20 = 10, 20 and 40, respectively, the parameter τe is 5, 10 and 20 times smaller than τL = 1/6RL. The ratio τe/τm grossly over-estimates what is considered to represent the time scale separation between the rate of global reorientation (RC) and the rate of local reorientation (RL) (note that 1/τm and 1/τe are global motion mode and main local motion mode eigenvalues, respectively, whereas RC = 1/6τm and RL=1/6τL are ‘bare’ diffusion constants for global and local motion, respectively). This may lead to inclusion of non-rigid residues into data sets used to determine τm from T1/T2,53 and improper usage of the Reduced Spectral Density55-57and model-independent58 approaches which are only valid when the local motion correlation time, τL, is very fast. It was observed by several authors14-16 that τe is nearly invariant as a function of temperature. This is not surprising since τe ∝ τL/c20, with both the numerator and the denominator decreasing with increasing temperature. While τL may exhibit Arrhenius-type temperature dependence, τe might not.
1c. General considerations
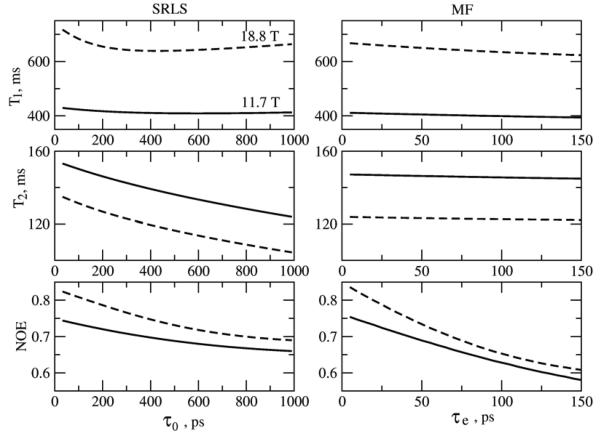
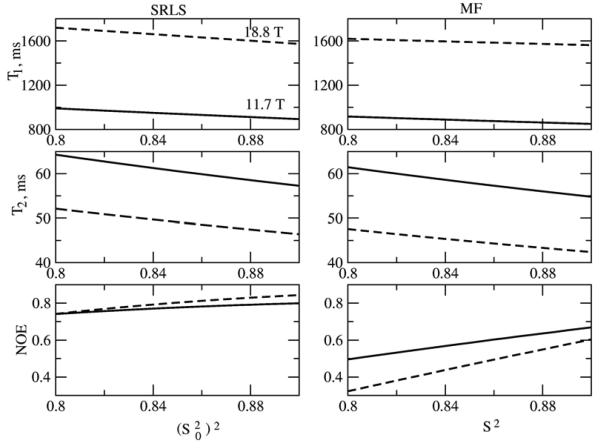
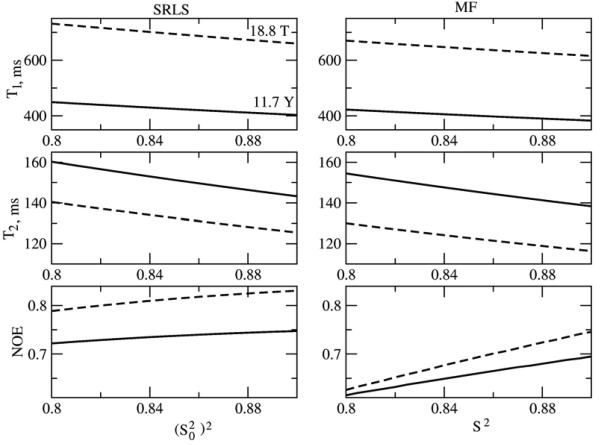
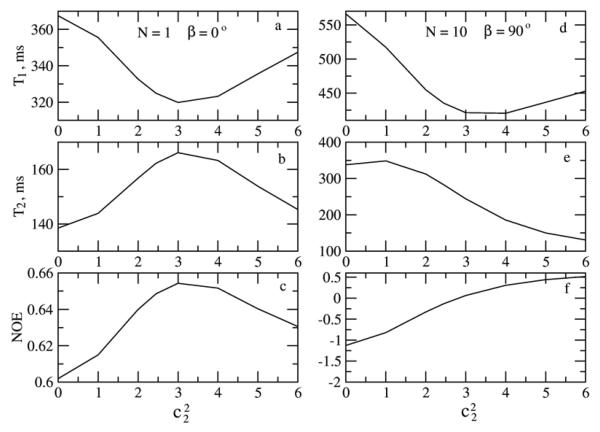
Illustrative simulated SRLS and MF T1, T2 and NOE values are shown in Figs. 5-8 for the parameter range where the original MF formula is typically applied. High ordering ((S20)2 = 0.85) and large time scale separation (τe/τm = 0.015 when τm = 5 ns, and τe/τm = 0.005 when τm = 15 ns) were used. In Figs. 5 and 7 c20 is fixed at 13.2, corresponding to (S20)2 = 0.85. In the SRLS calculations the local motion correlation time, τL = 1/6RL, is varied from 0 to 1000 ps (curves on the left). The corresponding MF effective local motion correlation time, τe = 2τL/c20 = τL/6.6, is varied from 0 to 150 ps (curves on the right). The global motion correlation times are τm = 5 and 15 ns, and the magnetic fields are 11.7 and 18.8 T. For fixed τm the parameter τL (τe) represents the time scale separation τL/τm (τe/τm). The SRLS and MF relaxation parameters in Figs. 5 and 7 were calculated using the same physical input (i.e, τL = 1/6RL in SRLS and τe = 2 τL/c20 in MF, with all the other parameters the same). It can be seen clearly that all the SRLS relaxation parameters depend significantly on τL/τm in ways which differ for low and high fields and small and large proteins (or high and low temperatures). On the other hand, in the parameter range considered, the MF T1 and T2 values vary to a small extent as a function of τe/τm, whereas the MF NOE's vary significantly in ways which differ from the variations in the SRLS NOE's.
Figure 5.
15N T1, T2 and NOE calculated with SRLS (JDD(ω) was calculated according to eq 10, and JCC(ω) as explained after eq 9) and MF (eq 16) for τm = 5 ns, 11.7 and 18.8 T, as a function of τL = τ0 = 1/6RL (left - SRLS) and corresponding τe = 2τ0/c20 (right – MF). The potential coefficient c20 = 13.2, corresponding to (S20)2 = 0.85, was used.
Figure 8.
15N T1, T2 and NOE calculated for τm = 15 ns, 11.7 and 18.8 T, as a function of the squared order parameter, (S20)2 (left - SRLS), and the generalized squared order, S2 (right - MF). The potential coefficient c20 = 13.2, corresponding to (S20)2 = 0.85, and was used. τ0 = 75 ps in SRLS, and τe = 75 ps in MF, were used.
Figure 7.
15N T1, T2 and NOE calculated for τm = 15 ns, 11.7 and 18.8 T, as a function of τL = τ0 = 1/6RL (left - SRLS) and corresponding τe = 2τ0/c20 (right - MF). The potential coefficient c20 = 13.2, corresponding to (S20)2 = 0.85, was used.
Table 8 shows the percent difference between 1/T1, 1/T2 and NOE shown in Figs. 5 and 7 calculated with SRLS for τL = 495 ps and MF for τe = 75 ps. These data illustrate clearly the field-dependence of the best-fit MF parameters. In this example the features which are likely to yield different results at different fields are the opposite trends in the 1/T1 discrepancy between SRLS and MF at 11.7 and 18.8 T for τm = 5 ns, and the very large field-dependence of the NOE discrepancy for τm = 15 ns. Clearly, fitting of combined multi-field data is expected to be problematic and in most cases impossible with MF, as often encountered in practice.
Table 8.
Percent difference ((varMF-varSRLS)/varSRLS), where ‘var’ represents 1/T1, 1/T2 or NOE from Figs. 5 and 7 for τL = 495 ps and the corresponding value of τe = 75 ps. The potential coefficient c20 = 13.2 ((S20)2 = 0.8) was used. The τm values and the magnetic field strengths are given in the Table.
| %diff versus SRLS | 1/T1,1/s | 1/T2, 1/s | NOE |
|---|---|---|---|
| 5 ns, 11.7 T | +0.8 | −6.2 | −4.3 |
| 5 ns, 18.8 T | −1.3 | −5.2 | −6.8 |
| 15 ns, 11.7 T | −5.9 | 0.0 | −4.8 |
| 15 ns, 18.8 T | −18.2 | +4.4 | −16.7 |
Another feature illustrated in Table 8 is the field-dependence of the deviation of the MF T1/T2 value from the SRLS T1/T2 value implying field-dependent τm values, as often encountered in MF analyses. The relaxation parameters shown in Figs. 5 and 7 were calculated with the proper analogy between SRLS and MF local motion correlation times, i.e., τL SRLS corresponds to τe = 2τL/c20 MF. The relaxation parameters shown in Figs. 6 and 8 were calculated with the improper analogy, i.e., τL SRLS the same as τe MF. This is done to show how misleading it is to consider τe as representing a bare (i.e., 1/6RL) local motion correlation time, although Lipari and Szabo6 indicated that this quantity is a composite. For example, one expects Arrhenius-type temperature dependence of τe, and obtains typically near temperature independence. Within the scope of the wobble-in-a-cone model τe depends on Dw and S2 (ref. 6). The physical parameter is the wobbling rate, Dw. If τs and Ss2, obtained from data dominated by mode-coupling, are used in this context the implications can be detrimental, as shown in section 5d.
Figure 6.
15N T1, T2 and NOE calculated with SRLS and MF for τm = 5 ns, 11.7 and 18.8 T, as a function of the squared order parameter, (S20)2 (left - SRLS) and the generalized squared order, S2 (right - MF). The potential coefficient c20 = 13.2, corresponding to (S20)2 = 0.85, and was used. τ0 = 75 ps in SRLS, and τe = 75 ps in MF, were used.
Figs. 5 and 7 show that SRLS relaxation rates calculated with τL agree reasonably with MF relaxation rates calculated with τe = 2τL/c20 for τL < 200 when τm = 5 ns, and for τL < 100 ps when τm = 15 ns. On the other hand, when τe = τL (Figs. 6 and 8) the agreement between the T1 and T2 values is reasonable (since in MF T1 and T2 depend only to a small extent on τe - see Figs. 5 and 7) but the NOE's differ significantly. This is precisely the empirical observation, which motivated the development of the extended MF formula8 commonly used in its reduced version. In the reduced extended MF formula a slow effective correlation time, τs, adjusts the NOE, whereas a scaling factor, Sf2, adjusts 1/T1 and 1/T2. This is related intimately to the ‘mode-independent’ form of this formula, with τm affecting predominantly the global motion term, τs affecting exclusively the local motion term, and neither affecting the weights of these terms. On the other hand, in SRLS the motional rates and the potential coefficients determine the weights of the various modes contributing to the spectral density, and the eigenvalues of the solution differ from the pure ‘mode-independent’ eigenvalues 1/τm and 1/τL. The MF parameterization of the spectral density impairs statistical properties of genuine fitting. For example, Vugmeyster et al.11 reported on a non-normal t-distribution in NOE values back-calculated using best-fit MF parameters obtained with models 1 and 2.
1d. τm determination
In MF the determination of the global diffusion tensor53 is implicitly based on the ‘Born-Oppenheimer’ type approximation inherent in factoring C(t) into CL(t)×CC(t) (eq 14). When CC(t) = exp(-t/τm), i.e., the protein is spherical, and CL(t) is given by the first term of eq 16 (model 1, where τL = 0), then deriving τm from T1/T2 is appropriate in SRLS, but in MF it is as inaccurate as implied by the omission of the D-to-CSA frame transformation. The various filtering procedures devised to extract from the complete data set the sub-set used for RC determination53 do not eliminate data, which correspond to the original MF formula (eq 16) (‘model 2’, where τL ≠ 0). The high sensitivity to τL of T1 and T2 for small proteins (Fig. 5), and T1 for large proteins (Fig. 7), indicates that significant errors will be introduced by including ‘model 2‘ data in the process of RC determination. When CC(t) corresponds to axial global diffusion and/or CL(t) corresponds to eq 19, the full SRLS time correlation function is to be used. Very precise τm values were reported recently by an MF study15 where C(t) was used in the fitting process, with CC(t) corresponding to axial global diffusion and CL(t) given by eqs 16 and 19. For the reasons outlined above the accuracy and precision of these results should be reassessed.
As shown below, when axial potentials are used model 1 is often selected instead of model 2 by force-fitting, yielding unduly high (S20)2 values. This has been documented in the literature by a recent SRLS application to nitroxide-labeled biomacromolecules which showed that force-fitting by using model 1 on a set of synthetic data which correspond to model 2, generates (S20)2 values which are too high and τm values which are too low.25 The discrepancies increase with increasing magnetic field.25 These are precisely the trends observed with MF analyses - higher S2 and lower τm at higher fields.
We found that a useful method for estimating the precision of τm is to first determine it from T1/T2 ratios (desirably acquired at low magnetic fields) of combination 1 fits, where τL → 0, and then scan the vicinity of this value. Illustrative calculations were carried out for residue 45 of VHHS acquired at 11.7 T, 295 K, fit by Vugmeyster et al.11 with MF model 1. The T1/T2-derived τm value is 2.5 ns. χ2 values obtained as a function of τm within a ±0.5 ns range centered at τm = 2.5 ns, using SRLS (black curve) and MF (red curve) are shown in Fig. 9. Let us assume that χ2 = 10 is the threshold (Vugmeyster et al.11 set χ2 = 25 as threshold for the site-specific fitting). It can be seen that in both cases practically identical (S20)2 (S2) values are obtained for τm within a range of 5-6% from the 2.58 (2.50) ns minimum. We believe that this is a realistic estimate of the precision with which τm can be currently determined in the BO limit for axial local ordering. Precision estimates of 0.2% (ref. 15) are highly over-rated. As shown in Fig. 9 about 4% accuracy is gained in this case by using SRLS instead of MF.
Figure 9.
χ2 probability distribution as a function of the global motion correlation time calculated with combination 1 SRLS (blue) and model 1 MF (red) for residue 45 of VHHS. The MF calculations used the same spectral densities as the SRLS calculations, except that the frame transformation D-to-CSA was omitted.
2. SRLS versus MF analysis in the mode-mixing regime
One of the greatest benefits of SRLS lies in the treatment of flexible residues where mode-mixing dominates the spectral density, as a consequence of local and global motions occurring on similar time scales. Allowing for rhombic local ordering shown previously to prevail at the N-H bond,26 and accounting for general features of local geometry,22-27 constitute additional significant advantages over MF. As pointed previously22,27 and mentioned above, the reduced extended MF formula is formally equivalent to JDD(ω) = A j0(ω), with SRLS fitting yielding A ∼ Sf2 ∼ 0.8 – 0.9. These spectral densities are typically inaccurate because terms, which contribute 10 – 20% have been omitted. Let us illustrate this quantitatively. The value of A = (1.5 cos2βMD - 0.5)2 = 0.8 is obtained for βMD = 15.4°, and A = 0.9 obtains for βMD = 10.7°. The values of B = 3 sin2βMD cos2βMD (C = 0.75 sin4βMD) corresponding to these angles are 0.20 (0.004) and 0.1 (0.0009), respectively. B is clearly not very small, but B j1(ω) could become negligible if j1(ω) were much smaller than j0(ω), in view of the imposed condition that RL∥ >> RL⊥. This is not borne out by the SRLS analysis of the experiment, since if it were, the fitting scheme would have returned A = 1, corresponding to βMD = 0°. By analogy, the validity of ‘reducing’ the extended MF formula, based on the condition that τs >> τf, is not borne out by the experiment, since if it were, the MF fitting scheme would have returned Sf2 = 1.
The values of j1(ω) and j2(ω) are, indeed, much smaller than j0(ω) for all ω values when RL∥ >> RL⊥, in the absence of mode-mixing. The presence of mode-mixing invalidates the relations j1(ω) << j0(ω) and j2(ω) << j0(ω) in the high ω regime. This is illustrated in Fig. 10, where we show the SRLS functions jK(ω) calculated using as input the best-fit parameters obtained by fitting with SRLS the 15N relaxation data of residue 124 of RNase H, acquired at 11.7 T, 300K. This residue pertains to the flexible loop αD/β5 and was fit previously with model 5 by Mandel et al.12 It can be seen that in the low frequency region j0(ω) >> j1(ω) and j2(ω). However, in the frequency range comprising the ω values relevant for the NOE (ωH + ωN and ωH - ωN) the j1(ω) values exceed the corresponding j0(ω) and j2(ω) values. Clearly even when RL∥ >> RL⊥ (or τs >> τf in MF) the term B j1(ω) cannot be ignored in eq 12 for the βMD = 10 - 20° geometry when mode-mixing is important. For the very same reason the incorrect eq 19 is further impaired when its last term is omitted.
Figure 10.
jK(ω) functions obtained with c20 = 3.2 ((S20)2 = 0.4), τ0/τm = 0.45, βMD = 16.3° and N = RL /RL∥ ⊥ = 916. The high-frequency region is shown in panel ‘a’ and the low-frequency region in panel ‘b’.
In principle setting B j1(ω) and C j2(ω) equal to zero implies the βMD = 0° geometry which corresponds to A = 1. In practice the fitting schemes returns A ≠ 1, indicating that force-fitting has occurred. The βMD = 90° geometry (which is approximately correct as the angle between N-H and Cαi-1-Cαi is 101.3°) corresponds to A = 0.25, B = 0 and C = 0.75. As shown below, this materializes by fitting when the potential is allowed to be rhombic and RL∥/RL⊥ is allowed to be arbitrary instead of being forced to be very high.
In the perturbation limit the ordering is very small. Motion about ZM is prohibited physically as the N-H bond is attached to the protein backbone. The highly plausible local diffusion/local ordering axes Cαi−1-Cαi and Ni-Cαi are tilted at approximately 90° from the N-H bond. Therefore the βMD = 90° geometry is actually implied in this limit. Outside the perturbation limit, ZM may be tilted with respect to the N-H bond. In order to assign physical meaning to the tensor RL we will assume, based on stereo-chemical considerations, that the βMD = 90° is preserved in the general case.
The experimental data of residue 124 of RNase H are further used for illustrative purposes as follows. They are shown in Table 9a along with the NMR relaxation data back-calculated with MF and SRLS using the best-fit parameters (given in Table 9b) obtained with the respective fitting processes. Clearly both SRLS and MF reproduce the experimental data. The specific J(ω) values entering the expressions for T1, T2 and NOE are shown in Table 9b. It can be seen that MF can fit the experimental data as well as SRLS, but this requires that the relevant JDD(ω) values be under-estimated, and the relevant JCC(ω) values over-estimated. This is a clear example of force-fitting. The different best-fit parameters can be further rationalized by examining Table 9c, where we show the dominant eigenvalues and their weights, which are clearly different in SRLS and MF. Thus, a fictitious physical situation characterized by MF eigenvalues and weights of C(t) and best-fit parameters of J(ω) obtained with force-fitting can reproduce technically very well the experimental data.
Table 9a.
Best-fit T1, T2 and NOE values corresponding to the best-fit parameters shown in Table 9b under the heading ‘input’. The χ2 values were practically zero for both SRLS and MF.
| T1 [ms] | T2 [ms] | NOE | |
|---|---|---|---|
| EXP | 633.8±10.1 | 112.3±3.1 | 0.5343±0.031 |
| SRLS | 633.8 | 112.3 | 0.5344 |
| MF | 633.8 | 112.3 | 0.5343 |
Table 9b.
Best-fit J(ω) values corresponding to the best-fit parameters listed under the heading ‘input’, used to calculate the data of Table 9a. The SRLS input set includes RC = 0.47 (τL⊥ = 4.36 ns, τm = 9.28 ns)), c20 = 3.04 ((S20)2 = 0.37) and (βMD = 16.6° (formally Sf2 = 0.770). N = RL∥/RL⊥ was fixed at the value of 1000. The MF input set includes τs/τm = 0.114, c20 = 10.2 derived from Ss2 = 0.806 (eqs 4 and 11), Sf2 = 0.809 and τf = 0. The formally22,27 analogous SRLS parameters are RC, c20, A = (P2(cos(βMD)))2 and RL∥ >> RL⊥, respectively. The units of J(ω) are ns.
| Input | Output | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c20 | (S20)2 | βMD | RC | JDD(0) | JDD(ωN) | JDD(ωH+ωN) | JDD(ωN) | JDD(ωH+ωN) | JCC(0) | JCC(ωN) |
| 3.01 | 0.368 | 16.6 | 0.47 | 2.61 | 0.318 | 1.09 | 0.0093 | 0.0081 | 1.96 | 0.241 |
| 10.2 | 0.806 | 15.0 | 0.11 | 2.48 | 0.303 | 1.08 | 0.0087 | 0.0074 | 2.48 | 0.303 |
Table 9c.
Significantly contributing SRLS eigenvalues and associated weights, and corresponding ‘independent’ MF eigenvalues and associated weights. The eigenvalues are given in units of RL, hence the ‘independent’ local motion eigenvalue is 6.
| SRLS | MF | |||||
|---|---|---|---|---|---|---|
| Eigenvalue | 2.13 | 10.12 | 24.52 | 14.13 | 0.678 | 6 |
| weight | 0.61 | 0.325 | 0.042 | 0.019 | 0.652 | 0.157 |
Note that the weights of the local and global motion terms in Table 9c, yielded by the reduced extended MF formula (eq 19 with the last term set equal to zero), do not sum up to unity. There is no requirement in the MF fitting schemes for normalization in the so-called ‘model 5’, which uses this formula. This is yet another aspect of the MF treatment which implies force-fitting of the experimental data. Additional confusion with regard to what the parameter τs represents is implied by the phenomenon of renormalization, which is important when the local potential is high. Since some Ss2 values are low and others are high, this complicates further the comparison among the τs values of different N-H sites. Finally, conformational entropy is often calculated from S2 instead of Ss2. The parameter Ss2 is formally equivalent to (S20)2, whereas S2 taken as Sf2×Ss2 includes the parameter Sf2 which is formally analogous to a geometric factor, P2(cos(βMD))2).22 Ss2 is highly inaccurate;19,20,22 S2 is qualitatively problematic.
We conclude this section by discussing the limit N = RL∥/RL⊥ >>1, which is the analogue of the MF restriction τs >> τf. The jK(ω) functions in Fig. 10 were obtained with N ∼ 1000. We show in Fig. 11 SRLS jK(ω) functions obtained with N = 1. Except for the value of N, the parameters used to calculate the Fig. 11 functions - RC = τL⊥/τm = 0.57, c20 = 4.04 ((S20)2 = 0.51), βMD = 20° and RL∥ >> RL⊥ - are quite similar to those used to calculate the Fig.10 functions. It can be seen that for a small time scale separation (τL⊥/τm = 0.57), moderate ordering ((S20)2 = 0.51) and the βMD = 20° geometry, the functions j1(ω) and j2(ω) are comparable in magnitude to j0(ω) over the entire range of ω values when N = 1 (Fig. 11). On the other hand, j0(ω) >> j1(ω), j2(ω) in the low frequency regime, whereas j1(ω) > j0(ω), j2(ω) in the high frequency regime when N >> 1 (Fig. 10). Thus, the parameter N affects the analysis significantly.
Figure 11.
SRLS spectral densities jK(ω) obtained with c20 = 4.04 ((S20)2 = 0.51), τL⊥/τm = 0.57, βMD = 20° and RL∥/RL⊥ >> 1 (blue curves). Reduced extended MF spectral density calculated with Ss2 = 0.51, Sf2 = 0.68 and τs′/τm = 0.57 (red curve). The coefficients of the SRLS jK(ω) functions shown in the expression of JDD(ω) are 0.68, 0.31 and 0.01 for K = 0, 1 and 2, respectively.
The restriction to high N = RL∥/RL⊥ was imposed in our first fitting scheme for SRLS (ref. 22) for practical reasons. RL∥ and RL⊥ represent the principal values of the diffusion tensor of ‘body 1’, i.e., the N-H bond. The inequality RL∥/RL⊥ >> 1 is clearly a simplifying approximation. As shown below, only by removing this restriction, and allowing for rhombic potentials, is a consistent physical picture obtained with data fitting. In SRLS the principal values of the local diffusion tensor comprise information on physically meaningful variations among the various N-H sites (examples of such variations among nitroxide-labeled sites in proteins appear in ref. 37). This information is lost when the restriction that N >> 1, whereby RL∥ is forced to be in the extreme motional narrowing limit, is imposed.
Within the scope of the formal (definitely not physical) analogy between SRLS and MF, RL∥/RL⊥ >> 1 in SRLS corresponds to τs/τf >> 1 in MF. Based on the results presented below, which indicate that RL∥/RL⊥ >> 1 is not to be imposed, the mathematical MF inequality τs/τf >> 1 constitutes an inappropriate oversimplification.
For comparison we also show in Fig. 11 the reduced extended MF spectral density obtained with the analogous parameters - τs′/τm = 0.57, Ss2 = 0.51, Sf2 = 0.68 ( Sf2 = A = 1.5 cos2(20°) - 0.5) and τf' = 0 (red curve). This function is clearly a poor approximation of JDD(ω) assembled according to eq 12 from the jK(ω) functions of Fig. 11, with the coefficients A = 0.68, B = 0.31 and C = 0.01, corresponding to the βMD = 20° geometry. This illustrates clearly the limited capabilities of MF, which cannot reproduce physical situations where RL∥/RL⊥ ∼ 1.
3. Conformational entropy derived in MF from S2.
In recent years squared generalized MF order parameters have been used extensively to derive thermodynamic quantities, notably configurational entropy.33-36 The logic behind this approach is as follows. S2 is determined with data fitting. Subsequently it is assumed that Peq is axially symmetric. Thereby the squared generalized order parameter becomes the square of an axial order parameter. This enables the derivation of the strength of an axial potential, the form of which must be guessed, based on equations similar to our eq 11. Since MF is an SRLS asymptote, the potential form given by eq 4 (with c22 set equal to zero) is appropriate. Employing other potential forms33-36 may increase the inaccuracy in the configurational entropy derived from the already inaccurate S2 value.
Large errors in the strength of the potential U/kBT = –c20 P2cos2βMD are illustrated in Table 7 in the best-case scenario where S2 is determined with models 1 or 2. As pointed out above, the errors in c20 are significantly larger than the errors in S2 due to the functional form of the (S20)2 versus c20 dependence (Fig.4). Table 7 shows that within the context of model 1 (or combination 1) the potential coefficient (c20) derived from S2 MF underestimates the SRLS potential coefficient on average by 23%, whereas in the context of model 2 (or combination 2) the potential coefficient derived from S2 MF overestimates the SRLS potential coefficient on average by 20%. Much larger inaccuracies in the potential underlying the calculation of configurational entropy are expected in the extended MF regime, where Ss2 MF and (S20)2 SRLS differ by factors of 3 - 4 (refs. 19, 20 and 22). When the local potential is rhombic, as it turns out to be at the N-H site,26 MF cannot provide the equilibrium probability distribution function, Peq = exp(U/kBT), since only one parameter, S2, is available while a rhombic potential is defined by at least two coefficients.
Contrary to MF, in SRLS the general rhombic form of the potential is implicit in the theory and its coefficients (c20 and c22) can be obtained directly with data fitting. The SRLS theory also emphasizes the relevance of the Euler angles ΩC′M. Peq, and therefore the thermodynamic quantities, are obtained straightforwardly in SRLS. Order parameters are thus not required to infer the potential in order to derive conformational entropy. They can be calculated independently, if so desired, using eqs 6-9.
4. Rhombic symmetry of the local potential/local ordering
4a. The rhombicity of the local ordering and the axiality of the global diffusion
A very large effect on the analysis not accounted for in MF is the rhombicity of the potential U/kBT(ΩCM). This is illustrated below in quantitative terms. The symmetry of the potential depends on the symmetry of local diffusion/local ordering frame, M, and the symmetry of the local director frame, C. In SRLS the local director is taken to be uniaxial for simplicity but the M frame is in general allowed to be rhombic. We found previously that the particular rhombic symmetry of the M frame is of the ‘nearly planar YM-XM’ type.26 Fig. 12 illustrates these frames in the context of the stereo-chemistry of the peptide plane. The C axis is considered to lie along the equilibrium Cαi−1-Cαi axis. The main ordering axis, YM, is parallel to the instantaneous orientation of the Cαi−1-Cαi axis. ZM is perpendicular to YM within the peptide plane and XM is perpendicular to both YM and ZM, i.e., perpendicular to the peptide plane. The axis XM lies along the symmetry axis of the lone electron pair of the nitrogen, assigning clear meaning to the ‘nearly planar YM-XM ordering’ symmetry, where |Sxx|, |Syy| >> |Szz|, Syy > 0 and |Syy| is slightly larger than |Sxx|. In the original MF formula, and in the extended MF formula as presented by its developers, C lies implicitly along the equilibrium N-H orientation. In a high ordering scenario the implied motion around N-H is not viable. On the other hand, motion around Cαi−1-Cαi, is definitely viable. Note that taking the local director to lie along the Cαi−1-Cαi axis within a presumed rigid peptide plane sets the angle βMD close to 90° for the N-H bond and close to 0° for the C′-Cα bond.
Figure 12.
Schematic illustration of high ‘nearly planar YM-XM ordering’ prevailing at the NH site, with YM as main ordering axis. The M frame denotes the rhombic local ordering/local diffusion frame. YM lies along the instantaneous orientation of the Cαi−1-Cαi axis (or the Ni-Cαi bond). XM lies along the symmetry axis of the lone pair of the nitrogen. The C frame denotes the uniaxial local director frame with ZC along the equilibrium orientation of the Cαi−1-Cαi axis (or the Ni-Cαi bond). Within the scope of high ordering YM is aligned preferentially along the C axis.
The rhombicity of the local ordering tensor, S, is outside the scope of MF. Since the above geometric considerations imply that rhombicity affects the experimental data, it must be absorbed by the best-fit MF parameters. We showed previously26 that the conformational exchange parameter, Rex, can absorb S rhombicity in the data fitting process. Another likely candidate, in particular in the BO limit where mode-mixing is limited, is RC axiality, as explored below.
The large effect of the symmetry of the local potential on the analysis is illustrated in Table 10 which shows NMR relaxation rates calculated for RC = 0.01, βMD = 0°, τm = 15 ns and axial or rhombic potentials on the order of 10×kBT. In the axial case c20 = 8 ((S20)2 = 0.754), whereas in the rhombic case c20 = 8 and c22 = 4. This is moderate rhombicity, corresponding to (Sxx - Syy)/Szz = 8.6%, where Sxx = −0.382, Syy = −0.454 and Szz = 0.836 are the principal values of the Cartesian ordering tensor. As shown in Table 10, potential (or ordering) rhombicity affects the NOE to a very large extent, amounting to 31.6 % (46.3 %) difference from the axial case at 11.7 T (18.8 T). The parameters used in Table 10 for the axial potential case are also used in Table 11 to illustrate the effect of global diffusion axiality with RC∥/RC⊥ = 1.2, which is a typical value for globular proteins.10 ‘%diff’ denotes the percent difference between corresponding variables calculated with βCC′ = 0° and βCC′ = 90°, where βCC′ denotes the angle between the (uniaxial) local director frame, C′, and the (axial) global diffusion frame, C. The effect illustrated in Table 11 is small relative to the large effect of moderate potential rhombicity on the NOE illustrated in Table 10. It is very likely that in many cases the rhombicity of the ordering tensor, S, was absorbed in the MF analyses by introducing RC axiality, in particular when the total time correlation function, C(t), rather than the time correlation function for global motion, CC(t), is used to determine RC. In the former case 15N T1, T2 and 15N-{1H} NOE enter the analysis, whereas in the latter case only 15N T1 and T2 enter the analysis.
Table 10.
Percent difference [var(axial) − var(rhombic)]/var(axial)×100 between 15N T1, T2 and NOE calculated with τm = 15 ns, RC = 0.01, and an axial (c20 = 8 and c22 = 0) or a rhombic (c20 = 8 and c22 = 4) potential. Calculations are shown for magnetic fields of 11.7, 14.1 and 18.8 T.
| 11.7 T | 14.1 T | 18.8 T | |
|---|---|---|---|
| T1 | −2.4 | −1.0 | +1.5 |
| T2 | −7.6 | −7.5 | −7.6 |
| NOE | +31.6 | +39.3 | +46.3 |
Table 11.
Percent difference [var(βCC'=0°) − var(βCC'=90°)]/var(βCC'=0°) ×100 between 15N T1, T2 and NOE calculated with τm(app) = 15 ns, RC(app) = 0.01, an axial potential given by c20 = 8, and a global diffusion anisotropy RC∥/RC⊥ = 1.2. Calculations are shown for magnetic fields of 11.7, 14.1 and 18.8 T.
| 11.7 | 14.1 | 18.8 | |
|---|---|---|---|
| T1 | +7.4 | +7.1 | +6.1 |
| T2 | −9.0 | −9.0 | −9.2 |
| NOE | −2.7 | −3.5 | −4.0 |
The data shown in Table 11 were obtained for a time scale separation of 0.01, which is quite large, and a potential strength of c20 = 8, which corresponds to the relatively high ordering of (S20)2 = 0.754. In this parameters range the effect of additional local motion eigenmodes on the correlation function is not very large.27 However, not accounting for it, and oversimplifying the local geometry, render the MF-based T1 and T2 values inaccurate by 7% and 9%, respectively. Interestingly, the field dependence of these discrepancies is small. This indicates that in those cases where MF analysis yields significantly field-dependent τm values either mode-mixing is pervasive in the experimental data, or the latter feature rhombic potentials.
It should be noted that in general all the parameters entering the Jx(ω) functions, including the global diffusion tensor, are to be determined in the same fitting process. The separate determination of RC in MF is implied by the mode-independence concept, the applicability of which to N-H bond dynamics we challenge herein.
4b. ‘Nearly planar YM-XM’ rhombic ordering
As pointed out above the ‘nearly planar YM-XM’ ordering symmetry with YM as main ordering axis represents realistic stereochemical and electronic properties of the N-H site in proteins. Let us investigate this symmetry in further detail. For convenience we use c20 = 2, and allow c22 to increase from 0 to 6, scanning thereby over a range of symmetries.
Table 12 shows potential coefficients and corresponding ordering tensor components in spherical tensor notation, S20 and S22, and in Cartesian tensor notation, Sxx, Syy and Szz. The numerical values of the Cartesian tensor components indicate clearly that the entry with c22 = 0 represents positive axial ZM ordering; the entry with c22 = 2.45 represents negative axial YM ordering; the entry with c22 = 3 represents rhombic negative YM ordering with ‘nearly planar YM-XM symmetry’; the entry with c22 = 4 represents positive rhombic XM ordering with ‘nearly planar YM-XM symmetry’; the entry c22 = 6 represents positive XM ordering and substantial negative YM ordering. The angle γMD was fixed in the calculations of this study, and all our previous studies, at 90°, in agreement with stereochemical considerations (Fig. 2b).
Table 12.
Potential coefficients c20 and c22 (eq 4) and corresponding principal values of the ordering tensor in spherical tensor notation, S20 and S22 (eq 11), and in Cartesian notation, according to ; and Szz = S20. Note that reversing the sign of c22 will cause the values of Sxx and Syy to be exchanged.
| c20 | c22 | S20 | S22 | Sxx | Syy | Szz |
|---|---|---|---|---|---|---|
| 2 | 0 | +0.440 | 0.000 | −0.220 | −0.220 | +0.440 |
| 2 | 2 | +0.265 | +0.368 | +0.093 | −0.358 | +0.265 |
| 2 | 2.45 | +0.188 | +0.460 | +0.188 | −0.376 | +0.188 |
| 2 | 3 | +0.088 | +0.572 | +0.306 | −0.394 | +0.088 |
| 2 | 4 | −0.082 | +0.750 | +0.500 | −0.418 | −0.082 |
| 2 | 6 | −0.183 | +0.878 | +0.739 | −0.446 | −0.293 |
The high sensitivity of the analysis to the symmetry of the local ordering (local potential) is illustrated in Fig. 13. We show the functions jKK′(ω) for all the relevant combinations of quantum numbers K and K′, as determined by the symmetry of the local potential. The potential is axial in Fig. 13a, with c20 = 1.5, and of the ‘nearly planar YM-XM ordering’ type in Figs. 13b (c20 = 2 and c22 = 3) and 13c (c20 = 2 and c22 = 3.25). To calculate the jKK′(ω) functions we used RC = 0.001, and to further calculate T1, T2 and NOE we used βMD = 0° in Fig. 13a and βMD = 90° in Figs. 13b,c, and the value of τm = 5.5 ns.
Figure 13.
Functions jK(ω) = jKK(ω) (Fig. 13a) and jK(ω) = jKK(ω) and jKK′(w) (Figs. 13b,c) calculated for RC = 0.001, and c20 and c22 as depicted in Figs. 13a-13c. The potential coefficients correspond to axial symmetry with c20 = 1.5 (Fig. 13a), and ‘nearly planar YM-XM symmetry’ with c20 = 2 and c22 = 3 in Fig. 13b and c20 = 2 and c22 = 3.25 in Fig. 13c. The black, red and green curves represent the functions jK(ω) with K = 0, 1 and 2, respectively. The blue, yellow and indigo curves in Fig. 13b and 13c represent the functions jKK′(ω) with KK′ = (2, 0) = (0, 2), KK′ = (2, −2) = (−2, 2) and KK′ = (1, −1) = (−1, 1), respectively. The NMR relaxation rates were calculated for βMD equal to 0° (Fig. 13a) or 90° (Figs. 13b, c), τm = 5.5 ns and a magnetic field of 11.7 T. Note that for βMD = 90° the contributions cross-terms include j20(ω) = j02(ω) and j2-2(ω) = j−22(ω)).
There are significant differences between the axial and rhombic potential scenarios. First of all, rhombic symmetry requires the cross terms j2-2(ω) = j−22(ω), j20(ω) = j02(ω) and j1-1(ω) = j−11(ω) in addition to the diagonal terms j0(ω), j1(ω) and j2(ω) required for axial symmetry. The NOE generated with the rhombic potential given by c20 = 2 and c22 = 3.25 (cf. Fig. 13) can also be reproduced with an axial potential given by c20 = 1.5 (with all the other input parameters being the same). However, T1 and T2 differ substantially indicating that a completely different set of input parameters featuring an axial potential would be required to reproduce satisfactorily the T1, T2 and NOE of Fig. 13c. This illustrates the need for force-fitting, with Fig.13c representing “model” experimental data. Figs. 13b and 13c show that the NMR relaxation rates are altered substantially when the rhombicity of the potentials changes moderately. Thus, an 8% increase in c22/c20 implies a 53% increase in the NOE, pointing out the very high sensitivity of the analysis to the precise form of the local potential.
The examples shown in Fig. 13 pertain to the large time scale separation regime where mode-mixing is limited. The effect of potential symmetry on the experimental variables for RC = 0.5, where mode-mixing is important, is illustrated in Fig. 14. The other parameters used include τm = 6.1 ns and c20 = 2. Isotropic RL (N = 1) and βMD = 0° were used in Figs. 14a - c, and axial RL (with N = 10) and βMD = 90° were used in Figs. 14d - f. The rhombic potential coefficient, c22, was varied from 0 to 6. Positive axial ZM ordering corresponds to c22 = 0 (Szz = 0.440, Sxx = Syy = −0.220), negative YM corresponds to c22 = 2.45 (Syy = −0.376, Sxx = Syy = 0.188), ‘nearly planar YM-XM ordering’ with YM as main (negative) ordering axis corresponds to c22 = 3 (Sxx = 0.306, Syy = −0.394 and Szz = 0.088), and rhombic XM ordering corresponds to c22 = 6 (Sxx = 0.629, Syy = −0.446 and Szz = −0.184).
Figure 14.
15T1, T2 and 15N-{1H} NOE calculated with τm = 6.1 ns, RC = 0.5 and a magnetic field of 11.7 T. Isotropic RL and βMD = 0° were used in panels a-c, and axial RL with N= RL∥/RL⊥ = 10 and βMD = 90° were used in panels d-f. The potential used was given by c20 = 2, with the rhombic coefficient, c22, varied from 0 to 6.
The ‘nearly planar YM-XM’ ordering symmetry with YM as main (but negative) ordering axis, which corresponds to c20 = 2 and c22 = 3, has unique features. For βMD = 0° the NOE and T2 assume their maximum values, whereas T1 assumes its minimum value for this symmetry. For βMD = 90°, T1 goes through a shallow minimum, whereas T2 and the NOE exhibit a maximum slope for this symmetry in the c22 range shown. Particularly noteworthy is the fact that the NOE is significantly higher when the symmetry of the local potential is rhombic instead of axial. By analogy with Fig. 14, the maximum NOE corresponding to rhombic ‘nearly planar YM-XM ordering’ is expected to be higher than the maximum NOE corresponding to axial ordering of similar magnitude. This turns out to be an important issue, discussed below in detail.
5. Examples of misleading force-fitting with MF
5a. ‘Missing’ contribution to 15N 1/T2.
Lee and Wand,9 who carried out a comprehensive MF analysis of multi-field Ubiquitin data acquired at 300K, reported on an apparently missing contribution to 1/T2 which led to large τm values despite having allowed for variations in the 15N CSA interaction. Only when T2 was excluded from the analysis was the expected value of τm = 4.1 ns recovered. As shown in Fig. 5, for τm = 5 ns mixed modes contribute to 1/T2 significantly already for τe on the order of 20 ps for S2 = 0.85 (Fig. 5). Quantitative estimates given in Table 8 show that even when the contribution of mixed modes is relatively small, the 1/T2's obtained with MF are 5-6% lower than the 1/T2's obtained with SRLS. Lower time scale separation and lower ordering, which imply larger effects of additional eigenmodes for local motion on the correlation functions, will increase further the difference between 1/T2 obtained with SRLS and 1/T2 obtained with MF. The data shown in Table 8 were obtained with axial potentials. For rhombic potentials the 1/T2 differences will be significantly larger since SRLS and MF differ, in general, to a larger extent when the potential is rhombic in SRLS and axial in MF (e.g., see Table 10).
Since MF does not account either for mode-mixing or potential rhombicity, the only way to render the fitting feasible is to exclude T2 from the analysis. SRLS-based data fitting with rhombic potentials and arbitrary RL∥/RL⊥ is expected to make more insightful fitting of complete multi-field data from Ubiquitin and other proteins.
5b. Low performance of the N-H bond as dynamic probe
The commonly used probe for studying protein backbone dynamics is the N-H bond. The experimental auto-correlated relaxation rates 15N T1, T2 and 15N-{1H} NOE are analyzed with data fitting. Recently the 13C′-13Cα bond has been suggested as a complementary dynamic probe (ref. 13 and papers cited therein). In this case the 13C′-13Cα dipolar-13C′ CSA cross-correlated relaxation rate, Γ, is measured. Since only a single relaxation rate is measured only model 1 cases, which feature τm and S2(C′-Cα) as free variables, can be treated. In such cases one can calculate S2(C′-Cα) from the expression of Γ using τm determined with 15N spin relaxation. Hence within the scope of MF combined N-H and C′-Cα analysis is relevant for rigid proteins where model 1 applies, assuming that the peptide plane is rigid.
The ribonuclease binase was studied earlier with 15N spin relaxation at 11.7 and 18.8 T,21 and recently with combined 15N-1H and C′-Cα spin relaxation at 11.7 T.13 In the earlier study the 18.8 T data could not be fit with MF, and the 11.7 T data yielded an altogether rigid backbone although other methods (X-ray crystallography, molecular dynamics and 13C′-13Cα cross-relaxation) indicated that the catalytic loops L2 and L5 are flexible.21 In reference 13 it was found that at 278 K S2(N-H) and S2(C′-Cα) are practically the same, in agreement with nearly rigid N-H and C′-Cα bonds. However, when the temperature was increased to 303 K, S2(C′-Cα) decreased by 10%, whereas S2(N-H) decreased by only 2%. This was considered contrary to the expectation that at the higher temperature the N-H bond should be more sensitive than the C′-Cα bond to the crankshaft motion38 occurring about an axis close to the Ni-Cαi bond, since N-H is perpendicular to this axis whereas C′-Cα is parallel to it. Similar results were obtained for Ubiquitin.
The unexpected temperature-independence of S2(N-H) is actually imprinted in the raw data. Fig. 15 shows the experimental NOE's obtained at 303 K, 11.7 and 18.8 T. The horizontal lines show the maximum ‘rigid sphere’ NOEs corresponding to τm values determined with T1/T2 analysis. It can be seen that most NOE's exceed the ‘rigid sphere’ value. This feature necessarily imposes on the fitting scheme model 1, where the ‘rigid sphere’ NOE value is obtained. The latter is independent of S2. T1 and T2 assume minimum values for S2 = 1. As shown in Table 13 the difference between the average experimental and minimum T1 and T2 values is approximately 8% at 278 K and 10% at 303 K. The S2 values yielded by the MF analysis are 0.903 at 278 K and 0.884 at 303 K, which differ from 1 by 10 and 12%, respectively. Hence the unexpectedly small temperature-dependence of S2(N-H) is to be assigned to issues related to the analysis, rather than issues related to the experimental data. A plausible interpretation is outlined below.
Figure 15.
Experimental NOE's of the ribonuclease binase acquired at 11.7 and 18.8 T, 303 K.21 The horizontal lines show the maximum NOE for 6.1 ns (11.7 T) and 5.5 ns (18.8 T), with the global motion correlation times determined based on T1/T2 ratios.
Table 13.
Maximum NOE (obtained with model 1) and minimum T1 and T2 (obtained with model 1 and S2 = 1) values corresponding to 11.7 T and 13. 44 (5.5) ns at 278 (303 K) (row 1); average experimental T1 and T2 and NOE values from ref. 13 (row 2); percent difference between corresponding data in rows 1 and 2 (row 3).
| 303 K | ||
|---|---|---|
| T1, ms | T2, ms | NOE |
| 368.5 | 117.2 | 0.771 |
| 408.2 ± 6.5 (1.6%) | 129.4 ± 2.5 (1.9%) | 0.771 |
| 10.8% | 10.4% | 0 |
| 278 K | ||
| T1, ms | T2, ms | NOE |
| 720.4 | 55.0 | 0.816 |
| 781.3 ± 18 (2.3%) | 59.2 ± 2.1 (3.6%) | 0.816 |
| 8.5% | 7.7% | 0 |
The maximum theoretical NOE value is determined for axial local potentials. However, we found previously that ‘nearly planar YM-XM ordering’ prevails at the N-H bond. Fig. 16 shows the effect of potential symmetry on the NOE. Calculations were performed for τm = 6.1 ns, τL⊥/τm = 0.5, RL∥/RL⊥ = 0.5, c20 = 2 and βMD = 0°. The rhombic potential coefficient, c22, was varied from 0 to 6, scanning thereby over the various symmetries of the coupling potential and the corresponding ordering tensor. As pointed out earlier c22 = 0 corresponds to axial ZM ordering; c22 = 2 corresponds to rhombic ZM ordering; c22 = 2.45 corresponds to axial perpendicular YM ordering; c22 = 3 corresponds to rhombic ‘nearly planar YM-XM ordering’ with YM as main ordering axis; c22 ~ 5 corresponds to rhombic XM ordering. It can be seen that the largest NOE value is obtained for c22 = 2 and c22 = 3, i.e., rhombic ‘nearly planar YM-XM ordering’, with YM as main ordering axis.
Figure 16.
15N-{1H} NOE calculated with τm = 6.1 ns, RC = 0.5, RL∥/RL⊥ = 0.5 and c20 = 2. The rhombic potential coefficient, c22, was varied from 0 to 6.
Figure 16 shows clearly that rhombic potentials can yield higher NOE's than axial potentials. Therefore if the theoretical spectral density used in the fitting scheme yields maximum NOE's corresponding to ‘nearly planar YM-XM ordering’, the experimental NOE's may not exceed the maximum NOE, and model 1 should not be imposed on the fitting scheme. Instead model 2, which yields significantly lower S2 values than model 1 (Table 7), may be selected at 303K.
Yet another illustrative example is presented in Table 14 which shows high c20 and c22 values, corresponding to high order parameters, S20 and S22, as appropriate for model 1. For the expected βMD= 90° geometry corresponding to the ‘nearly planar YM-ZM ordering’, the term τm/(1 + ω2τm2) in the combination 1 spectral density is multiplied by the coefficient [0.25 (S20)2 + 0.75 (S22)2], since (d200)2 = 0.25 and 2×[d220)2] = 0.75 (eq 20). Let us assume that this expression is mimicked by (S20)axial2 in an axial potential scenario. We calculated S20 and S22 in terms of the potential of eq 6 with c20 = 10 and c22 varied from 16 to 22. These potentials correspond to ‘nearly planar YM-XM ordering’. Table 14 also shows c20(axial), corresponding to (S20)axial2. It can be seen that for c22/c20 ≥ 1.6 the axial mimic (S20)axial2 corresponds to very high potentials. This agrees with the high S2(N-H) values obtained in reference 13 at the higher temperatures for binase and Ubiquitin.
Table 14.
Order parameters S20 and S22, and (S20)axial2 = 0.25 (S20)2 + 0.75 (S22)2, for c20 and c22 as given in the Table.
| c20 | c22 | S20 | S22 | (S20(axial))2 | c20(axial) | c22/c20 |
|---|---|---|---|---|---|---|
| 10 | 16 | −0.305 | 1.034 | 0.825 | 11.3 | 1.6 |
| 10 | 18 | −0.381 | 1.100 | 0.943 | 34.9 | 1.8 |
| 10 | 20 | −0.412 | 1.130 | 0.971 | > 50 | 2.0 |
| 10 | 22 | −0.432 | 1.147 | 1.005 | very high | 2.2 |
Combined analysis of N-H and C′-Cα bond dynamics can be carried out when the peptide plane is assumed to be rigid. The local ordering (associated with the M frame) is then a common property. Since the magnetic dipolar frames of 15N-1H and 13C′-13Cα differ, the local geometry (ΩMD) differs. However, for axial local ordering model 1 (combination 1) S2 ((S20)2) represents the mean square amplitude of all the local motions.25 In this case the angle βMD does not enter the model. For that a rhombic M frame is required (eq 20). Hence one cannot expect differential sensitivity of the N-H and C′-Cα bonds to the crankshaft motion unless the local ordering is allowed to be rhombic in the theoretical spectral density. As pointed out previously26 this ordering symmetry actually prevails at the N-H site. As shown herein, it can be treated with SRLS.
Unlike S2(N-H), which is determined with combined fitting of the relaxation quantities 15N T1, T2 and 15N-{1H} NOE, S2(C′-Cα) is calculated directly from the expression for the cross-correlated relaxation rate, Γ. This can be accomplished assuming that model 1 is valid, i.e., the spectral density is given by the first term of eq 16, and that τm is determined with N-H bond dynamics analysis. In this case S2(C′-Cα) is the only unknown parameter. The maximum Γ value for 11.7 T is −2.395 (−1.194) for τm = 13.44 ns (5.5 ns) whereas the corresponding experimental value is −1.992 (−0.885). Hence limitations implied by the experimental values exceeding maximum values implied by axial potentials, as in the N-H case, do not exist in this case. For rigid residues the 13C′-13Cα dipolar - 13C′ CSA cross-correlated relaxation rate depends to a large extent on JDD(0) and JCC(0). Therefore local motion effects are small, justifying the use of model 1. Since βMD is approximately 0° for C′-Cα, (S20)axial2 is approximately equal to (S20)2. For this reason at the higher temperatures the relatively accurate S2(C′-Cα) MF value is smaller than the force-fitted S2(N-H) MF value.
The 3D Gaussian Axial Fluctuations (GAF) model40 accounts quantitatively for the different geometric features at the N-H and C′-Cα sites. In this approach the S2 MF is expressed in terms of harmonic fluctuations around the Cαi-1-Cαi axis (σ2γ) and perpendicular to it (σ2αβ). 3D GAF was applied to Ubiquitin at 300 K.40 Molecular dynamics simulations showed that σ2γ > σ2αβ. MF fitting of the 15N relaxation data showed the same trend with even larger absolute magnitudes of σ2γ and σ2αβ. The S2(N-H) values obtained with the usual MF analysis were reproduced by the S2(N-H) values calculated with the 3D GAF model when (relatively small) contributions, which are usually ignored in 15N spin relaxation of 15N,13C-labeled proteins, were taken into account. S2(C′-Cα) values calculated with the 3D GAF model were found to higher than S2(N-H) as N-H senses the σ2γ fluctuations, which can be considered to represent the crankshaft motion, whereas C′-Cα senses the σ2αβ fluctuations. Thus, accounting for the asymmetry of the local ordering with 3D GAF yields S2(N-H) < S2(C′-Cα), as expected.
Instead of harmonic fluctuations σ2γ and σ2αβ (applicable to ‘rigid’ residues only) and a predetermined geometry, which is implicit in 3D GAF, SRLS allows for a rhombic ordering tensor with principal values S20 (eq 7) and S22 (eq 8) defined in terms of a rhombic potential (eqs 6 and 9). The SRLS treatment is not limited to ‘rigid’ residues, and the orientation of the M frame is not fixed. The perpendicular orientations are not considered equivalent and the magnitude of the local asymmetry is quantified through c20 and c22, or S20 and S22. The local motion is treated as diffusive, which is reasonable (although improved modeling can be introduced to account for any inertial effects).23
Calculations using our fitting scheme for SRLS, which allows for rhombic ordering, were carried out for the average values of 15N T1, T2 and NOE acquired at 303K and 11.7 T, and for combined 11.7 and 18.8 T data of residue 16 acquired at 303 K. In the former case we fixed the ratio RL∥/RL⊥ at 1000 and the angle βMD at 90°, allowing c20, c22 and RC to vary. In the latter case we allowed c20, c22, RC, RL∥/RL⊥ and βMD to vary. The results are shown in Table 15. S2(C′-Cα) = 0.806 was derived in ref. 13 from the experimental cross-correlated relaxation rate, Γ, measured at 11.7 T and 303 K, as outlined above. Based on eq 11 S2(C′-Cα) = 0.806 corresponds to c20 = 10.2. The components of the Cartesian ordering tensor are Sxx = Syy = −0.449, Szz = 0.898. It can be seen that the magnitude of the local potentials at N-H and C′-Cα sites is similar. The local geometry is different with βMD = 90° for the N-H bond and βMD = 0° for the C′-Cα bond. Since RL∥/RL⊥ and βMD were fixed in the fitting of the average 15N relaxation data (as only 3 data points were available), and rhombic potentials could not be used to treat C′-Cα bond dynamics (as only one data point was available), we regard the data in Table 15 as interim results. However, they can be used for illustrative purposes. If the peptide plane is rigid it is expected to determine the same tensor components permuted from ZM ordering for C′-Cα to YM ordering for N-H. Thus, Szz = 0.898 is to be compared with Syy = 0.761 (results for the average data). If (Syy)2 is considered to represent the crankshaft fluctuations then 0.58 for N-H is to be compared with 0.20 for C'-Cα. Clearly proper analysis bears out the higher sensitivity of N-H bond dynamics as compared to C′-Cα bond dynamics to the crankshaft fluctuations.
Table 15.
Best-fit parameters for N-H site dynamics obtained with rhombic potentials for the average experimental data of binase at 11.7 T, 303 K (ref. 13) and for the combined 11.7 and 18.8 T data of binase residue 16 at 303 K (data from ref. 21 kindly provided by Prof. E. Zuiderweg). τm = 5.5 ns was used.
| c20 | c22 | RC | βMD° | RL∥/RL⊥ | Sxx | Syy | Szz | |
|---|---|---|---|---|---|---|---|---|
| Av | 5.5 | 9.7 | 0.97 | 90_fixed | 1000_fixed | −0.470 | 0.761 | −0.291 |
| res 16 | 5.0 | 10.0 | 1.0 | 90.0 | 30.0 | −0.65 | 0.75 | −0.10 |
It is concluded that the effects of potential rhombicity, mixed modes and the D-to-CSA tilt must be accounted for in the theoretical spectral density in order to obtain physically insightful information. This is outside the scope of model-free and can only be accomplished with SRLS. As shown above, combined 15N and 13C spin relaxation analysis is expected to be useful within the scope of SRLS analysis. As shown below, binase is not a singular case but a representative case.
Figure 17 shows the experimental NOE's of oxidized flavodoxin acquired at 11.7 T, 300 K.17 Similar to binase, most NOE's exceed the maximum NOE calculated for axial potentials, depicted by the horizontal line. 15N spin relaxation data of oxidized flavodoxin data could not be fit with the standard MF fitting scheme. Fitting became possible only after reducing the experimental NOE's globally by 10%, rendering them smaller than the maximum NOE. Even then model 1 was used predominantly, the S2 values were very high, and τm was 7.6 ns, which is about 2 ns shorter than expected for a bare sphere with a molecular weight of nearly 20 kDa, corresponding to oxidized flavodoxin. Clearly the experimental data were force-fitted similar to the binase case. SRLS-based fitting with axial potentials gave similar results, in support of the assessment that rhombic potentials are to be used.
Figure 17.
Experimental NOE's of oxidized flavodoxin acquired at 11.7, 303K (ref. 17). The horizontal line shows the maximum NOE for 7.6 ns, with the global motion correlation time determined based on T1/T2 ratios.
The enzyme ribonuclease H (RNase H) was studied with 15N spin relaxation at 285, 300 and 310 K, 11.7 T.12 The experimental NOE's are shown in Fig. 18. It can be seen that quite a few NOE's exceed the maximum NOE values depicted by horizontal lines. The average slope d(1–S)/dT determined with MF analysis was 5.9×10−4 K−1, to be compared to 8×10−4 K−1 for binase, considered to be low. Analysis with SRLS, using the equivalents of the MF models 1-5 (ref.46) as implemented in our 2D grid-based fitting scheme for SRLS featuring axial potentials,22 yielded 6.4×10−4 K−1 for the β2 strand of RNase H. Thus, the detrimental features of model 1 analysis with axial potentials outlined for binase recur with RNase H.
Figure 18.
Experimental NOE's of Ribonuclease H acquired at 285, 300 and 310 K, 11.7 T.12 The horizontal lines show the maximum NOE for the τm values shown, which were determined based on T1/T2 ratios.
15N relaxation data of RNase H were also acquired at 11.7, 14.1 and 18.8 T, 300 K (ref 57). The NOE's obtained for the rigid part of the protein backbone are shown in Fig. 19. NOE's exceeding the maximum NOE are pervasive at 18.8 T, where the local motion contributes significantly to the spectral density. Hence valuable information will be lost with force-fitted MF analysis.
Figure 19.
Experimental NOE's of RNase H acquired at 11.7, 14.1 and 18.8 T, 300 K.59 The horizontal lines show the maximum NOE for the τm = 9.28 ns determined based on T1/T2 ratios.
A small value of d(1–S)/dT was also reported for Ubiquitin. The experimental NOE's acquired for Ubiquitin by Lee and Wand9 are shown in Fig. 20. It can be seen that similar to binase and RNase H, many NOE's exceed the maximum NOE.
Figure 20.
Experimental NOE's of Ubiquitin acquired at 11.7, 14.1 and 18.8 T, 298 K.9 The horizontal line shows the maximum NOE's for 4.1 ns.
Figures 15 - 20, as well as the last column of Table 5, indicate that in many cases MF fitting schemes select model 1 by force-fitting, yielding inaccurate S2 values. When the main effect is the omission of the D-to-CSA frame transformation, as for the Villin Headpiece (Table 5), (S20)2 is underestimated by S2. When the main effect is over-simplified symmetry of the local ordering, (S20)2 is overestimated by S2 (Table 14). Thus, the S2 profile over the protein backbone may become qualitatively inaccurate.
5c. Limited information on main-chain conformational entropy from N-H bond dynamics
Similar to its low sensitivity to temperature variations, S2(N-H) was also found to exhibit low sensitivity to ligand binding. The parameters S2(N-H) and S2(C′-Cα) of ligand-free and ligand-bound Ca2+-Calmodulin (CaM) were derived in reference 18. The experimental data were acquired at 11.7 T, 308K, and the MF analysis used predominantly model 1. Except for the central linker and several loops, S2(N-H) changed very little whereas S2(C′-Cα) changed significantly upon ligand binding. As in the temperature-dependent study, it was concluded that the N-H bond does not sense (in this case ligand-binding-dependent) backbone fluctuations sensed by the C′-Cα bond. This is important for conformational entropy derivation from S2 MF in the context of complex formation.
Similar to the apparently low sensitivity of S2(N-H) of binase, RNase H and Ubiquitin to temperature changes, the apparently low sensitivity of S2(N-H) to ligand binding to CaM stems from force-fitting the experimental data with axial potentials, instead of physically sounder rhombic potentials. Proper interpretation requires a very efficient SRLS fitting scheme featuring rhombic potentials, which is currently being developed. We illustrate below the need for model generality using our SRLS fitting scheme featuring axial potentials. Table 16 features results obtained for the domain residues 30, 100 and 135 of CaM (associated with high experimental NOE's) and for the central linker residues 77 and 80 (associated with low experimental NOE's). τm = 7.5 ns for the free form and τm = 8.3 ns for the bound form were used.35,60 SRLS combinations 1 and 2, which correspond to MF models 1 and 2, did not yield acceptable results. SRLS combination 5, with N fixed at the value of 1, gave the results shown in Table 16. This combination differs from model 2 MF in allowing the orientation of the ordering tensor, βMD, to vary (recall that (1.5 cos2βMD - 0.5)2 is formally equivalent to Sf2 in MF). For all the residues examined the local potential and the local ordering are high for both Calmodulin forms, as found in reference 60, whereas βMD is small for the free form and on the order of 25° for the bound form. Thus, ligand binding changes the orientation of the ordering tensor preserving the magnitude of its principal value. This could not be determined with MF because in models 1 and 2 the angle βMD is zero.
Table 16.
Best-fit parameters obtained by subjecting the domain residues 30, 100 and 135, which feature high experimental NOE's, and residues 77 and 80 of the central linker region which feature low experimental NOE's, to SRLS analysis using combination 5 ((S20)2, RC and βMD varied) with N = RL∥/R⊥ = 1. The symbols ‘f’ and ‘b’ denote the ligand-free and ligand-bound forms, respectively.
| c20 | (S20)2 | RC | βMD° | χ2* | |
|---|---|---|---|---|---|
| 30_f | 19.3 | 0.897 | 0.05 | 0.01 | 4.9 |
| 30_b | 18.3 | 0.892 | 0.06 | 20.0 | 5.0 |
| 100_f | 16.4 | 0.879 | 0.07 | 2.6 | 0.016 |
| 100_b | 15.7 | 0.874 | 0.05 | 22.5 | 25.2 |
| 135_f | 21.4 | 0.907 | 0.01 | 0.03 | 0.3 |
| 135_b | 16.4 | 0.879 | 0.06 | 22.2 | 13.1 |
| 77_f | 8.6 | 0.771 | 0.06 | 8.0 | 0.0 |
| 77_b | 8.4 | 0.765 | 0.03 | 24.8 | 7.9 |
| 80_f | 8.7 | 0.773 | 0.06 | 0.0 | 18.2 |
| 80_b | 8.2 | 0.760 | 0.03 | 28.0 | 21.6 |
Note that a threshold value of χ2 = 25 was also used by Vugmeyster et al.11
As shown in Fig. 11 MF does not have the capability to fit flexible N-H sites where N ∼ 1 because it does not consider the spectral density components j1(ω) and j2(ω). The fact that the MF scheme converged to models 1 and 2 instead of model 5 is therefore assignable to force-fitting. Residues 77 and 80 of the central linker are associated with an average c20 value of 8.5 (corresponding to an average (S20)2 value of 0.77), whereas residues 30, 100 and 135 are associated with c20 ∼ 17.9 ((S20)2 ∼ 0.9). Hence axial-potential-based SRLS fitting differentiates between the central linker and the domains of CaM. These results will change when the potential will be allowed to be rhombic and N will be allowed to vary. However, Table 16 illustrates clearly the fact that sensitivity to ligand binding can be borne out by properties of the ordering tensor other than S2, which is the only property measured when models 1 or 2 MF are used. The local ordering is characterized by a tensor, not merely by a scalar, S2.
5d. Calmodulin: the detection of an incorrect phenomenon
Ca+2-ligated Calmodulin is made of an N-terminal domain and a C-terminal domain connected by a helical linker, which is flexible in the middle. In the crystal CaM adopts an elongated dumb-bell structure61 with the N-, and C-terminal regions of the helical linker parallel to one another (Figs. 21 and 22). Since the middle linker region is flexible the N-, and C-domains may adopt various relative orientations in solution. The helical target peptide, essential for CaM recognition and regulation, binds in-between the domains. Hence molecular shape, linker flexibility, and domain mobility are related to function, and deriving a reliable dynamic picture is important.
Figure 21.
Ribbon diagram of Ca2+-ligated Calmodulin reproduced from reference 64. The coordinates of the crystal structure of Babu et al.61 (PDB accession number 3CLN) were used. The data depicted define the global diffusion tensor as determined in ref. 64. ‘N’ and ‘C’ denote the N-, and C-terminal domains of Ca2+-ligated Calmodulin.
Figure 22.

Ribbon diagram of the crystal structure of Babu et al.61 (PDB accession number 3CLN) and corresponding inertia frame, I. The global diffusion frame, D, shown in Fig. 21 is also depicted.
Several experimental and theoretical methods, including NMR spin relaxation, have been used to study CaM flexibility. The NMR-based dynamic picture changed as data were acquired at an increasing number of magnetic fields and temperatures. The first 15N spin relaxation study of Ca+2-saturated Drosophila CaM data acquired at 11.7 T, 35°, was carried out in 1992.62 These data were analyzed with the original MF formula. The assumption that Ca2+-CaM is nearly spherical in solution (also found in references 18 and 60) was corroborated by comparing N-H orientations in solution and in the crystal structure,61 and by the nearly flat T1/T2 profile. Somewhat different isotropic correlation times on the order of 6-8 ns were assigned to the N-, and C-domains. Except for the flexible residues 78-81 of the central linker and two loops, the CaM backbone was found to be quite rigid, with S2 ∼ 0.85 and τe < 100 ps.
At low magnetic fields the local motion makes a relatively small contribution to the spectral density. If the spectral density used to calculate the NMR variables is appropriate, the addition of higher field data will merely increase accuracy and precision. If it is not, then inconsistencies will arise because local motion effects will be parameterized in different ways at different magnetic fields. The Ca+2-free Xenopus CaM study of Tjandra et al.63 identified such inconsistencies when 11.7 T and 14.1 T data were analyzed in concert. The inconsistencies detected could be reconciled by using the reduced extended MF formula (eq 19 with τf′ set equal to zero) instead of the original MF formula (eq 15). With Sf2 fixed at 0.85 and uniform parameters within each domain, the fitting yielded τm = 12 ns, Ss2 ∼ 0.7 and τs ∼ 3 ns. The parameters Ss2 and τs were interpreted within the scope of wobble-in-a-cone motions of the two domains. The vertex angle of the cones was approximately 30°, which is incompatible with isotropic τm. Semi-quantitative arguments in support of an elongated solution structure with N = RC∥/RC⊥ ∼ 1.6 were invoked.
15N spin relaxation data of Ca+2-saturated Xenopus CaM were acquired by Baber et al.64 at 8.5 (except for NOE's), 14.1 and 18.8 T, 308 K. The analysis was similar to that of Tjandra et al.63 The larger data set available made possible determination of the global diffusion tensor, RC, and removal of the restrictions that τf = 0 and Sf2 = 0.85. Chang et al.65 extended the experimental data set of Baber et al.64 so that 15N T1, T2 and NOE's became available at 8.5, 14.1 and 18.8 T, at 294, 300, 308 and 316 K. These data were analyzed in concert assuming that (1) Sf2, τf and N = RC∥/RC⊥ are the same for all the residues within a given domain and are independent of temperature, and (2) the temperature dependence of τm(app) (with τm(app) = 1/6D(app), D(app) = 1/3(2D⊥ + D∥)) is controlled by the Stokes-Einstein formula. A sudden decrease (increase) in Ss2 (τs) was observed when the temperature was increased from 308 to 316 K, interpreted as ‘melting’ of residues 74-77 of the central linker, considered important from a biological point of view.
Since data acquired at several magnetic fields and several temperatures are analyzed in concert, the analysis is particularly prone to force-fitting. It will be shown below that qualitatively erroneous results are obtained in the case of Ca2+-Calmodulin.
Global diffusion
The axial global diffusion tensor, RC, was determined together with the site-specific parameters using the total time correlation function, C(t) (eq 13), with CL(t) given by the extended MF formula, and the coordinates of the elongated dumb-bell shaped crystal structure.61 In most MF studies RC is determined separately based on CC(t). The C(t)-based fitting of the combined multi-field multi-temperature data yielded Θ = 68°, as shown in Fig. 21, with the global diffusion axis, C, along the symmetry axis of the molecule. Accordingly, the principal axis of the inertia tensor, I, must be tilted at 68° from C. This disagrees with the orientation of the inertia tensor in the crystal structure, shown in Fig. 22. Clearly the latter orientation of the inertia tensor is correct, hence the orientation of the global diffusion tensor is likely in error.
Fig. 23 shows the experimental T1/T2 data acquired at 8.5, 14.1 and 18.8 T, and 294 and 316 K and filtered according to traditional criteria.64,65 The width of the distribution divided by the average error is 6 (4), 8.5 (9.0) and 13.0 (14.0) for 8.5, 14.1 and 18.8 T at 294 K (316 K). It is obvious that the distribution in T1/T2 is significantly smaller at 8.5 T, in agreement with the 11.7 T data of Barbato et al.62 and Wand and co-workers.18,60 Isotropic global diffusion analysis with the program QUADRIC66 yielded the τm values depicted in Fig. 23. The shape of the distribution in T1/T2 values is both field-, and temperature-dependent, although it has been assumed that RC is temperature-independent except for τm(app), which does not affect the shape of the T1/T2 distribution. It is very likely that the structured T1/T2 profiles at the higher fields represent mixed-mode contributions and unaccounted for geometric effects. If this is not the case then analyses based on single-field data and the concerted analysis should yield the same results. This test is carried out below.
Figure 23.
Experimental T1/T2 ratios at 8.5, 14.1 and 18.8 T, 294 and 316 K, from reference 65. The isotropic global diffusion correlation times, τm, determined with the program QUADRIC,66 are also shown.
Using the filtered T1/T2 data of Chang et al.65 we determined the axial global diffusion tensor RC at each magnetic field and temperature separately with the program QUADRIC.66 This corresponds to using CC(t) instead of C(t). In Fig. 24 we show D(app) = 1/3 (D∥ + 2 D⊥) (RC(app) in our notation) as a function of P2(cos(βCC′)) at 8.5, 14.1 and 18.8 T, and at the temperatures of 294 and 316 K.
Figure 24.
Analysis of the experimental data shown in Fig. 23 with the program QUADRIC66 assuming axial global diffusion (RC). The resulting parameters, which define the RC tensor, are also shown.
The spread of points about the theoretical straight lines (obtained with linear regression) in Fig. 24 is invariably large, indicating that theory and data are incompatible. There are very few points corresponding to βCC′ = 0, in disagreement with the purported solution structure (Fig. 21). The largest spread of points is obtained for 8.5 T, 316 K, although χ2 assumes the smallest value (χ2 = 2) in this case. This is certainly not expected for models matching the data to which they are applied, but can occur when force-fitting sets in. All four parameters defining the global diffusion tensor are field-dependent. In all the cases except for 8.5 T, 316 K, the angle Θ of the individual analyses is much closer to 0° (coincident inertia and diffusion frames) than to the non-physical angle of 68° yielded by the concerted multi-field multi-temperature MF analysis. The contradictions between the raw data, the single-field analysis and the concerted MF analysis are substantial. For example, the raw T1/T2 profile at 8.5 T, 316 K is nearly flat (Fig. 23), while the RC tensor illustrated in Fig. 24 is closest to the axial tensor yielded by the concerted analysis. As shown in Fig. 23, isotropic RC analysis also yielded inconsistent τm values. Inaccuracies in CL(t) must have been clearly absorbed by CC(t). Therefore the local motion parameters must be highly inaccurate, as demonstrated below.
Local motion
Fig. 25 reproduces the Ss2 and τs temperature-dependent profiles obtained by Chang et al.65 The squared generalized order parameter Ss2 shows very limited temperature dependence between 294 and 308 K and decreases abruptly upon increasing the temperature to 316 K. The slow local motion correlation time, τs, is temperature-independent between 294 and 308 K and increases abruptly upon increasing the temperature to 316 K. Within the scope of the cone model used by Chang et al.65 the correlation time for slow local motion, τs, depends analytically on Ss2 and Dw. The respective expression is used to show that the abrupt increase in τs is due to the abrupt decrease in Ss2, while Dw increases with temperature. However, inspection of the absolute values of Dw shows that 1/6Dw is equal to 8.3 (6.8) ns for the N-domain (C-domain), while the apparent global motion correlation time is 6.88 ns. This is not tenable physically, nor consistent with the basic MF mode-independence assumption of time scale separation between the global and local motions.
Figure 25.
Best-fit Ss2 and τs values obtained with the extended MF formula as outlined in ref. 65 for the N-domain (open squares) and C-domain (solid circles). Additional best fit parameters are Sf2 ∼ 0.86, τf ∼ 15 ps and global diffusion parameters D∥/D⊥ = 1.62, Θ ∼ 68°, Φ ∼ 94° for the N-domain and 146° for the C-domain. The τm(app) values are 11.55, 9.87, 8.12 and 6.88 ns at 294, 300, 308 and 316 K.
The discontinuities in Ss2 and τs between 308 and 316 K in Fig. 25 results from the force-fitting process veering into a different region of the parameter space at 8.5 T and 316 K. Inspection of the experimental data presented in Figures 26-28 shows that the T2's at 8.5 T, 316 K, are outliers. Fig. 24 shows that the graph obtained for 8.5 T, 316 K is an outlier. Inspection of Figs. 5-8, where SRLS and MF are compared for corresponding parameter values near the BO limit, shows that for τm = 15 ns the corresponding NMR variables are comparable in magnitude. On the other hand, for τm = 5 ns T2 obtained with MF is significantly higher than T2 obtained with SRLS. This supports the assertion that artificial results can be obtained by force fitting large data sets covering extensive parameter ranges. In the case under consideration the experimental data acquired at low fields and high temperatures do not accommodate the force-fitted parameters, which fit all the other data. Therefore discontinuities in Ss2 and τs, which are merely technical in nature, ensue.
Figure 26.
Longitudinal 15N T1 relaxation times of Ca2+-calmodulin at 294 K (black), 300 K (red), 308 K (green) and 316 K (blue), and 8.5, 14.1 and 18.8 T.65 The vertical dashed lines depict the central linker (residues 74 – 78).
Figure 28.
Steady-state 15N-{1H} NOE's of Ca2+-calmodulin at 294 K (black), 300 K (red), 308 K (green) and 316 K (blue), and 8.5, 14.1 and 18.8 T.65 The vertical dashed lines depict the central linker (residues 74 – 78).
SRLS analysis
The Calmodulin data were analyzed separately for each temperature and magnetic field using our fitting scheme for SRLS based on axial potentials. We assumed that in view of large-amplitude domain motion RC is on average isotropic, and used the τm(app) values of Chang et al.65 Isotropic RC is consistent with the Calmodulin studies of Barbato et al.62 and Wand and co-workers,18,60 and with other 15N spin relaxation studies of proteins exhibiting large-amplitude domain motion.19,21 As mentioned above, this SRLS fitting scheme assumes implicitly that RL∥≫RL⊥, in analogy with τs ≫ τf in MF. The SRLS fitting scheme selected primarily combination 5, which corresponds to model 5 MF. The average (S20)2 (the formal analogue of Ss2 in MF) and τL⊥ (the formal analogue of τs in MF) values for each magnetic field are shown as a function of temperature in Fig. 29.
Figure 29.
Best-fit (S20)2 and τL⊥ values obtained with combination 5 SRLS by averaging over the results obtained for the individual residues using τm values of 11.55, 9.87, 8.12 and 6.88 ns at 294, 300, 308 and 316 K. βMD was on average 15°. Experimental data from ref. 65 were used.
It can be seen that τL⊥ SRLS differs in magnitude from τs MF (cf. Fig. 25). While τs shows the non-physical temperature dependence illustrated in Fig. 25, τL⊥ shows physically reasonable temperature dependence illustrated in Fig. 29. (S20)2 SRLS is approximately half of Ss2 MF and decreases monotonically with increasing temperature. No sudden change is observed between 308 and 316K in either parameter. The inconsistencies among (S20)2 and τL⊥ values obtained at different magnetic fields are expected to be eliminated in future work, where rhombic potentials will be used.
Dynamic picture according to MF.65
CaM is an elongated dumb-bell shaped molecule. The N-, and C-terminal domains wobble within cones with vertex angles increasing suddenly from 22.5° (27°) to 27° (37°) for the N-domain (C-domain) when the temperature is increased from 308 to 316 K. This corresponds to a squared generalized order parameter, Ss2, decreasing from 0.77 (0.68) to 0.68 (0.53) for the N-domain (C-domain). The average wobbling rate, Dw, is 1.7×107 (2.0 107) s−1 at 295 (316 K) for the N-domain, which translates into correlation times, 1/6Dw, of 9.8 (8.3) ns. The value of Dw is 2.0 107 (2.45 107) s−1 at 295 (316 K) for the C-domain, which translates into correlation times of 8.3 (6.8 ns). The correlation time for global motion is 11.55 (6.88) ns at 295 (316 K). Hence, at 316 K the correlation time for local motion, 1/6Dw, is larger than the correlation time for global motion, τm, for the N-domain, and equal to τm for the C-domain. Motion about the N-H bond is on the order of 20 ps and Sf2 is on the order of 0.85 throughout the temperature range investigated.
The abrupt change in best-fit parameters values upon increasing the temperature from 308 to 316 K is interpreted as ‘melting’ of residues 74-77. This process is purported to have biological implications for target peptide binding by prolonging the flexible part of the central linker by 50%. Note that the experimental data of residues 74-77 (as well as many other CaM residues) are not observed experimentally at 316 K (Figs. 26-28, residues demarcated by the dashed vertical lines).
Dynamic picture according to the current SRLS analysis
CaM is on average spherical in solution due to large-amplitude nanosecond segmental motions of its N-, and C-terminal domains. This is physically plausible, consistent with the T1/T2 profiles at 11.7 T which are determined predominantly by the global motion, and the quantitative analysis by Barbato at al.62 and Wand and co-workers.18,60 Average spherical shapes in solution were also determined with 15N spin relaxation for AKeco19 and Binase,21 which similar to CaM, feature large-amplitude domain or loop motions in solution. Both AKeco and Binase have elongated shapes in the crystalline state, similar to CaM..
Domain motion is expected to occur on the same time scale as the global motion, implying mode-mixing. This is accounted for by the SRLS analysis, which yields τL⊥ on the order of 3 – 6 ns in the temperature range of 294 – 316 K. As expected, τL⊥ decreases monotonically with increasing temperature. (S20)2 is on order of 0.2-0.35 in this temperature range and decreases monotonically with increasing temperature. No discontinuity is exhibited by either the τL⊥ or the (S20)2 temperature profiles.
The results shown in Fig. 29 are interim results because the analysis used oversimplified axial symmetry for the local orienting potential/local ordering, and assumed implicitly that RL∥/RL⊥ ≫ 1. The implication of removing these restrictions are illustrated and discussed in the next section.
6. Reliable fitting; the mixed mode concept
The SRLS approach is a general one, although we have implemented it with several simplifying assumptions. In our current fitting scheme the local and global diffusion tensors are allowed to be axially symmetric, and the local ordering tensor (or local coupling/mixing/orienting potential) is allowed to be rhombic. The magnetic tensors have arbitrary symmetry and orientation. If one starts with the original MF limit, where the magnetic tensors are collinear, their frame is the same as the local ordering frame, and the global and local diffusion tensors are isotropic, one can then systematically lower symmetries until the complexity of the model matches the integrity of the data. In this case reliable fitting, which extracts the dynamic information inherent in the experimental data, can be accomplished. Table 17 illustrates the last part of such a process, where SRLS spectral densities are upgraded to include more detailed features in a stepwise fashion.
Table 17.
Best-fit parameters obtained with the SRLS combination 5 (SRLS_5) and SRLS combination 6 (SRLS_6) using the axial-potential 2D grid-based fitting scheme. SRLS_rh represents the calculation carried out with the fitting scheme allowing for rhombic potentials and arbitrary N = RL∥/RL⊥ values, with the jKK'(ω) functions calculated on the fly. The combined 15N relaxation data of residue 124 of RNase H acquired at 11.7, 14.1 and 18.8 T, 300 K, were used. τm = 9.28 ns was used. RC is the same as τL/τm, and N = RL∥/RL⊥. Sxx, Syy and Szz are the components of the Cartesian ordering tensor.
| RC | c20 | c22 | Sxx | Syy | Szz | βMD° | N | |
|---|---|---|---|---|---|---|---|---|
| SRLS_5 | 0.44 | 3.2 | 0.0 | −0.315 | −0.315 | 0.63 | 16.1 | 1000(fixed)(χ2 = 15.9) |
| SRLS_6 | 0.45 | 3.2 | 0.0 | −0.315 | −0.315 | 0.63 | 16.3 | 916(χ2 = 15.8) |
| SRLS_rh | 0.23 | 4.8 | 10.1 | −0.469 | 0.799 | −0.330 | 99.5 | 40.0(χ2 = 12.3) |
The example considered is residue 124 of RNase-H, which pertains to the flexible loop αD/β5. The 15N relaxation data acquired at 11.7 T, 300 K, were fit previously with MF model 5.12 The global motion correlation time of RNase H was determined to be τm = 9.28 ns at 300 K, and it has been ascertained that the protein is spherical within a good approximation. We subjected the combined 11.7, 14.1 and 18.8, 300K, data of this residue (kindly provided by Prof. A. G. Palmer III of Columbia U.), to SRLS analysis.
The first row of Table 17 shows the best-fit parameters obtained with SRLS combination 5 (SRLS_5), which is formally analogous with MF model 5. In this scenario the local ordering is axially symmetric and N = RL∥/RL⊥ ≫1. The potential is small (c20 = 3.2), the corresponding squared order parameter is small ((S20)2 = 0.47), the D-to-M tilt is small (βMD = 16.1°), and there is only a modest time scale separation between the global and perpendicular component, RL⊥ (i.e., RC = 0.44 in units of RL⊥). Similar values were obtained previously for the flexible residues of a large number of proteins and can be considered typical. This modest time scale separation between the global and local motion (RC) is expected. Quite unexpectedly, the local potential is weak (c20 = 3.2) for the tightly packed globular proteins, i.e., the local ordering is low ((S20)2 = 0.40). Also, the diffusion tilt is on the order of 16° instead of being on the order of 90°, corresponding to preferred ordering around Cαi−1-Cαi of Ni−Cαi. No improvement was achieved by allowing N to vary (SRLS_6), indicating that N ≫ 1 is not the main reason for these difficult-to-reconcile results. On the other hand, significant improvement was achieved by allowing the potential to be rhombic, in addition to allowing N to vary, as shown by the last row of Table 17.
Let us compare the results obtained for axial (SRLS_6) and rhombic (SRLS_rh) potentials. The value of N = 40 corresponds to τL∥ = 53 ps whereas N = 916 corresponding to τL∥ ∼ 0. The ordering is high in the rhombic case, as implied by c22 = 10.1, and low in the axial case. The ratio c22/c20 obtained with SRLS_rh corresponds to ‘nearly planar YM-XM ordering’, as expected This is borne out clearly by the values of the components of the Cartesian ordering tensor shown in Table 17. The diffusion tilt is βMD ∼ 90° in the rhombic case, compatible with crankshaft fluctuations38 or peptide-plane reorientation about the Cαi−1-Cαi axis or the Ni-Ci bond, small-amplitude N-H wobbling motion and/or nitrogen pyramidalization,39 which are illustrated in Fig. 30. The rate of this motion, RL⊥ = 0.23×9.23 = 2.1 ns, shows that, as expected, loops move on the same time scale as the entire molecule. Unlike the axial scenario the rhombic scenario is consistent and physically appropriate.
Figure 30.
Various local motion modes including the anti-correlated Φi and Ψi−1 crankshaft motion (upper left), peptide-plane motion about Cαi−1-Cαi (upper right), nitrogen pyramidalization (lower left) and fast small-amplitude fluctuations (lower right).
Data fitting with rhombic potentials was also carried out for residues 45 and 47 of Adenylate kinase from E. coli (AKeco), which are representative of the mobile domain AMPbd of AKeco. The results are shown in Table 18. The symmetry of the rhombic potential is of the same type as found for residue 124 of RNase H, and the angle βMD is also close to 90°. However, RC is on the order of 0.8 for the AKeco residues 45 and 47 as compared to 0.2 for residue 124 of RNase-H, and τL∥ ∼ 1.5 ns for the AKeco residues 45 and 47 as compared to τL∥ = 75 ps for residue 124 of RNase-H. This indicates significantly stronger dynamical coupling and smaller local diffusion anisotropy for mobile domains than for flexible loops.
Table 18.
Best-fit parameters obtained with the SRLS combination 6 featuring rhombic potentials using Adenylate kinase data acquired at 14.1 and 18.8 T, 303 K.19 τm = 15.1 ns was used. RC = τL∥/τm. Sxx, Syy and Szz are the components of the Cartesian ordering tensor.
| res | c20 | c22 | RC | Sxx | Syy | Szz | βMD° | N |
|---|---|---|---|---|---|---|---|---|
| 46 | 5.7 | 10.5 | 0.82 | −0.465 | 0.827 | −0.361 | 101.4 | 9.6 |
| 47 | 4.3 | 10.3 | 0.73 | −0.470 | 0.761 | −0.291 | 100.7 | 6.3 |
The SRLS version which allows for rhombic potentials and axial local diffusion clearly yields a consistent physical picture. It appears that this is as much as one can extract from 15N relaxation data in proteins. Note that nine data points, acquired at three magnetic fields were used in these calculations. The rhombic potential coefficient, c22, is the only extra parameter in SRLS as compared to extended MF. On the other hand, for flexible residues which are typically of biological interest, SRLS features a single local motion while MF features two local motions (fast and slow), so the MF concept is actually a more compounded one.
The meaning of mixed modes
Let us consider a cylinder diffusing freely in an isotropic medium. The diffusion rates are RL∥ and RL⊥, with N = RL∥/RL⊥ determined by its shape. The solution of the diffusion equation yields eigenvalues τK−1 = 6 RL⊥ + K2 (RL∥ – RL⊥), with K = 0, 1 and 2. Let us now consider the same cylinder diffusing in the presence of a locally orienting potential. This is a reasonable model for an N-H bond attached physically to the protein, with the local potential representing the restrictions imposed on its motion by the immediate protein environment. The protein itself is reorienting at a slower rate with respect to a fixed lab frame. When the local and global motions do not occur on a greatly separated time scale, and the local potential is neither very low nor very high, the potential couples or mixes the motions of the N-H bond and the protein. The local ordering can be expressed (as usually done for restricted motions in liquids) in terms of an ordering tensor with principal values defined in terms of the orienting potential.
This is the two-body problem solved by the SRLS model. A Smoluchowski equation of the form of eq 1 is solved where the SRLS diffusion operator Γ̂ can be written in either of the two equivalent forms given by eq 2 or eq 3. In eq 2 the orientation of each body is referred to the lab (inertial) frame, but with a potential coupling them, which depends on their relative orientations. Simple products of basis functions of the two rotators (N-H body and protein), corresponding to their free diffusion (i.e., zero potential coupling them), are utilized to provide a matrix representation of Γ̂. This is a convenient basis set when the potential is relatively small, i.e., weak coupling. In eq 3 only the global motion of the protein is referred to the lab frame, whereas the local motion of the N-H bond is referred to the local director frame in the protein. This latter scenario thus describes the local motion in relative coordinates. Then product basis functions for the overall motion and the relative motion are used to provide the matrix representation of Γ̂. This is a more natural choice when the potential coupling is large. Since these two approaches are mathematically equivalent, one may use either choice. In our past work we have utilizes eq 2, whereas in the newer work we have reported in this paper we utilized eq 3.
The eq 2 perspective on mixed modes means that as a coupling potential is added, the new eigenmodes of Γ̂ become linear combinations of the product functions of the two free rotors. This is a point of view where there are two sources of “mixed-modes”: the first results from the coupling between the two rotors, so that the motion of the internal rotor becomes more that of its motion relative to the protein. This is a feature that exists even when there is time-scale separation, i.e., RC/RL ⪡ 1. The second arises when RC/RL ∼ 1, so there is no longer a significant time-scale separation. In that case the diffusive reorientation of the internal rotor becomes a mixture of the global and local motions. That is, an observer that detects just the 15N label on a particular N-H bond can no longer distinguish between a local and a global mode of motion. Thus, these modes become mixed. In the case, wherein eq 3 and its convenient basis set are used, the intuitive picture changes somewhat, but the final analysis must remain equivalent. In simple mathematical terms, this means that the eigenvalues of Γ̂ are unchanged, but the eigenmodes are represented in (or referred to) the different basis sets, and appear different, although (again) they must be equivalent. Here, for very high ordering and RC/RL ⪡ 1, the eigenmodes can be represented by the overall motion and by the relative internal motion with eigenvalues given (for axial potentials) by eq 18, and eigenfunctions given elsewhere (refs. 29 and 48), yielding simple limiting correlation functions. As the coupling potential is reduced (but RC/RL ⪡ 1), then the correlation functions for the relative motion (i.e., for the D2MK(ΩCM)) become more complex, involving several eigenmodes of this motion. Again, as RC/RL⊥ → 1, there must be “mixed modes” of the two coupled dynamic processes.
We illustrate some of these concepts with a relevant computational study presented by Polimeno and Freed.23 This was performed using the basis set appropriate for eq 2, i.e., the global and internal rotors both referred to the lab frame. Table 19 shows the eigenvalues (in units of RL) and corresponding weights of the two isotropic rotors coupled by an axial potential whose strength is given by c20. Values of RC/RL of 1.0, 0.1 and 0.01 are considered, and c20 ranges from 0 to 4. When c20 = 0, the motions are uncoupled, so only the motion of the local rotor relative to the lab frame is relevant. It corresponds to eigenvalue 6 (or more precisely 6RL) with weight 1.00, independent of RC/RL. For the case of RC/RL = 0.01 there is good time-scale separation of the motions. Thus, as c20 is increased the global motion retains the eigenvalue 0.06, but its relative weight in the correlation function C0(t) increases roughly according to (S20)2. The local motion is represented by several eigenvalues with the (typically two) major ones given in Table 19. The results for RC/RL = 0.1 are qualitatively similar. For RC/RL = 1 the results are quite different. As c20 is increased from 0, two main mixed modes appear, one of which decreases from the value of 6.00, while the other increases from this value, and the former becomes relatively more important in C0(t). We can intuitively suggest that the former represents a mixed mode, wherein as the protein reorients in one sense (e.g., clockwise), the internal rotor is attempting to reorient in the opposite (e.g., counterclockwise) sense; the latter mixed mode would correspond to more additive reorientational diffusion (i.e., both are in the same sense).
Table 19.
Dominant eigenvalues and respective weights (given in parenthesis) in the C0(t) correlation function for two isotropic rotators, RC and RL = 1, mixed by an axial potential of strength 0 ≤ c20 ≤ 4, and time scale separation given by RC = 1.0, 0.1 and 0.01. The (S20)2 values corresponding to the c20 values are also presented.
| RC | ||||
|---|---|---|---|---|
| c20 | (S20)2 | 1.0 | 0.1 | 0.01 |
| 0 | 0 | 6.00(1.00) | 6.00(1.00) | 6.00(1.00) |
| 1 | 0.049 | 4.79(0.60) | 0.59(0.06) | 0.06(0.05) |
| 7.57(0.39) | 5.94(0.65) | 5.57(0.32) | ||
| 7.33(0.15) | 6.04(0.27) | |||
| 2 | 0.193 | 3.90(0.71) | 0.58(0.23) | 0.06(0.19) |
| 10.0(0.27) | 6.55(0.57) | 6.04(0.26) | ||
| 8.10(0.11) | 6.68(0.36) | |||
| 3 | 0.366 | 3.41(0.79) | 0.56(0.41) | 0.06(0.37) |
| 13.43(0.17) | 7.95(0.37) | 7.20(0.15) | ||
| 9.47(0.12) | 8.02(0.33) | |||
| 4 | 0.506 | 3.19(0.85) | 0.55(0.56) | 0.06(0.51) |
| 10.06(0.22) | 10.0(0.30) | |||
| 11.54(0.14) |
Finally, a comment on the tensors RC and RL which constitute input values in a given SRLS calculation, or best-fit parameters in fitting actual experiments, is in order. These quantities may represent more complex local and global rotators, and not just the RC and RL tensors corresponding to simple rotators. One can suggest tentative interpretations, as was done in a recently published SRLS application to nitroxide-labeled T4 Lysozyme.37 In that paper RL∥ was interpreted as motion around a specific bond of the nitroxide tether, whereas RL⊥ was associated with motion around the symmetry axis of the helix comprising the nitroxide-labeled residue. For N-H bond motion we associated at this stage of our studies RL⊥ with motion around the Cαi−1-Cαi axis and RL∥ with motion around an axis perpendicular to Cαi−1-Cαi within the peptide plane.
Table 20 illustrates the high sensitivity of the eigenvalues and weights comprised in the time correlation functions CKK′(t) to the symmetry of the coupling potential. The parameters used include RC = 0.001 and c20 = 2, with c22 = 0 for axial ordering and 3 for rhombic ‘nearly planar YM-XM ordering’. For axial ordering only C0(t) is relevant. For the βMD = 90° geometry only the correlation functions C0(t), C2(t) and C2–2(t) = C−22(t) contribute to the measurable spectral density. The dominant eigenvalues and their eigenmode composition depend to a large extent on the symmetry of the coupling potential. The individual Lorentzians in a given function jK1K1(ω), obtained by Fourier transformation of the corresponding time correlation function, are multiplied by d2KK′ to yield the measurable spectral density. The d2KK′ values are also presented in Table 20. The additional contributions, comprising a large number of eigenvalues with small individual weights, are not shown. It can be seen clearly that potentials of similar magnitude but different symmetry are associated with a different composition of dynamic modes. Obviously potential symmetry is a very influential component, to which the experimental data are highly sensitive.
Table 20.
Dominant eigenvalues and respective weights (given in parenthesis) of the CK(t) and CKK'(t) components which contribute to the measurable spectral density for βMD = 0° in the axial case given by RC = 0.001, c20 = 2 and c22 = 0, and βMD = 90° in the rhombic case given by RC = 0.001, c20 = 2 and c22 = 3. The respective irreducible ordering tensor components, S20 and S22, are also presented.
| c22 | S20 | S22 | eigenvalue(weight) | K | K' | d2KK' |
|---|---|---|---|---|---|---|
| 0.0 | 0.439 | 0.0 | 6.58(0.415) | 0 | 0 | 1.0 |
| 0.006(0.193) | ||||||
| 5.95(0.193) | ||||||
| 8.52(0.161) | ||||||
| .............. | ||||||
| 3.0 | 0.088 | 0.572 | 5.40(0.349) | 0 | 0 | 0.25 |
| 4.44(0.237) | ||||||
| 4.36(0.232) | ||||||
| 8.31(0.114) | ||||||
| 0.006(0.008) | ||||||
| ............... | ||||||
| 4.36(0.214) | 2 | 2 | 0.75 | |||
| 1.85(0.209) | ||||||
| 5.70(0.184) | ||||||
| 0.006(0.098) | ||||||
| ............... | ||||||
| 4.36(0.428) | 2 | −2 | 0.75 | |||
| 0.006(0.163) | ||||||
| 14.40(0.125) | ||||||
| 5.40(0.120) | ||||||
| 4.44(0.08) | ||||||
| ............... |
As indicated, we have developed recently a fitting scheme for SRLS where the spectral densities jK(ω) and jKK′(ω) are calculated on the fly. This fitting scheme allows for rhombic potentials, and arbitrary axiality of the local and global diffusion tensors. Efforts to improve the computational efficiency of this scheme are underway.
IV. Conclusions
Model-free is a very simplified approach for analyzing spin relaxation data in proteins, such that the quality of the experimental data and their variations (e.g., with magnetic field) are frequently beyond its capabilities. Small data sets (three data points at a given field in the case of N-H bond dynamics) can usually be force-fitted with good statistics but inaccurate best-fit parameters, which are obtained through parameterization of the experimental spectral densities. When larger data sets acquired at several magnetic fields, temperatures, states of complex formation, etc., are subjected to MF analysis, or when N-H and C′-Cα bond dynamics are analyzed in concert, force-fitting is so pervasive that functional dynamics may be missed, qualitatively erroneous results may be derived, and incorrect phenomena may be detected. Conformational entropy derived from parameterizing entities is inaccurate. Entropy profiles over the protein backbone may even be qualitatively inaccurate.
On the other hand, the experimental data can be analyzed significantly more reliably with SRLS spectral densities, which constitute generalized forms of the MF formulae. The main aspects which greatly improve the analysis include non-symmetric (e.g., axial) local motion, rhombic local ordering, rigorous account of mode-coupling, and proper treatment of general features of local geometry. The dynamic picture emerging, which differs significantly from the MF picture, is physically insightful, consistent and comprehensive. Conformational entropy can be derived with SRLS in a straightforward manner from experimentally determined local potentials of arbitrary symmetry.
Figure 27.
Transverse 15N T2 relaxation times of Ca2+-calmodulin at 294 K (black), 300 K (red), 308 K (green) and 316 K (blue), and 8.5, 14.1 and 18.8 T.65 The vertical dashed lines depict the central linker (residues 74 – 78).
Acknowledgments
This work was supported by the Israel Science Foundation grant number 279/03 and the Binational Science Foundation 2000399 to E.M., and the Damadian Center for Magnetic Resonance research at Bar-Ilan University, Israel.
References
- 1.(a) Kay LE. Nat. Struct. Biol. 1998;5:513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]; (b) Ishima R, Torchia DA. Nat. Struct. Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]; (c) Palmer AG., II Annu. Rev. Biophys. Biomol. Struct. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 2.(a) Case D. Acc. Chem. Res. 2002;35(325):325–33. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]; (b) Bruschweiler R. Curr. Opin. Struct. Biol. 2003;13:175–183. doi: 10.1016/s0959-440x(03)00036-8. [DOI] [PubMed] [Google Scholar]
- 3.Palmer AG., III Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 4.Peng JW, Wagner G. In: Methods in Enzymology. James TL, Oppenheimer NJ, editors. Vol. 239. Academic Press; New York: 1994. pp. 563–595. [DOI] [PubMed] [Google Scholar]
- 5.Abragam A. Principles of Nuclear Magnetism. Oxford University Press (Clarendon); London: 1961. [Google Scholar]
- 6.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 7.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 8.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 9.Lee AL, Wand J. J. Biomol. NMR. 1999;13:101–112. doi: 10.1023/a:1008304220445. [DOI] [PubMed] [Google Scholar]
- 10.Osborne MJ, Wright PE. J. Biomol. NMR. 2001;19:209–230. doi: 10.1023/a:1011283809984. [DOI] [PubMed] [Google Scholar]
- 11.Vugmeyster L, Trott O, McKnight CJ, Raleigh DP, Palmer AG., III J. Mol. Biol. 2002;320:841–854. doi: 10.1016/S0022-2836(02)00537-5. [DOI] [PubMed] [Google Scholar]
- 12.Mandel AM, Akke M, Pamer AG., III Biochemistry. 1996;35:16009–16023. doi: 10.1021/bi962089k. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, Cai S, Zuiderweg ERP. J. Am. Chem. Soc. 2003;125:8639–8643. doi: 10.1021/ja034077+. [DOI] [PubMed] [Google Scholar]
- 14.Zeeb M, Jacob M, H., Schindler T, Balbach J. J. Biomol. NMR. 2003;27:221–234. doi: 10.1023/a:1025449611201. [DOI] [PubMed] [Google Scholar]
- 15.Korchuganov DS, Gagnidze IE, Tkach EN, Schulga AA, Kirpichnikov MP, Arseniev AS. J. Biomol. NMR. 2004;30:431–442. doi: 10.1007/s10858-004-4242-7. [DOI] [PubMed] [Google Scholar]
- 16.Chang S-L, Tjandra N. J. Magn. Res. 2005;174:43–53. doi: 10.1016/j.jmr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P, Dayie K, Wagner G. J. Mol. Biol. 1997;272:443–455. doi: 10.1006/jmbi.1997.1266. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Frederick KK, Igumeniva TI, Wand AJ, Zuiderweg ERP. J. Am. Chem. Soc. 2005;127:828–829. doi: 10.1021/ja045743p. [DOI] [PubMed] [Google Scholar]
- 19.Tugarinov V, Shapiro Yu. E., Liang Z, Freed JH, Meirovitch E. J. Mol. Biol. 2002;315:171–186. doi: 10.1006/jmbi.2001.5231. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro Yu. E., Kahana E, Tugarinov V, Liang Z, Freed JH, Meirovitch E. Biochemistry. 2002;41:6271–6281. doi: 10.1021/bi012132q. [DOI] [PubMed] [Google Scholar]
- 21.Pang Y, Buck M, Zuiderweg ERP. Biochemistry. 2002;41:2655–2666. doi: 10.1021/bi011657f. [DOI] [PubMed] [Google Scholar]
- 22.Tugarinov V, Liang Z, Shapiro Yu. E., Freed JH, Meirovitch E. J. Am. Chem. Soc. 2001;123:3055–3063. doi: 10.1021/ja003803v. [DOI] [PubMed] [Google Scholar]
- 23.Polimeno A, Freed JH. Adv. in Chem. Phys. 1993;83:89–210. [Google Scholar]
- 24.Polimeno A, Freed JH. J. Phys. Chem. 1995;99:10995–11006. [Google Scholar]
- 25.Liang Z, Freed JH. J. Phys. Chem. B. 1999;103:6384–6396. 1999. [Google Scholar]
- 26.Meirovitch E, Shapiro Yu.E., Tugarinov V, Liang Z, Freed JH. J. Phys. Chem. B. 2003;107:9883–9897. [Google Scholar]
- 27.Meirovitch E, Shapiro YE, Liang Z, Freed JH. J.Phys. Chem. B. 2003;107:9898–9904. [Google Scholar]
- 28.Polimeno A, Moro GJ, Freed JH. J. Chem. Phys. 1996;104:1090–1104. [Google Scholar]
- 29.Polnaszek CF, Freed JH. J. Phys. Chem. 1975;79:2283–2306. [Google Scholar]
- 30.Lin WJ, Freed JH. J. Phys. Chem. 1979;83:379–401. [Google Scholar]
- 31.Millet O, Muhandiram DR, Skrynnikov NS, Kay LE. J. Am. Chem. Soc. 2002;124:6439–6448. doi: 10.1021/ja012497y. [DOI] [PubMed] [Google Scholar]
- 32.Choy W-Y, Kay LE. J. Biomol. NMR. 2003;25:325–333. doi: 10.1023/a:1023065310430. [DOI] [PubMed] [Google Scholar]
- 33.Akke M, Bruschweiler R, Palmer AG., III J. Am. Chem. Soc. 1993;115:9832–9833. [Google Scholar]
- 34.(a) Yang D, Kay LE. J. Mol. Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]; (b) Yang D, Mok Yu-K., Forman-Kay JD, Farrow NA, Kay LE. J. Mol. Biol. 1997;272:790–804. doi: 10.1006/jmbi.1997.1285. [DOI] [PubMed] [Google Scholar]; (c) Kay LE, Muhandiram DR, Farrow NA, Aubin Y, Forman-Kay JD. Biochemistry. 1996;35:361–368. doi: 10.1021/bi9522312. [DOI] [PubMed] [Google Scholar]; (d) Yang D, Mok Y-K, Forman-Kay JD, Farrow NA, Kay LE. J. Mol. Biol. 1997;272:790–804. doi: 10.1006/jmbi.1997.1285. [DOI] [PubMed] [Google Scholar]
- 35.(a) Li Z, Raychaudhuri S, Wand AJ. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee AL, Kinnear SA, Wand AJ. Nat. Struct. Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]; (c) Lee AL, Wand AJ. Nature. 2001;411:501–504. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 36.Massi F, Palmer AG., III J. Am. Chem. Soc. 2003;125:11158–11159. doi: 10.1021/ja035605k. [DOI] [PubMed] [Google Scholar]
- 37.Liang Z, Lou Y, Freed JH, Columbus L, Hubbell WL. J. Phys. Chem. B. 2004;108:17649–17659. [Google Scholar]
- 38.Brunne RM, Berndt KD, Guntert P, Wuthrich K, van Gunsteren WF. Proteins. 1995;23:49–62. doi: 10.1002/prot.340230107. [DOI] [PubMed] [Google Scholar]
- 39.Palmo K, Mannfors B, Mirkin NG, Krimm S. Biopolymers. 2003;68:383–394. doi: 10.1002/bip.10254. [DOI] [PubMed] [Google Scholar]
- 40.Lienin SF, Bremi T, Brutscher B, Bruschweiler R, Ernst RR. J. Am. Chem. Soc. 1998;120:9870–9879. [Google Scholar]
- 41.Moro G, Freed JH. J. Chem. Phys. 1981;74:3757–3773. [Google Scholar]
- 42.Moro G, Freed JH. Math Studies Series. In: Cullum J, Willough R, editors. Large-Scale Eigenvalue Problems. Vol. 127. North Holland: 1986. pp. 143–160. [Google Scholar]
- 43.Freed HJ, Nayeem A, Rananavare SB. In: The Molecular Dynamics of Liquid Crystals. Luckhurst GR, Veracini CA, editors. Kluwer Academic Publishers; The Netherlands: 1994. pp. 271–312. Chapter 12. [Google Scholar]
- 44.Fushman D, Tjandra N, Cowburn D. J. Am. Chem. Soc. 1998;120:10947–10952. [Google Scholar]
- 45.Cornilescu G, Bax A. J. Am. Chem. Soc. 2000;122:10143–10154. [Google Scholar]
- 46.Mandel AM, Akke M, Palmer AG., III J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 47.Fushman D, Cahill S, Cowburn D. J. Mol. Biol. 1997;266:173–194. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 48.Freed JH. J. Chem. Phys. 1977;66:4183–4199. [Google Scholar]
- 49.Polnaszek CF, Bruno GV, Freed JH. J. Chem. Phys. 1973;58:3185–3199. [Google Scholar]
- 50.Kinosita K, Kawato S, Ikegami A. Biophys. J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipari G, Szabo A. Biophys. J. 1980;30:489–506. doi: 10.1016/S0006-3495(80)85109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.(a) Vold RL. In: The Molecular Dynamics of Liquid Crystals. Luckhurst GR, Veracini CA, editors. Kluwer Academic Publishers; The Netherlands: 1994. [Google Scholar]; (b) Dingemans T, Photinos DJ, Samulski ET, Terzis AF, Wutz C. J. Chem. Phys. 2003;118:7046–7061. [Google Scholar]
- 53.(a) Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]; (b) Pawley NH, Wang C, Koide S, Nicholson LK. J. Biomol. NMR. 2001;20:149–165. doi: 10.1023/a:1011249816560. [DOI] [PubMed] [Google Scholar]
- 54.Damberg P, Jarvet J, Graslund A. J. Am. Chem. Soc. 2004;127:1995–2005. doi: 10.1021/ja045956e. [DOI] [PubMed] [Google Scholar]
- 55.Peng JW, Wagner G. J. Magn. Res. 1992;98:308–332. [Google Scholar]
- 56.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 57.Ishima R, Nagayama K. J. Magn. Res. Ser.B. 1995;108:73–76. [Google Scholar]
- 58.Fushman D, Cowburn D. J. Am. Chem. Soc. 1998;120:7109–711. [Google Scholar]
- 59.Kroenke CD, Rance M, Palmer AG., III J. Am. Chem. Soc. 1999;121:10119–10125. [Google Scholar]
- 60.Lee A, Sharp KA, Kranz JK, Song X-J, Wand AJ. Biochemistry. 2002;41:13814–13825. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 61.Babu YS, Bugg CE, Cook WJ. J. Mol. Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 62.Ikura M, Barbato G, Kay LE, Pastor RW, Bax A. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 63.Tjandra N, Kuboniwa H, Ren H, Bax A. Eur. J. Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 64.Baber JL, Szabo A, Tjandra N. J. Am. Chem. Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 65.Chang S-L, Szabo A, Tjandra N. J. Am. Chem. Soc. 2003;125:11379–11384. doi: 10.1021/ja034064w. [DOI] [PubMed] [Google Scholar]
- 66.Lee LK, Rance M, Chazin WJ, Palmer AG., III J. Biomol. NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]