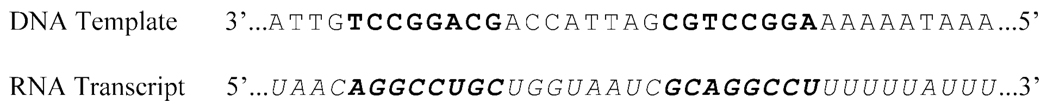Abstract
Nuclear magnetic resonance spectroscopy and proton exchange have been used to characterize two RNA-DNA hybrids from the tR2 intrinsic transcription terminator site of phage λ. The hybrids have the same base sequence: 5’-GGCGCAGGCC(T/U)(T/U)CC-3’/5’-GGAAGGCC(T/U)GCGCC-3’, but differ from each other by an interchange of DNA and RNA strands. The opening of single base pairs in the two hybrids is characterized by measuring the rates of exchange of imino protons with solvent protons as a function of the concentration of a proton acceptor (ammonia base) at 10 °C. The free energy change in the opening reaction provides a measure of the stability of the base pair, while the rates of opening and closing define the base-pair dynamics. The results demonstrate that, within the same base sequence context, dA-rU base pairs are less stable than dT-rA base pairs. The differences in stability are enhanced when two dA-rU base pairs are located next to each other in the hybrid structure. For the G-C base pairs, the rates of opening and closing, and the stability are affected by the base sequence context and by the nature of the sugar moiety attached to the guanine. The dominant feature of the base sequence is the proximity of the dA-rU base pair, which destabilizes the G-C base pair when the guanine is located on the DNA strand. Two G-C base pairs (namely, those in the 4th and 10th positions) exhibit large differences in their opening and closing rates between the two hybrids, while maintaining the same stability. These results provide the first demonstration that, for RNA-DNA hybrid structures with the same base sequence, the opening dynamics and the stability of individual base pairs is strongly influenced by the chemical nature of each strand.
RNA-DNA hybrid structures play important roles in the transmission and processing of biological information (1). The hybrid between template DNA and messenger RNA is the key intermediate in the expression of genetic information. Hybrid structures between RNA and DNA strands serve as initiation sites for the synthesis of Okazaki fragments in DNA replication and as sites for the elongation of telomeric repeats by telomerase. The complementarities between bases in the RNA-DNA hybridization reaction is also the basis for a variety of applications, such as identification of specific of sites genomes by in situ hybridization, the antisense technology, and microarray gene expression profiling.
The numerous biological roles of RNA-DNA hybrids have prompted great interest in characterization of the structures and the physical properties of these duplexes. Crystallographic, spectroscopic and molecular dynamics studies have demonstrated that the structures of hybrids encompass a large domain in the conformational space, close to the canonical A-form, but often incorporating features of the B-form (2–11). Thermodynamic analyses have revealed that the stability of hybrid structures is strongly influenced by the base sequence (12–14). Furthermore, the stability depends on the nature of the bases in each strand. This property has been first demonstrated for RNA-DNA hybrid duplexes with polypurine and polypyrimidine strands: the hybrid with a polypurine DNA strand is significantly less stable than the hybrid with a polypurine RNA strand (15–17). Molecular dynamics simulations have shown that the nature of each strand also affects the fluctuations of the bases and the phosphodiester backbone: the fluctuations are enhanced when the polypurine sequence is present in the DNA strand (11). Together, these strand-specific alterations in stability and molecular flexibility may contribute to the recognition and cleavage of RNA-DNA hybrids by RNase H (10, 11).
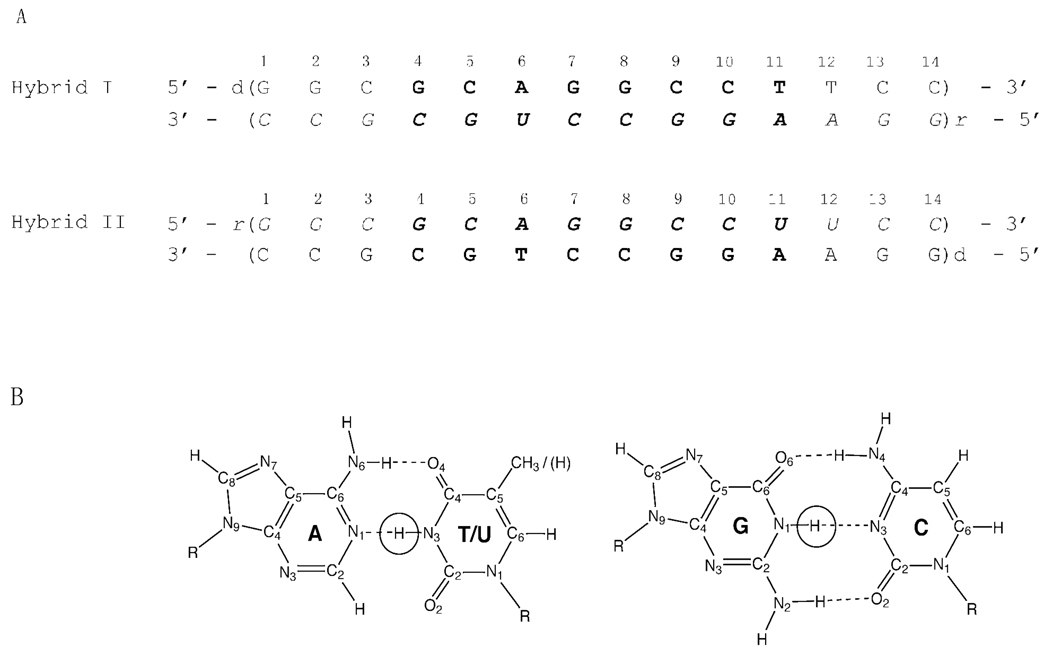
The work presented in this paper addresses the question of the stability and dynamics of individual base pairs in RNA-DNA hybrids of mixed, but homologous, base sequences. The two hybrids investigated correspond to RNA-DNA hybrids formed during transcription at the intrinsic transcription termination site tR2 of phage λ (18). Intrinsic transcription terminators are sites on bacterial genomes where the nascent RNA transcript is spontaneously released from the DNA template, thereby ending transcription. Termination of transcription at these sites does not require any auxiliary proteins (e.g., rho protein). Instead, the process relies upon structural and energetic signals encoded in the DNA template, the RNA transcript and the RNA-DNA hybrids formed during transcription. The tR2 site of phage λ has been a reference model system for investigations of intrinsic transcription termination and its molecular mechanisms (18–21). This site contains two structural elements that are conserved in all intrinsic transcription terminators: two G/C-rich sequences with dyadic symmetry, and an A-rich tract where termination of transcription occurs (Figure 1). The RNA-DNA hybrids that form when the two G/C-rich sequences are transcribed are homologous to each other, i.e., the base sequence of the RNA transcript in the upstream hybrid is the same as that of the DNA template in the downstream hybrid, and the base sequence of the RNA transcript in the downstream hybrid is the same as that of the DNA template in the upstream hybrid (with thymine in DNA and uracil in RNA). The RNA-DNA hybrids investigated in the present work (Figure 2A) reproduce these two hybrids: the base pairs in their central positions (4th to 11th) correspond to the hybrids formed by the G/C-rich sequences at the tR2 transcription termination site. Two and three G-C/C-G base pairs are added at the ends of each hybrid to minimize the effects of fraying upon the central base pairs of interest. The base sequences and the design of the two hybrids allows us to define how the interchange of RNA and DNA strands affects the dynamic and energetic properties of individual base pairs in the same base sequence context.
Figure 1.
Base sequences of the DNA template and the messenger RNA transcript in the tR2 intrinsic transcription termination site of phage λ. The bases in the two GC-rich dyadic sequences are shown in bold.
Figure 2.
A. The base sequences and numbering of base pairs in the RNA-DNA hybrids investigated in the present work. The DNA strands are in regular fonts and the RNA strands are in italic fonts. The base pairs from the two GC-rich dyadic sequences are shown in bold. In hybrid I, the base pairs are numbered in the 5’->3’ direction of the DNA strand. To emphasize the similarity in base sequence, in hybrid II, the base pairs are numbered in the opposite direction (5’->3’ in the RNA strand).
B. The structures of the G-C and A-T/A-U base pairs. The imino proton in each base pair is circled.
EXPERIMENTAL PROCEDURE
Materials
The DNA strands were synthesized on an automated DNA synthesizer (Applied Biosystems) using the solid-support phosphoramidite method. The DNA was purified by HPLC on a semi-preparative column (Hamilton PRP-1) in 50 mM triethylamine acetate buffer with a gradient of 5–32% acetonitrile over 46 minutes at 60 °C. The RNA strands were synthesized by in vitro transcription using T7 RNA polymerase (22). 15N-GTP was purchased from Spectra Stable Isotopes (Columbia, MD). The RNA was purified by electrophoresis on a 20% polyacrylamide gel. For both DNA and RNA strands the counterions were replaced with Na+ ions by repeated centrifugation in Centricon YM-3 tubes (Millipore) using 0.5 M NaCl, followed by centrifugation against water. The final NMR samples contained 0.5 to 1 mM RNA-DNA duplex in 100 mM NaCl and 0.5 mM EDTA in 90%H2O/10%D2O at pH 8.3±0.1 (at 10 °C). All samples also contained 1 mM triethanolamine, which was used to determine the pH of the samples directly in the NMR tube as we have described (23).
Methods
NMR Experiments
The NMR experiments were performed at 10 °C on a Varian INOVA 500 spectrometer operating at 11.75 T. The exchange rates of imino protons were measured by transfer of magnetization from water. The water proton resonance was selectively inverted using a Gaussian 180° pulse (5.8 ms) followed by a variable delay for the exchange of magnetization between water and imino protons. A gradient of 0.21 G/cm was applied during the exchange delay to prevent the effects of radiation damping upon the recovery of water magnetization to equilibrium. A second selective pulse on water was applied to bring the water magnetization back onto the z-axis before observation. Twenty-five values of the exchange delay in the range from 1 to 800 ms were used in each experiment. The exchange rates were calculated from the dependence of the intensity of the imino proton resonance on the exchange delay as we described (24). For the unlabeled samples the observation was with the Jump-and-Return pulse sequence (25). For the 15N-labeled samples the observation was with the one-dimensional (1D) version of the fast HSQC (fHSQC) pulse sequence (26) to edit the resonances of protons attached to 15N. A modified fHSQC pulse sequence (27) was used to filter out the resonances of protons attached to 15N and retain all other proton resonances. The highest exchange rates that can be accurately measured by these methods is ~ 70–80 s−1. The 1H-1H NOESY spectra were obtained using the Watergate pulse sequence (28) with a mixing time of 250 ms.
Optical Melting
The optical melting curves were obtained on a Beckman DU650 spectrophotometer at 260 nm. The concentration of each RNA-DNA hybrid was 1.5 µM in 100 mM NaCl.
Imino Proton Exchange in Nucleic Acid Duplexes
The exchange of imino protons in nucleic acids is a two-step process. In the first step, the base containing the imino proton (Figure 2B) flips out of the structure into an open state in which the barriers to exchange are removed. In this open state, the imino proton is accessible to proton acceptors, and the hydrogen bond in which the proton participates breaks. The second step is the actual transfer of the proton to an acceptor, such as OH− and NH3. The exchange rate observed experimentally is given as (24, 29):
| (1) |
where kop and kcl are the rates of opening and closing, respectively, of the base containing the imino proton, and kex,open is the rate of exchange from the open state. The rates of opening and closing define the equilibrium constant of the opening reaction:
| (2) |
This equilibrium constant is related to the free energy change in the opening reaction by
| (3) |
where T is the absolute temperature and R is the universal gas constant.
The rate of exchange from the open state depends on the concentration of proton acceptor B as:
| (4) |
where kB is the rate constant for the transfer of the imino proton to proton acceptor B in isolated nucleotides, and α is a factor that accounts for differences in proton transfer rate between isolated nucleotides and open base pairs. Previous studies have shown that, for nucleic acid duplexes, α has a value close to unity (30). The rate constant kB is calculated from the pKa value of the imino group (pKNH) and the pKa of the proton acceptor (pKB) as (31):
| (5) |
where kcoll is the rate of diffusion-controlled collision between the imino group and the proton acceptor in the open state of the base pair.
The equation describing the dependence of the exchange rate observed experimentally on the concentration of proton acceptor is obtained by inserting Eq. 4 in Eq.1 (with αα=1):
| (6) |
Two kinetic regimes for imino proton exchange can be distinguished depending on how the rate of exchange from the open state compares with the rate of closing. When the concentration of proton acceptor is high enough to make the exchange from the open state very fast ( kex,open ≫kcl , EX1 regime) the exchange is limited by the rate of base opening. In this case, Eq. 6 becomes:
| (7) |
At low concentrations of proton acceptor, the rate of exchange from the open state is much smaller than the rate of base-pair closing ( kex,open ≪ kcl , EX2 regime). In this regime, the observed exchange rate depends linearly on the concentration of proton acceptor:
| (8) |
In the present work we have used ammonia base (NH3) as the imino proton acceptor in the exchange. Previous work from this and other laboratories has shown that, due to its small size and its lack of charge, ammonia base is the acceptor of choice for proton exchange studies of nucleic acids (24, 29). The rate constant for proton transfer to NH3 was calculated from Eq. 5 as 8.8×108 M−1 s−1 for both guanine and uracil, and 4.1×108 M−1 s−1 for thymine at 10 °C. The concentration of ammonia base NH3 was calculated from the total ammonia concentration C0 and the pH as:
| (9) |
The pH was measured at each ammonia concentration, directly in the NMR tube, using the proton resonances of triethanolamine (23). The pK value of ammonia at 10 °C is 9.73 (32).
RESULTS
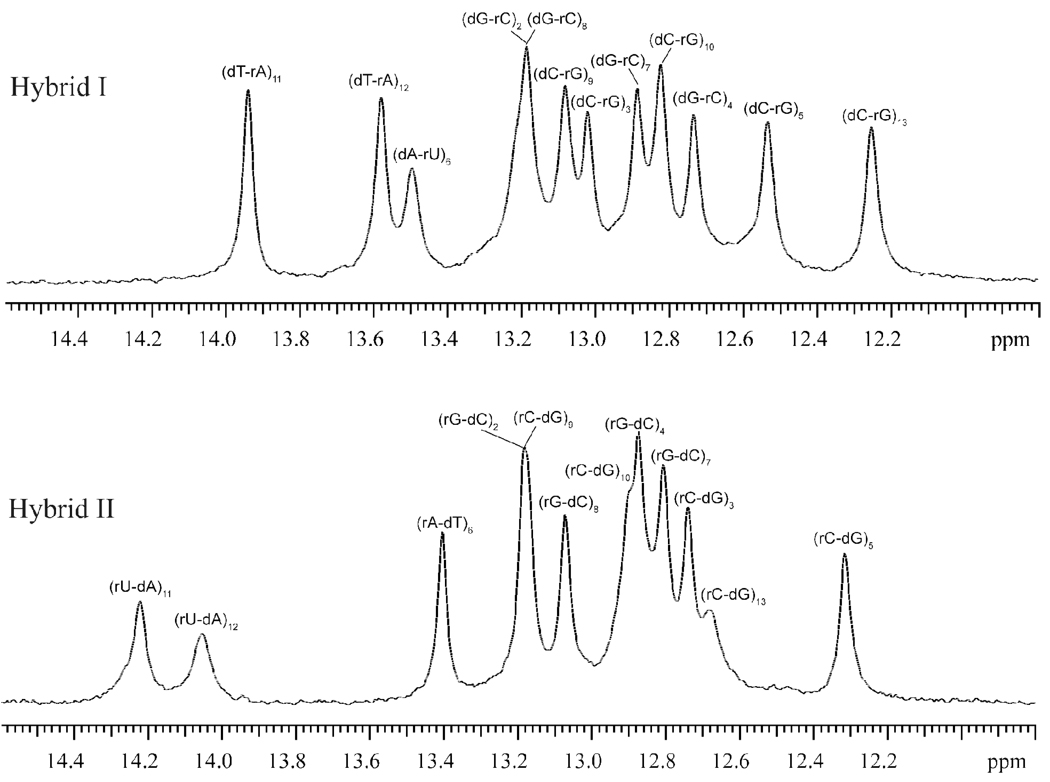
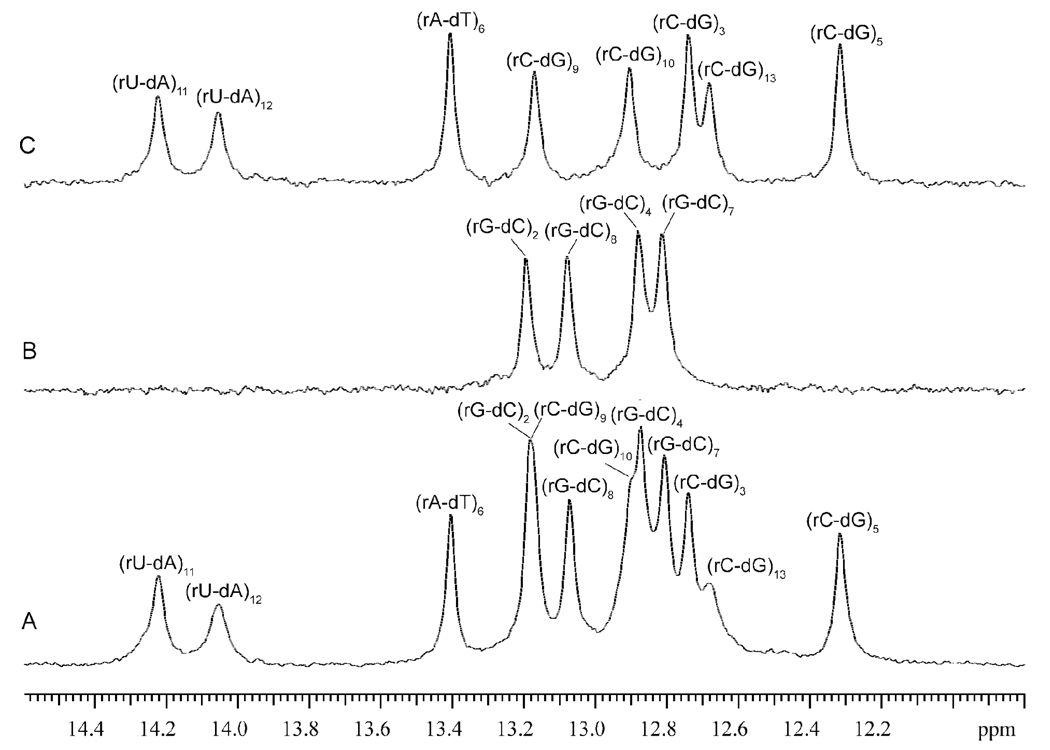
Our characterization of base-pair dynamics and energetics in the two RNA-DNA hybrids relies upon the exchange of imino protons with solvent protons. The location of imino protons in the structures of the base pairs is shown in Figure 2B. The NMR resonances of these protons in the two RNA-DNA duplexes investigated are shown in Figure 3. The resonances were assigned to specific protons using 15N-editing and 1H-1H NOESY, as detailed in Supporting Information. In the NMR spectrum of hybrid II, four imino proton resonances overlap: the resonance from the (rG-dC)2 base pair overlaps that from the (rC-dG)9 base pair, and the resonance from the (rG-dC)4 base pair overlaps that from the (rC-dG)10 base pair. To remove this overlap and observe each resonance separately we have used a sample of hybrid II in which the guanines in the RNA strand were labeled with 15N. 15N-editing of the spectrum (panel B in Figure 4) allowed specific observation of the imino proton resonances of RNA guanines, including those from (rG-dC)2 and (rG-dC)4. 14N-editing of the same spectrum (panel C in Figure 4) gave the imino proton resonances from DNA guanines [including those from (rC-dG)9 and (rC-dG)10] as well as those from thymines and uracils. In the NMR spectrum of hybrid I (Figure 3), the imino proton resonance from (dG-rC)2 overlaps that from (dG-rC)8. This overlap cannot be removed by the 15N-/14N-editing procedure used for hybrid II because, in hybrid I, the guanines in positions 2 and 8 belong to the same strand (i.e., DNA strand).
Figure 3.
The NMR resonances of imino protons in the RNA-DNA hybrids investigated. Solvent conditions: 100 mM NaCl, 0.5 mM EDTA and 1 mM triethanolamine at pH 8.3±0.1 and at 10 °C.
Figure 4.
15N-/14N-editing of the NMR resonances of imino protons for a sample of hybrid II in which the RNA strand was labeled with 15N at guanines. (A) All imino proton resonances. (B) 15N-editing: the observed resonances originate from guanine imino protons in the RNA strand. (C) 14N-editing: the observed resonances originate from imino protons of uracils/thymines and of guanines in the DNA strand.
The rates of exchange of imino protons in the two hybrids were measured as a function of the concentration of ammonia (from 0 to 600 mM total ammonia concentration). Increasing ammonia concentration does not affect the conformation of the two RNA-DNA hybrids, as shown in Supporting Information. The dependence of the imino proton exchange rates on the concentration of ammonia base is shown in Figure 5 and Figure 6. Figure 5 does not include the G-C base pairs in positions 2 and 8 because, as explained above, the imino proton resonances of these base pairs in hybrid I are not resolved in the NMR spectra. For the terminal G-C base pairs (i.e., positions 1 and 14), the imino proton resonances are broadened beyond detection (Figure 3) by fraying at the ends of the duplex structures.
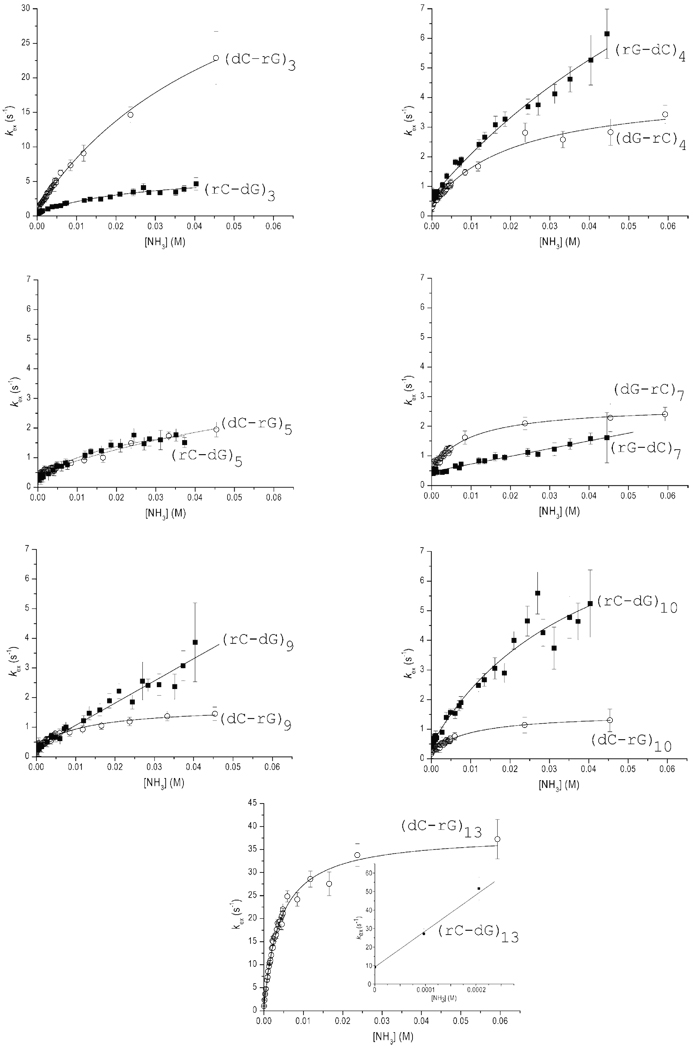
Figure 5.
Dependence of the exchange rates of selected guanine imino protons in the RNA-DNA hybrids investigated on the concentration of ammonia base at 10 °C. Empty circles represent exchange rates in hybrid I. Solid squares represent exchange rates in hybrid II. For (rG-dC)7, (rG-dC)9 and (rC-dG)13 in hybrid II the lines represent least-squares fits to Eq. 8. For the other imino protons the curves represent nonlinear least-squares fits to Eq. 6.
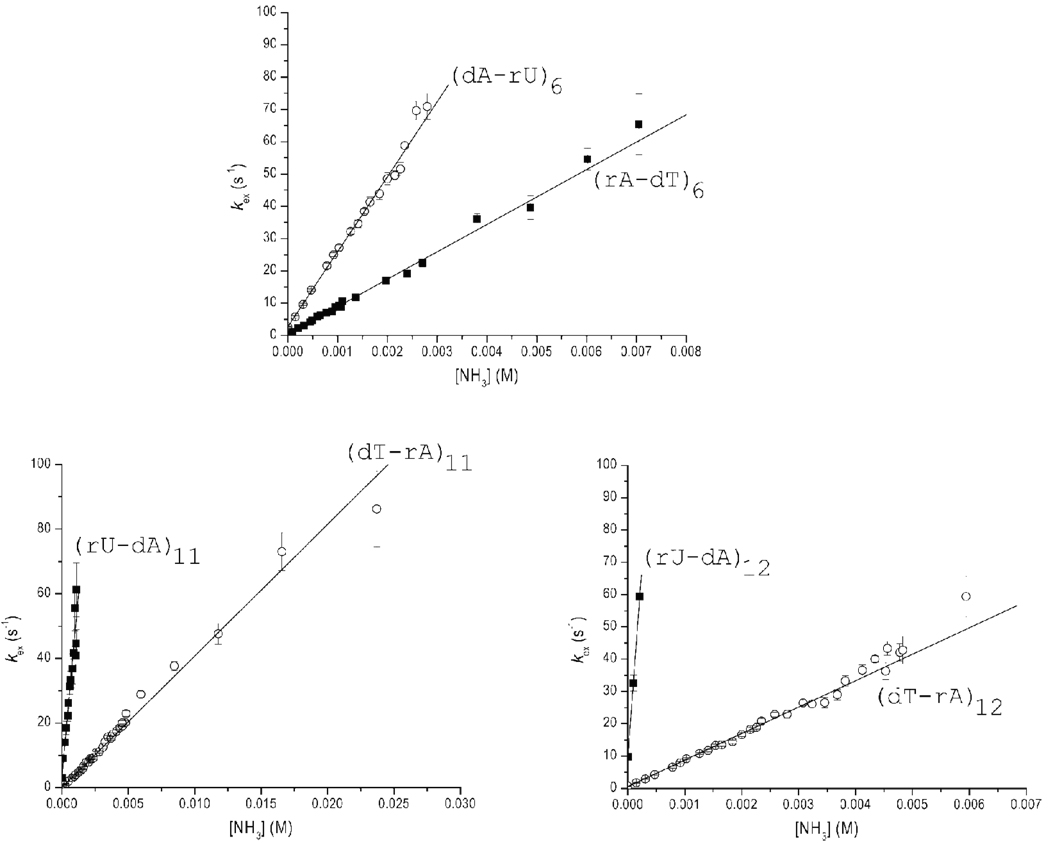
Figure 6.
Dependence of the exchange rates of thymine/uracil imino protons in the RNA-DNA hybrids investigated on the concentration of ammonia base at 10 °C. Empty circles represent exchange rates in hybrid I. Solid squares represent exchange rates in hybrid II. The lines represent least-squares fits to Eq. 8.
The dependence of the imino proton exchange rates on ammonia base concentration was analyzed according to the exchange model presented in the Experimental Procedures section. The results of this analysis are summarized in Table 1. For several G-C base pairs, such as those in positions 3, 4, 5, and 10, the exchange at the highest ammonia concentration reaches, or closely approaches, the EX1 regime. For these base pairs, the exchange rates were fitted as a function of ammonia base concentration to Eq. 6. The fits provided the rates of opening and closing of each base pair (kop and kcl, respectively). The equilibrium constant of the opening reaction (Kop) was calculated from the rates based on Eq. 2. For other G-C base pairs the exchange did not reach the EX1 regime even at the highest ammonia concentration investigated, e.g., (rG-dC)7, (rG-dC)9 and (rG-dC)13 in hybrid II. Similarly, for the A-T/A-U base pairs the exchange did not reach the EX1 regime (Figure 6) because the exchange rates are very high even at low ammonia concentrations. As the ammonia concentration is increased, the exchange rates become higher than ~ 70–80 s−1, and cannot be measured by the NMR transfer of magnetization technique. When the EX1 regime was not reached, only the equilibrium constants of the opening reaction could be obtained by fitting the exchange rates as a function of ammonia base concentration to Eq. 8. The opening equilibrium constants for each base pair in the two hybrids are compared in Table 1 and Figure 7.
Table 1.
Base-Pair Opening Parameters in the RNA-DNA hybrids investigated at 10 °C
| Hybrid I | Hybrid II | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Base pair | kop (s−1) |
kcl (×10 −7 s−1) |
Kop (×106) |
ΔGop (kcal/mol) | Base pair | kop (s−1) |
kcl (×10 −7s−1) |
Kop (×106) |
ΔGop (kcal/mol) |
| (dG-rC)2 | c | c | c | c | (rG-dC)2 | 33.2 ± 1.4 | 0.65 ± 0.08 | 5.1 ± 0.6 | 6.85 ± 0.07 |
| (dC-rG)3 | 48 ± 2 | 4.8 ± 0.3 | 1.0 ± 0.1 | 7.77 ± 0.05 | (rC-dG)3 | 6.6 ± 0.7 | 2.4 ± 0.5 | 0.27 ± 0.06 | 8.50 ± 0.13 |
| (dG-rC)4 | 4.3 ± 0.3 | 1.8 ± 0.2 | 0.24 ± 0.03 | 8.59 ± 0.07 | (rG-dC)4 | 18.8 ± 3.9 | 10 ± 3 | 0.19 ± 0.06 | 8.71 ± 0.19 |
| (dC-rG)5 | 5.1 ± 1.4 | 8.1 ± 2.7 | 0.06 ± 0.02 | 9.33 ± 0.22 | (rC-dG)5 | 2.6 ± 0.2 | 2.0 ± 0.5 | 0.13 ± 0.03 | 8.92 ± 0.14 |
| (dA-rU)6 | a | a | 26.4 ± 0.3 | 5.93 ± 0.01 | (rA-dT)6 | a | a | 21.8 ± 0.4 | 6.04 ± 0.01 |
| (dG-rC)7 | 2.7 ± 0.1 | 0.9 ± 0.1 | 0.32 ± 0.03 | 8.42 ± 0.05 | (rG-dC)7 | b | b | 0.030 ± 0.002 | 9.75 ± 0.03 |
| (dG-rC)8 | c | c | c | c | (rG-dC)8 | b | b | 0.035 ± 0.002 | 9.65 ± 0.03 |
| (dC-rG)9 | 1.7 ± 0.1 | 1 ± 0.1 | 0.16 ± 0.02 | 8.79 ± 0.08 | (rC-dG)9 | b | b | 0.085 ± 0.003 | 9.16 ± 0.02 |
| (dC-rG)10 | 1.5 ± 0.1 | 0.9 ± 0.1 | 0.18 ± 0.02 | 8.74 ± 0.06 | (rC-dG)10 | 9.9 ± 1.8 | 3.5 ± 1.2 | 0.3 ± 0.1 | 8.49 ± 0.22 |
| (dT-rA)11 | a | a | 10.5 ± 0.1 | 6.45 ± 0.01 | (rU-dA)11 | a | a | 51 ± 2 | 5.56 ± 0.02 |
| (dT-rA)12 | a | a | 21.1 ± 0.2 | 6.06 ± 0.01 | (rU-dA)12 | a | a | 270 ± 3 | 4.62 ± 0.01 |
| (dC-rG)13 | 38.3 ± 0.8 | 0.37 ± 0.03 | 10.4 ± 0.8 | 6.46 ± 0.04 | (rC-dG)13 | a | a | 221 ± 13 | 4.74 ± 0.03 |
The exchange rates in the EX1 regime are too fast to be measured by NMR.
The EX1 regime could not be reached at the highest ammonia concentrations.
Imino proton resonance is not resolved in the NMR spectrum (Figure 3).
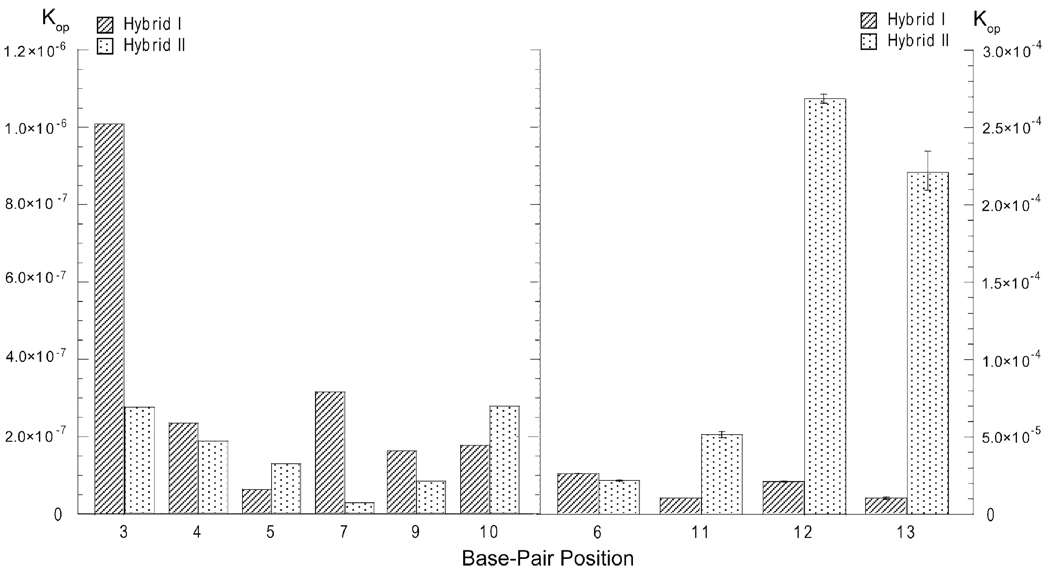
Figure 7.
Comparison of the equilibrium constants for base-pair opening in the two RNA-DNA hybrids investigated. The left panel shows the base pairs whose opening equilibrium constants are ≤ 1·10−6 (i.e., positions 3, 4, 5, 7, 9, and 10). The right panel shows the base pairs whose opening equilibrium constants are ≥ 5·10−6 (i.e., positions 6, 11, 12, and 13).
DISCUSSION
The results obtained in the present work show that the imino proton exchange rates for individual base pairs in hybrid I differ from those of the homologous base pairs in hybrid II. These differences depend on the nature of the base pair (e.g., A-T or A-U) as well as on the location of the base pair in the structure.
For the A-T/A-U base pairs (Figure 6), the exchange rates of uracil imino protons are consistently higher than those of thymine protons. At any concentration of ammonia base, the exchange rate for (dA-rU)6 is 2.6-fold higher than that for (rA-dT)6, the exchange rate for (rU-dA)11 is 10-fold higher than that for (dT-rA)11 and the exchange rate for (rU-dA)12 is 27-fold higher than that for (dT-rA)12. These differences in exchange rates are due to two factors: the different pKa values of uracil and thymine, and the stabilities of dA-rU and rA-dT base pairs. The pKa values of the two bases at 10 °C are: pKa = 9.58 for uracil and pKa = 10.07 for thymine (33). The lower pKa of uracil promotes the exchange of the imino proton from the open state of a dA-rU base pair. As a result, the rate constant for transfer of the imino proton from the open state (Eq. 5) is higher for uracil than for thymine: kB = 8.8×108 M−1s−1 for uracil and kB = 4.1×108 M−1s−1 for thymine (at 10 °C, with kcoll = 1.9×109 M−1s−1 and pKa = 9.73 for ammonia (32)). Hence, the differences in pKa values account for a ~2-fold increase in the exchange rates of the uracil imino protons relative to those of thymine imino protons. The remaining enhancements in the exchange rates of uracil protons are due to the lower stabilities of dA-rU base pairs. As shown in Table 1 and in Figure 7, the opening equilibrium constants (Kop) for dA-rU base pairs are higher than those for dT-rA base pairs: 21% higher in position 6, 5-fold higher in position 11, and 13-fold higher in position 12. Accordingly, the free energy changes required for the opening of dA-rU base pairs are lower than the corresponding ones for rA-dT base pairs by 0.11 to 1.44 kcal/mol (Table 1). These findings therefore demonstrate that, within the same base sequence context, dA-rU base pairs are less stable than dT-rA base pairs. These differences in stability are largest when two dA-rU base pairs are located next to each other in the structure (i.e., positions 11 and 12 in the two hybrids investigated).
For the G-C base pairs the imino proton exchange rates are influenced by the nature of the base pair, i.e., dG-rC versus rG-dC, as well as by the base sequence. A feature of the base sequence that clearly affects the exchange rates of G-C imino protons is the proximity of A-U/A-T base pairs. As Figure 5 shows, the exchange rates for the G-C base pairs in positions 10 and 13 are higher in hybrid II than in hybrid I. Similarly, the exchange rates for the G-C base pair in position 7 are higher in hybrid I than in hybrid II. Hence, the exchange rates increase when the G-C base pair is located next to an A-U base pair. The only exceptions to this trend are the G-C base pairs in position 5: their exchange rates in the two hybrids are similar (Figure 5). This observation suggests that an A-U base pair affects its neighboring G-C base pair only when the guanine is located on the DNA strand, i.e, “across” the helix from the uracil. Another factor that influences the exchange rates of imino protons in G-C base pairs is the nature of the sugar moiety attached to guanine (i.e., ribose, r, or deoxyribose, d). This can be seen for positions 3 and 4 where the rG bases in the RNA strand have higher exchange rates than the dG bases in the DNA strand. The base sequence context can, however, override this effect of the sugar. For example, in position 9, the rC-dG base pair in hybrid II has a higher exchange rate than the dC-rG base pair in hybrid I. This reversal in exchange rates may be due to the fact that, in hybrid II, the (dG-rC)9 base pair is located close to the destabilizing (dA-rU)11 and (dA-rU)12 base pairs.
The exchange rates for G-C base pairs differ between the two hybrids due to the kinetics of base-pair opening as well as the energetic stability of the base pairs. For some G-C base pairs kinetic effects alone can account for the differences in exchange rates. In these cases, the rate of opening (kop) increases without significant changes in the equilibrium constant of the opening reaction (Kop). The G-C base pairs in positions 4 and 10 illustrate this case. The kop rates of these base pairs are larger in hybrid II relative to hybrid I [i.e., 4-fold in position 4 and 7-fold in position 10] but the Kop values are the same, within experimental errors. The Kop values are the same because the rates of closing of the base pairs (kcl) also change, in concert with the opening rates (Eq. 2 and Table 1). Hence, the G-C base pairs in positions 4 and 10 in hybrid II are kinetically destabilized relative to the same base pairs in hybrid I; their energetic stabilities are, however, the same as those in hybrid I. For other G-C base pairs, the differences in exchange rates originate from changes in both, the kinetic and the energetic parameters of the opening reaction. For example, the kop rate for (dC-rG)3 in hybrid I is 7-fold higher than that for (rC-dG)3 in hybrid II. The corresponding increase in Kop is only 4-fold because the kcl rate is also increased ~2-fold (Table 1). Another example is that of the G-C base pairs in position 9. The kop rate for (rC-dG)9 in hybrid II could not be determined because the exchange of the imino proton did not reach the EX1 regime at the highest ammonia concentrations investigated. Nevertheless, the data shown in Figure 5 clearly indicate that the kop rate for this base pair should be higher than ~ 4 s−1. This represents an increase of at least 2-fold relative to the kop rate of (dC-rG)9 in hybrid I. In spite of this increase of the opening rate the equilibrium constant Kop for (rC-dG)9 in hybrid II is 2-fold smaller than that for (dC-dG)9 in hybrid I. These opposite changes in kop and Kop are due to the fact that the kcl rate for (rC-dG)9 in hybrid II is at least 4-fold higher than that for (dC-dG)9 in hybrid I. In summary, for G-C base pairs, a difference in stability between the two hybrids is observed when the change in opening rate exceeds, or is smaller than, the change in closing rate. When the change in opening rate parallels that in closing rate the stability of the G-C base pair remains unaffected.
Differences in the stability of homologous RNA-DNA hybrids have been previously observed in optical melting experiments (12, 13, 15–17, 34). In contrast to our present approach, which provides the stability of individual base pairs, optical melting experiments measure the overall stability of the hybrid duplex structure. In these previous studies, the largest differences in stability were observed for RNA-DNA hybrids that contained all purine (R) and all pyrimidine (Y) bases in each strand. Specifically, the hybrids rR-dY (i.e., purines in the RNA strand and pyrimidines in the DNA strand) are more stable than the homologous dR-rY hybrids (13, 15–17). Kool and coworkers have elucidated the structural origin of these energetic differences for a RNA-DNA hybrid containing an adenine-rich strand and a thymine/uracil-rich strand (34). Using appropriate chemical modifications of the hybrid, they have shown that each C5 methyl group of thymine enhances the overall stability of the hybrid by an average of 0.17 to 0.27 kcal/mol (at 37 °C) relative to the homologous hybrid in which thymine is replaced by uracil. The origin of this effect is the methyl-induced enhancement of the stacking of thymine onto neighboring bases (11, 34). The ribose 2’-OH group also has a stabilizing effect but only when placed in the purine-rich strand; for example, substitution of a dA-rU base pair with a rA-dU base pair stabilizes the structure by 0.32 kcal/mol (34). These findings offer important insights into the structural origin of the differences in stability between dA-rU and rA-dT base pairs observed in the present work. They suggest that rA-dT base pairs are stabilized relative to dA-rU base pairs due to the presence of the C5 methyl group in thymine and to the location of the 2’-OH group in the purine.
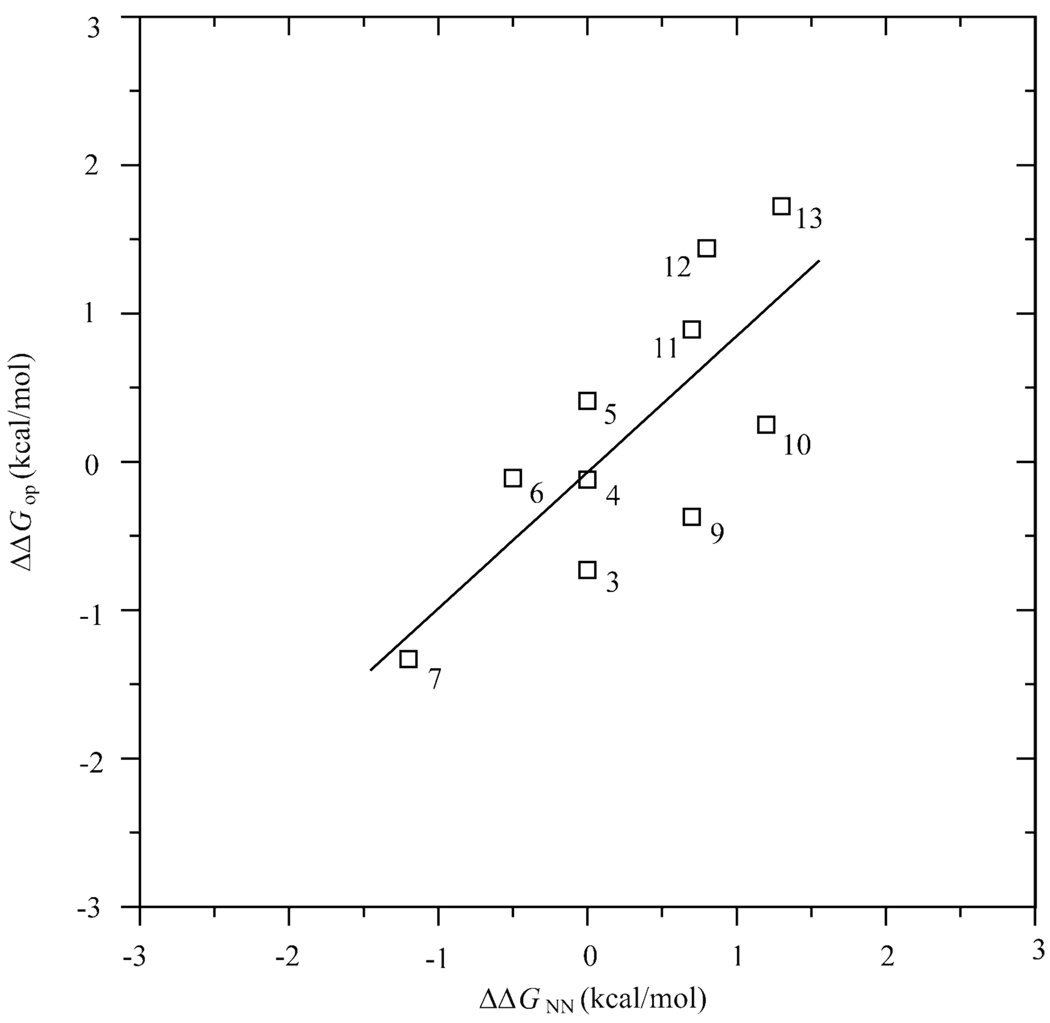
For homologous RNA-DNA hybrids of mixed base sequences, such as those investigated in the present work, the overall stabilities of the structures do not follow simple trends like those discussed above for rR-dY and dR-rY hybrids. Instead, in these cases, the stabilities are strongly dependent on the particular base sequence of the two homologous hybrids (12–14). This dependence has been rationalized using the nearest-neighbor (NN) model for stability of duplex nucleic acids. Sugimoto and coworkers have determined the free energies of stabilization for all 16 NN base-pair sets present in RNA-DNA duplexes using optical melting data for 41 different base sequences (35). These NN sets predict that the overall stability of hybrid I investigated here is ~ 4 kcal/mol higher then that of hybrid II. This prediction is in agreement with the stabilization free energies that we obtained from the melting curves of the two hybrids, namely, ΔG0 = −16.8 kcal/mol (Tm = 63 °C) for hybrid I, and ΔG0 = −13.9 kcal/mol (Tm = 59 °C) for hybrid II. Do the NN sets of free energies also predict the differences in opening free energies of individual base pairs observed in the present work? The answers to this question are shown in Table 2. For most base pairs the predicted values differ from the experimentally determined opening free energy changes both, in the magnitude of the differences and in their direction. In spite of these discrepancies, a correlation is observed between the opening free energies obtained in the present work and the NN free energies (Figure 8). This observation suggests that the discrepancies between the free energy differences predicted by the NN model and the differences in base-pair opening free energies are likely due to the fact that the former are obtained by averaging the melting data over a large number of base sequences (i.e., 41 sequences in the study by Sugimoto and coworkers (35)). In contrast, the proton exchange measurements probe the stability of each base pair in a given base sequence context. Hence, the proton exchange data reflect local changes in the structure induced by the nature of the base pair and by the base sequence. Such structural changes can have significant energetic consequences by affecting, for example, the stacking of a given base pair to its near neighboring base pairs. Characterization of these local structural changes, and their relationships to the energetic differences observed in the present work, awaits the determination of the structures of the two RNA-DNA hybrids, currently in progress in our laboratory.
Table 2.
Comparison of the differences in base-pair opening free energies (ΔΔGop) in the two RNA-DNA hybrids investigated to the differences in the NN free energies (ΔΔGNN) predicted by duplex melting data
| Position | Base pair | ΔΔGop (kcal/mol)1 | ΔΔGNN (kcal/mol)2 |
|---|---|---|---|
| 3 | C-G | −0.73 | 0 |
| 4 | G-C | −0.12 | 0 |
| 5 | C-G | 0.41 | 0 |
| 6 | A-U/T | −0.11 | −0.5 |
| 7 | G-C | −1.33 | −1.2 |
| 8 | G-C | - | - |
| 9 | C-G | −0.37 | 0.7 |
| 10 | C-G | 0.25 | 1.2 |
| 11 | T/U-A | 0.89 | 0.7 |
| 12 | T/U-A | 1.44 | 0.8 |
| 13 | C-G | 1.72 | 1.3 |
ΔΔ Gop was calculated from the values reported in Table 1, as ΔΔGop = ΔGop(Hybrid I) – ΔGop(Hybrid II).
ΔΔGNN was calculated as ΔΔGNN = ΔGNN(Hybrid I) – ΔGNN(Hybrid II).
The ΔGNN value for each base pair was calculated as the sum of the NN free energies for the base pair of interest and the two base pairs adjacent to it (35).
Figure 8.
Correlation between the differences in base-pair opening free energies (ΔΔGop ) in the two RNA-DNA hybrids investigated and the differences in the NN free energies (ΔΔGNN) predicted by duplex melting data (35). The line represents the least-squares fit ΔΔGop (−0.1 ± 0.2)+(0.9 ± 0.3)· ΔΔGNN. The position of each base pair in the two hybrids is indicated for each point.
Supplementary Material
Acknowledgments
We thank Dr. Smita Patel (UMDNJ-Robert Wood Johnson Medical School) for a gift of T7 RNA polymerase.
Abbreviations used
- NMR
nuclear magnetic resonance
- d
deoxyribose
- r
ribose
- R
purine
- Y
pyrimidine
- GTP
guanosine triphosphate
- EDTA
ethylenediaminetetraacetic acid
- HSQC
heteronuclear single quantum coherence spectroscopy
- fHSQC
fast HSQC
Footnotes
Supporting Information Available
Assignments of NMR resonances to specific imino protons in the two RNA-DNA hybrids using 15N-editing and 1H-1H NOESY, and the imino proton resonances of hybrid II at different concentrations of ammonia. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by a grant from the National Institutes of Health (GM077188)
REFERENCES
- 1.Rich A. Discovery of the Hybrid Helix and the First DNA-RNA Hybridization. J. Biol. Chem. 2006;281:7693–7696. doi: 10.1074/JBC.X600003200. [DOI] [PubMed] [Google Scholar]
- 2.Hall KB. NMR spectroscopy of DNA/RNA hybrids. Curr. Opin. Struct. Biol. 1993;3:336–339. [Google Scholar]
- 3.Fedoroff OY, Salazar M, Reid BR. Structure of a DNA:RNA Hybrid Duplex. Why RNase H Does Not Cleave Pure RNA. J. Mol. Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 4.Sanghani SR, Lavery R. Theoretical studies of DNA-RNA hybrid conformations. Nucleic Acids Res. 1994;22:1444–1449. doi: 10.1093/nar/22.8.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton NC, Finzel BC. The Structure of an RNA/DNAHybrid: A Substrate of the Ribonuclease Activity of HIV-1 Reverse Transcriptase. J. Mol. Biol. 1996;264:521–533. doi: 10.1006/jmbi.1996.0658. [DOI] [PubMed] [Google Scholar]
- 6.Fedoroff OY, Ge Y, Reid BR. Solution Structure of r(gaggacug):d(CAGTCCTC) Hybrid: Implications for the Initiation of HIV-1 (+)-Strand Synthesis. J. Mol. Biol. 1997;269:225–239. doi: 10.1006/jmbi.1997.1024. [DOI] [PubMed] [Google Scholar]
- 7.Gyi JI, Lane AN, Conn GL, Brown T. Solution Structures of DNA-RNA Hybrids with Purine-Rich and Pyrimidine-Rich Strands: Comparison with the Homologous DNA and RNA Duplexes. Biochemistry. 1998;37:73–80. doi: 10.1021/bi9719713. [DOI] [PubMed] [Google Scholar]
- 8.Xiong Y, Sundaralingam M. Crystal structure of a DNA-RNA hybrid duplex with a polypurine RNA r(gaagaagag) and a complementary polypyrimidine DNA d(CTCTTCTTC) Nucleic Acids Res. 2000;28:2171–2176. doi: 10.1093/nar/28.10.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopka ML, Leavelle L, Won Han G, Ng H-L, Dickerson RE. An Unusual Sugar Conformation in the Structure of an RNA/DNA Decamer of the Polypurine Tract May Affect Recognition by RNase H. J. Mol. Biol. 2003;334:653–665. doi: 10.1016/j.jmb.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 10.Noy A, Perez A, Marquez M, Luque FJ, Orozco M. Structure, Recognition Properties, and Flexibility of the DNA·RNA Hybrid. J. Am. Chem. Soc. 2005;127:4910–4920. doi: 10.1021/ja043293v. [DOI] [PubMed] [Google Scholar]
- 11.Priyakumar UD, MacKerell AD., Jr Atomic Detail Investigation of the Structure and Dynamics of DNA·RNA Hybrids: A Molecular Dynamics Study. J. Phys. Chem. B. 2008;112:1515–1524. doi: 10.1021/jp709827m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall KB, McLaughlin LW. Thermodynamic and Structural Properties of Pentamer DNA·DNA, RNA·RNA, and DNA·RNA Duplexes of Identical Sequence. Biochemistry. 1991;30:10606–10613. doi: 10.1021/bi00108a002. [DOI] [PubMed] [Google Scholar]
- 13.Ratmeyer L, Vinayak R, Zhong YY, Zon G, Wilson WD. Sequence Specific Thermodynamic and Structural Properties for DNA·RNA Duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 14.Lesnik EA, Freier SM. Relative Thermodynamic Stability of DNA, RNA, and DNA:RNA Hybrid Duplexes: Relationship with base Composition and Structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 15.Martin FH, Tinoco IJ. RNA-DNA hybrid duplexes containing oligo (dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts RW, Crothers DM. Stability and Properties of Double and Triple Helices: Dramatic Effects of RNA or DNA Backbone Composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 17.Gyi JI, Conn GL, Lane AN, Brown T. Comparison of the Thermodynamic Stabilities and Solution Conformations of DNA-RNA Hybrids Containing Purine-Rich and Pyrimidine-Rich Strands with DNA and RNA Duplexes. Biochemistry. 1996;35:12538–12548. doi: 10.1021/bi960948z. [DOI] [PubMed] [Google Scholar]
- 18.Wilson KS, von Hippel PH. Stability of Escherichia coli Transcription Complexes Near an Intrinsic Terminator. J. Mol. Biol. 1994;244:36–51. doi: 10.1006/jmbi.1994.1702. [DOI] [PubMed] [Google Scholar]
- 19.von Hippel PH. An Integrated Model of the Transcription Complex in Elongation, Termination, and Editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 20.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied Force Reveals Mechanistic and Energetic Details of Transcription Termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datta K, von Hippel PH. Direct Spectroscopic Study of Reconstituted Transcription Complexes Reveals That Intrinsic Termination is Driven Primarily by Thermodynamic Destabilization of the Nucleic Acid Framework. J. Biol. Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Russu IM. Sequence-Dependence of the Energetics of Opening of AT Base Pairs in DNA. Biophys. J. 2004;87:1–7. doi: 10.1529/biophysj.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russu IM. Probing Site-Specific Energetics in Proteins and Nucleic Acids by Hydrogen Exchange and NMR Spectroscopy. Methods Enzymol. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 25.Plateau P, Gueron M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 26.Mori S, Abeygunawardana C, Johnson MO, van Zijl PCM. Improved Sensitivity of HSQC Spectra of Exchanging Protons at Short Interscan Delays Using a new Fast HSQC (FHSQC) Detection Scheme That Avoids Water Saturation. J. Magn. Reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Jiang L, Michalczyk R, Russu IM. Structural Energetics and Base-Pair Opening Dynamics in Sarcin-Ricin Domain RNA. Biochemistry. 2006;45:13606–13613. doi: 10.1021/bi060908n. [DOI] [PubMed] [Google Scholar]
- 28.Lippens G, Dhalluin C, Wieruszeski J-M. Use of a water flip-back pulse in the homonuclear NOESY experiment. J. Biomol. NMR. 1995;5:327–331. doi: 10.1007/BF00211762. [DOI] [PubMed] [Google Scholar]
- 29.Gueron M, Leroy J-L. Studies of Base Pair Kinetics by NMR Measurement of Proton Exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 30.Gueron M, Charretier E, Hagerhorst J, Kochoyan M, Leroy JL, Moraillon A. Applications of Imino Proton Exchange to Nucleic Acid Kinetics and Structures. In: Sarma RH, Sarma MH, editors. Structure & Methods. Albany, NY: Adenine Press; 1990. pp. 113–137. [Google Scholar]
- 31.Benight AS, Schurr JM, Flynn PF, Reid BR, Wemmer DE. Melting of a Self-complementary DNA Minicircle. Comparison of Optical Melting Theory with Exchange Broadening of the Nuclear Magnetic Resonance Spectrum. J. Mol. Biol. 1988;200:377–399. doi: 10.1016/0022-2836(88)90248-3. [DOI] [PubMed] [Google Scholar]
- 32.Weast RC. CRC Handbook of Chemistry and Physics. 67th ed. ed. Boca Raton, FL: CRC Press; 1987. [Google Scholar]
- 33.Ts'o POP. Basic Principles in Nucleic Acid Chemistry. Vol. I. New York, NY: Academic Press; 1974. [Google Scholar]
- 34.Wang S, Kool ET. Origins of the Large Differences in Stability of DNA and RNA Helices: C-5 Methyl and 2'-Hydroxyl Effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 35.Wu P, Nakano S, Sugimoto N. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. 2002;269:2821–2830. doi: 10.1046/j.1432-1033.2002.02970.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.