Abstract
We have cloned and characterized a chick homologue of the human vitronectin receptor α subunit (αv) whose primary sequence is 83% identical with its human counterpart but less than 40% identical with any other known integrin α subunit. Comparison of the chick and human sequences reveals several highly conserved regions, including the cytoplasmic domain. The putative ligand binding domain contains αv-specific residues that may contribute to ligand binding specificity. These are concentrated in three regions that are located before and between the first three Ca2+ binding domains. Polyclonal antibodies raised against two peptides deduced from the putative cytoplasmic and extracellular domains of the chick αv sequence recognize specifically integrin heterodimers in chick embryo fibroblasts. At least three putative β subunits coimmunoprecipitate with the chick αv subunit. In addition to a protein with the same molecular weight as β3 (94K), protein bands of Mr 84K and 110K are also coprecipitated. By successive immunodepletions, we demonstrate that this latter Mr 110K subunit is β1, which appears to be one of the αv-associated subunits in chick embryo fibroblasts.
The integrin family of cell-surface receptors mediates multiple interactions between cells and extracellular matrix proteins that are involved in cellular adhesion [reviewed by Hynes (1987), Ruoslahti and Pierschbacher (1987), Buck and Horwitz (1987), and Hemler (1990)]. Integrin receptors are heterodimers composed of two noncovalently associated subunits named α and β. The integrin family has been divided into three main subfamilies according to their β subunit. The β1 subfamily includes the fibronectin, collagen, and laminin receptors, or VLA receptors, the β2 subfamily includes three leukocyte adhesion molecules, and the β3 subfamily contains the vitronectin receptor and the GPIIb/IIIa (αIIb/βIIIa) platelet receptor. Recently the integrin receptor family complexity has been increased by the discovery and characterization of new β subunits that are associated with some α subunits in certain tissues and cell lines. The α6 subunit can be associated with either a β1 in VLA-6 (Sonnenberg et al., 1988a) or a β4 subunit (Kajiji et al., 1989; Sonnenberg et al., 1988b). Similarly, α4 has been described associated on lymphocytes either with β1 in VLA-4 (Hemler et al., 1987) or with a novel βP subunit (Holzmann & Weissman, 1989). The αv subunit of the vitronectin receptor can form heterodimers either with a β3 subunit in many cell types, or with a βX subunit in a carcinoma cell line (Cheresh et al., 1989a), or with a βS subunit in an osteosarcoma cell line (Freed et al., 1989).
The α subunits have a major role in determining the ligand specificity. The α1–α6 subunits associate with the β1 subunit to form αβ1 heterodimers in the VLA-1 to -6 series but constitute receptors with different binding characteristics [reviewed by Hemler (1990)]. Different αβ1 heterodimers bind specific ligands (collagen, laminin, fibronectin, VCAM-1), different combinations of these ligands, and even different cell binding domains on the same ligands (Wayner et al., 1989; Elices et al., 1989; Guan & Hynes, 1990).
To determine the conservation of functional domains in an integrin α subunit throughout evolution, we cloned and sequenced the chick homologue of the human vitronectin receptor α subunit (αv) to compare it with its human equivalent. Additionally, chick embryo fibroblasts were analyzed, and multiple αv-associated β subunits were identified including β3 and β1, increasing the number of known αv-containing heterodimers.
Experimental Procedures
Isolation of cDNA Clones
An E10 chicken embryo cDNA library in λgt10 was obtained from Dr. C. Nottenburg, and an El3 chicken brain cDNA library in λgtl0 was a gift from Dr. B. Ranscht. The libraries were plated on Escherichia coli Y1089 and screened according to the plate hybridization method of Benton and Davis (1977). The filters were screened at low stringency according to a modified procedure of Church and Gilbert (1984). Prehybridizations were performed overnight at 43 °C in 0.2 M sodium phosphate, pH 6.8, 7% SDS, 15% formamide, 1 mM EDTA, and 1% bovine serum albumin. Hybridizations were carried out in a fresh solution of the same composition at 40 °C for 36 h with the denatured probe at about l06 cpm/mL. Probes were made from gel-purified restriction fragments by using the random priming method of Feinberg and Vogelstein (1983) with a kit from United States Biochemical Corp., using 50 μCi of [α-32P]dCTP (Amersham Inc.). The filters were washed 4 × 15 min, 45 °C, in 2× SSC and 0.1% SDS (1× SSC is 150 mM sodium chloride and 15 mM sodium citrate, pH 7.0) before autoradiography. The phage library screenings involving high-stringency hybridization were performed as described by Maniatis et al. (1982).
DNA Sequencing
cDNAs were subcloned in M13mp18 (Yanisch-Perron et al., 1985) and sequenced by the dideoxy chain termination method (Sanger et al., 1977) from their extremities. The nucleotide sequence displayed in Figure 2 has been determined on both strands by reconstituting a linear sequence from overlapping stretches. The coding strand was sequenced by using the 5′ ends of the cDNA clones d10, d14, 1.4, 1.1, 1.3, 1.7, d15, and 1.5 (Figure 1d,e) and subclones constructed with the EcoRI, HindIII, and BglII restriction sites (Figure 1c). Two specific oligonucleotides were used to initiate sequencing reactions at positions 748 and 2611 (Figure 2). The noncoding strand was sequenced by using the 3′ ends of the cDNA clones 1.7, 1.1, and 1.8 (Figure 1d,e) and subclones constructed with the HindIII and BglII restriction sites (Figure 1c) and PvuII at position 1782. Specific oligonucleotides initiating sequences at positions 3330, 2815, 2365, 1026, and 675, respectively (Figure 2), were used to fill the gaps in the noncoding strand.
FIGURE 2.

cDNA and deduced amino acid sequence of the chick αv subunit and comparison with the human αv subunit amino acid sequence. Amino acids are abbreviated with the single-letter code. The amino acids in the mature human αv sequence that are different from the residues in the chick αv sequence are mentioned below the corresponding chick residues. Nonconservative substitutions are indicated by an asterisk. Amino acid substitutions within the following groups were considered as conservative: A, S, T; M, I, L, V; D, E; F, Y, W; R, K; N, Q. The three amino acids residues in the human αv sequence that do not have chick equivalents are listed with a (#). The four putative metal binding (I-IV) and transmembrane (TM) domains are underlined. The arrows pointing upward indicate the two potential cleavage sites: after the signal peptide (between residues −1 and 1) and within the extracellular domain (between residues 857 and 858). The 15 potential N-glycosylation sites are followed by a (@).
FIGURE 1.
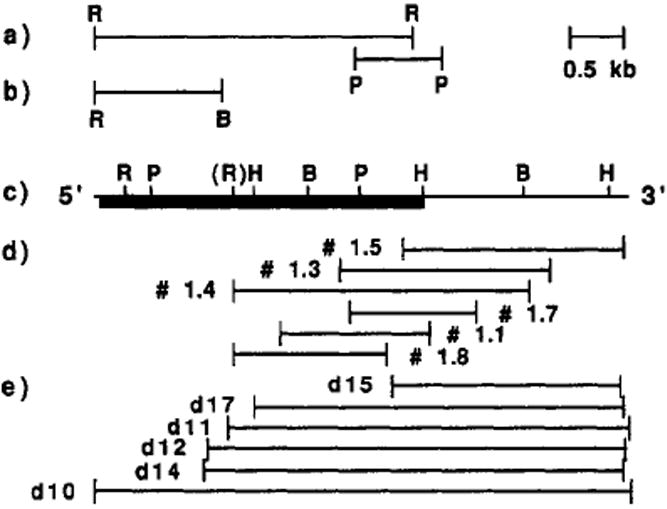
Restriction map of the probes and purified cDNAs. (a) Map of the EcoRI (R) and PstI (P) restriction fragments and EcoRI–BglII (B) fragment in (b), from human αv cDNAs used as probes for screening at low stringency. (c) Restriction map of the purified chick cDNAs. The box indicates the location of the main open reading frame. (EcoRI, R; BglII, B; PstI, P; HindIII, H). (d) Map of the six chick embryonic (E10) cDNAs (1.1, 1.3, 1.4, 1.5, 1.7, 1.8) purified with the probes mentioned in (a). (e) Map of six cDNAs (d10, d11, d12, d14, d15, and d17) purified from a chick brain (E13) cDNA library.
The nucleotide and amino acid sequence analyses were performed with the PCGENE program (Intelligenetics Corp., Mountain View, CA).
RNA Hybridization Analysis
RNA was purified from chick embryo fibroblasts according to the guanidinium/cesium chloride method described by Maniatis et al. (1982). Poly-(A+) RNA was then purified with oligo(U)–Sepharose 4B (Pharmacia) using the manufacturer’s recommendations. Two micrograms of poly(A+) RNA was electrophoresed through a 0.8% agarose/l6% formaldehyde gel (Maniatis et al., 1982) and transferred by capillary action to a Biotrans nylon membrane, 0.2 μm (ICN). RNA blots were prehybridized overnight at 42 °C in 5× SSPE, 50% formamide, 5× Denhardt’s solution, 10 mg/mL carrier DNA, and 10 mg/mL yeast tRNA [20× SSPE is 3.6 M NaCl, 0.2 M NaH2PO4, and 20 mM EDTA, pH adjusted to 7.4; 1× Denhardt’s solution is 0.02% BSA, 0.02% poly(vinylpyrrolidone), and 0.02% Ficoll type 400]. The probe was 50 ng of the 5-kb-long d10 cDNA that was gel-purified and random-labeled as described above for the isolation of clones. The RNA blot was washed 4 × 30 min with 1 × SSC/0.1% SDS at room temperature and then 2 × 15 min with 0.1 × SSC/0.1% SDS at 65 °C. RNA sizes were determined by running in parallel a RNA ladder of known molecular weight markers (Bethesda Research Laboratories) and by hybridizing the RNA blot with probes specific for transcripts of known sizes, including actin (1.75 kb) and the β1 integrin subunit (3.75 kb).
Production of Antipeptide Sera
Oligopeptides were synthesized on solid phase at the Howard Hughes Medical Institute by Dr. C. Turk. Their sequences are CKRVRPPQEEQEREQLQPHENGEGTSEA for the C peptide and CKIKISAPKEDEKNETFSREDNRNHRIS for the L peptide. These correspond to N-terminal cysteines added to amino acids 989–1015 and 829–855 in the primary sequence of the chick αv sequence (see Figure 2). The peptides were coupled to sulfo-MBS (PIERCE)-activated KLH (Calbiochem), using a ratio of 7.5 mg of peptide/mg of KLH. The amount of free sulfhydryl-containing peptide was determined by Ellman’s method (1959) with DTNB, as well as by quantifying the disappearance of free peptide during the coupling reaction. Rabbits were immunized with 1 mg of KLH–peptide complex in Freund’s complete adjuvant and then boosted with 0.5 mg of antigen in Freund’s incomplete adjuvant. Immunizations were performed either at the UCSF animal care facility or at Caltag Laboratories (South San Francisco, CA). Sera collected after the third boost were highly reactive to the peptide alone by ELISA or to the native protein by immunoprecipitation. The polyclonal antibodies that react with the β1 subunit were raised against the last 23 C-terminal residues of this subunit as described by Tomaselli et al. (1988). The LM609 monoclonal antibody that recognizes specifically the αvβ3 complex was generously provided by Dr. D. Cheresh.
Cells
Chick embryo fibroblasts (CEFs) were prepared from E9 or E10 embryos (Feather Hill, Petaluma, CA) according to Rein and Rubin (1968). All experiments described here were performed with confluent or subconfluent secondary cultures.
Cell-Surface Iodination and Immunoprecipitations
CEFs were surface labeled according basically to Tomaselli et al. (1988). Briefly, about l07 cells were collected and iodinated with 1.0 mCi of Na125I in 1 mL of PBS with lactoperoxidase (PBS is 0.2 g/L KCl, 0.2 g/L KH2PO4, 2.16 g/L Na2HPO4–7H2O, and 8 g/L NaCl). The reaction was stopped by successive washes of cold KI (5 mM) in PBS. Cells were lysed on ice 30 min in 1 mL of extraction buffer and then centrifuged 20 min at 12000g. The extraction buffer is PBS, 1 mM CaCl2, 1 mM MgCl2, 1 mM PMSF, and 1% Triton X-100. The supernatant was preincubated 2 times successively with 100 μL of Sepharose Cl-4B 5 min and centrifuged 1 min at l0000g each time. The resulting supernatant was used immediately for immunoprecipitation or kept at 4 °C a few days or frozen at −80 °C.
Immunoprecipitations were performed by mixing (5–10) × l06 cpm of surface-labeled CEF extract with 10 μL of serum in a total of 0.5 mL. This mixture was incubated overnight at 4 °C on a shaker, then added to a 75 μL of protein A–Sepharose (Pharmacia), incubated 45 min at 4 °C, and then centrifuged for 1 min at 10000g. The pellet was washed 5–7 times with extraction buffer. Finally, 50 μL of electrophoresis sample buffer (4% SDS, 150 mM Tris-HC1, pH 6.8, 20% glycerol, and bromophenol blue) was added to the washed pellet with or wtihout β-mercaptoethanol for SDS–PAGE analysis in reducing or nonreducing conditions. The samples were boiled 5 min and loaded on 6% SDS–PAGE gels, with a 5% stacking gel (Laemmli, 1970). Molecular weights were determined by comparison with molecular weight standards purchased from Bio-Rad Corp. (myosin, 200K; β-galactosidase, 116K; phosphorylase B, 97K; BSA, 66K).
In immunodepletion experiments, extracts of surface-labeled CEFs were mixed 7 times successively with 10-μL aliquots of integrin β1-specific plyclonal antibodies followed by incubation with protein A–Sepharose and centrifugation. Subsequently, the extract was incubated sequentially with two aliquots of protein A–Sepharose and was centrifuged after each incubation. Finally, 10 μL of αv-specific (anti-C peptide) serum was used to form an immunoprecipitate. A positive control was performed in parallel, with seven precipitations using nonimmune rabbit serum before the final αv-specific immunoprecipitation.
Results
Isolation of cDNA Clones
The cDNA clone VNR-2 and the 0.8-kb PstI restriction fragment of VNR-8 (Figure 1a) contain the whole coding sequence of the human vitronectin receptor α subunit (Fitzgerald et al., 1987). These restriction fragments were used to screen a chick embryonic cDNA library at low stringency (Tm −47 °C). Six overlapping clones have been purified and sequenced from their extremities. None of them contained the 5′ end of the main open reading frame (ORF). Figure 1d illustrates the alignment of these clones relatively to the main ORF (Figure 1c). In order to obtain the missing 5′ part of the coding sequence, another cDNA library (chick brain E13) was screened both at low stringency with an EcoRI–BglII 1.2-kb restriction fragment (Figure 1b) from the 5′ end of the human VNR-2 clone and at high stringency with the most 5′ cDNA (1.4) isolated during the first screening. Six cDNAs that were positive with the 1.4 cDNA probe were purified (Figure 1e). One of them, d10, was 5 kb long and positive with both probes. It contains the 5′ end of the coding sequence as well as 32 base pairs of the 5′-untranslated sequence and 1.9 kb of 3′-untranslated sequence.
Main Coding Sequence Analysis
The nucleotide sequence was determined by reconstituting a continuous sequence on both strands from overlapping partial sequences obtained from 10 different cDNAs and their subclones. This nucleotide sequence and the deduced amino acid sequence of the main open reading frame are presented in Figure 2. The cDNA sequence includes 3102 nucleotides of open reading frame and 32 and 361 nucleotides of 5′- and 3′-untranslated regions, respectively. Two nucleotide substitutions in individual cDNAs have been found at positions 1252 and 2087. In the first case, the substitution eliminates an EcoRI site found in the two most 5′ clones from the E10 library in all the cDNAs purified from the E13 library that overlap this site. Both substitutions affect third codon bases that do not change the encoded amino acid.
The translation of the main open reading frame (Figure 2) defines a 1034 amino acid long polypeptide that shares 83% identity with the sequence of the human vitronectin receptor αv subunit (Suzuki et al., 1986; Fitzgerald et al., 1987) and is less than 40% identical with that of any other integrin α subunit published so far. Therefore, this sequence appears to be the chick homologue of the human αv subunit.
The different human amino acids are listed in Figure 2 below the chick residues, and the nonconservative changes are also highlighted with an asterisk. The N-terminus of the chick αv sequence starts with a 21-residue-long putative signal sequence. Although it is less than 25% conserved with its human equivalent, sequence analysis indicates that it has the features of a signal sequence including appropriate amino acids at positions −1 and −3 (von Heijne, 1986). After cleavage of this peptide, the 1015 amino acid long mature polypeptide has a calculated molecular weight of 112 502. Fifteen potential N-glycosylation sites (Asn-X-Thr/Ser) are present (Figure 2). Of these, 80% (12) are conserved in the human sequence. Assuming an average contribution of 2500 daltons per glycosylation site, we obtain an expected molecular weight of 150 002. This chick αv sequence contains all of the specific features typical of an integrin α subunit. In the N-terminal half of the sequence, there are seven repeats, the last four of which have a putative Ca2+ binding domain (DxD/NxDD/GxxD). In the C-terminal portion are a highly hydrophobic and presumably transmembrane domain followed by a short cytoplasmic domain. Out of 32 residues, 30 of this cytoplasmic tail are identical in the human αv sequence (Figure 2). The 18 cysteines of the human αv sequence are conserved in the chick subunit, but surprisingly, an additional 19th cysteine residue is present in the latter sequence at position 317 between the second and third Ca2+ binding domains. Like the human αv subunit, the chick sequence has a putative protease cleavage site in the extracellular domain between residues 857 and 858, in a segment relatively close to the transmembrane domain. With the possible exception of α4, this cleavage site (RR.D) satisfies the consensus observed so far among the cleaved integrin α subunits, that is, K/R-R-E/D (Takada et al., 1989). Analysis of the chick αv protein presented below provides evidence that this potential cleavage site is actually used to generate a large extracellular fragment linked by a disulfide bond to a small transmembrane fragment containing the cytoplasmic tail. The C-terminal domain of the large fragment is comparatively poorly conserved between the chick and human αv subunits, with only 36% identical residues in the last 25 amino acids. This chick sequence segment is hydrophilic and is also highly antigenic (see below).
The cDNA clones extending the furthest 3′ (d10 and d11, Figure 1e) exhibit a stretch of poly(A) at their 3′ end. The absence of a consensus polyadenylation site (AATAAA) in the immediate 5′ vicinity of this site suggests that this may not be the true polyadenylation site of the mRNA, but rather an internal A-rich sequence. In this case, part of the poly(A) stretch may not be encoded in the mRNA but may instead reflect a sequence complementary to the poly(dT) primer used in the construction of the library.
RNA Blot Hybridization
When RNA blots of chick embryo fibroblast poly(A+) RNA were hybridized with d10 cDNA as a probe, multiple transcripts were detected (Figure 3). The main transcript is about 9 kb long, and two minor products are 6.6 and 5.6 kb long. The presence of a major mRNA of 9 kb suggests also that the main polyadenylation site is several kilobases further 3′ than the d10 cDNA 3′ end. It is presently unclear whether the existence of several poly-(A+) transcripts reveals the presence of alternatively spliced transcripts, different initiation sites, or alternative polyadenylation sites. The oligo(A) stretch found about 2 kb beyond the coding sequence at the 3′ end of some purified cDNAs (e.g., d10 and d 11, Figure 1e) is compatible with the latter hypothesis.
FIGURE 3.
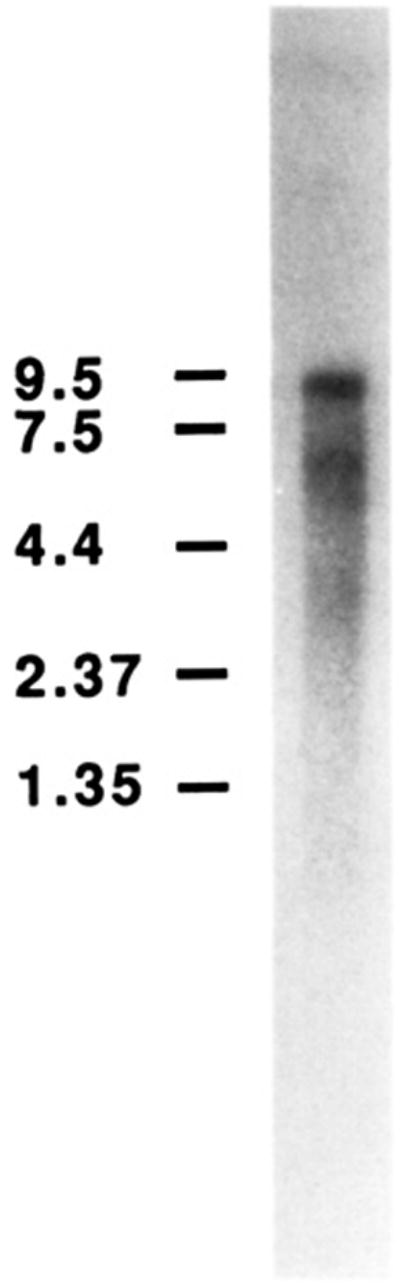
RNA hybridization analysis. Chick embryo fibroblast poly(A+) RNA was electrophoresed through a denaturing agarose gel, transferred to a membrane, and probed with the chick αv subunit d10 cDNA. The numbers at left indicate RNA size standards in kilobases.
αv-Specific Antisera
To analyze the protein product encoded by this chick αv cDNA, we coupled synthetic peptides derived from this sequence to keyhole lipet hemocyanin (KLH) to generate polyclonal antisera in rabbits. We chose two sequences which were both hydrophilic and located in a C-terminus position. The first peptide (C) was the cytoplasmic domain, and the second peptide (L) was the predicted C-terminus of the large extracellular fragment expected to be generated during integrin maturation by proteolytic cleavage. While extracellular, the latter sequence is not very well conserved with its human counterpart (9 out of 27 residues are conserved) and therefore seemed likely to be antigenic. Both peptides proved to be highly antigenic in rabbits. In fact, as illustrated in Figure 4, immune sera to both peptides precipitated a specific set of proteins from surface-labeled chick embryo fibroblasts (lanes 2 and 4) that are not precipitated with preimmune sera (lanes 1 and 3).
FIGURE 4.
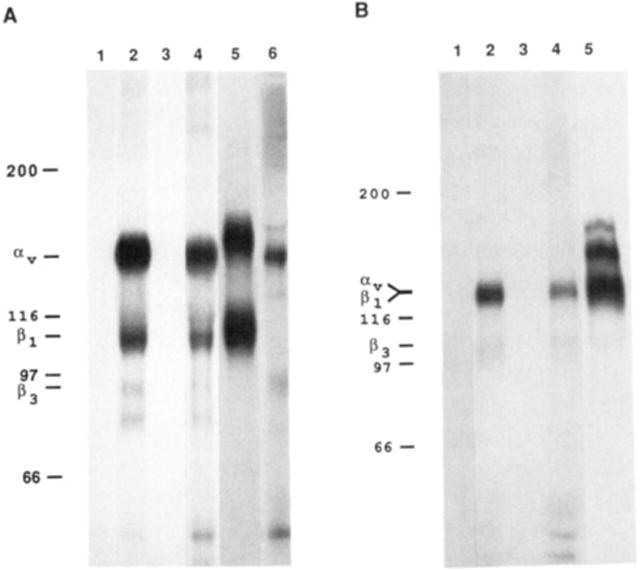
Immunoprecipitation of chick embryo fibroblasts (CEFs). Surface-labeled CEF extracts were immunoprecipitated with (1) preimmune and (2) immune sera raised against the C peptide of the chick αv subunit, (3) preimmune and (4) immune sera raised against the L peptide of the chick αv subunit, and (5) integrin β1-specific serum and (6) αvβ3-specific monoclonal antibody LM609. After protein A–Sepharose precipitation, the samples were washed and electrophoresed in nonreducing (A) and reducing (B) conditions. The gels were then dried and autoradiographed. The numbers at left indicate molecular weight markers × 10−3.
By comparing in Figure 4 lanes 2 and 4, we can see that the immune sera raised against the C and L peptides immunoprecipitate the same set of integrin subunits migrating at Mr 150K, 110K, 94K, and 84K in nonreducing conditions.
The Mr 150K band, which migrates at Mr 127K in reducing conditions, has a molecular weight consistent with that predicted by glycosylation of the chick αv amino acid sequence. Its calculated molecular weight is 150K, and the expected fragment sizes after proteolytic cleavage at position 857 are 122K and 28K. After reduction and denaturation, the αv subunit migrated with a molecular weight close to that predicted for the large fragment, indicating that the two fragments were linked by a disulfide bond. The short, transmembrane fragment is too small to be visualized in Figure 4.
The three lower molecular weight protein bands appear to be multiple β subunits coprecipitating with the avian αv subunit. The larger and most abundant of these migrates at Mr 110K when nonreduced, and comigrates with αv at Mr 127K in reducing conditions. This band migrates with precisely the same mobility as the lower band immunoprecipitated in lanes 5A and 5B with a β1 subunit specific serum. The upper bands in lanes 5A and 5B appear to be mixtures of the β1-associated α subunits. The second band which coprecipitates with the αv subunit has the same mobility as the β3 subunit that is visualized in lane 6, using the αvβ3 complex specific monoclonal antibody (LM609). Finally, a lower band of 84 kDa in nonreducing conditions indicates that a third β subunit is probably coprecipitating with some αv subunits. This could be βS or a novel β subunit. This band is unlikely to be a breakdown product of β1 or β3 because it is not present in immunoprecipitates of β1 or αvβ3 heterodimers (Figure 4, lanes 5 and 6).
The αv subunit was first characterized in association with a β3 subunit (Pytela et al., 1985; Suzuki et al., 1986), but more recently two other β subunits have been shown to form heterodimers with αv. A βX subunit has been found in some carcinoma cells, which is slightly larger than β3 (Cheresh et al., 1989a), and a βS subunit in osteosarcomas that migrates below the β3 subunit (Freed et al., 1989). A possible existence of αvβ1 heterodimers was suggested by vitronectin–integrin binding that could be inhibited by the β1-specific CSAT monoclonal antibody (Buck & Horwitz, 1987).
In order to further investigate the presence of αvβ1 heterodimers, immunodepletions using a β1-specific polyclonal antibody were performed as illustrated in Figure 5. Extracts from surface-labeled chick embryo fibroblasts (CEFs) were depleted 7 times successively, resulting in a virtually complete depletion of β1-containing heterodimers (Figure 5, lanes 1–7). When the β1-depleted extract was mixed with αv-specific antibodies to immunoprecipitate the remaining αv-containing heterodimers, the amount of protein migrating with the mobility of the β1 subunit was strikingly diminished (lane 8) after the β1 depletions, compared to a control (lane 9) depleted 7 times with normal rabbit serum. This indicates that one β subunit precipitated by the αv-specific antiserum is indeed β1. As shown in lane 8, even after extensive depletions with β1, there is still a faint band located at approximately the same position as β1, at about 110 kDa. It is therefore possible that another β subunit, that comigrates with the β1 subunit in nonreducing conditions, coprecipitates with the αv subunit in CEFs. Alternatively, it is possible that the β1-specific polyclonal antibody is unable to completely deplete the β1 heterodimers due to a limited affinity of the antibodies. However, the latter possibility seems unlikely because the same β1-specific antiserum was able to deplete completely another αβ1 heterodimer (data not shown).
FIGURE 5.
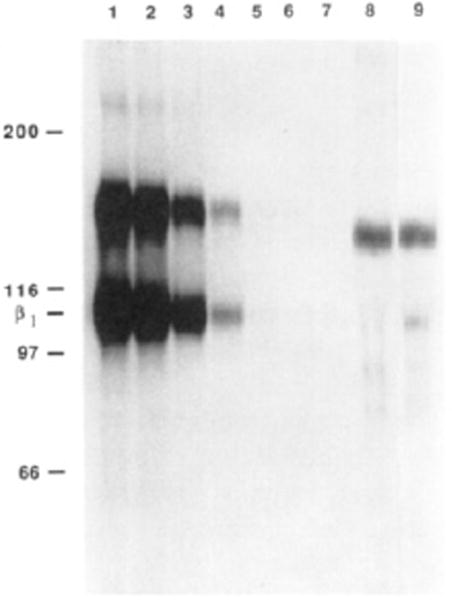
Immunodepletion with anti-β1 polyclonal antibodies. Surface-labeled CEF extracts were immunodepleted 7 times with successive doses of anti-β1 subunit serum (lanes 1–7). The depleted extract was then mixed with the serum raised against the C peptide derived from the chick αv subunit cytoplasmic domain (lane 8). An identical CEF extract sample was depleted 7 times with normal rabbit serum and finally was depleted 7 times with normal rabbit serum and finally immunoprecipitated (lane 9) with the same αv subunit specific serum used in lane 8.
Inspection of Figure 5 indicates that the amount of αv in lane 8 was not strongly reduced by depletion with the β1 antibody compared to controls, as would be expected if the majority of the subunits was present in αvβ1 heterodimers. The most likely explanation for this is the presence of a large proportion of monomeric αv subunits in the extract, dissociated from β subunits after cellular lysis. Consistent with this possible explanation, a high apparent ratio of αv subunits compared to β subunits was seen in each experiment in which an αv-specific serum was used for immunoprecipitation, in contrast to the ratio observed with a β1-specific serum (Figures 4 and 5).
As another demonstration that αv associates with β1, a gel was overloaded with an excess of αv-specific antibodies and their immunoprecipitated products. After electrophoresis, we were able to detect the presence of β1 subunits in the immunoprecipitate by using the β1-specific monoclonal antibody W1B10 in an antigen blot (data not shown).
Discussion
We have cloned and analyzed chick cDNAs that are homologous to the human vitronectin receptor α subunit (αv). We have also raised specific antibodies to the subunit encoded by these cDNAs and analyzed the heterodimeric receptors that it forms in CEFs by immunoprecipitation and SDS–PAGE.
The chick amino acid sequence deduced from the purified cDNA clones is very similar to the human αv subunit; 83% of the 1015 amino acids are identical. An additional 6% of the residues have been substituted by a homologous amino acid. This high sequence conservation between two α subunits is much higher than the similarities observed between different integrin α subunits in the same organism (<45%). Moreover, this amino acid conservation is comparable to the 85% conservation that has been observed between the chick and human integrin β1 subunits (Tamkun et al., 1986; Scott Argraves et al., 1987). We therefore assume that the subunit described in the present paper is the chick homologue of the vitronectin receptor αv subunit (Suzuki et al., 1986; Fitzgerald et al., 1987).
Analysis of the different structural domains of this αv subunit indicates that the putative transmembrane and cytoplasmic domains of the chick and human αv subunits are particularly well conserved (95%), as they are in the β1 subunits (100% conserved) of the same two species. This high conservation between cytoplasmic domains from homologous subunits in different organisms contrasts with the diversity observed between cytoplasmic domains from different subunits in the same organism [cf. Marcantonio and Hynes (1988) and Takada et al. (1989)]. This suggests that the cytoplasmic domains of the α subunits are functionally important, as it has been directly demonstrated for the β1 subunit [e.g., see Solowska et al. (1989)].
The putative Ca2+ binding domains of the integrin α subunits are also likely to have important functions. They are highly conserved (91%) between the human and chick αv subunits. Association of the α and β subunits has been shown to require Ca2+ in the platelet heterodimer GPIIb/IIIa, also named αIIbβ3 (Fitzgerald & Phillips, 1985). In addition, integrity of integrin heterodimers is necessary for the ligand binding activity of avian β1 integrins (Buck et al., 1986). Calcium is required for the binding of the vitronectin receptor to peptide ligands (Cheresh et al., 1987). Binding of the I domain containing integrins α2β1, αLFA-1β2, and αMAC-1β2 requires Mg2+ [cf. Santoro (1986) and Dransfield and Hogg (1989)].
Several integrin receptors recognize an Arg-Gly-Asp (RGD)-containing sequence in different extracellular matrix components (Pierschbacher et al., 1984; Ruoslahti & Pierschbacher, 1987). Vitronectin, for example, contains a cellular attachment site that contains a single RGD sequence (Suzuki et al., 1985). Two RGD binding sites of the vitronectin receptor α subunit have been recently mapped by photoaffinity labeling between amino acids 139 and 349. This interval overlaps and precedes the N-terminal side of the first three Ca2+ binding domains (Smith & Cheresh, 1990). By comparing the chick and human αv sequences within the 139–349 interval, we can identify residues that did not diverge during evolution of these two species and thus might be involved in the ligand binding specificity of these receptors. To eliminate residues not specific for αv, we have also compared the same region to the sequence of the α5 subunit of the fibronectin receptor. The α5 subunit is more similar in amino acid sequence to αv than any other known integrin α subunit, but α5-containing heterodimers do not bind vitronectin or fibrinogen, in contrast to at least some αv-containing heterodimers (Pytela et al., 1985; Cheresh et al., 1989b). The result of such a comparison is presented in Figure 6. It appears that residues specific to the two αv sequences are scattered throughout the 139–349 interval, but three regions (A, B, and C) mentioned in Figure 6 have particularly high numbers of them. In the A region, which is located N-terminal to the first Ca2+ binding domain (residues 186–204), 11 out of 19 residues (58%) are conserved between the 2 αv subunits only; 2 others (11%) are also present in the FNR α5 subunit. Intriguingly, this is the location of an alternatively spliced exon in the Drosophila integrin α homologue PS2 (Brown et al., 1989). The B and C regions, residues 247–262 and 320–334, respectively, are located between the first three Ca2+ binding domains. Fifty percent of the residues of the B region and 59% of the residues in the C region are specific to the two compared αv sequences. In addition, 31% of the residues in the B region and 18% of the C region are also shared with the α5 subunit. Interestingly, in comparison to the chick and human αv subunits, α5 and all other sequenced α subunits have additional amino acids inserted in the C region (Takada et al., 1989).
FIGURE 6.
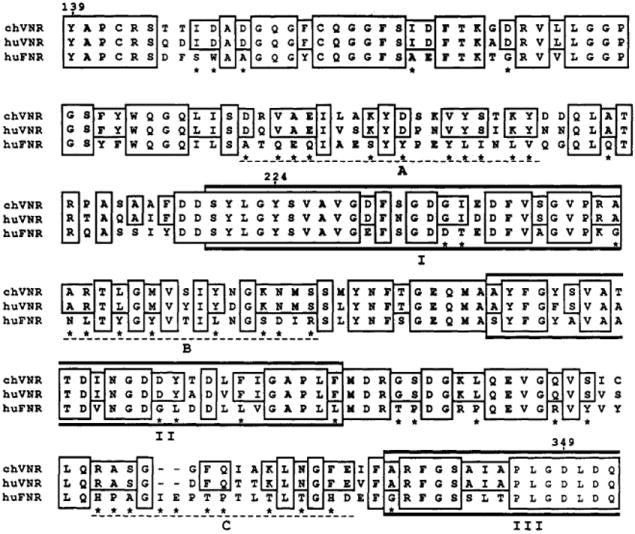
RGD binding domains comparison. Amino acid comparison of the RGD binding domain from the human αv subunit (huVNR) and its chick counterpart (chVNR), with the homologous sequence from the α5 subunit of the human fibronectin receptor (huFNR). Boxes indicate identical residues between chick and human vitronectin receptors (two upper row boxes) or between the three compared α subunits (three row boxes). Nonconservative substitutions between the two αv and the α5 sequences are indicated by an asterisk, using the rule defined in the Figure 2 legend. The three main regions, A, B, and C, containing vitronectin receptor specific sequences are underlined by dashed lines. Amino acid numbers correspond to the human sequence (Fitzgerald et al., 1987).
Thus, αv-specific residues do exist in the RGD binding site interval (residues 139–349). Forty percent of them are scattered, but 60% are concentrated in three regions that are either preceding and/or interspersed between the three first Ca2+ binding domains. If the ligand binding domain is restricted to the RGD binding site and its vicinity, these αv-specific residues are likely to have an important role in determining the binding properties of αv-containing heterodimers. In addition to the α subunits, there is also evidence that the β chains participate in the ligand binding specificity. Most notably, a carcinoma cell line (UCLA-P3) expresses a vitronectin receptor composed of an αv subunit associated with a βX subunit, also called β5 (Cheresh et al., 1989a). This αvβX receptor exhibits a different ligand binding specificity than the αvβ3 heterodimer, that is most likely due to the different β subunits in these two heterodimers.
Evidence in this report indicates that αv can form heterodimers with multiple β subunits in CEFs. Several lines of evidence show that one of the β subunits associated with αv is the β1 integrin subunit. After coimmunoprecipitation with an αv subunit, this β subunit migrates with a mobility identical with β1 either in reducing or in nonreducing conditions (Figure 4). Moreover, it can be specifically immunodepleted with β1-specific polyclonal antibodies (Figure 5). In addition to this β1 subunit, two smaller subunits were coimmunoprecipitated with αv-specific antibodies. One of these has the electrophoretic mobility expected for the β3 integrin subunit, but the other one (84 kDa nonreduced) is smaller than any αv-associated β subunit described so far (Cheresh et al., 1989a; Freed et al., 1989). This small subunit may be another known αv-associated β subunit (β5 or βS) that migrates faster in CEFs due to a difference in glycosylation. Alternatively, it may be a novel β subunit.
Since the completion of this work, two reports have appeared that demonstrate association of the αv subunit with the integrin β1 subunit in two human cell lines, 293 and IMR 32 (Bodary & McLean, 1990; Vogel et al., 1990). There is no agreement in these reports on the ligand binding specificity of αvβ1 heterodimers. The evidence presented indicates that this receptor functions as a fibronectin, but not vitronectin, receptor in the neuroblastoma cell line IMR 32 and as a vitronectin, but not fibronectin, receptor in the embryonic kidney cell line 293. One possible reconciliation of these results is that the ligand binding specificity of αvβ1 heterodimers is determined, in part, by cell-specific factors, as has been demonstrated for α2β1 integrins (Languino et al., 1989; Elices & Hemler, 1989). Such factors could include differences in splicing, glycosylation, or accessory proteins. Taken together with the results in the present paper, the data indicate that the αv subunit associates with multiple β subunits, including β1, in both avian and mammalian species. They also indicate that the ligand binding specificity of the αvβ1 heterodimer must be determined in each cell type in which it is expressed. Clearly, further studies on factors modulating its specificity are needed.
Acknowledgments
We acknowledge the generous gifts of antibodies LM609 and W1B10 from Dr. David Cheresh and Dr. Rick Horwitz. We also thank Dr. Larry Fitzgerald for giving us samples of human αv cDNA clones, Dr. Carol Nottenburg for the E10 chick cDNA library, Dr. Barbara Ranscht for the E13 chick brain cDNA library, and Ella Wetzel for her help with poly(A+) RNA purification and hybridization. This work has benefited from stimulating conversations with Drs. David Cheresh, Kevin Tomaselli, and Karla Neugebauer. We thank Drs. Kevin Jones and Mike Ignatius for helpful comments on the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant NS21824 to L.F.R. and by the Howard Hughes Medical Institute. B.B. was supported successively by a young researcher fellowship and by Fellowship 83.668.0.88, both from the Fonds National Suisse de la Recherche Scientifique. L.F.R. is an investigator of the Howard Hughes Medical Institute.
References
- Benton WD, Davis RW. Science. 1977;196:180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bodary SC, McLean JW. J Biol Chem. 1990;265:5938–5941. [PubMed] [Google Scholar]
- Brown NH, King DL, Wilcox M, Kafatos FC. Cell. 1989;59:185–195. doi: 10.1016/0092-8674(89)90880-5. [DOI] [PubMed] [Google Scholar]
- Buck CA, Horwitz AF. J Cell Sci Suppl. 1987;8:231–250. doi: 10.1242/jcs.1987.supplement_8.13. [DOI] [PubMed] [Google Scholar]
- Buck CA, Shea E, Duggan K, Horwitz AF. J Cell Biol. 1986;103:2421–2428. doi: 10.1083/jcb.103.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh DA, Pytela R, Pierschbacher M, Klier FG, Ruoslahti E, Reisfeld RA. J Cell Biol. 1987;105:1163–1173. doi: 10.1083/jcb.105.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh DA, Smith JW, Cooper HM, Quaranta V. Cell. 1989a;57:59–69. doi: 10.1016/0092-8674(89)90172-4. [DOI] [PubMed] [Google Scholar]
- Cheresh DA, Berliner SA, Vicente V, Ruggeri ZM. Cell. 1989b;58:945–953. doi: 10.1016/0092-8674(89)90946-x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield I, Hogg N. EMBO J. 1989;8:3759–3765. doi: 10.1002/j.1460-2075.1989.tb08552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elices MJ, Hemler ME. Proc Natl Acad Sci U S A. 1989;86:9906–9910. doi: 10.1073/pnas.86.24.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elices MJ, Osborn L, Takada Y, Crouse C, Luhowskyj S, Hemler ME, Lobb RR. Cell. 1989;60:577–584. doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fitzgerald LA, Phillips DR. J Biol Chem. 1985;260:11366–11374. [PubMed] [Google Scholar]
- Fitzgerald LA, Poncz M, Steiner B, Rall SC, Jr, Benett JS, Phillips DR. Biochemistry. 1987;26:8158–8165. doi: 10.1021/bi00399a021. [DOI] [PubMed] [Google Scholar]
- Freed E, Gaillit J, van der Geer P, Ruoslahti E, Hunter T. EMBO J. 1989;8:2955–2965. doi: 10.1002/j.1460-2075.1989.tb08445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Hynes R. Cell. 1990;60:53–61. doi: 10.1016/0092-8674(90)90715-q. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Annu Rev Immunol. 1990;8:365–400. doi: 10.1146/annurev.iy.08.040190.002053. [DOI] [PubMed] [Google Scholar]
- Hemler ME, Huang C, Takada Y, Schwarz L, Strominger JL, Clabby ML. J Biol Chem. 1987;262:11478–11485. [PubMed] [Google Scholar]
- Holzmann B, Weissman IL. EMBO J. 1989;8:1735–1741. doi: 10.1002/j.1460-2075.1989.tb03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- Kajiji S, Tamura R, Quaranta V. EMBO J. 1989;8:673–680. doi: 10.1002/j.1460-2075.1989.tb03425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Languino RR, Gehlsen KR, Wayner E, Carter WG, Engvall E, Ruoslahti E. J Cell Biol. 1989;109:2455–2462. doi: 10.1083/jcb.109.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. [Google Scholar]
- Marcantonio EE, Hynes RO. J Cell Biol. 1988;106:1765–1772. doi: 10.1083/jcb.106.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- Pytela R, Pierschbacher MD, Ruoslahti E. Proc Natl Acad Sci U S A. 1985;82:5766–5770. doi: 10.1073/pnas.82.17.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Rubin H. Exp Cell Res. 1968;49:666–678. doi: 10.1016/0014-4827(68)90213-9. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E, Pierschbacher MD. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SA. Cell. 1986;46:913–920. doi: 10.1016/0092-8674(86)90073-5. [DOI] [PubMed] [Google Scholar]
- Scott Argraves W, Suzuki S, Arai H, Thompson T, Pierschbacher MD, Ruoslahti E. J Cell Biol. 1987;105:1183–1190. doi: 10.1083/jcb.105.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JW, Cheresh DA. J Biol Chem. 1990;265:2168–2172. [PubMed] [Google Scholar]
- Solowska J, Guan JL, Marcantonio EE, Trevithick JE, Buck CA, Hynes RO. J Cell Biol. 1989;109:853–861. doi: 10.1083/jcb.109.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Modderman PW, Hogervorst F. Nature. 1988a;336:487–489. doi: 10.1038/336487a0. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Hogervorst F, Osterop A, Veltman FEM. J Biol Chem. 1988b;263:14030–14038. [PubMed] [Google Scholar]
- Suzuki S, Oldberg A, Hayman, Pierschbacher MD, Ruoslahti E. EMBO J. 1985;4:2519–2524. doi: 10.1002/j.1460-2075.1985.tb03965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Scott Argraves W, Pytela R, Arai H, Krusius T, Pierschbacher MD, Ruoslahti E. Proc Natl Acad Sci U S A. 1986;83:8614–8618. doi: 10.1073/pnas.83.22.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Elices MJ, Crouse C, Hemler M. EMBO J. 1989;8:1361–1368. doi: 10.1002/j.1460-2075.1989.tb03516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO. Cell. 1986;46:271–282. doi: 10.1016/0092-8674(86)90744-0. [DOI] [PubMed] [Google Scholar]
- Tomaselli KJ, Damsky CH, Reichardt LF. J Cell Biol. 1988;107:1241–1252. doi: 10.1083/jcb.107.3.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel BE, Tarone G, Giancotti FG, Gaillit J, Ruoslahti E. J Biol Chem. 1990;265:5934–5937. [PubMed] [Google Scholar]
- von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayner EA, Garcia-Pardo A, Humphries MJ, McDonald JA, Carter WG. J Cell Biol. 1989;109:1321–1330. doi: 10.1083/jcb.109.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


