Abstract
An endogenous 0.6-kb rice DNA transposon, nDart1-0, was found as an active nonautonomous element in a mutable virescent line, pyl-v, displaying leaf variegations. Here, we demonstrated that the active autonomous element aDart in pyl-v corresponds to Dart1-27 on chromosome 6 in Nipponbare, which carries no active aDart elements, and that aDart and Dart1-27 are identical in their sequences and chromosomal locations, indicating that Dart1-27 is epigenetically silenced in Nipponbare. The identification of aDart in pyl-v was first performed by map-based cloning and by detection of the accumulated transposase transcripts. Subsequently, various transposition activities of the cloned Dart1-27 element from Nipponbare were demonstrated in Arabidopsis. Dart1-27 in Arabidopsis was able to excise nDart1-0 and Dart1-27 from cloned sites, generating footprints, and to integrate into new sites, generating 8-bp target site duplications. In addition to Dart1-27, Nipponbare contains 37 putative autonomous Dart1 elements because their putative transposase genes carry no apparent nonsense or frameshift mutations. Of these, at least four elements were shown to become active aDart elements in transgenic Arabidopsis plants, even though considerable sequence divergence arose among their transposases. Thus, these four Dart1 elements and Dart1-27 in Nipponbare must be potential autonomous elements silenced epigenetically. The regulatory and evolutionary implications of the autonomous Dart1 elements and the development of an efficient transposon-tagging system in rice are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0410-x) contains supplementary material, which is available to authorized users.
Keywords: Autonomous elements, DNA transposon, Epigenetic silencing, hAT superfamily, Transposition activity, Rice
Introduction
Considerable attention has recently been given to DNA transposons causing mutable alleles in plants for utilizing transposon tagging as a functional genomic tool to elucidate the function of genes of interest (Feschotte et al. 2002; Kunze and Weil 2002; May and Martienssen 2003; Chopra et al. 2006; Johzuka-Hisatomi et al. 2008). Rice (Oryza sativa L.) is an important staple food for more than one half of the world’s population, and it has become the first crop plant for which the 389-Mb genome of a variety, cv. Nipponbare, has been sequenced (International Rice Genome Sequencing Project 2005). However, only a few active endogenous DNA transposons causing mutable alleles have been reported in rice. For example, the mutable slg (slender glume) allele, which shows a variegated glume shape, was found to be caused by the integration of 430-bp mPing of Tourist-like miniature inverted-repeat transposable elements (MITEs) belonging to the PIF/Harbinger superfamily into the Rurm1 (Rice ubiquitin-related modifier-1) locus (Nakazaki et al. 2003). The mutable virescent (or pale-yellow-leaf variegated, pyl-v) and thumbelina-mutable (thl-m) alleles, which display leaf variegation and mutable gibberellin-insensitive dwarfism, were caused by the integration of the same 607-bp nonautonomous element nDart1-0 belonging to the hAT superfamily into the OsClpP5 and GID1 (GIBBERELLIN INSENSITIVE DWARF1) genes encoding the P5 subunit of the ATP-dependent caseinolytic protease and the soluble gibberellin receptor, respectively (Tsugane et al. 2006), whereas the mutable albino allele due to chlorophyll deficiency was caused by the insertion of a 607-bp nDart1-0-related element, nDart1-3, into the gene for Mg-protoporphyrin IX methyltransferase (Fujino et al. 2005). In the mutable pyl-v allele, somatic excision of nDart1-0 from OsClpP5 in the presence of an active autonomous element, aDart, results in the pyl-v leaf variegation phenotype, a dark-green sector consisting of somatically reverted cells due to nDart1-0 excision in a pale-yellow background comprising cells having nDart1-0 inserted into OsClpP5 in the homozygous condition (Tsugane et al. 2006). Plants containing the pyl-v allele without an active aDart element display pale-yellow leaves without variegation; this has been termed as the pale-yellow leaf-stable (pyl-stb) phenotype. In addition, the 670-bp nonautonomous element dTok0 of the hAT superfamily found within the FON1 (FLORAL ORGAN NUMBER1) gene exon 1 was also shown to be an active transposon, even though the fon1 mutant line containing dTok0 gave phenotypic revertants only rarely (Moon et al. 2006). Among these active rice DNA transposons, nDart1-0 and its relatives appear to be more suitable than the others for transposon tagging in rice because (1) their transposition can be controlled under natural growth conditions, i.e., the transposition of nDart1-0 can be induced by crossing with a line containing an active aDart element and stabilized by segregating aDart, and (2) nDart1-0 and its relatives are often found at GC-rich regions in the genome and tend to integrate into promoter proximal genic regions (Tsugane et al. 2006; Takagi et al. 2007; K. Takagi and M. Maekawa unpublished data).
In the sequenced Nipponbare genome containing no active aDart elements, the nDart-related elements can be classified into three subgroups of about 0.6-kb nonautonomous elements (nDart1-3, nDart1-101, and nDart1-201) and four subgroups of elements longer than 2 kb, which comprise epigenetically silenced inactive iDart and genetically defective dDart elements (iDart1/dDart1, iDart2/dDart2, dDart3, and iDart4) on the basis of their lengths and sequence characteristics (Tsugane et al. 2006). According to the 4.0 pseudomolecules of the Nipponbare genome (International Rice Genome Sequencing Project 2005), the small nDart1 elements of 0.6 kb and longer iDart1/dDart1 elements are 18 and 53 copies, respectively, and both elements are distributed throughout the Nipponbare chromosomes (Takagi et al. 2007; Johzuka-Hisatomi et al. 2008). Interestingly, nDart1-0, which was originally identified as an element causing the pyl-v allele in the mutable virescent line and subsequently found to be the most active element in the nDart1-3 subgroup, is not present in the Nipponbare genome (Tsugane et al. 2006; Johzuka-Hisatomi et al. 2008). Because only a fraction of 0.6-kb nDart1 elements belonging to the nDart1-3 subgroup in the pyl-v plant can be transposed in the presence of an active aDart element and because the excision of the same nDart1-3 subgroup elements can be induced by treating Nipponbare with 5-azacytidine (5-azaC), Nipponbare must contain an epigenetically silenced autonomous element or elements that can become aDart by 5-azaC treatment (Tsugane et al. 2006). Among the 53 iDart1/dDart1 elements in Nipponbare, 38 iDart1 elements are putative autonomous element silenced epigenetically because their putative transposase genes carry no apparent nonsense or frameshift mutations (Tsugane et al. 2006; Takagi et al. 2007). It is, thus, highly likely that an active aDart element is similar in structure to one of these 38 iDart1 elements.
In the F2 population from a cross between pyl-stb and pyl-v, the ratio of pyl-v to pyl-stb was 3:1, indicating that the pyl-v plant contains only one copy of the active aDart element (Tsugane et al. 2006; M. Maekawa unpublished data). We subsequently exploited the capability of the active aDart element to excise the nonautonomous nDart1-0 element from the OsClpP5 gene in the pyl-stb tester plant to explore the presence and distribution of an aDart element in 49 japonica and 51 indica varieties; several japonica varieties were found to contain aDart (Nishimura et al. 2008). We show here that, by map-based cloning, aDart in the mutable pyl-v plant coincides with one of the 38 iDart1 elements, iDart1-27, residing on chromosome 6 in Nipponbare and that the transcripts of the accumulated transposase gene in the pyl-v leaves are predominantly from Dart1-27. Moreover, iDart1-27 from Nipponbare can be converted into an active aDart element by cloning into Escherichia coli and subsequently introduced into Arabidopsis by Agrobacterium-mediated transformation; Dart1-27 in Arabidopsis can mobilize nDart1-0 and transpose by itself from the introduced vectors into various sites in the Arabidopsis genome, and the generation of footprints and 8-bp target site duplications can be observed upon excisions and insertions of the elements, respectively. The results demonstrate that Dart1-27 fulfills the molecular criteria for the autonomous element. Consequently, Dart1-27 in pyl-v and Nipponbare is an active and an epigenetically silenced autonomous element, respectively. We also showed that not only Dart1-27 but also at least four other Dart1 elements could render active autonomous elements in the introduced Arabidopsis plants, indicating that they are potential autonomous elements silenced epigenetically in Nipponbare. Moreover, cloned Dart1-27 derivatives from Nipponbare were shown to behave as an active aDart element mobilizing nDart1-0 from OsClpP5 when they were introduced into pyl-stb plants by Agrobacterium-mediated transformation. These findings would facilitate the development of an efficient gene-tagging system in rice and shed light on epigenetic regulatory and evolutionary aspects of autonomous elements in the nDart/aDart system.
Materials and methods
Nucleic acid procedures
All nucleic acid procedures, including the preparation of genomic DNA and RNA, PCR amplification, and Southern blot hybridization and RACE analyses were performed as previously described (Tsugane et al. 2006; Nishimura et al. 2008; Yamauchi et al. 2008). The list of primers used for PCR and RT-PCR amplifications, probes for Southern blot hybridization, and RACE analysis is found in Supplementary Table S1.
Mapping of an active aDart element in the pyl-v line
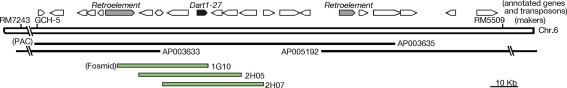
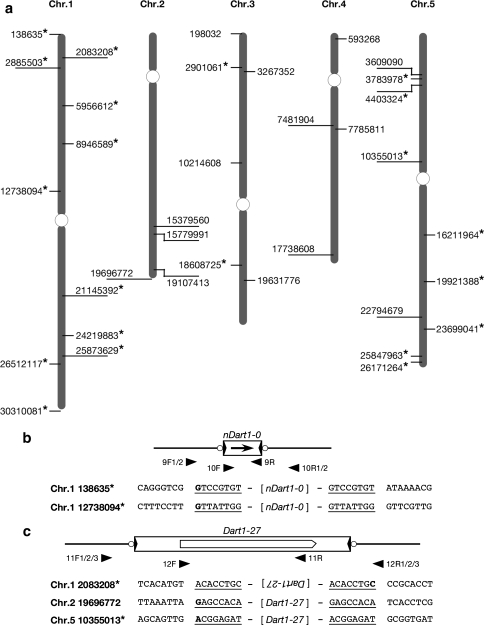
To map an active aDart element in the pyl-v mutant line with the background of a temperate japonica line T-65 (Tsugane et al. 2006), we first obtained 21 pyl-stb segregants out of 669 F2 plants from the cross between a pyl-v line and the indica line Kasalath, which carries no aDart element (Tsugane et al. 2006; Nishimura et al. 2008), and used 57 SSR markers for genome-wide coverage (Maekawa et al. 2005) for rough mapping. Because aDart appeared to reside in the region near the RM30 marker in the long arm of chromosome 6 (Fig. 1), 550 pyl-stb segregants out of 10,370 F2 plants from the cross between the pyl-v line and Kasalath were subsequently employed for fine mapping using simple sequence repeat (SSR) markers RM5509 and RM7243 (McCouch et al. 2002) and sequence length polymorphism (SLP) marker GCH-5 on the basis of the available rice genome sequence data (Nishimura et al. 2008). PCR amplification with primers Dt1-F (1F) and Dt1-R (1R) (Fig. 2a, Supplementary Table S1) was used to detect the presence or absence of a Dart1 element in the pyl-v line at the corresponding Dart1-27 site in Nipponbare.
Fig. 1.
Mapping of an active aDart element in the pyl-v line. The aDart element was mapped at the 259-kb region between the SSR markers RM7243 and RM5509 on chromosome 6 and subsequently located within the 170-kb region between the SLP marker GCH-5 and RM5509, which was covered by three PAC clones and contained 23 annotated genes, including Dart1-27 in the Nipponbare genome. The horizontal pentagonals represent the annotated genes, and three Fosmid clones contained Dart1-27 and its adjacent segment from the pyl-v genome
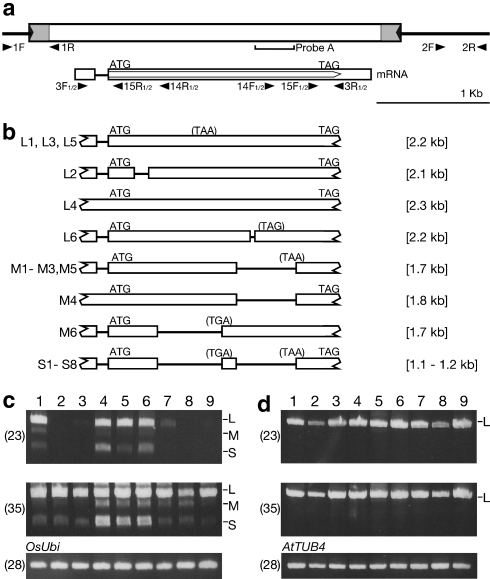
Fig. 2.
Transcripts of the Dart1 transposase genes detected in rice and transgenic Arabidopsis plants. a Structure of Dart1-27 in the pyl-v genome. The large horizontal filled arrowheads and the shadowed areas at both ends of the transposon Dart1-27 box indicate the terminal inverted repeats and subterminal regions, respectively. The transposase gene carries an approximately 92-bp intron in its 5′-untranslared region, and its mRNA is indicated under the Dart1-27 box; the open pentagonal arrow in the 2nd exon of the mRNA boxes represents the coding region of the transposase gene. Probe A was used for screening Fosmid clones in Fig. 1, and the horizontal arrowheads with the codes (numerals with either F or R) represent primers for PCR or RT-PCR amplification and RACE analysis (Supplementary Table S1). b Schematic representation of the alternatively spliced transposase transcripts observed in rice and transgenic Arabidopsis plants. Three different transcripts in sizes (L, M, and S) were observed in rice, and their splicing patterns are categorized. The premature stop codons are indicated in parentheses, and the sizes of the RT-PCR amplified fragments were indicated in brackets. For detailed structural information concerning the individual transcript, see Supplementary Table S2. c Transcripts of the Dart1 transposase genes in rice by RT-PCR analysis. Lane 1, pyl-v; lane 2, pyl-stb; lane 3, Nipponbare; lanes 4–6, three different pyl-stb plants carrying a single copy of Dart1-27* generated by transforming pZEN12; lanes 7–9, three different pyl-stb plants carrying a single copy of Dart1-27D generated by transforming pZEN13. OsUbi (Os03g0234200) was used as an internal control. d Transcripts of the Dart1 transposase genes in Arabidopsis by RT-PCR analysis. Vectors used for transformation are lane 1, pZEN2; lane 2, pZEN3; lane 3, pZEN4; lane 4, pZEN5; lane 5, pZEN6; lane 6, pZEN7; lane 7, pZEN8; lane 8, pZEN9; lane 9, pZEN10. ATTUB4 (M21415) was used as an internal control. The numerals in parentheses indicate the cycles of PCR amplification
A Fosmid library from the pyl-v plants was prepared using a kit for CopyControl Cloning Systems (Epicentre Technologies, Madison, WI, USA). Out of about 140,000 clones bearing average 37-kb genomic segments, 216 clones containing Dart1 sequences were screened by colony hybridization with Probe A containing the dimerization-domain sequence of the putative transposase gene (Fig. 2a). Subsequently, we identified three Fosmid clones (1G10, 2H05, and 2H07) that carry the 0.79-kb segment adjacent to the Dart1-27 element by PCR analysis with the primers DtAJ-F (2F) and DtAJ-R (2R) (Figs. 1, 2a, Supplementary Table S1).
Characterization of Dart1 transcripts
The 5′- or 3′-RACE analysis for Dart1 transcripts was performed with a GeneRacer kit (Invitrogen, Carlsbad, CA, USA) with primers 5RC1-R (14R1) and 5RC2-R (15R1) or 3RC1-F (14F1) and 3RC2-F (15F1), respectively, for two consecutive amplifications, and RT-PCR analysis was carried out with Thermoscript reverse transcriptase (Invitrogen) for first-strand cDNA synthesis and with PrimeSTAR GXL Taq polymerase (Takara Bio, Ohtsu, Japan) for subsequent PCR amplification with primers Dt2-F1 (3F1) and Dt2-R1 (3R1) (Fig. 2a, Supplementary Table S1), as described previously (Tsugane et al. 2006; Yamauchi et al. 2008). For characterizing Dart1-52 transcripts, primers 5RC(1-51)1-R (14R2), 5RC(1-51)2-R (15R2), 3RC(1-51)1-F (14F2), 3RC(1-51)2-F (15F2), Dt2-F2 (3F2), and Dt2-R2 (3R2) were used in the 5′- or 3′-RACE and RT-PCR analyses (Fig. 2a, Supplementary Table S1).
Vector construction
A 0.7-kb fragment containing the entire nDart1-0 element was prepared from the pyl-stb genome by PCR amplification using LA Taq polymerase (Takara Bio) with primers Clp-F and Clp-R (Supplementary Table S1): initial denaturation (94°C for 1 min), 10 cycles of denaturation (94°C for 30 s), annealing (74°C for 30 s), and extension (72°C for 1 min), followed by a reduction of the annealing temperature in a 1°C decrement per cycle, then 30 cycles of reactions with an altered annealing condition (62°C for 30 s), and a final extension (72°C for 7 min). The resultant fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced to confirm that it contained the nDart1-0 sequence without any sequence alterations.
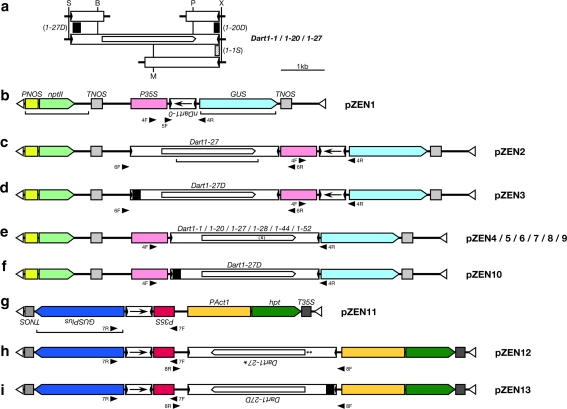
Fragments of about 3.7 kb containing six different Dart1 elements, Dart1-1,Dart1-20,Dart1-27,Dart1-28, Dart1-44, and Dart1-52, were first obtained from the appropriate BAC or PAC clones (AC105744, AC084817, AP003635, BX000499, AP004458, and AP002863, respectively) derived from Nipponbare by two consecutive PCR amplifications using KOD-Plus-polymerase (Toyobo, Osaka, Japan) with appropriate primers (Supplementary Table S1). The first PCR reaction consisted of initial denaturation (96°C for 5 min), 10 cycles of denaturation (96°C for 1 min), annealing (74°C for 30 s), and extension (72°C for 2.5 min), followed by a reduction of the annealing temperature in a 1°C decrement per cycle, then 50 cycles of reactions with an altered annealing condition (65°C for 30 s), and a final extension (72°C for 5 min). For the subsequent nested PCR amplification, 35 cycles of reactions, rather than 50 cycles, were performed before the final extension (72°C for 5 min). The amplified fragments were cloned either into pCR4 Blunt-TOPO-X, a pCR4 Blunt-TOPO derivative carrying an XhoI site next to the unique NotI site, or into pCR-BluntII-TOPO (Invitrogen). The cloned fragments were sequenced to examine whether they contained the anticipated sequences. While Dart1-28, Dart1-44, and Dart1-52 carried the expected sequences, Dart1-1 was found to contain a small substitution, and both Dart1-20 and Dart1-27 bore small deletions. In order to distinguish these altered Dart1 elements from those with anticipated sequences, we tentatively call these altered Dart1 elements Dart1-1S,Dart1-20D, and Dart1-27D in the present study (Fig. 3a).
Fig. 3.
Structures of the T-DNA regions in the plasmid vectors used. a Construction of cloned Dart1 elements. Because the amplified Dart1-1, Dart1-20, and Dart1-27 fragments cloned were found to contain a small substitution or deletions, represented by small hatched and black boxes below and above the Dart1 map, respectively, these DNA rearranged regions were substituted by appropriate fragments derived from PCR-amplified fragments containing proper sequences drawn at the top and bottom (see Materials and methods). Restriction cleavage sites are S, SpeI; B, BstBI; M, MunI; P, PstI; and X, XhoI. b–i T-DNA structures of the vectors used for transformation. The structures of the Dart1 elements and primers used are drawn as in Fig. 2a, and the arrows within the nDart1-0 boxes indicate the orientation of nDart1-0. The x in parentheses within the transposase coding region in (e) indicates the premature stop codon present in Dart1-44 carried by pZEN8. The brackets under nptII, Dart1-27, and GUSPlus represent the probes used for Southern blot hybridization. The horizontal arrowheads with the codes represent primers for PCR amplification (Supplementary Table S1)
To obtain the proper Dart1 elements, we tried to replace the segments containing small rearrangements by PCR-amplified fragments with proper sequences. For Dart1-1S and Dart1-20D containing small rearrangements at their 3′- terminal regions, 1.8- and 0.8-kb PCR-amplified fragments containing the 3′- termini of Dart1-1 and Dart1-20 were prepared, and the segments of Dart1-1S and Dart1-20D with small rearrangements were substituted by the 1.6-kb MunI–XhoI and 0.3-kb PstI–XhoI fragments to yield Dart1-1 and Dart1-20, respectively (Fig. 3a). The sequences of the reconstructed Dart1-1 and Dart1-20 elements were found to be identical to those in the Nipponbare genome. Similarly, the 0.8-kb fragment containing the 5′-terminus of Dart1-27D was prepared by PCR amplification, and its 0.7-kb SpeI–BstBI fragment was replaced by the corresponding fragment of Dart1-27 (Fig. 3a).
For Arabidopsis transformation, we constructed vectors based on pBI121 (Jefferson et al. 1987). The 0.7-kb fragment containing nDart1-0 was cut out from the pGEM-T Easy vector and cloned into pBI121 in front of the GUS gene encoding β-glucuronidase by blunt-end ligation to yield pZEN1 (Fig. 3b). The 3.7-kb fragment containing either Dart1-27 or Dart1-27D was cut out from pCR4 Blunt-TOPO-X and cloned into pZEN1 to yield pZEN2 and pZEN3, respectively (Fig. 3c, d). To examine the excision activity of Dart1 elements, the fragments bearing six different Dart1 elements, Dart1-1,Dart1-20,Dart1-27, Dart1-28, Dart1-44, and Dart1-52, were cut out from the vectors mentioned above and cloned into pBI121 to yield six vectors, pZEN4 to pZEN9 (Fig. 3e). pZEN10 having Dart1-27D inserted between the 35S promoter and the GUS reporter gene was constructed in the same way (Fig. 3f).
For rice transformation, we constructed vectors based on pCAMBIA 1305.1 (CAMBIA, Canberra, Australia). The region containing the 35S promoter and the hpt gene for hygromycin B resistance on the pCAMBIA vector was replaced by the segment carrying the rice Actin 1 promoter with its intron fused with the modified hpt gene from pRIT1 (Terada et al. 2002), and the 0.7-kb fragment containing nDart1-0 was inserted between the 35S promoter and the GUSPlus gene to yield pZEN11 (Fig. 3g). The 3.7-kb fragment containing either Dart1-27 or Dart1-27D was cloned into pZEN11 to yield pZEN12 and pZEN13, respectively (Fig. 3h, i). Unfortunately, the inserted Dart1-27 sequence on pZEN12 was found to carry two point mutations; a single bp deletion at position 649 and G to A substitution at position 663. Because the Dart1 transcripts comprise 5′-untranslated exon 1 and exon 2, which contains the entire coding region of the Dart1 transposase, and because the two point mutations found in the reconstructed Dart1-27 element are within the 92-bp intron at positions 585-686, the mutations are expected to be spliced out in the mature transcripts (Fig. 2b, Supplementary Table S2; Nishimura et al. 2008). Therefore, the reconstructed element, which we named Dart1-27* (Fig. 3h), is likely to produce transposase proteins identical to those of the wild-type Dart1-27 element. Although the origin of these two point mutations remains obscure, Dart1-27* must have been present as a minor contaminant in the reconstructed Dart1-27 element carried by pCR4 Blunt-TOPO-X.
Plant transformation and transposition of nDart1-0 and its relatives
For the transformation of Arabidopsisthaliana ecotype Col-1 plants, Agrobacterium strain GV3101 (pMP90) harboring an appropriate pZEN vector was used for floral dip transformation (Clough and Bent 1998). For the transformation of rice pyl-stb plants, the high-speed transformation procedure (Toki et al. 2006) was employed with the following modifications. After cocultivation with Agrobacterium strain EHA105 harboring an appropriate pZEN vector, vancomycin (200 mg/L) was used for washing to remove the bacteria, and cefatoxim (400 mg/L) and vancomycin (100 mg/L) were added to the N6D and ReIII media instead of carbenicillin. About five or six calli were usually obtained from one seed infected with the appropriate Agrobacterium, and approximately four to six plants were usually regenerated from each callus.
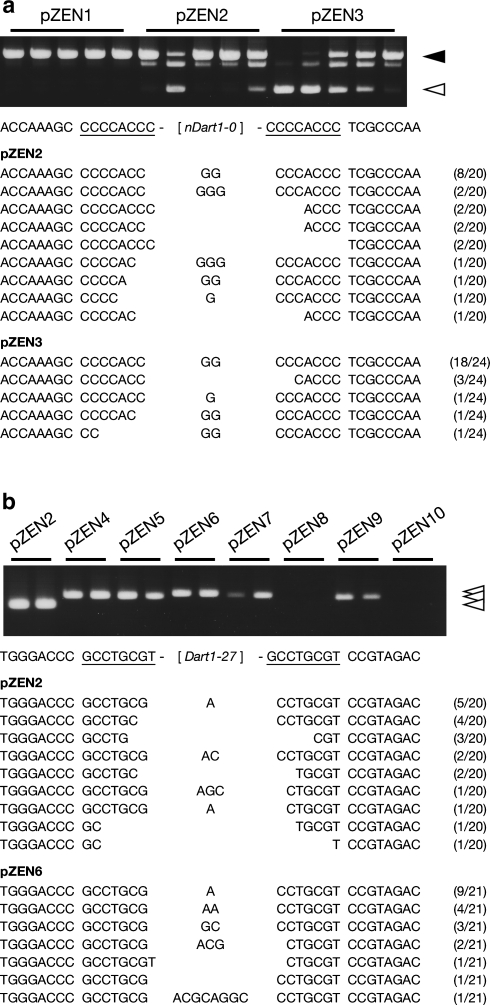
Excisions of the nDart1-0 and Dart1 elements from the GUS gene were examined by assaying the GUS activity (Fig. 4; Ishikawa et al. 2002) and by detecting the appearance of PCR-amplified fragments (Fig. 5) with appropriate primers (Supplementary Table S1) from the GUS gene having nDart1-0/Dart1 excised. To determine the footprints generated by the transposon excisions (Fig. 5, Supplementary Figs. S1, S2, and S3), the PCR-amplified fragments were cloned into pCR4-TOPO (Invitrogen) and sequenced.
Fig. 4.
Excision activities of the introduced nDart1-0 and Dart1 elements in transgenic Arabidopsis plants. GUS-positive patterns in leaves (a), inflorescences (b), and roots (c)
Fig. 5.
Excisions of nDart1-0 and Dart1-27 from the introduced vectors and generations of footprints upon excisions of the elements in transgenic Arabidopsis plants. a Excisions of nDart1-0. The filled and open arrowheads point to the PCR-amplified bands with and without the nDart1-0 insert, respectively, in the PCR analysis. The target site duplication (TSD) sequences of nDart1-0 originated from the pyl-stb plant are underlined. b Excisions of Dart1-27 and other Dart1 elements examined. The open arrowheads point to the PCR-amplified bands without the Dart1 inserts in the PCR analysis. The TSD sequences of Dart1-27 originated from Nipponbare are underlined. The numerals in the fractional expressions in the parentheses at the rightmost column indicate the occurrences of the footprint sequences observed. Footprints generated by the excisions of other Dart1 elements examined are shown in Supplementary Fig. S1
To determine the nDart1-0 and Dart1-27 integration sites on the Arabidopsis genome, a transposon display for the nDart1/Dart1 elements was employed (Takagi et al. 2007), and the sequences obtained by direct sequencing of appropriate PCR-amplified fragments were compared with the Arabidopsis pseudomolecules in the Arabidopsis Information Resource (TAIR) 8.0 database (http://www.arabidopsis.org/).
Results
Identification of aDart in the pyl-v plant
Segregation data indicated that the pyl-v plant contains only one copy of the active aDart element (Tsugane et al. 2006; M. Maekawa unpublished data). Fine mapping revealed that aDart in the pyl-v line is located within the 170-kb region between markers GCH-5 and RM5509 on chromosome 6 (Fig. 1). The annotations of The Rice Annotation Project (2007, see also http://rapdb.dna.affrc.go.jp/) and Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) predicted 23 putative genes, including one copy of the Dart1 element, Dart1-27, and two retroelements longer than 2 kb in the region of the Nipponbare genome. The presence of one Dart1 element in the pyl-v line that corresponds to Dart1-27 in Nipponbare was confirmed by the appearance of the 0.37-kb PCR-amplified fragment derived from the 5′-terminal junction of the Dart1 element with primers Dt1-F (1F) and Dt1-R (1R) (Fig. 2a). Indeed, all of the 817 pyl-v segregants analyzed in the F2 population from the cross between the pyl-v line and the indica line Kasalath, which contains no Dart1 element corresponding to Dart1-27 (Nishimura et al. 2008), displayed the 0.37-kb PCR-amplified band derived from the Dart1 element, whereas none of the 550 pyl-stb segregants examined in the same F2 population exhibited the 0.37-kb band (data not shown). Moreover, we examined whether the pyl-v plant carries an additional Dart1 insertion by comparing PCR-amplified fragments covering the entire 170-kb region from pyl-v and Nipponbare; no additional insertion of an element of about 3.5 kb could be found in the 170-kb region of the pyl-v plant compared with Nipponbare, although small insertions/deletions of less than about 0.2 kb might be present (data not shown). It is, thus, highly likely that aDart in the mutable pyl-v line resides in the Dart1-27 locus on chromosome 6. To obtain aDart in the pyl-v line, we screened the Fosmid library prepared from the pyl-v plants and isolated three Fosmid clones (Fig. 1) containing the Dart1 element together with the adjacent 0.79-kb segment, detected by PCR analysis with primers DtAJ-F (2F) and DtAJ-R (2R), from the segment adjacent to Dart1-27 (Fig. 2a). Subsequently, we determined the entire sequence of the Dart1 element on these three Fosmid clones, which was found to be identical to Dart1-27 in Nipponbare. The results indicate that Dart1-27 in the pyl-v line acts as the active aDart element, whereas Dart1-27 in Nipponbare is epigenetically silenced, even though their sequences are identical to each other and reside at the identical site on chromosome 6.
Transcripts from the Dart1 elements in pyl-v, pyl-stb, and Nipponbare
Although one of the full-length cDNA clones in Nipponbare (The Rice Full-Length cDNA Consortium 2003) contains the entire transposase coding sequence of Dart1-52 (AK072732), its 171-bp 5′-untranslated region (5′-UTR) is derived from a gene residing at 4.4-kb upstream of Dart1-52 according to The Rice Annotation Project (2007). We first employed 5′-RACE analysis to examine whether such aberrant splicing often occurred in the leaves of pyl-v, pyl-stb, and Nipponbare (Fig. 2a). No such aberrant splicing could be detected, but we found that a 92-bp intron was present immediately upstream of the ATG initiation codon of the putative transposase and that the spliced transcripts contained 175-bp 5′-UTR derived from the Dart1 elements in the plants examined. Since 3′-RACE analysis revealed that most of the transcripts carried 170-bp 3′-UTR (Fig. 2a), we employed RT-PCR analysis with primers Dt2-F1 (3F1) and Dt2-R1 (3R1) to examine whether the transcripts from Dart1-27 were accumulated in the leaves of pyl-v and whether Dart1-related transcripts were detectable in the leaves of pyl-stb and Nipponbare. The RT-PCR analysis with 23 cycles of amplification indicated that long Dart1 transposase transcripts were abundantly expressed in pyl-v (Fig. 2b, c), and sequencing analysis revealed that they comprised predominantly the Dart1-27 transcripts (Supplementary Table S2), supporting the mapping data in Fig. 1 that aDart in pyl-v must be Dart1-27. Although no transcripts were detectable in pyl-stb and Nipponbare, two additional smaller transcripts could be detected in pyl-v (Fig. 2). While the longest (L) transcripts in pyl-v contained the Dart1-27 transcripts having intron 1 at 5′-UTR spliced, the middle (M) and the shortest (S) transcripts were mixtures that had one or two additional introns spliced, respectively, although the exact splicing sites were often different (Fig. 2b, c). In the Dart1-27 S1 and S2 transcripts detected in the pyl-v plant, for example, their 3′ splice sites in intron 2 differ by 7 bp, whereas their 5′ splice sites are identical (Supplementary Table S2). The RT-PCR analysis with 35 cycles of amplification showed that these three transcripts, different in size, were observed not only in pyl-v but also in pyl-stb and Nipponbare. The major transcripts detected in pyl-stb or Nipponbare were derived from Dart1-28, together with the minor transcripts from Dart1-18, Dart1-30, and Dart1-34, implying that the residual expression of these Dart1 elements in both pyl-stb and Nipponbare would be too weak to act on the nDart1-3 subgroup elements including nDart1-0 at OsClpP5 to lead to their excision even though some of them might encode an active transposase. It is noteworthy that no transcripts from Dart1-27 could be detected in pyl-stb and Nipponbare. We also noticed that all the shorter transcripts characterized in pyl-v were derived from Dart1-27, indicating that some of the abundantly expressed transcripts must have undergone further splicing in rice.
Transposition activities of the Dart1 elements of Nipponbare from introduced vectors in Arabidopsis
The mapping data (Fig. 1) and the results of the Dart1 transcripts accumulated in the leaves (Fig. 2b, c) indicated that Dart1-27 in pyl-v acts on nDart1-0 as an active aDart element, whereas Dart1-27 in Nipponbare appears to be epigenetically silenced. The results also indicated that the active transposase is likely to be carried by the longest transcripts. To confirm these notions, we first cloned both Dart1-27 from Nipponbare and nDart1-0 from pyl-stb into pBI121 to yield pZEN2, in which the excision of nDart1-0 would result in the activation of the GUS gene (Fig. 3c). Subsequently, we introduced pZEN2 into Arabidopsis to examine whether the cloned Dart1-27 from Nipponbare was able to act on the introduced nDart1-0 element and to show the excision capability of nDart1-0. We also included pZEN3 carrying Dart1-27D with a 192-bp deletion at the 5′-subterminal region (Fig. 3d) in order to examine the transposition activities. Because the putative transposase genes of the 38 Dart1 elements, including Dart1-27 in Nipponbare, carry no apparent nonsense or frameshift mutations (Tsugane et al. 2006; Takagi et al. 2007), these 38 putative autonomous elements are likely candidates for potential autonomous elements silenced epigenetically. To test whether at least a few of them can indeed behave as an active aDart element in the introduced Arabidopsis plants, we cloned several of their representatives into pBI121 to yield pZEN4 to pZEN10, in which the excision of Dart1 would result in the GUS activation (Fig. 3e, f) and, subsequently, introduced them into Arabidopsis to examine their excision capability. Among the 38 Dart1 elements, we chose four elements in addition to Dart1-27 for the following reasons (Supplementary Table S3); in Dart1-1, its subterminal regions are the most homologous to those of nDart1-0; in Dart1-20, its putative promoter and intron 1 regions contain 22-bp and 11-bp deletions, respectively; in Dart1-28, its intron 1 of the longest transcripts detected in pyl-stb and Nipponbare is either 117 bp (instead of 92 bp because of a single G to T transversion at the 3′ splice site and the removal of the common ATG initiation codon, resulting in the possible utilization of the inframe ATG codon 63-bp downstream of the common ATG) or unspliced, and its putative transposase gene contains an additional 6-bp deletion (Fig. 2b, c, Supplementary Tables S2 and S3); and, in Dart1-52, its aberrant transcript in Nipponbare (AK072732) was reported in the full-length cDNA clones (The Rice Full-Length cDNA Consortium 2003). We also noticed that Dart1-52 carries 49 insertions/deletions compared with Dart1-27; therefore, the putative transposase of Dart1-52 shows only 75% identities in the amino acid sequence to that of Dart1-27 (Supplementary Table S3). In addition, we chose Dart1-44, whose putative transposase gene contains a 6-bp deletion and a premature TAA stop codon; it encodes only a polypeptide of 441 amino acids, while Dart1-27 encodes a putative transposase of 726 amino acids (Supplementary Table S3).
The excision of the nDart1-0 and Dart1 elements from GUS in the introduced Arabidopsis plants can be monitored by detecting GUS-positive spots and sectors in leaves, inflorescences, and roots (Table 1), and representative GUS-positive spots and sectors are shown in Fig. 4. Among the variegations detected in these tissues, the most frequently observed variegation was GUS-positive sectors (Table 1 III). To confirm the excisions of these elements, the PCR-amplified bands containing footprints generated by the excisions were subsequently characterized (Fig. 5, Supplementary Fig. S1). Not only Dart1-27 in pZEN2 but also Dart1-27D in pZEN3 was able to act on nDart1-0 and lead the nDart1-0 excisions from GUS on the introduced vectors, indicating that both elements can produce active transposases. Anticipated footprints generated by the nDart1-0 excisions were observed in individual clones of the PCR-amplified excision bands, even though the transgenic plants transformed with pZEN2 produced faint excision bands (Fig. 5a). The excision of Dart1-27 in pZEN6 also occurred, producing GUS-positive spots and sectors, whereas that of Dart1-27D in pZEN10 did not (Fig. 4, Table 1). About half of the plants with pZEN10 produced a small blurred GUS-positive spot or spots at the styles in young flower buds (Fig. 4b), and only one plant with pZEN10 also gave a tiny spot at the top of a root tip (data not shown); however, no PCR-amplified excision bands could be detected (Fig. 5b, Table 1). Although the significance of these rare GUS-positive spots observed in plants with pZEN10 remains to be elucidated, the results suggested that the 192-bp segment in the 5′-subterminal region deleted in Dart1-27D contains a cis-element that is necessary for its own transposition. As expected, the Dart1-27 excisions could be detected to generate footprints (Fig. 5b).
Table 1.
Dart1-mediated excision activities in transgenic Arabidopsis plants
| Vectors | Plants examined | Leaves | Inflorescences | Roots | ||||
|---|---|---|---|---|---|---|---|---|
| Variegation detected (%) | [I/II/III/IV/V] | Excision detected (%) | Variegation detected (%) | [I/II/III/IV/V] | Variegation detected (%) | [I/II/III/IV/V] | ||
| pZEN1 | 5 | 0 (0) | [0/0/0/0/0] | 0 (0) | 0 (0) | [0/0/0/0/0] | 0 (0) | [0/0/0/0/0] |
| pZEN2 | 9 | 5 (56) | [0/0/3/2/0] | 6 (67) | 6 (67) | [0/0/5/0/1] | 8 (89) | [0/0/8/0/0] |
| pZEN3 | 9 | 7 (78) | [0/0/5/1/1] | 7 (78) | 7 (78) | [0/0/6/0/1] | 7 (78) | [3/0/4/0/0] |
| pZEN4 | 13 | 12 (92) | [0/0/12/0/0] | 13 (100) | 13 (100) | [0/0/13/0/0] | 13 (100) | [0/0/12/1/0] |
| pZEN5 | 14 | 13 (93) | [0/0/11/0/2] | 8 (57) | 12 (86) | [0/0/11/1/0] | 12 (86) | [0/0/11/1/0] |
| pZEN6 | 16 | 15 (94) | [0/2/13/0/0] | 16 (100) | 15 (94) | [0/0/15/0/0] | 14 (88) | [0/0/11/3/0] |
| pZEN7 | 10 | 9 (90) | [0/0/1/4/4] | 10 (100) | 9 (90) | [0/0/7/1/1] | 6 (60) | [0/0/6/0/0] |
| pZEN8 | 5 | 0 (0) | [0/0/0/0/0] | 0 (0) | 0 (0) | [0/0/0/0/0] | 0 (0) | [0/0/0/0/0] |
| pZEN9 | 12 | 10 (83) | [0/0/5/2/3] | 11 (92) | 11 (92) | [0/0/7/4/0] | 12 (100) | [0/0/11/1/0] |
| pZEN10 | 9 | 0 (0) | [0/0/0/0/0] | 0 (0) | 5a (56) | [0/0/5/0/0] | 1b (11) | [0/0/1/0/0] |
Excisions of nDart1-0 in pZEN2 and pZEN3 were mediated by the transposase encoded by Dart1-27 and Dart1-27D, respectively, whereas those of Dart1-1, Dart1-20, Dart1-27, Dart1-28, and Dart1-52 in pZEN4, pZEN5, pZEN6, pZEN7, and pZEN9, respectively, were promoted by their own transposases. GUS-positive patterns detected are I, entirely stained; II, entirely stained and sectors; III, sectors; IV, sectors and spots; V, spots
Superscript letters a and b in pZEN10 indicate a small blurred spot(s) that appeared at the styles (Fig. 4b) and a tiny spot at the top of a root tip (see text), respectively
Among the other Dart1 elements tested, Dart1-1 in pZEN4, Dart1-20 in pZEN5, Dart1-28 in pZEN7, and Dart1-52 in pZEN9 were all shown to be excised from the vectors, whereas Dart1-44 in pZEN8 failed to be excised (Figs. 4, 5b, Table 1). Expected footprints were observed in individual clones of the PCR-amplified excision bands (Supplementary Fig. S1). The results indicated that at least four Dart1 elements tested, Dart1-1, Dart1-20, Dart1-28, and Dart1-52, in addition to Dart1-27, are all potential autonomous elements silenced epigenetically in Nipponbare, whereas Dart1-44, which contains a premature stop codon, is a defective element, designated as dDart1-44. Southern blot analysis with the nptII region as a probe indicated that these transgenic Arabidopsis plants carried an average of 4.6 copies (within a range of 1–12 copies) of the transgenes in their genomes (data not shown). Interestingly, only the longest Dart1 transcripts were accumulated in the transgenic Arabidopsis plants into which these Dart1 elements were introduced (Fig. 2b, d). The accumulation of the longest Dart1 transcripts, including the defective dDart1-44 element, was confirmed by sequencing of individual clones (Supplementary Table S2). To determine whether the Dart1 elements that are capable of being excised by themselves can promote the transposition of nDart1-0, the transgenic plants bearing the individual Dart1 element were crossed with the plant containing nDart1-0 originated from pZEN1; then, F1 hybrids carrying both Dart1 and nDart1-0 were examined to determine whether the excision of nDart1-0 from pZEN1 could take place and produce PCR-amplified fragments containing footprints. As expected, elements Dart1-1, Dart1-27, Dart1-28, and Dart1-52 were all able to excise nDart1-0, whereas dDart1-44 was not (Supplementary Fig. S2). Moreover, Dart1-27 could act on dDart1-44 and excise dDart1-44 from GUS on the introduced pZEN8 vector to generate footprints.
We also examined whether both nDart1-0 in pZEN2 and Dart1-27 in pZEN6 were shown to integrate into new sites in the Arabidopsis genome. A comparison of the bands detected in the Southern blot analysis using the transposase-coding region of Dart1-27 as a probe with those using the GUS-coding region as a probe indicated that no clear additional insertion bands of Dart1-27 were detected (data not shown), probably because the cells containing a somatic integration of Dart1-27 in the genome were too few to be detectable as bands. We, thus, employed a transposon display for analyzing the integrations of nDart1-0 and Dart1-27 separately (Takagi et al. 2007). As Fig. 6a shows, the results clearly demonstrated that both nDart1-0 and Dart1-27 were integrated into various sites in the unique regions of the genome and that some of them were inserted within the genic regions (Supplementary Table S4). Furthermore, the anticipated 8-bp target site duplications were found in these elements examined (Fig. 6b, c). Based on these results, we can conclude that (1) Dart1-27 in pyl-v is the active autonomous aDart element, whereas Dart1-27 in Nipponbare is silenced epigenetically; (2) several other Dart1 elements, Dart1-1, Dart1-20, Dart1-27, Dart1-28, and Dart1-52, described here, as well as Dart1-26 described previously (Nishimura et al. 2008), are all potential autonomous elements silenced epigenetically in Nipponbare; (3) Dart1-44 containing a premature stop codon is a defective element, dDart1-44; and (4) the length of their putative transposases can vary from 703 amino acids in Dart1-28 or 711 amino acids in Dart1-52 to 726 amino acids in Dart1-27, Dart1-1, and Dart1-20. The putative transposases in Dart1-1 and Dart1-20 were different from that in Dart1-27 by six and eleven amino acid substitutions, respectively (Supplementary Table S3).
Fig. 6.
Integrations of nDart1-0 and Dart1-27 into the transgenic Arabidopsis genome. a Localization of nDart1-0 and Dart1-27 in the Arabidopsis genome. The short and long bars represent nDart1-0 and Dart1-27, respectively, and the leftward and rightward bars indicate the downward and upward orientations of the inserted elements on the map, respectively. The numerals are according to the Arabidopsis pseudomolecules in the Arabidopsis Information Resource (TAIR) 8.0 database (http://www.arabidopsis.org/). The asterisks indicate the integrations of the genic regions consisting of putative coding regions including introns and 0.5-kb segments upstream of the coding regions (Supplementary Table S4). b TSDs generated by nDart1-0 integrations. c TSDs generated by Dart1-27 integrations. The small open circles adjacent to the transposon boxes indicate TSDs. The horizontal arrowheads with the codes under maps indicate primers used to determine TSDs (Supplementary Table S1). Newly generated TSDs are underlined. The nucleotides in bold correspond to the positions in the Arabidopsis chromosomes shown in (a)
Excisions of nDart1-0 from either the endogenous OsClpP5 gene or the GUS gene on the introduced vectors promoted by newly introduced Dart1-27 derivatives in the rice pyl-stb line
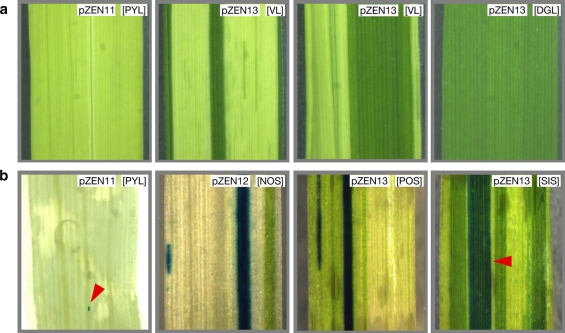
Both Dart1-27 from Nipponbare and its deletion derivative Dart1-27D produce active transposases and promote nDart1-0 excisions in Arabidopsis. We were interested in determining whether both excisions of nDart1-0 from the endogenous OsClpP5 gene and from the newly introduced GUS gene can often occur simultaneously when a vector bearing Dart1-27 together with nDart1-0 within GUS is introduced into the pyl-stb plant. To this end, we cloned both Dart1-27 from Nipponbare and nDart1-0 from OsClpP5 in pyl-stb into a pCAMBIA derivative carrying the rice Actin 1 promoter-driven hpt gene to yield pZEN12, in which the excision of nDart1-0 would result in GUS activation (Fig. 3h). Unfortunately, the cloned element, Dart1-27*, was later found to contain two point mutations within intron 1. In addition, we also constructed pZEN13 carrying Dart1-27D (Fig. 3i). Approximately 20% of the regenerants obtained exhibited albino phenotype, and we used the remaining apparently normally regenerated plants for further investigation. Of 63 and 99 transgenic pyl-stb plants transformed with pZEN12 and pZEN13 examined, 60 and 96 plants at the 7- to 10-leaf stages exhibited leaf variegations or an entirely dark-green leaf due to the nDart1-0 excision from OsClpP5, and 57 and 85 plants displayed GUS-positive variegations, respectively; typical examples are shown in Fig. 7. In almost all such variegated leaves, we detected PCR-amplified fragments containing footprints generated by the nDart1-0 excisions from OsClpP5 as well as from GUS (Supplementary Fig. S3A and S3B). The results clearly indicate that the transposases produced from either pZEN12 or pZEN13 act not only on nDart1-0 in the GUS gene carried by the newly introduced vectors but also on nDart1-0 in the endogenous OsClpP5 gene in the genome and lead these nDart1-0 elements to be excised with comparable efficiencies. In addition, Dart1-27* could be excised from the introduced pZEN12 vector in rice, but Dart1-27D could not (Supplementary Fig. S3C).
Fig. 7.
Excisions of nDart1-0 from the OsClpP5 and GUS genes. Excisions of nDart1-0 from the OsClpP5 gene resulting in dark-green spots/sectors in a pale-yellow background (a) and those from both OsClpP5 and GUS genes leading to both dark-green and GUS-positive spots/sectors (b). Introduction of pZEN11 occasionally produced a minute GUS-positive spot pointed by the red arrowhead in [PYL]. Abbreviations in brackets are PYL pale-yellow leaf, VL variegated leaf, DGL dark-green leaf, NOS nonoverlapping sector(s), POS partially overlapping sector(s), SIS superimposed sector
Thus, more than 90% of transgenic plants examined showed phenotypes conferred by the nDart1-0 excisions. Southern blot analysis with the GUS region as a probe indicated that these transgenic rice plants carried an average of 3.2 copies (within the range of 1–18 copies) of the transgenes in their genomes (data not shown). Out of these transgenic plants with pZEN12 and pZEN13, 43 and 57 plants displayed leaf variegations and GUS-positive spots and/or sectors, and 38 and 52 plants exhibited dark-green spots/sectors partially overlapping with GUS-positive spots/sectors, respectively (Fig. 7b [POS]). Only two plants transformed with pZEN13 displayed a dark-green sector superimposed with a GUS-positive sector (Fig. 7b [SIS]), suggesting that the simultaneous excisions of nDart1-0 from OsClpP5 and GUS occurred rarely. We also noticed that only a few transgenic plants transformed with the control vector pZEN11 exhibited either a dark-green-leaf variegation or minute GUS-positive spots (Fig. 7b [PYL]), probably because stresses caused during Agrobacterium-mediated transformation, including callus formation, were likely to activate potential autonomous elements silenced epigenetically (Kaeppler et al. 2000; Cheng et al. 2006; Slotkin and Martienssen 2007).
Three transcripts with different sizes were observed in the leaves of the transgenic pyl-stb plants transformed with pZEN12 and pZEN13; the longest (L) transcripts contained transcripts having intron 1 spliced, whereas the middle (M) and the shortest (S) transcripts were mixtures that had an additional one or two introns spliced, and most of them were the Dart1-27 transcript derivatives (Fig. 2b, c, Supplementary Table S2). Interestingly, the Dart1-27 transcripts accumulated in three independent transgenic pyl-stb plants carrying only one copy of Dart1-27D were all considerably less abundant than those in three independent transgenic plants with one copy of Dart1-27* (Fig. 2c). Such a reduction of the Dart1-27 transcripts accumulated in the pyl-stb plants with pZEN13 bearing Dart1-27D might provide an explanation for detecting the residual transcripts from Dart1-18 and Dart1-20 in addition to those from Dart1-27 (Supplementary Table S2). Nevertheless, the reduced amounts of the Dart1-27 transcripts appeared to be sufficient to promote the excision of nDart1-0 from the GUS and OsClpP5 genes in the transgenic pyl-stb plants carrying only one copy of Dart1-27D (Supplementary Fig. S3).
Discussion
Among the active nonautonomous DNA transposable elements conferring mutable traits in rice reported, i.e., 430-bp mPing (Nakazaki et al. 2003), 607-bp nDart1-0 (Tsugane et al. 2006), and 670-bp dTok0 (Moon et al. 2006), nDart1-0 is unique in the copy numbers of the nonautonomous elements relative to their putative autonomous elements; the Nipponbare genome carries a number of nonautonomous mPing and dTok elements with only a few putative autonomous elements (Jiang et al. 2003; Kikuchi et al. 2003; Moon et al. 2006), whereas it contains 13 nonautonomous nDart1-3 subgroup elements with 38 putative autonomous Dart1 elements (Tsugane et al. 2006; Takagi et al. 2007; Johzuka-Hisatomi et al. 2008). For mPing, two elements, Ping and Pong, are thought to be autonomous elements (Jiang et al. 2003; Kikuchi et al. 2003), and Ping was recently reported to be actively transposed into a new site in rice (Ohmori et al. 2008). In addition, both Ping and Pong were shown to promote the transposition of mPing in Arabidopsis (Yang et al. 2007). We showed here that Dart1-27 in the mutable pyl-v line is the active autonomous element and that at least five elements, Dart1-1, Dart1-20, Dart1-27, Dart1-28, and Dart1-52, are potential autonomous elements, which are likely to be epigenetically silenced in Nipponbare, because all of them were able to behave as active autonomous elements in transgenic Arabidopsis plants. Because 5-azacytidine treatments of Nipponbare seeds were shown to activate potential autonomous elements silenced epigenetically (Tsugane et al. 2006), they are likely to be hypermethylated. Indeed, preliminary bisulfite sequencing data indicated that the 5′ region of Dart1-27 in both pyl-stb and Nipponbare was found to be more heavily methylated than that in the pyl-v plant, implying that the DNA methylation states of the potential autonomous elements appear to determine whether they are active aDart elements or epigenetically silenced autonomous elements (C.-H. Eun unpublished data). The DNA methylation states of the potential autonomous Dart1 elements in Nipponbare were expected to be erased by cloning of these elements into Escherichia coli and subsequently introduced into Agrobacterium for plant transformation. Since bisulfite sequencing results indicated that no apparent DNA methylation was detected at the 5′ region of Dart1-27* carried by pZEN12 in Agrobacteria, which was to be introduced into rice (data not shown), potential autonomous Dart1 elements cloned and introduced into Arabidopsis plants in Fig. 3 must have been much less DNA methylated than the corresponding elements in Nipponbare. Such hypomethylation states of the newly introduced Dart1 elements in the transgenic Arabidopsis plants must be one of the main reasons why they were able to act as active Dart1 elements (Figs. 4, 5, 6, Supplementary Fig. S1, Table 1). Of 100 rice cultivars comprising 49 japonica and 51 indica varieties examined, only seven and one japonica varieties were found to bear a single aDart element, whose sequences are identical to Dart1-27 and Dart1-26, respectively (Nishimura et al. 2008; K. Tsugane unpublished data), and these two elements exhibit 98% identities in DNA sequence. It would be intriguing to elucidate the molecular mechanisms whereby the Dart1-27 or Dart1-26 element in specific varieties is able to escape from epigenetic gene silencing associated with DNA methylation whereas the others are silenced.
The length of putative transposases in the active aDart elements (Fig. 3, Table 1) in the introduced Arabidopsis plants can vary from 703 amino acids in Dart1-28 and 711 amino acids in Dart1-52 to 726 amino acids in Dart1-27, Dart1-1, and Dart1-20. Interestingly, the transposase of the Dart1-52 is more closely related to that of Dart2-1, which contains no apparent premature stop codon in its transposase gene and belongs to another subgroup of Dart elements (Dart2) found in Nipponbare, than that of Dart1-27; the putative transposases between Dart1-52 and Dart2-1 showed 99% identities in amino acid sequence, whereas those between Dart1-52 and Dart1-27 showed only 75% (Tsugane et al. 2006). Because Dart1-52 could excise by itself from GUS in transgenic Arabidopsis plants and also promote the nDart1-0 excision from GUS by crossing a Dart1-52-expressing Arabidopsis plant with a pZEN1-transformed Arabidopsis plant containing nDart1-0 in the GUS gene, it is conceivable that Dart2-1 would act as an active autonomous element if its transposase gene were expressed. Presumably, autonomous Dart1 elements bear a considerable sequence divergence of transposases, which is one of the dynamic features of the transposase genes in regulation as well as an evolution characteristic of the nDart/aDart system in rice. Although it would be intriguing to compare the relative activities of these divergent transposases, our experimental design could not allow quantitative analysis to assess the activities of individual elements. Nonetheless, we can speculate that most of the remaining 32 putative autonomous elements in Nipponbare are likely to be potential autonomous elements silenced epigenetically, even though considerable divergence of their transposase sequences arose among these elements.
In transgenic Arabidopsis plants transformed with pZEN4 to pZEN10, the transposase genes in the cloned Dart1 elements can be transcribed either from their own promoters or the 35S promoter originated from the pBI121 (Fig. 3e, f). Preliminary results indicate that the transcriptions of these transposase genes were initiated from their own promoters in all 22 transgenic plants examined, which were transformed with either one of seven different pZEN vectors, and that only 6 of 22 transgenic plants introduced with one of four different pZEN vectors displayed additional transposase transcripts initiated by the integrated 35S promoter (K. Tsugane and C.-H. Eun unpublished data). Although the molecular mechanisms of the observed promoter selection remain to be studied, the results appear to indicate that the transposase promoter was more frequently used for the expression of the transposase gene than the 35S promoter under our experimental conditions. Because some of the transgenic Arabidopsis plants transformed with an identical vector contain only the transcripts initiated from the transposase promoter while others had the mixture of two different transcripts initiated from either one of these two promoters, the activity of the 35S promoter appears to be affected by the integration sites of the vector rather than the vector itself. Since the transgenic plants examined carried multiple copies of the transgene, it is also conceivable that the activity of the 35S promoter may vary from one transgene to another. Consistently with this notion, the transformation of Arabidopsis with the 35S promoter-driven GUS gene is known to result in a highly variable GUS activity pattern (Butaye et al. 2005). We noticed that splicing of intron 1 occurred in all of the transposase transcripts initiated from either the 35S promoter or their own promoters, even though the 3′ spliced site of intron 1 in Dart1-28 differed from that in other Dart1 elements (Supplementary Table S2) because of a single base substitution at the 3′ splice site (Supplementary Table S3).
The Dart1 transcripts expressed in rice underwent complicated posttranscriptional processing to produce a mixture of smaller M and S transcripts due to the splicing of variable introns 2 and 3 (Fig. 2, Supplementary Table S2). Although the biological significance of these smaller transcripts in rice remains obscure, one can speculate that truncated transposase proteins encoded by the smaller transcripts may act as repressors of transposition, as is the case for the P transposable elements in Drosophila (Rio 2002). Alternatively, the variable splicing processes in rice may have something to do with the degradation of the transcripts associated with nonsense-mediated mRNA decay (Lejeune and Maquat 2005). Nonetheless, the generation of the small transcripts in rice appears to be passive processes of the abundantly accumulated transcripts, because they were derived predominantly from Dart1-27 in the pyl-v line as well as the transgenic pyl-stb plants transformed with pZEN12, in which the longest Dart1-27 transcripts were extensively accumulated. While both 5′ and 3′ splice sites of intron 1 are common among the transcripts of all Dart1 elements examined, except for Dart1-28 containing a single base substitution at the 3′ splice site (Supplementary Table S3), the splice sites of introns 2 and 3 varied even in the same elements, including Dart1-27 (Fig. 2, Supplementary Table S2). These variable splice sites fit well with the consensus sequence for splicing in rice (Campbell et al. 2006), and these small transcripts appear to be generated by rice-specific posttranscriptional processing because no such processing could be observed in transgenic Arabidopsis plants (Fig. 2d, Supplementary Table S2).
While no apparent difference was observed between the amounts of the overexpressed transcripts from Dart1-27 and those from Dart1-27D in Arabidopsis (Fig. 2d), the Dart1-27 transcripts accumulated in three independent transgenic rice pyl-stb plants with only one copy of Dart1-27D were all considerably less abundant than those in three independent transgenic pyl-stb plants with one copy of Dart1-27* (Fig. 2c). The 192-bp segment in the 5′-subterminal region deleted in Dart1-27D is likely to contain a cis-element partially enhancing the expression of the transposase gene in rice, in addition to a cis-element necessary for its own transposition in Arabidopsis and rice (Fig. 4, Supplementary Fig. S3C, Table 1). However, we could not disregard an alternative remote possibility, namely that two point mutations within intron 1 in Dart1-27* affect its expression in rice. It appears to be paradoxical that the reduced amounts of the Dart1-27 transcripts were found to be sufficient to promote the excision of nDart1-0 from the GUS and OsClpP5 genes in the transgenic pyl-stb plants carrying only one copy of Dart1-27D (Fig. 2c, Supplementary Fig. S3), whereas the comparable amounts of the Dart1 transcripts containing the transcripts from the autonomous Dart1-28 element in pyl-stb (Fig. 2c, Supplementary Table S2) could not mediate the excision of nDart1-0 from the OsClpP5 gene (Tsugane et al. 2006). Presumably, newly produced Dart1-27 transcripts from the Dart1-27D transgene accounted for a significant portion of the Dart1 transcripts in the transgenic pyl-stb plants and were sufficient to promote the nDart1-0 excision. Indeed, major transcripts in a transgenic pyl-stb plant with only one copy of Dart1-27D were found to be the Dart1-27 transcripts (Supplementary Table S2). An additional possible explanation is that the putative Dart1-27 transposase may be more efficiently produced and/or more active than the putative Dart1-28 transposase, because Dart1-28 carries base alterations in its promoter region and at the 3′ splice site of intron 1, which results in the elimination of the normal initiation codon for the Dart1 transposase, and its putative transposase gene contains a 6-bp deletion (Supplementary Table S3).
Although foreign elements, such as T-DNA or the maize DNA transposons, Ac/Ds and En/Spm, as well as the endogenous retrotransposon, Tos17, have been systematically employed for gene tagging in rice, tissue cultures are absolutely necessary to either introduce these foreign elements into rice calli or activate dormant Tos17 in the genome (Hirochika et al. 2004; Leung and An 2004; Upadhyaya 2007); thus, somaclonal variations associated with tissue cultures are inevitable (Kaeppler et al. 2000). In this respect, the generation of albino, a good indicative of somaclonal variation, by high-speed rice transformation (Toki et al. 2006) is much higher than the conventional rice transformation (Hiei et al. 1994); by our hands in pyl-stb with the T-65 background, approximately 20% of the regenerants obtained by the high-speed procedure exhibited an albino phenotype, whereas about 2% of those obtained by the conventional method were albino (Z. Shimatani unpublished data). We noticed that the regeneration of albino tends to occur in the later stage of regenerants obtained through multiple shoots. Perhaps, high-speed transformation is more suitable for application to complementation analysis of rice mutants than to reversed genetic analyses, including gene tagging, in order to minimize the occurrence of somaclonal variations.
While endogenous active DNA transposons, which are free from somaclonal variation because no tissue culture is involved in generating insertion mutants, have been extensively used for gene tagging in maize, snapdragon, petunia, and morning glories (May and Martienssen 2003; Chopra et al. 2006), only a few active endogenous DNA transposons, mPing/Ping, nDart1, and dTok, have been identified in rice (Nakazaki et al. 2003; Fujino et al. 2005; Moon et al. 2006; Naito et al. 2006; Tsugane et al. 2006; Ohmori et al. 2008). Among the active nonautonomous DNA elements, nDart1 appears to be the most suitable for gene tagging because nDart1 seems to have a tendency to integrate more into GC-rich promoter proximal genic regions than mPing (Naito et al. 2006; Takagi et al. 2007; K. Takagi and M. Maekawa unpublished data) and because dTok was reported to transpose rarely (Moon et al. 2006). Our results in Arabidopsis (Fig. 6a, Supplementary Table S4) are consistent with this notion. Moreover, we are currently generating a large number of mutant lines tagged by nDart1 and its relatives, and they are free from somaclonal variation because no tissue culture is involved in the generation of such mutants (Tsugane et al. 2006; Johzuka-Hisatomi et al. 2008; M. Maekawa unpublished data). We have unambiguously demonstrated here that the active autonomous element in the pyl-v line is Dart1-27 on chromosome 6 and that the rice genome contains many potential autonomous Dart1 elements silenced epigenetically. These features would certainly provide important and necessary information that would facilitate an effective and somaclonal variation-free gene tagging system in rice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1. Generations of footprints upon excisions of the Dart1 elements examined from the introduced vectors in Arabidopsis. Symbols are as in Fig. 5. TSDs flanking Dart1-20 and Dart1-52 were lost during the construction of pZEN5 and pZEN9, respectively. (EPS 851 kb)
Supplementary Fig. S2. Generations of footprints upon excisions of nDart1-0 and dDart1-44 from the introduced vectors in Arabidopsis when one of the autonomous Dart1 elements was introduced by the cross indicated. TSD flanking dDart1-44 was lost during the construction of pZEN8. Symbols are as in Fig. 5. (EPS 491 kb)
Supplementary Fig. S3. Generations of footprints upon excisions of nDart1-0 from OsClpP5 and GUS and those of Dart1-27* from pZEN12 in rice. Symbols are as in Fig. 5, and the asterisks indicate the samples obtained from transgenic pyl-stb plants carrying only one copy of Dart1-27* or Dart1-27D. (TIFF 684 kb)
Acknowledgments
We thank Miwako Matsumoto, Miki Shimamoto, Hisayo Asao, Yoko Kobayashi, Seiko Nakano, and Kazue Hiramatsu for technical assistance, and Toshihiko Komari and Yikoh Hiei for discussion about somaclonal variation. We also thank Seiichi Toki for instructing their high-speed rice transformation procedure. This work was supported by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) from Bio-oriented Technology Research Advancement Institution (BRAIN) in Japan and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Z. S., K. Takagi, and Y. J.-H. are recipients of a fellowship from the Japan Society for the Promotion of Science for Young Scientists, and C.-H. E. is a recipient of a fellowship awarded by the Japan Society for the Promotion of Science for Foreign Researchers.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0410-x) contains supplementary material, which is available to authorized users.
References
- Butaye KMJ, Cammue BPA, Delaure SL, De Bolle MFC (2005) Approaches to minimize variation of transgene expression in plants. Mol Breed 16:79–91 [DOI]
- Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR (2006) Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7:327 [DOI] [PMC free article] [PubMed]
- Cheng C, Daigen M, Hirochika H (2006) Epigenetic regulation of the rice retrotransposon Tos17. Mol Gen Genomics 276:378–390 [DOI] [PubMed]
- Chopra S, Hoshino A, Boddu J, Iida S (2006) Flavonoid pigments as tools in molecular genetics. In: Glotewold E (ed) The science of flavonoids. Springer, New York, pp 147–173
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed]
- Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341 [DOI] [PubMed]
- Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Gen Genomics 273:150–157 [DOI] [PubMed]
- Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, Sundaresan V, Leung H (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54:325–334 [DOI] [PubMed]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282 [DOI] [PubMed]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800 [DOI] [PubMed]
- Ishikawa N, Johzuka-Hisatomi Y, Sugita K, Ebinuma H, Iida S (2002) The transposon Tip100 from the common morning glory is an autonomous element that can transpose in tobacco plants. Mol Genet Genomics 266:732–739 [DOI] [PubMed]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167 [DOI] [PubMed]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907 [DOI] [PMC free article] [PubMed]
- Johzuka-Hisatomi Y, Maekawa M, Takagi K, Eun C–H, Yamauchi T, Shimatani Z, Ahmed N, Urawa H, Tsugane K, Terada R, Iida S (2008) Homologous recombination-dependent gene targeting and an active DNA transposon nDart-promoted gene tagging for rice functional genomics. In: Hirano H-Y, Hirai A, Sano Y, Sasaki T (eds) Rice Biology in the Genomics Era: Biotechnology in Agriculture and Forestry, vol 62. Springer, Berlin, pp 81–94
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188 [DOI] [PubMed]
- Kikuchi K, Terauchi K, Wada M, Hirano H-Y (2003) The plant MITE mPing is mobilized in anther culture. Nature 421:167–170 [DOI] [PubMed]
- Kunze R, Weil CF (2002) The hAT and CACTA superfamilies of plant transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington, DC, pp 565–610
- Lejeune F, Maquat LE (2005) Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17:309–315 [DOI] [PubMed]
- Leung H, An G (2004) Rice functional genomics: large-scale gene discovery and applications to crop improvement. Adv Agron 82:55–111 [DOI]
- Maekawa M, Takamure I, Ahmed N, Kyozuka J (2005) Bunketsu-waito, one of tillering dwarf, is controlled by a single recessive gene in rice (Oryza sativa L.). Breed Sci 55:193–196 [DOI]
- May BP, Martienssen RA (2003) Transposon mutagenesis in the study of plant development. Crit Rev Plant Sci 22:1–35 [DOI]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207 [DOI] [PubMed]
- Moon S, Jung K-H, Lee D-E, Jiang W-Z, Koh HJ, Heu M-H, Lee DS, Suh HS, An G (2006) Identification of active transposon dTok, a member of the hAT family, in rice. Plant Cell Physiol 47:1473–1483 [DOI] [PubMed]
- Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103:17620–17625 [DOI] [PMC free article] [PubMed]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T (2003) Mobilization of a transposon in the rice genome. Nature 421:170–172 [DOI] [PubMed]
- Nishimura H, Ahmed N, Tsugane K, Iida S, Maekawa M (2008) Distribution and mapping of an active autonomous aDart element responsible for mobilizing nonautonomous nDart1 transposons in cultivated rice varieties. Theor Appl Genet 116:395–405 [DOI] [PubMed]
- Ohmori Y, Abiko M, Horibata A, Hirano H-Y (2008) A transposon, Ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant Cell Physiol 49:1176–1184 [DOI] [PubMed]
- Rio DC (2002) P transposable elements in Drosophila melanogaster. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington, DC, pp 484–518
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285 [DOI] [PubMed]
- Takagi K, Ishikawa N, Maekawa M, Tsugane K, Iida S (2007) Transposon display for active DNA transposons in rice. Genes Genet Syst 82:109–122 [DOI] [PubMed]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20:1030–1034 [DOI] [PubMed]
- The Rice Annotation Project (2007) Curated genome annotation of Oryzasativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17:175–183 [DOI] [PMC free article] [PubMed]
- The Rice Full-Length cDNA Consortium (2003) Collection, mapping, and annotation of over 28, 000 cDNA clones from japonica rice. Science 301:376–379 [DOI] [PubMed]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976 [DOI] [PubMed]
- Tsugane K, Maekawa M, Takagi K, Takahara H, Qian Q, Eun C-H, Iida S (2006) An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J 45:46–57 [DOI] [PubMed]
- Upadhyaya NM (ed) (2007) Rice functional genomics – challenges, progress and prospects. Springer, New York
- Yamauchi T, Moritoh S, Johzuka-Hisatomi Y, Ono A, Terada R, Nakamura I, Iida S (2008) Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. J Plant Physiol 165:1774–1782 [DOI] [PubMed]
- Yang G, Zhang F, Hancock CN, Wessler SR (2007) Transposition of the rice miniature inverted repeat transpoisable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA 104:10962–10967 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary Fig. S1. Generations of footprints upon excisions of the Dart1 elements examined from the introduced vectors in Arabidopsis. Symbols are as in Fig. 5. TSDs flanking Dart1-20 and Dart1-52 were lost during the construction of pZEN5 and pZEN9, respectively. (EPS 851 kb)
Supplementary Fig. S2. Generations of footprints upon excisions of nDart1-0 and dDart1-44 from the introduced vectors in Arabidopsis when one of the autonomous Dart1 elements was introduced by the cross indicated. TSD flanking dDart1-44 was lost during the construction of pZEN8. Symbols are as in Fig. 5. (EPS 491 kb)
Supplementary Fig. S3. Generations of footprints upon excisions of nDart1-0 from OsClpP5 and GUS and those of Dart1-27* from pZEN12 in rice. Symbols are as in Fig. 5, and the asterisks indicate the samples obtained from transgenic pyl-stb plants carrying only one copy of Dart1-27* or Dart1-27D. (TIFF 684 kb)