Abstract
IscU is a scaffold protein that functions in iron-sulfur cluster assembly and transfer. Its critical importance has been recently underscored by the finding that a single intronic mutation in the human iscu gene is associated with a myopathy resulting from deficient succinate dehydrogenase and aconitase [Mochel, F., Knight, M. A., Tong, W. H., Hernandez, D., Ayyad, K., Taivassalo, T., Andersen, P. M., Singleton, A., Rouault, T. A., Fischbeck, K. H., and Haller, R. G. (2008) Am. J. Hum. Genet. 82, 652–660]. IscU functions through interactions with a chaperone protein HscA and a cochaperone protein HscB. To probe the molecular basis for these interactions, we have used NMR spectroscopy to investigate the solution structure of IscU from Escherichia coli and its interaction with HscB from the same organism. We found that wild-type apo-IscU in solution exists as two distinct conformations: one largely disordered and one largely ordered except for the metal binding residues. The two states interconvert on the millisecond time scale. The ordered conformation is stabilized by the addition of zinc or by the single site IscU mutation, D39A. We used apo-IscU(D39A) as a surrogate for the folded state of wild-type IscU and assigned its NMR spectrum. These assignments made it possible to identify the region of IscU with the largest structural differences in the two conformational states. Subsequently, by following the NMR signals of apo-IscU(D39A) upon addition of HscB, we identified the most perturbed regions as the two N-terminal β-strands and the C-terminal α-helix. On the basis of these results and analysis of IscU sequences from multiple species, we have identified the surface region of IscU that interacts with HscB. We conclude that the IscU:HscB complex exists as two (or more) distinct states that interconvert at a rate much faster than the dissociation of the complex and that HscB binds to and stabilizes the ordered state of apo-IscU.
Iron-sulfur (Fe-S) proteins play indispensable roles in electron transfer, catalysis, and gene regulation (1). The prosthetic group of Fe-S proteins, the FenSn cluster, comes in a variety of sizes and geometries and includes 2Fe-2S, 3Fe-4S, and 4Fe-4S clusters (2). The functional importance of Fe-S proteins spurred investigations of cluster assembly mechanisms and led to the discovery of the Nif (nitrogen fixation), Isc (iron-sulfur cluster), and Suf (sulfur utilization factor) assembly systems (2). Of these three systems, the Isc system is responsible for generalized Fe-S cluster biosynthesis in most organisms (2, 3).
The Isc operon of E. coli encodes several protein products: IscR, IscS, IscU, IscA, HscB, HscA, and Fdx (2). Of these components, IscU is particularly important because it acts as a scaffold on which the cluster is assembled and from which the cluster is subsequently transferred to a recipient apo-protein (4). It has been reported that the Asp39Ala (D39A) substitution in Azotobacter vinelandii IscU (5), or in the corresponding IscU variants from Aquifex aeolicus (6), Schizosaccharomyces pombe (7), and Homo sapiens (8) stabilizes the cluster-coordinating complex of IscU. This stabilization has been attributed to a decrease in solvent accessibility of the cluster (9, 10).
In vitro experiments have shown that holo-IscU is able to transfer its cluster to an apo-protein but that the presence of HscA and HscB greatly enhance the rate and efficiency of this process (11, 12). HscA and HscB are specialized versions of an Hsp70-type chaperone and an Hsp40-type cochaperone, respectively. The nature of the nucleotide (ATP or ADP) bound to HscA allosterically regulates the affinity of the chaperone for IscU (13), and the ATPase activity of HscA is highly enhanced by its interaction with HscB and IscU (14). The differential affinity of HscA for IscU, and the role of HscB in enhancing the binding of IscU to HscA and increasing the ATPase activity of HscA appear to regulate the cycle of cluster transfer from IscU to a protein such as apo-ferredoxin (11, 12).
Our understanding of Fe-S cluster transfer has also been aided by structural investigations of IscU, HscA, HscB, and complexes of these proteins. The NMR solution structures of Zn-bound IscU from Haemophilus influenzae (15) and Mus musculus (PDB 1wfz) indicated that the Zn-bound protein has a compact structural core that consists of two α-helices (α2 and α5) packed against a three-stranded anti-parallel β-sheet (β1-β3)on one side and two short α-helices (α3 and α4) on the other. The NMR solution structure of Zn-bound H. influenzae IscU (15) shows the Zn atom coordinated by two cysteines located in the loop region (C37 and C63) and one histidine and one cysteine at the edge of helix α5 (H105 and C106). In the absence of zinc, H. influenzae IscU was found to be much less structured (15). Consistent with an inherent structural flexibility, the recent crystal structure of holo-IscU from A. aeolicus revealed a different conformation for each of the three IscU monomers within the trimeric holo-complex (16). In addition, apo-SufU of Thermotoga maritima (an IscU counterpart in the Suf system) was reported to have a molten globule-like tertiary structure consisting of multiple conformations (17, 18). The best model for the interaction between IscU and HscA comes from the X-ray structure of the substrate binding domain (SBD) of HscA co-crystallized with ELPPVKIHC (19), an IscU-derived peptide that was shown to interact with HscA (20, 21). The crystal structure revealed that the ELPPVKIHC peptide binds to the hydrophobic cleft of HscA(SBD) in a fully extended conformation, implying a significant conformational change in IscU as a result of the binding interaction with HscA (19). The binding interaction between apo-IscU and HscB involves the formation of a 1-to-1 complex with a dissociation constant of ≅13 µM (14). A recent NMR spectroscopic study proposed that the binding site for IscU is located on one face of HscB’s C-terminal domain and contains several highly-conserved hydrophobic as well as acidic residues (22). Furthermore, alanine shaving experiments suggested that the hydrophobic residues (L92, M93, and F153 in E. coli HscB) may play an important role in strengthening the binding interaction (22).
We report here the results from a complementary investigation of E. coli apo-IscU and its interaction with E. coli HscB. Our study indicates that apo-IscU exists in solution in two conformational states, one largely disordered and one ordered with the exception of the metal binding region. The ordered structure resembles that stabilized by the addition of Zn2+. A single site mutation IscU(D39A) leads to the stabilization of the ordered state. Our results identify residues of IscU that are perturbed upon the addition of the cochaperone HscB and provide information on the nature of the IscU:HscB complex.
Experimental Procedures
Materials
BL21 competent cells were obtained from Novagen (Madison, WI), and strain JM15 [CGSC strain # 5042 (23)] was obtained from the E. coli Genetic Stock Center (EGSC) of Yale University (New Haven, CT). The QuikChange II Site-Directed Mutagenesis Kit was purchased from Stratagene (Cedar Creek, TX). 15NH4Cl and [U-13C]-D-glucose were purchased from Cambridge Isotope Laboratories (Andover, MA). Other chemicals, including natural abundance amino acids and chemicals for buffers, were purchased from Sigma-Aldrich Corp. (St. Louis, MO). DE-52 anion exchange resin was purchased from Whatman Inc. (Florham Park, NJ), DEAE Bio-Gel anion exchange gel was purchased from Bio-Rad Laboratories (Hercules, CA), and the HiLoad 16/60 Superdex 75 prep grade gel filtration column was purchased from GE Healthcare Bio-Sciences Corp. (Piscataway, NJ).
Site-Directed Mutagenesis
To express IscU(D39A), the QuikChange technique was used to introduce the appropriate mutation into the pTrcIscU expression vector (14). The presence of the mutation in the new expression vector [henceforth called pTrcIscU(D39A)] was subsequently confirmed by DNA sequencing.
Expression and Purification of Proteins
HscB was expressed and purified as described previously (14). [U-15N]- and [U-13C,U-15N]-labeled samples of IscU and IscU(D39A) were prepared as follows. A colony of BL21 cells transformed with the pTrcIscU [or pTrcIscU(D39A)] plasmid was used to inoculate 10 mL of TB liquid medium containing 100 µg of ampicillin/mL. The cells were grown for ∼12 h at 37 °C, and a 10 µL inoculum was transferred to 50 mL TB liquid medium containing 100 µg of ampicillin/mL, which was subsequently grown for ∼12 h at 37 °C. Cells from this 50 mL culture were used to inoculate 500 mL M9 medium containing 100 µg of ampicillin/mL and supplemented with either 0.5 g 15NH4Cl and 2.5 g glucose (for production of [U-15N]IscU) or 0.5 g 15 NH4Cl and 1.5 g [U-13C]-D-glucose (for production of [U-13C,U-15N]IscU). Gene expression was induced at Abs600 ≈ 1 by adding IPTG to a final concentration of 0.4 mM. Protein production was allowed to proceed for ∼ 3 h, after which cells were harvested and stored at –80 °C. Protein purification was carried out as described previously (14), except that a HiLoad 16/60 Superdex 75 gel filtration column was employed for the final purification step in place of reversed phase chromatography. The elution buffer for this step consisted of 20 mM Tris·HCl pH 8.0, 1 mM DTT, 0.5 mM EDTA, and 150 mM NaCl.
IscU(D39A) was further characterized to determine whether it serves as a suitable model for investigating the interaction between wild-type IscU and HscB. The affinity of apo-IscU(D39A) for HscB was determined directly using isothermal titration calorimetry. Figure S1 shows that apo-IscU(D39A) forms a 1:1 complex with HscB with binding affinity ≅10 µM and thermodynamic parameters similar to those observed with apo-IscU (14, 21). We also measured the ability of apo-IscU(D39A) to stimulate the ATPase activity of HscA, a sensitive probe for monitoring the binding interactions between HscA, IscU and HscB (14, 21). As shown in Figure S2, apo-IscU(D39A) acts synergistically with HscB to stimulate HscA ATPase activity in a manner similar to that observed for wild-type IscU (14) indicating that apo-IscU(D39A) interacts with HscB and HscA in a physiologically relevant manner.
Isotopic Labeling of Proteins
To produce proteins selectively reverse-labeled with an amino acid of natural abundance, 500 mL of M9 growth medium containing 100 µg of ampicillin/mL was initially supplemented with 0.5 g 15NH4Cl and 1 g glucose. When the cell culture reached Abs600≈1, the following additions were made: IPTG (final concentration 0.4 mM), 1 g glucose, and an unlabeled amino acid (0.28 g glycine to produce [U-15N,14N-Gly]IscU, 0.2 g L-arginine·HCl to produce [U-15N,14N-Arg]IscU, or 0.2 g L-lysine·HCl to produce [U-15N, 14N-Lys]IscU) (24). To prepare [U-15N,14N-Cys]IscU, a Cys-auxotrophic strain of JM15 cells (23) was transformed with the pTrcIscU(D39A) plasmid. Initial cell growth was carried out as described above for BL21 cells, but later steps were modified as follows. Cells from the 50 mL TB were used to inoculate a 125 mL TB medium, which was subsequently grown overnight at 37 °C. Cells from the 125 mL TB medium were transferred to 500 mL M9 medium supplemented with 0.5 g 15NH4Cl and 1 g glucose. Following incubation for ∼2 h, the following was added: IPTG (to a final concentration of 0.4 mM), 1 g glucose, and 0.03 g of L-cysteine. After further incubation for ∼3 h, the cells were harvested and stored, and proteins were purified as described above.
NMR Samples
The solvent used for NMR samples contained 20 mM Tris·HCl pH 8.0, 0.5 mM EDTA, 5–10 mM DTT, 150 mM NaCl, 7% D2O, 0.7 mM DSS, and 0.02% sodium azide. A sample of 0.8 mM [U-15N]apo-IscU was employed to obtain 2D NMR spectra while a sample of 2.0 mM [U-13C,U-15N]apo-IscU was used to collect 3D NMR spectra. To prepare Zn2+ bound IscU, 0.8mM [U-15N]IscU was exchanged into 20 mM Tris·HCl pH 8.0, 3 mM ZnCl2, 5–10 mM DTT, 150 mM NaCl, 7% D2O, 0.7 mM DSS, and 0.02% sodium azide. The concentration of [U-13C,U-15N]apo-IscU(D39A) used for resonance assignments was 2.5 mM. The concentration of [U-15N]apo-IscU(D39A) used for 15N-relaxation and 1H-15N heteronuclear NOE measurements was 0.6–0.7 mM. Chemical shift perturbation experiments started with a sample of 0.6–0.8 mM [U-15N]IscU(D39A) to which was added aliquots of 2.5 mM unlabeled HscB.
NMR Spectroscopy
Unless specified otherwise, all NMR spectra were acquired at 25 °C with 600 MHz or 800 MHz Varian Unity-Inova spectrometers equipped with a z-gradient cold probe. NMRPipe (25) was used to process the raw NMR data and Sparky (26) was used for data analysis. The 2D exchange experiment (27, 28) with apo-IscU was collected on a 750 MHz Bruker DMX spectrometer equipped with a CryoProbe. The mixing time used to detect chemical exchange was set to 60 ms. The following data sets were collected and used to assign the resonances from [U-13C,U-15N]IscU(D39A): 2D 15N-HSQC, 3D HNCO, 3D CBCA(CO)NH, 3D HNCACB, 3D HN(CO)CA, 3D HNCA, and 2D 15N-NOESY-HSQC (τm=100ms). 2D 15N-HSQC spectra of selectively reverse-labeled IscU(D39A) ([U-15N, 14N-Arg]IscU(D39A), [U-15N, 14N-Gly]IscU(D39A), [U-15N, 14N-Lys]IscU(D39A), and [U-15N, 14N-Cys]IscU(D39A)) were also used to assign ambiguous resonances. Chemical shift assignments were used as input for the PECAN web server (29) to predict the secondary structure of IscU(D39A). Resonance assignment of apo-IscU was achieved with the help of the apo-IscU(D39A) assignments and by collecting and analyzing 2D 15N-HSQC, 3D HNCO, 3D CBCA(CO)NH, 3D HNCACB, and 3D HNCA spectra. The tendency of a certain residue to form a secondary structural element in apo-IscU was predicted by comparing the difference between the secondary 13Cα and 13Cβ chemical shifts of apo-IscU with the difference between 13Cα and 13Cβ chemical shifts of a random coil (for Gly residues, only 13Cα chemical shifts were compared) (30, 31).
R1, R2 relaxation and 1H-15N heteronuclear NOE data sets of [U-15N]apo-IscU(D39A) were acquired at 25 °C on a 600 MHz Varian Unity-Inova spectrometer equipped with a z-gradient cold probe. R1 relaxation rate experiments utilized relaxation delays of 100, 200, 300, 400, 600, 800, and 1200 ms; R2 relaxation rate experiments utilized relaxation delays of 10, 30, 50, 70, 90, 110, and 130 ms. The 1H excitation time in the 1H-15N heteronuclear NOE experiments was set to 3 s, and the intensities of each peak in spectra collected with and without the 1H excitation were compared to determine the NOE value. Two replicate sets of R1, R2, and 1H-15N NOE data were collected, standard deviations were estimated from their differences, and the replicate values were averaged to yield the reported values.
For chemical shift perturbation measurements, HscB was titrated into a solution of [U-15N]IscU(D39A) and a 2D 15N-TROSY-HSQC spectrum was recorded after the addition of each aliquot. Titrations were performed at 25 °C with spectra recorded on a 600 MHz Varian Unity-Inova spectrometer equipped with a z-gradient cold probe. The final molar ratio of HscB:IscU was 2. HscB-induced changes in amide peak positions of IscU, ΔδHN (in ppm), were calculated according to Eq. 1 (32),
| (1) |
where ΔδH and ΔδN are the chemical shift changes in the proton and nitrogen dimensions, respectively.
Isothermal Titration Calorimetry
ITC measurements were carried out at 25 ºC in 50 mM HEPES pH 7.5, 150 mM NaCl, and 4 mM TCEP using a VP-ITC isothermal titration calorimeter (MicroCal LLC). The injection syringe contained 3.84 mM apo-IscU or 3.95 mM apo-IscU(D39A), and the sample cell contained 0.27 mM HscB. All solutions were thoroughly degassed by stirring under vacuum before use. Each titration was started with an initial injection of 3 µL, followed by 34 injections of 8 µL with a 240 s gap between consecutive injections. The ORIGIN software supplied with the instrument was used to integrate the peaks of the thermograms and to fit each isotherm by nonlinear regression analysis to yield binding constants (Ka), enthalpies of binding (ΔH), and stoichiometries (n). Entropies of binding (ΔS) were calculated from the Ka and ΔH values by standard thermodynamic relationships. Estimated errors in Ka, ΔH, and n were generated automatically by the software. Because the heat produced upon injection of buffer into buffer was small (< 0.2 kcal/mol), the data presented in the text were not corrected for this effect. The heat of dilution of IscU was approximated from the heats of the last few injections in each series, and the data presented in the text are corrected for this effect.
ATPase Assays
ATPase rates were determined at 23 °C in HKM buffer (50 mM HEPES pH 7.3, 150 mM KCl, 10 mM MgCl2) containing 1 mM ATP by measuring phosphate released using the EnzCheck coupled enzyme phosphate assay kit as described previously (14).
Sequence alignment
TBLASTN was used to search for amino acid sequences of homologs to E. coli IscU. In addition to IscU homologs, these initial searches also identified many NifU-/SufU-type proteins. To screen out the latter, sequences lacking the 99LPPVK103 (or a highly-similar) HscA recognition motif of E. coli IscU were discarded. The final 131 sequences were divided into 7 groups: 9 vertebrates, 20 fungi/metazoa, 11 other eukaryotes, 11 α-proteobacteria, 28 β-proteobacteria, 46 γ-proteobacteria, and 6 other prokaryotes. Sequences of each group were aligned separately with ClustalW2 (33) using default parameters, and the alignment result within each group was analyzed to identify identical residues, and positions that had ‘conserved substitutions’ or ‘semi-conserved substitutions’ as classified by ClustalW2 (33). The global sequence conservation pattern among all 131 sequences was evaluated by comparing the results of each group alignment and identifying residues that were identical, >90% similar, or >80% similar among all groups.
Results
Two Interconverting Conformational States of Apo-IscU
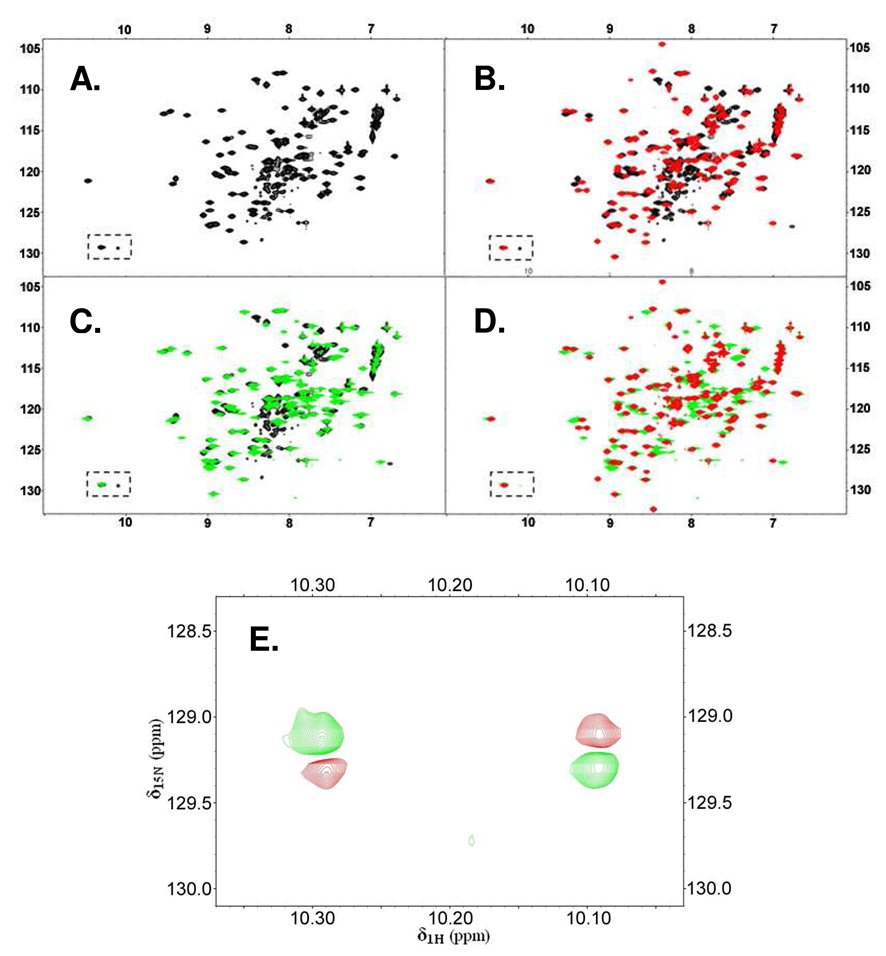
Initial evidence for two conformational states of apo-IscU that interchange slowly on the NMR time scale came from the observation of two 15N-HSQC cross peaks corresponding to the side chain of the single tryptophan residue in the protein (δ1H 10.3 ppm, δ15N 129.1 ppm and δ1H 10.1 ppm, δ15N 129.3 ppm; boxed signals of Figure 1A). In addition, the 15N-HSQC spectrum of apo-IscU showed two sets of signals; one had sharp and well-dispersed peaks, and the other had broad peaks that were overlapped in the center of the spectrum. While the former is the characteristic of folded proteins, the latter is more typical of disordered proteins. The addition of ZnCl2 caused the broad and overlapped signals in the center of the HSQC spectrum to disappear while most sharp and dispersed peaks were retained without significant peak shifts (Figure 1B). A similar observation was noted previously for H. influenzae IscU (15).
Figure 1.
Evidence from 2D 15N-HSQC NMR spectra that E. coli apo-IscU exists in solution as two slowly interconverting conformational states and that the equilibrium is shifted either by zinc ion binding or by the single site mutation D39A. The boxed region in (A – D) contains resonances from the single tryptophan residue Trp76. (A) Spectrum of E. coli apo-IscU. Because IscU contains only a single Trp, the presence of cross peaks in the boxed spectral region indicates the existence of two different conformations. (B) Spectrum of E. coli IscU in the presence of 3 mM ZnCl2 [red] overlaid with the spectrum of apo-IscU [black]. Zinc binding to IscU results in the disappearance of the minor Trp side chain peak as well as the cluster of broad peaks present in the central region of the spectrum of apo-IscU. (C) Spectrum of E. coli apo-IscU(D39A) [green] overlaid with the spectrum of apo-IscU [black]. As with zinc binding, the D39A mutation leads to the disappearance of the minor Trp side chain signal and the cluster of broad and overlapped peaks in the central region of the spectrum of apo-IscU. (D) Spectrum of E. coli IscU in the presence of 3 mM ZnCl2 [red] overlaid with the spectrum of E. coli apo-IscU(D39A) [green]. The close correspondence between two spectra indicates that Zn-IscU and apo-IscU(D39A) are structurally similar. (E) Two-dimensional 1H-15N exchange spectrum (600 MHz NMR spectrometer) of apo-IscU. Cross peaks assigned to Trp76 (green) are connected by exchange cross peaks (red). This result demonstrates that these two peaks originate, not from two covalently distinct protein species, but from two interchanging conformations.
Unexpectedly, we found that the D39A substitution, which stabilizes the Fe-S cluster in holo-IscU (5–8), also stabilizes the more structured state of IscU. In the 2D 15N-HSQC spectrum of [U-15N]apo-IscU(D39A) the peaks corresponding to the less structured state became very weak in intensity compared to those of the more structured conformation (Figure 1C). However, most cross peaks showed only small shifts in positions compared to apo-IscU. Thus, the D39A substitution shifts the equilibrium between the ordered and disordered states of IscU rather than altering its secondary or tertiary structure.
The extensive congruence between the spectra of Zn-IscU and apo-IscU(D39A) (Figure 1D) indicates that the zinc-bound state of IscU and the ordered state of apo-IscU stabilized by the D39A substitution have similar folds. The major differences between two spectra correspond mainly to the peaks of apo-IscU(D39A) assigned (see below) to residues located in loop regions (C37, G38, V40–L43, G62, C63, S69–S71, V73, T74, K89, and N90) or in a portion of helix α5 (C106–L109, E111–A113, K115, and A117). These are residues involved in metal binding or that become ordered in Zn-IscU.
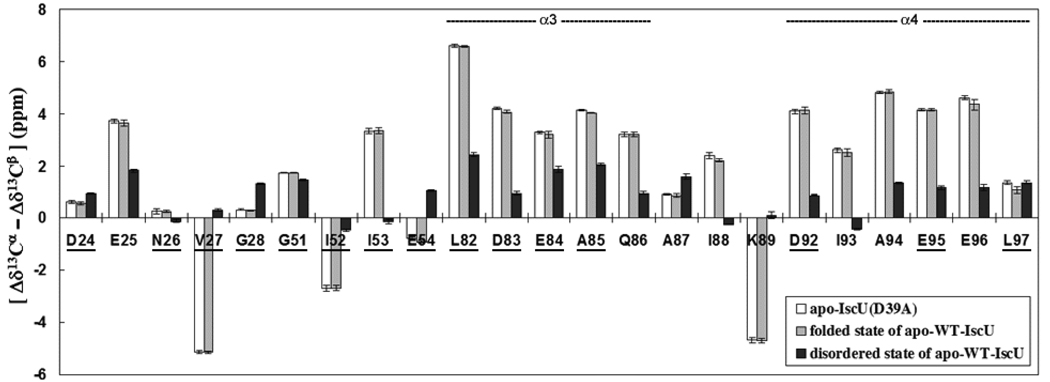
A 2D exchange spectrum (27, 28) of apo-IscU (Figure 1E) revealed the presence of exchange cross peaks linking the two Trp side chain peaks. This result confirmed that these two peaks originate from two exchanging conformations, rather than two covalently distinct protein species, and indicated that the two states have lifetimes in the ms–s range. The 2D exchange spectrum enabled us to identify other, linked pairs of signals in the 2D 15N-HSQC spectrum of [U-15N]apo-IscU (Table S1), that could be used to assign the backbone amide signals of residues D24–G28, G51–E54, L82–K89, and D92–L97 in both states (see below). We used the 13Cα and 13Cβ chemical shifts of these residues to analyze the secondary structure of the two conformational states (Figure 2). It has been shown that the difference between the secondary chemical shifts of 13Cα and 13Cβ [Δδ13Cα – Δδ13Cβ] is a reliable indicator of secondary structural elements (30, 31). Our results indicate that residues E25, V27, I52, I53, L82–Q86, I88, K89 and D92–E96 form ordered secondary structural elements in the folded conformation whereas these structural elements are only partially populated in the disordered conformation.
Figure 2.
Comparison of the predictor of secondary structure [Δδ13 Cα – Δδ13Cβ] (30, 31) for corresponding residues in: [white bars] apo-IscU(D39A), [gray bars] the ordered state of apo-IscU, and [black bars] the disordered state of apo-IscU. Positive values of [Δδ13 Cα – Δδ13 Cβ] indicate α-helix and negative values indicate β-strand. Values near zero indicate random coil. Note the close agreement between the [Δδ13Cα – Δδ13Cβ] values for residues in the ordered state of apo-IscU and those for apo-IscU(D39A). Comparison of the [Δδ13Cα – Δδ13Cβ] values for the ordered and disordered forms of IscU indicate that helix α3 in the disordered state is populated at about 33–50% and that helix α4 in the disordered state is populated at about 25–33%. The results suggest that the two conformational states of apo-IscU interconvert by means of an order-disorder transition affecting the stability of helices α3 and α4. Residues of apo-IscU(D39A) that show minimal chemical shift changes upon formation of the HscB complex are underlined. This result shows that the ordered state of apo-IscU is stabilized upon complex formation and that the underlined residues probably do not contact HscB. Only residues Q86 (part of helix α3) and I93, A94, and E95 (part of helix α4) exhibit shifts upon complex formation.
Resonance Assignments of Apo-IscU(D39A)
Owing to the diminished intensity of the broad and overlapped set of peaks, the NMR spectra of apo-IscU(D39A) were much easier to interpret than those of apo-IscU. Therefore, we concentrated first on assigning spectra of apo-IscU(D39A). Resonance assignments were achieved by employing samples of apo-IscU(D39A) that were uniformly 13C/15N labeled and selectively reverse-labeled. These efforts led to resonance assignments for residues K6–D9, R15–N22, D24–A34, C37–K59, G62, C63, S69–L99, V102–I104, and C106–K128. Notably, most of the unassigned residues were located in the N-terminal region or in loop regions observed to be involved in metal coordination in the zinc complex (15). We used the PECAN web server (29) to predict the secondary structural elements of apo-IscU(D39A) from its assigned chemical shifts and amino acid sequence. The results matched closely the secondary structure elements determined from the solution structure of H. influenzae Zn-IscU (15), except for the absence of the N-terminal helix a1 of Zn-IscU. We have used the designators for the corresponding secondary structural elements in H. influenzae Zn-IscU (15) in describing those in E. coli apo-IscU(D39A): strand β1, G28–G33; strand β2, V40–V47; strand β3, E54-K59; helix α2, S69–W76; helix α3, L82–Q86; helix α4, N90–L97; and helix α5, I104–K124.
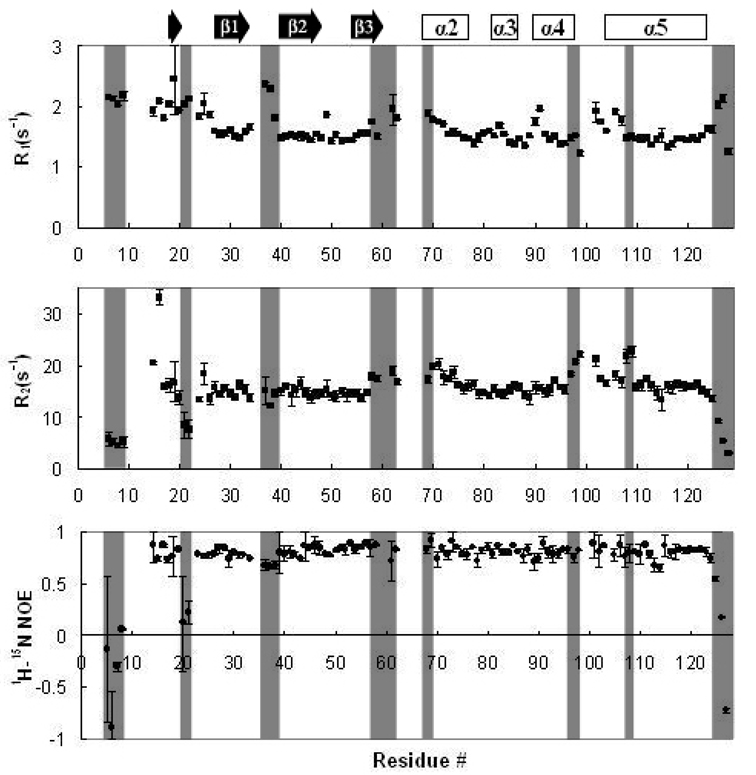
Backbone Dynamics of Apo-IscU(D39A)
We measured 15N R1 and R2 relaxation rates and 1H-15N heteronuclear NOEs to investigate the backbone dynamics of apo-IscU(D39A) (Figure 3). The mean values of R1, R2 and the heteronuclear NOE were 1.64 (±0.26) s−1, 15.37 (±3.90) s−1, and 0.73 (±0.29), respectively. The N- and C-terminal residues, and some residues in loop regions including K6–D9, D21, N22, C37, G38, and A39 showed distinctively higher R1 and lower 1H-15N NOE values characteristic of ps–ns dynamics; the dynamic properties of these residues could account for the failure to detect signals from other residues in these regions (M1–E5, H10–N13, N23, and A36). Both the R1 and R2 values for residues F58–K59, G62, C63, and S69–S71 were slightly higher than average, which implies that the loop between strand β3 and helix α2 undergoes simultaneous ps–ns and µs–ms dynamics. It is likely that the dynamic properties of this region are responsible for the failure to observe signals from residues T60, Y61, and G64–A68. Residues L97–L99, I108, and L109 had distinctively high R2 values without either an increase in R1 values or a decrease in 1H-15N NOE, suggesting that they are affected by a slow (µ–ms) chemical exchange process. It is of interest that this dynamic region includes the conserved 99LPPVK103 motif known to interact with HscA (20, 21).
Figure 3.
R1, R2 relaxation rates and 1H-15N heteronuclear NOE results for E. coli apo-IscU(D39A) plotted as a function of the peptide sequence. Data were collected at 600 MHz (1H) at 25 °C. Blank regions correspond to residues whose signals were either not observed or not assigned. Error bars indicate the standard deviation from two replicate data sets acquired under the same conditions. Some of the residues showing distinctive relaxation rates or NOE values are shaded for comparison. Secondary structural elements predicted from the assigned chemical shifts are indicated at the top of the figure.
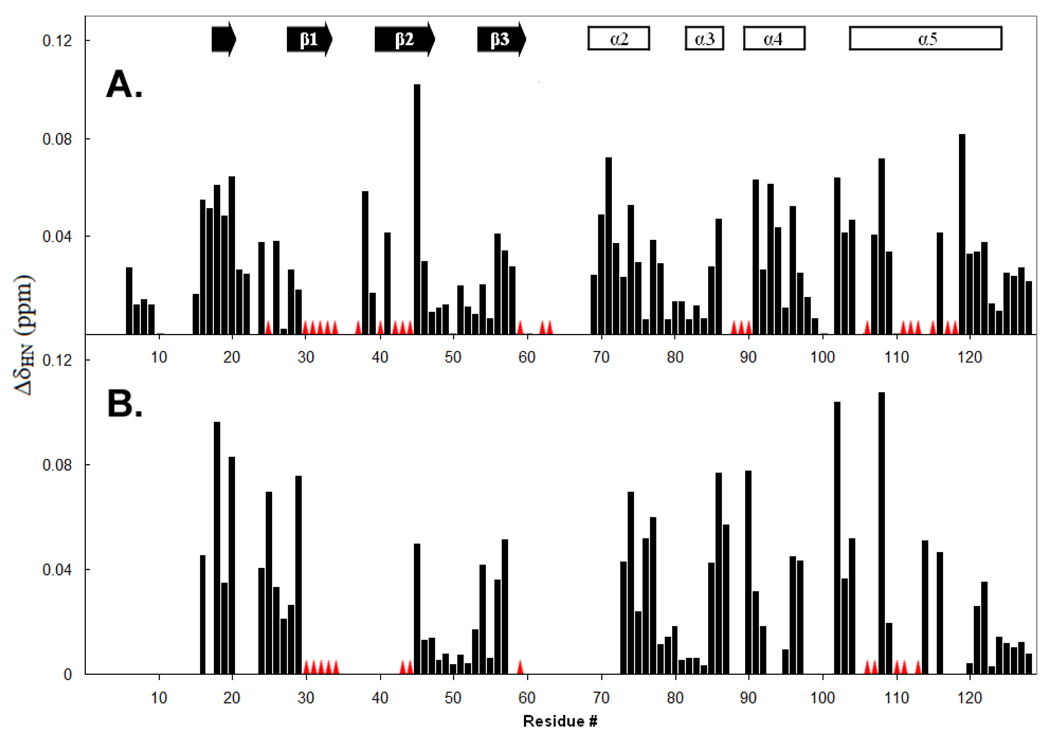
Perturbations in the Chemical Shifts of Apo-IscU(D39A) upon Addition of HscB
To probe the effect of HscB on IscU, we collected a series of 2D 15N-TROSY-HSQC spectra of samples containing different molar ratios of [U-15N]apo-IscU(D39A) and HscB. Peaks corresponding to residues K6–D9, R15, D21, N22, D24, N26–S29, A39, K46–D49, G51–D55, R57, F58, S69, L72, V73, E75–A85, D92, E95, L97–L99, L109, D120–K128 showed only minor chemical shift perturbations (< 0.04 ppm; Figure 4A), suggesting that HscB has minimal effect on these residues. Peaks corresponding to N16–F20, G38, M41, I45, A56, S70, S71, T74, Q86, T91, I93, A94, E96, V102–I104, S107, I108, A116, A119 showed larger chemical shift perturbations (> 0.04 ppm; Figure 4A), indicative of a more pronounced environmental change in the presence of HscB. A fraction of peaks, corresponding to residues E25, G30–A34, C37, V40, K42–Q44, K59, G62, C63, I88–N90, C106, E111–A113, K115, A117, and I118, showed only minor chemical shift perturbations before broadening beyond detection at substoichiometric amounts of HscB (Figure 4A). These peaks disappeared from the HSQC spectrum at an HscB:IscU ratio of ∼0.4.
Figure 4.
Chemical shift perturbations (Δδhn) plotted as a function of the IscU peptide sequence of (A) apo-IscU(D39A) upon the addition of 2 equivalents HscB and (B) apo-IscU upon the addition of 2.2 equivalents of HscB. Blank regions correspond to residues whose signals were not observed or not assigned. Red triangles indicate residues for which chemical shift perturbation could not be determined, either because the signals broadened out completely or shifted discontinuously to unknown locations. Secondary structural elements predicted from the assigned chemical shifts are indicated at the top of the figure.
To exclude the possibility that the observed chemical shift perturbations are caused by a nonspecific protein-protein interaction, we collected 2D 15N-TROSY-HSQC spectra of [U-15N]apo-IscU(D39A) mixed with various amounts of HscB(L92A,M93A,F153A). This HscB mutant was shown previously to be defective in achieving maximal stimulation of HscA ATPase activity in the presence of IscU, and also increased the concentration of IscU required for half-maximal stimulation from ≅ 3 to 24 µM (22). Indeed, not only did we observe no line broadening effect in the spectrum of [U-15N]apo-IscU(D39A) mixed with 3 equivalents of HscB(L92A,M93A,F153A), but chemical shift changes were also negligible (data not shown). Based on these results, we conclude that the observed chemical shift perturbation and line broadening effects noted earlier arise from a specific interaction between apo-IscU(D39A) and HscB.
Resonance Assignments of Apo-IscU Complexed with HscB
To ascertain whether apo-IscU(D39A) and apo-IscU behave similarly when interacting with HscB, we also conducted a titration experiment using [U-15N]apo-IscU and HscB. Although we were unable to achieve complete resonance assignments for apo-IscU, because of severe spectral overlaps and ambiguities arising from the presence of two distinct conformations, we could assign some signals by comparing their positions with peaks from apo-IscU(D39A). We were then able to partially extend these assignments with additional NMR experiments. The chemical shift perturbation pattern observed for the assignable resonances of apo-IscU in the presence of 2.2 equivalents of HscB was similar to that obtained for apo-IscU(D39A) (Figure 4B). Backbone amide signals from residues N16, G18, F20, D24, E25, S29, I45, E54, R57, V73, T74, W76, V77, A85-A87, N90, E96, L97, V102-I104, I108, I114, and A116 exhibited shifts > 0.036 ppm while those from G30–A34, L43, Q44, K59, C106, S107, A110, E111, and A113 suffered extreme line-broadening.
Discussion
The Isc system is an important and complex multi-protein system. Its importance is linked to its highly-conserved, ‘house-keeping’ Fe-S cluster assembly and transfer activity, while its complexity derives from the involvement of several proteins (CyaY, IscR, IscS, IscU, IscA, HscB, HscA and ferredoxins) and the challenges of determining the mechanisms for Fe-S cluster assembly and transfer (2). In these mechanisms, an understanding of the versatile and dynamic functions of IscU has remained particularly elusive. Previous studies suggested that IscU, as a scaffold protein in the ‘generalized’ Fe-S cluster biosynthesis system, needs to have considerable flexibility in order to be capable of transferring Fe-S clusters efficiently to a wide range of apo-proteins (17, 18); on the other hand, IscU needs to have a stable structural fold in order to maintain meaningful interactions with various protein components of the Isc system (e.g. IscS, HscB, and HscA). The solution structures of the non-physiological, Zn-bound forms of IscU from H. influenzae (15) and M. musculus (PDB 1wfz) gave static pictures of the protein structure and did not explain the reported versatility of IscU. Rather, the recent crystal structure of holo-IscU from A. aeolicus (16) and the solution structural study of T. maritima apo-SufU (17, 18), a homolog of IscU, provided consistent evidence for structural flexibility.
The present study of E. coli apo-IscU gives more direct evidence for the dynamic nature of IscU. Our NMR results indicate that IscU in solution populates two major conformations that interchange slowly. One conformation is largely disordered, and the other is largely structured except for the metal-binding regions, which remain dynamically disordered such that their NMR signals were not observed. From this observation arises an intriguing hypothesis that two different conformations of IscU may serve different functions. It would be reasonable to speculate that the ordered conformation is appropriate to scaffold the Fe-S cluster and that the disordered conformation serves to facilitate cluster transfer.
We also found that both the binding of zinc ion and the D39A mutation shift the equilibrium toward the structured conformation. IscU(D39A) has been found to reconstitute a more stable holo-complex than IscU (5–8), but with a decreased rate of cluster transfer to apo-ferredoxin (7, 9). It was suggested that the stabilization of the Fe-S cluster assembled on IscU(D39A) was a consequence of its being less solvent-accessible than that on holo-IscU (9, 10). Our results suggest that the increased stability may result from the fact that the D39A mutation energetically stabilizes the structured state of IscU that is required for the holo-complex. As discussed above, it is tempting to speculate that the slow step of the cluster transfer reaction is the opening up of IscU to allow release of the cluster, and that this opening is accelerated by an order-disorder transition in IscU. Because the order-disorder transition is diminished in IscU(D39A), this could explain the lower transfer rate observed with this mutant (7, 9).
Signals showing exchange cross-peaks in the exchange spectrum of apo-IscU are localized primarily to helices α3 and α4. Our results indicate that helices α3 and α4 undergo conformational exchange between an ordered secondary structure and a random coil-like conformation (Figure 2). By contrast, the neighboring region, including helix α2 and the adjacent loop containing the 99LPPVK103 motif, are only slightly affected by the presence of HscB, suggesting that this region remains flexible and accessible after formation of the IscU . HscB complex. Thus the 99LPPVK103 motif of IscU, which is known to bind HscA (20, 21), appears to be accessible. The crystal structure of the complex between HscA and ELPPVKIHC suggested, however, that IscU must undergo a conformational change in order to form the complex (19). This result raises the possibility that the order-disorder transition of helices α3 and α4 of IscU, which are adjacent to the binding motif, is somehow coupled to the binding interaction with HscA.
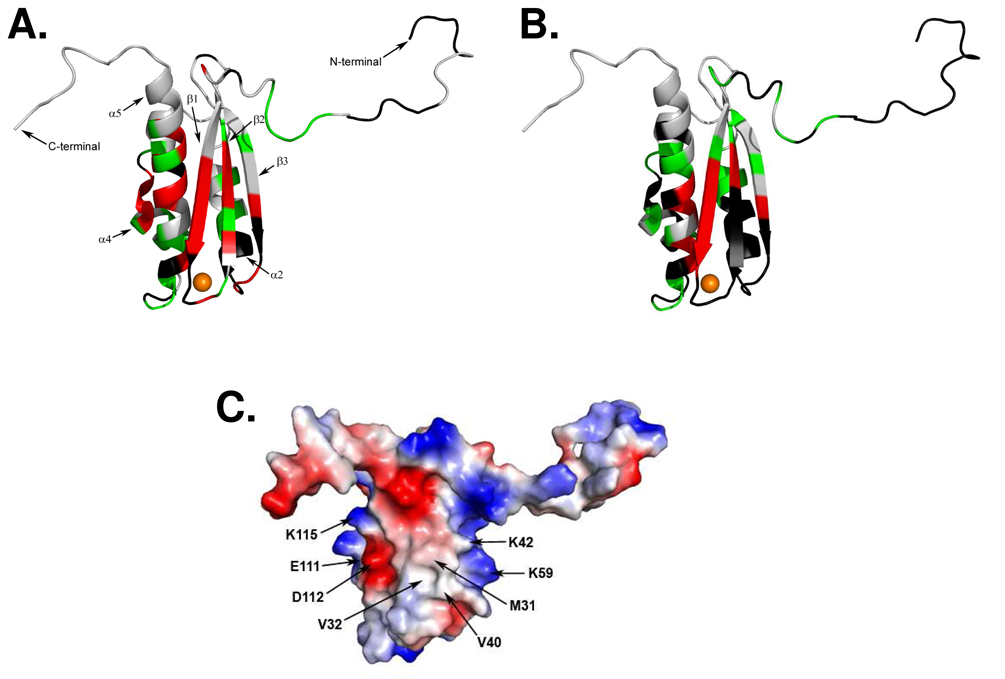
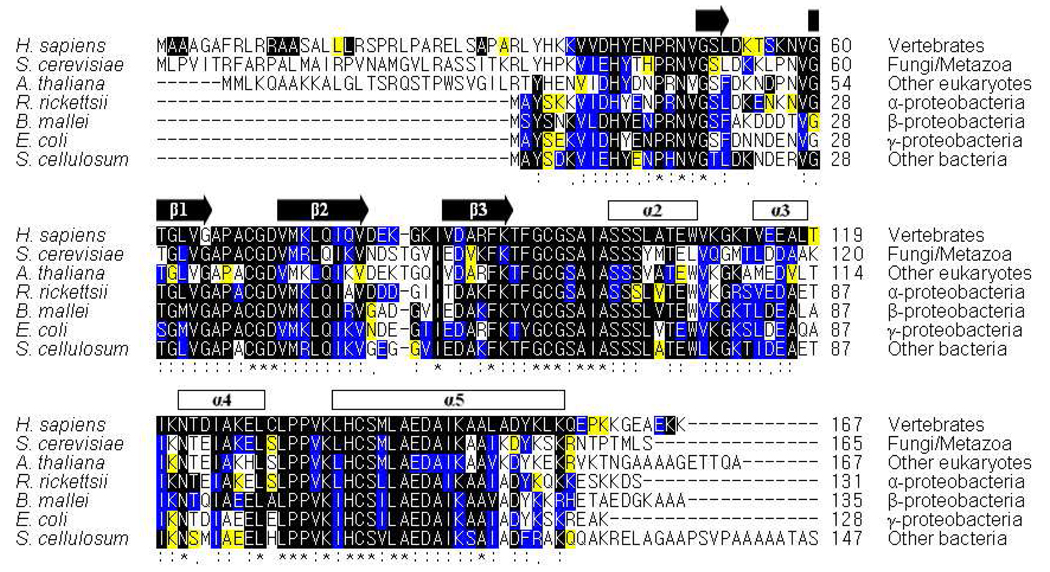
Our NMR results indicate that the residues of apo-IscU(D39A) most perturbed in the presence of HscB are localized primarily in the first two β-strands (β1 and β2) and the last α-helix (α5) of the scaffold protein (Figure 5A). The same pattern of perturbed residues is seen also for the structured form of apo-IscU (Figure 5B), although fewer assignments are available. A comprehensive sequence analysis of IscU homologs shows that these regions are highly conserved from proteobacteria to vertebrates (Figure 6).
Figure 5.
Putative surface of apo-IscU that interacts with HscB. (A) Chemical shift perturbations of apo-IscU(D39A) resulting from the addition of HscB mapped onto the structure of H. influenzae Zn-IscU [PDB 1r9p] (15). Red indicates residues whose signals broadened beyond detection; green indicates residues with large (Δδhn> 0.04 ppm) chemical shifts changes; and black indicates residues whose signals could not be assigned or followed during the titration. The zinc atom is colored orange. (B) Chemical shift perturbations of apo-IscU resulting from the addition of HscB mapped onto the structure of H. influenzae Zn-IscU (15). Red indicates residues whose signals broadened beyond detection; green indicates residues with large (Δδhn> 0.036 ppm) chemical shifts changes; and black indicates residues whose signals could not be assigned or followed during the titration. (C) Surface electrostatic potential of H. influenzae Zn-bound IscU. Neutral potential is shaded white, negative surface potential is colored red, and positive surface potential is colored blue. Residues proposed to be involved in the binding interaction with HscB are labeled.
Figure 6.
Sequence alignment of IscU homologs. A total of 131 aligned sequences were divided into 7 groups: 9 sequences of vertebrates, 20 of fungi/metazoa, 11 of other eukaryotes, 11 of α-proteobacteria, 28 of β-proteobacteria, 46 of γ-proteobacteria, and 6 of other prokaryotes. For simplicity, the figure shows representative aligned sequences from each of the 7 groups. Sequence conservation within each group is represented by three colors: ‘black’ indicates total conservation within the group; ‘blue’ indicates only conserved substitutions within the group; and ‘yellow’ indicates only semi-conserved substitutions within the group. The level of global conservation in the 131 sequences is represented by three symbols: ‘*’ indicates residues identically conservation in all sequences; ‘:’ indicates >90% similarity in all sequences; and ‘.’ indicates >80% similarity in all sequences. The secondary structural elements predicted from the present study of E. coli IscU(D39A) are indicated at the top of the sequence alignment.
Our chemical shift perturbation results indicated that several hydrophobic residues in the first two β-strands of apo-IscU(D39A), namely M31, V32, V40, and L43, were highly perturbed upon addition of HscB (Figure 4A). Because the hydrophobic side chains of M31 and V40 are solvent exposed in the solution structure of H. influenzae Zn-IscU (Figure 5C), we speculate that these two residues are involved in a hydrophobic interaction in the IscU:HscB complex. The NMR signals of a number of highly conserved charged residues of apo-IscU were also perturbed in the presence of HscB. These include K42 and K59 in the β2/β3 strands, which are located at the boundary of the hydrophobic patch with their charged side chains exposed to solvent (Figure 5C), as well as residues E111, D112 and K115 in α5 (Figure 5C). Whereas M31 and V40 might provide stability to the complex, the charged residues may be important for conferring specificity and/or to strengthen the interaction. Consistent with these results, the proposed binding site of IscU on HscB includes a highly conserved, solvent-exposed hydrophobic patch as well as an acidic triad of glutamates (22).
The results (Figure 4) indicate that binding to HscB stabilizes the ordered state of apo-IscU. Signals from most of the residues that exhibited large chemical shift differences between the ordered and disordered forms of apo-IscU remained at their ordered positions [equivalent to those of apo-IscU(D39A) upon addition of HscB (Figure 2)].
A challenging aspect of the apo-IscU:HscB interaction is the excessive line broadening that affects a large number of HSQC cross peaks (Figure 4). Broadening occurs at substoichiometric concentrations of added HscB, and the peaks do not sharpen upon saturation of apo-IscU with bound HscB. For these reasons, the effect cannot be explained by simple association-dissociation kinetics. Explanation of the effect requires a dynamic structural rearrangement of the bound complex at a rate much faster that the dissociation rate for the complex. At substoichiometric concentrations of added HscB, signals from nuclei with appreciably different chemical shifts in the distinct bound states are saturated as a result of the interconversion, and this saturation is transferred to the pool of uncomplexed IscU. The rearrangement may involve shuffling of hydrophobic residues in the contact region between the two proteins or shifting of the complex between two (or more) orientations.
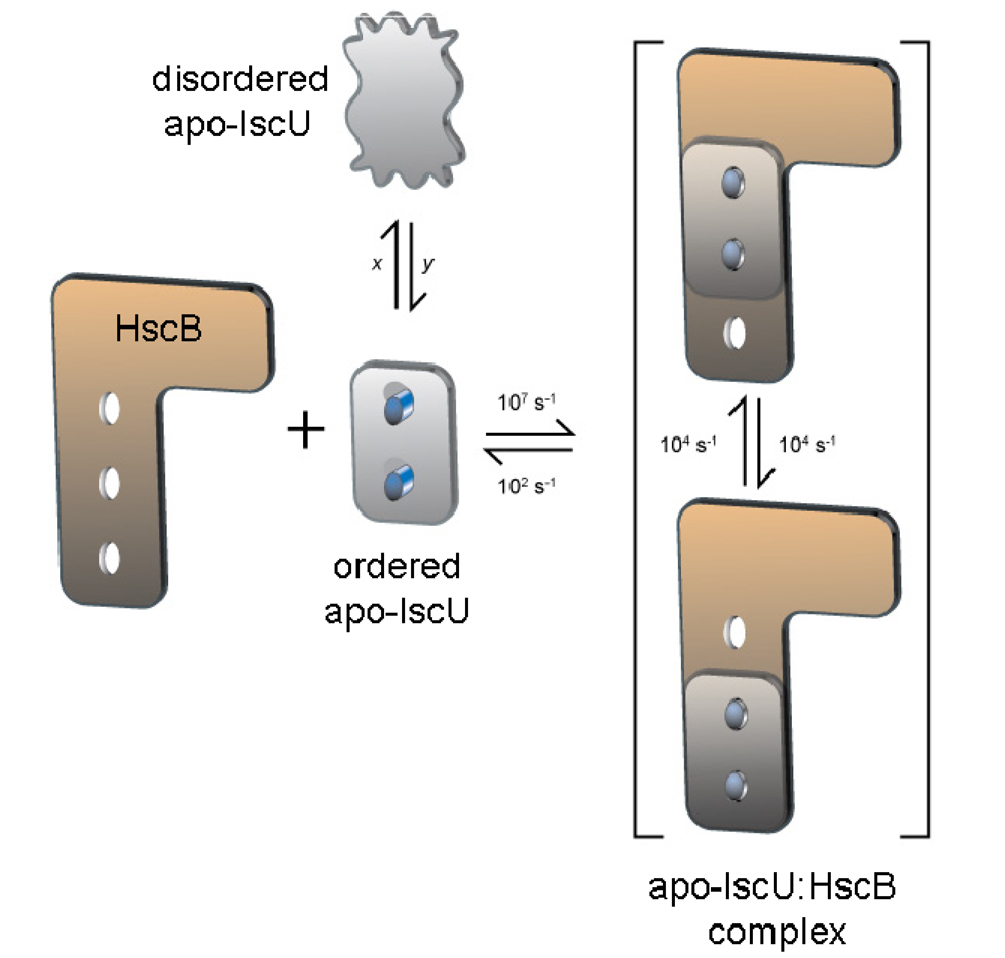
The schematic diagram presented in Figure 7 illustrates our finding that HscB binds to the ordered state of apo-IscU and depicts the simplest kind of rearrangement that would explain the substoichiometric broadening of NMR signals from apo-IscU reported here as well as those from HscB as reported earlier (22). The rates shown are approximations based on the known dissociation constant (≅10 µM), an assumed association rate of 107 s−1, and assumed chemical shift differences of about 0.5 ppm (300 / 400 s−1 for 1H, respectively at 600 / 800 MHz) for the affected peaks in the interconverting states. This model can be tested and the rates refined by future NMR experiments that we are planning to undertake.
Figure 7.
Cartoon depicting the dynamic interactions between apo-IscU and HscB that explain the extreme broadening of signals from IscU (HscB) upon the addition of substoichiometric amounts of HscB (IscU). Apo-IscU exists as an equilibrium between a disordered and ordered state. The rates x and y are of similar magnitude, because the two states have nearly equal populations. Individually, x and y must be slower than 10 s−1 to explain the observation of separate signals for the disordered and ordered state, but faster than 0.3 s−1 to account for the observation of 2D exchange peaks. The ordered state of apo-IscU binds to HscB, but the complex formed undergoes a dynamic rearrangement that is faster than the off-rate for dissociation of the complex. The change occurring within the complex could involve the relative displacement of the two proteins, as depicted here, or a rearrangement of hydrophobic groups in the contact area without a relative displacement. The NMR signals of IscU and HscB that become excessively broadened upon complex formation must have appreciable chemical shift differences in the two bound states. The rates shown are crude estimates (see text).
Supplementary Material
Abbreviations
- DTT
dithiothreitol
- DSS
2,2-dimethyl-2-silapentane-5-sulfonic acid
- HscB/A
heat shock cognate protein B/A
- Hsp
heat shock protein
- HSQC
heteronuclear single quantum coherence
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- IscR/S/U/A
iron-sulfur cluster protein R/S/U/A
- IscU
unless noted otherwise, IscU from Escherichia coli
- IscU(D39A)
IscU mutant in which aspartate at residue 39 is replaced by alanine)
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- PECAN
protein energetic conformational analysis from NMR chemical shifts
- SBD
substrate binding domain
- TROSY
transverse relaxation optimized spectroscopy
- [U-15N]
uniformly 15N-labeled
Footnotes
This work was supported by NIH grant R01 GM58667 (JLM) and GM 54624 (LEV). NMR data were collected at the National Magnetic Resonance Facility at Madison (NMRFAM) with support from NIH grants P41 RR02301 (JLM) and P41 GM66326 (JLM).
1H, 13C, and 15N resonance assignments and 15N R1, R2, and heteronuclear NOE values of apo-IscU(D39A) were deposited to the BioMagResBank as entry 7432. 1H and 15N resonance assignments of IscU(D39A) in the complex with HscB were deposited as entry 15967.
Supporting Information Available
Table S1 lists the chemical shifts of exchange cross peaks observed in the 2D exchange spectrum of [U-15N]apo-IscU. Figure S1 provides evidence that IscU(D39A) forms a 1:1 complex with HscB with binding affinity ∼10 µM and thermodynamic parameters similar to those observed with wild-type IscU. Figure S2 presents assay results showing that apo-IscU(D39A) acts synergistically with HscB to stimulate HscA ATPase activity in a manner similar to that observed for wild-type IscU. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 3.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 4.Bonomi F, Iametti S, Ta DT, Vickery LE. Multiple turnover transfer of [2Fe2S] clusters by the iron-sulfur cluster assembly scaffold proteins IscU and IscA. J. Biol. Chem. 2005;280:29513–29518. doi: 10.1074/jbc.M504344200. [DOI] [PubMed] [Google Scholar]
- 5.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 6.Shimomura Y, Kamikubo H, Nishi Y, Masako T, Kataoka M, Kobayashi Y, Fukuyama K, Takahashi Y. Characterization and crystallization of an IscU-type scaffold protein with bound [2Fe-2S] cluster from the hyperthermophile. Aquifex aeolicus, J. Biochem. (Tokyo) 2007;142:577–586. doi: 10.1093/jb/mvm163. [DOI] [PubMed] [Google Scholar]
- 7.Wu G, Mansy SS, Wu SP, Surerus KK, Foster MW, Cowan JA. Characterization of an iron-sulfur cluster assembly protein (ISU1) from Schizosaccharomyces pombe. Biochemistry. 2002;41:5024–5032. doi: 10.1021/bi016073s. [DOI] [PubMed] [Google Scholar]
- 8.Foster MW, Mansy SS, Hwang J, Penner-Hahn JE, Surerus KK, Cowan JA. A mutant human IscU protein contains a stable [2Fe-2S]2+ center of possible functional significance. J. Am. Chem. Soc. 2000;122:6805–6806. [Google Scholar]
- 9.Wu SP, Wu G, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 10.Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis: toward an understanding of cellular machinery and molecular mechanism. Acc. Chem. Res. 2004;37:719–725. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- 11.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47:12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 13.Silberg JJ, Hoff KG, Tapley TL, Vickery LE. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem. 2001;276:1696–1700. doi: 10.1074/jbc.M009542200. [DOI] [PubMed] [Google Scholar]
- 14.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J. Mol. Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura Y, Wada K, Fukuyama K, Takahashi Y. The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Bertini I, Cowan JA, Del Bianco, C., Luchinat C, Mansy SS. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J. Mol. Biol. 2003;331:907–924. doi: 10.1016/s0022-2836(03)00768-x. [DOI] [PubMed] [Google Scholar]
- 18.Mansy SS, Wu SP, Cowan JA. Iron-sulfur cluster biosynthesis. Biochemical characterization of the conformational dynamics of Thermotoga maritima IscU and the relevance for cellular cluster assembly. J. Biol. Chem. 2004;279:10469–10475. doi: 10.1074/jbc.M312051200. [DOI] [PubMed] [Google Scholar]
- 19.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J. Mol. Biol. 2004;342:1265–1278. doi: 10.1016/j.jmb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 20.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J. Biol. Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 21.Hoff KG, Cupp-Vickery JR, Vickery LE. Contribution of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J. Biol. Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
- 22.Füzéry AK, Tonelli M, Ta DT, Cornilescu G, Vickery LE, Markley JL. Solution structure of the iron-sulfur cluster cochaperone HscB and its binding surface for the iron-sulfur assembly scaffold protein IscU. Biochemistry. 2008;47:9394–9404. doi: 10.1021/bi800502r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones-Mortimer MC. Positive control of sulphate reduction in Escherichia coli. Isolation, characterization and mapping of cysteineless mutants of E. coli K12. Biochem. J. 1968;110:589–595. doi: 10.1042/bj1100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods. Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- 25.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 26.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2008. [Google Scholar]
- 27.Wider G, Neri D, Wüthrich K. Studies of slow conformational equilibria in macromolecules by exchange of heteronuclear longitudinal 2-spin-order in a 2D difference correlation experiment. J. Biomol. NMR. 1991;1:93–98. [Google Scholar]
- 28.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 29.Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J. Biomol. NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Eghbalnia HR, Markley JL. Probabilistic approach to determining unbiased random-coil carbon-13 chemical shift values from the protein chemical shift database. J. Biomol. NMR. 2006;35:155–165. doi: 10.1007/s10858-006-9022-0. [DOI] [PubMed] [Google Scholar]
- 32.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 33.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.