Abstract
The ubiquitin-mediated degradation of mitotic cyclins is required for cells to exit from mitosis. Previous work with cell-free systems has revealed four components required for cyclin-ubiquitin ligation and proteolysis: a nonspecific ubiquitin-activating enzyme E1, a soluble fraction containing a ubiquitin carrier protein activity called E2-C, a crude particulate fraction containing a ubiquitin ligase (E3) activity that is activated during M-phase, and a constitutively active 26S proteasome that degrades ubiquitinated proteins. Here, we identify a novel approximately 1500-kDa complex, termed the cyclosome, which contains a cyclin-selective ubiquitin ligase activity, E3-C. E3-C is present but inactive during interphase; it can be activated in vitro by the addition of cdc2, enabling the transfer of ubiquitin from E2-C to cyclin. The kinetics of E3-C activation suggest the existence of one or more intermediates between cdc2 and E3-C. Cyclosome-associated E3-C acts on both cyclin A and B, and requires the presence of wild-type N-terminal destruction box motifs in each cyclin. Ubiquitinated cyclins are then rapidly recognized and degraded by the proteasome. These results identify the cyclosome-associated E3-C as the component of the cyclin destruction machinery whose activity is ultimately regulated by cdc2 and, as such, the element directly responsible for setting mitotic cyclin levels during early embryonic cell cycles.
Full text
PDF




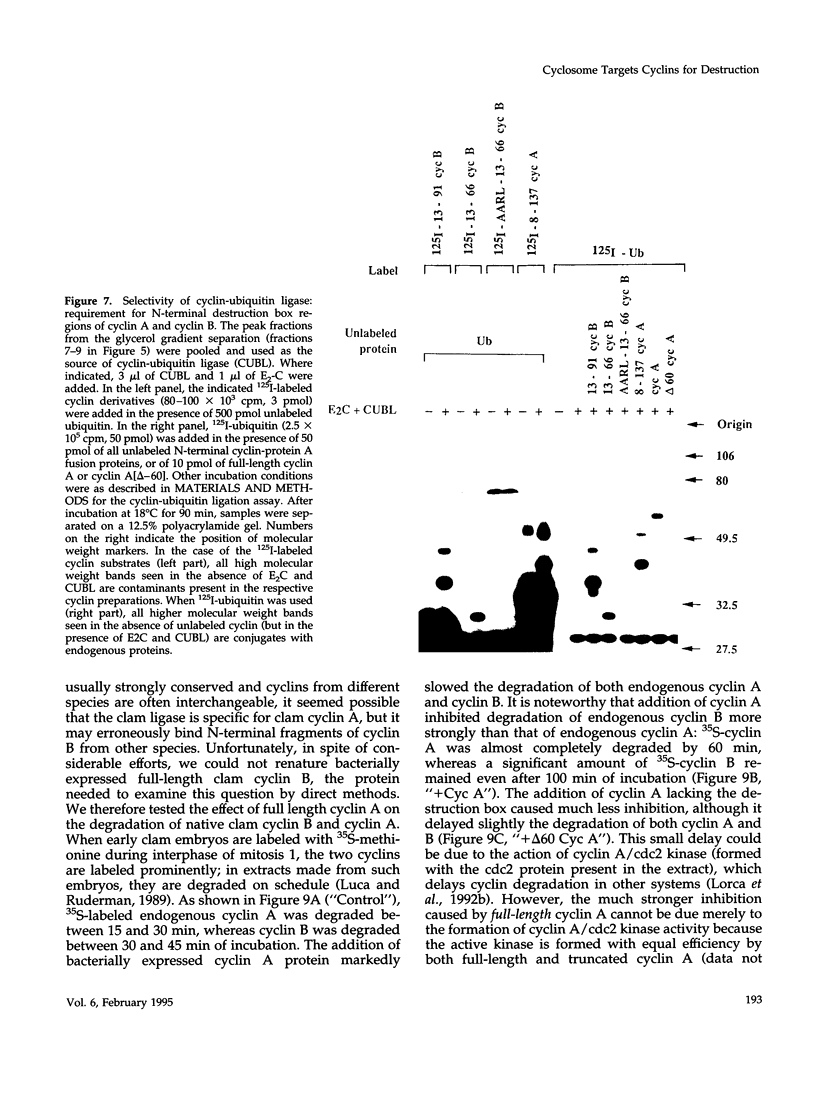




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., DiGiuseppe J. A., Bercovich B., Orian A., Richter J. D., Schwartz A. L., Brodeur G. M. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J., Madura K., Bartel B., Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Rosenthal E. T., Youngblom J., Distel D., Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983 Jun;33(2):389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Félix M. A., Labbé J. C., Dorée M., Hunt T., Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990 Jul 26;346(6282):379–382. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- Gallant P., Nigg E. A. Cyclin B2 undergoes cell cycle-dependent nuclear translocation and, when expressed as a non-destructible mutant, causes mitotic arrest in HeLa cells. J Cell Biol. 1992 Apr;117(1):213–224. doi: 10.1083/jcb.117.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiara J. B., Richardson H. E., Sugimoto K., Henze M., Lew D. J., Wittenberg C., Reed S. I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991 Apr 5;65(1):163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Heller H., Hershko A. A ubiquitin-protein ligase specific for type III protein substrates. J Biol Chem. 1990 Apr 25;265(12):6532–6535. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ganoth D., Pehrson J., Palazzo R. E., Cohen L. H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991 Sep 5;266(25):16376–16379. [PubMed] [Google Scholar]
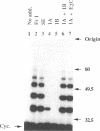
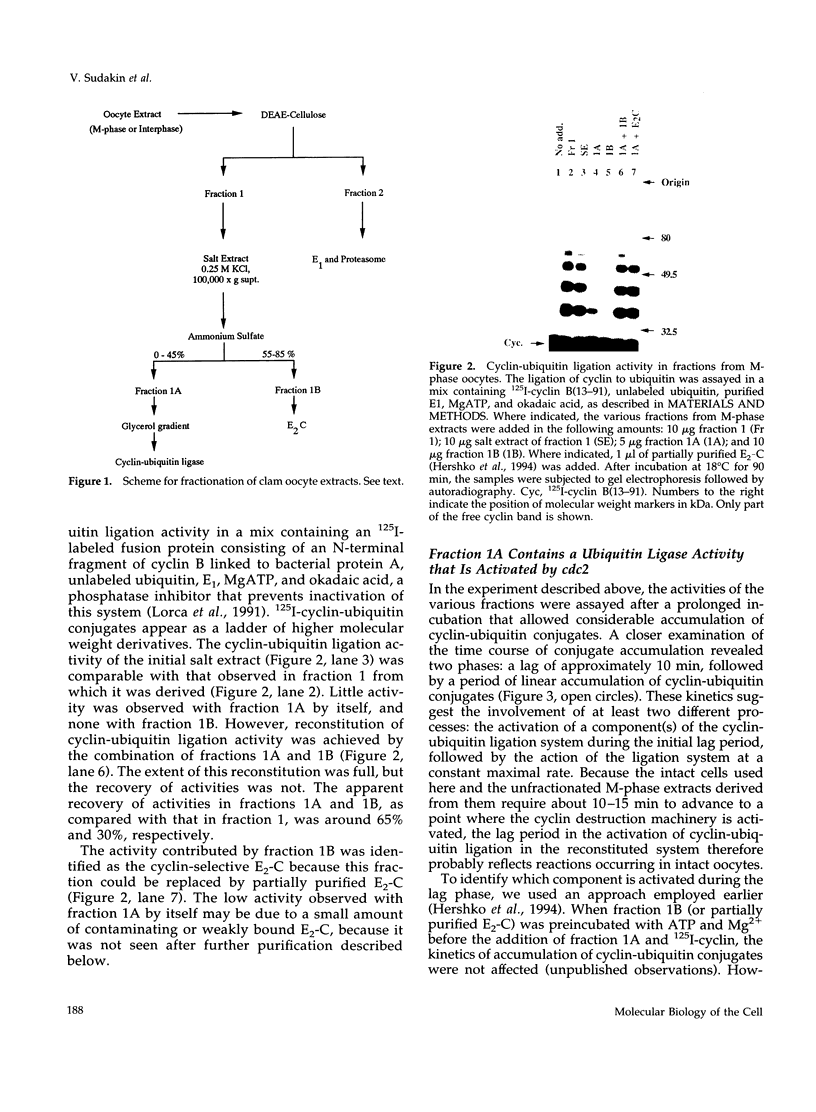
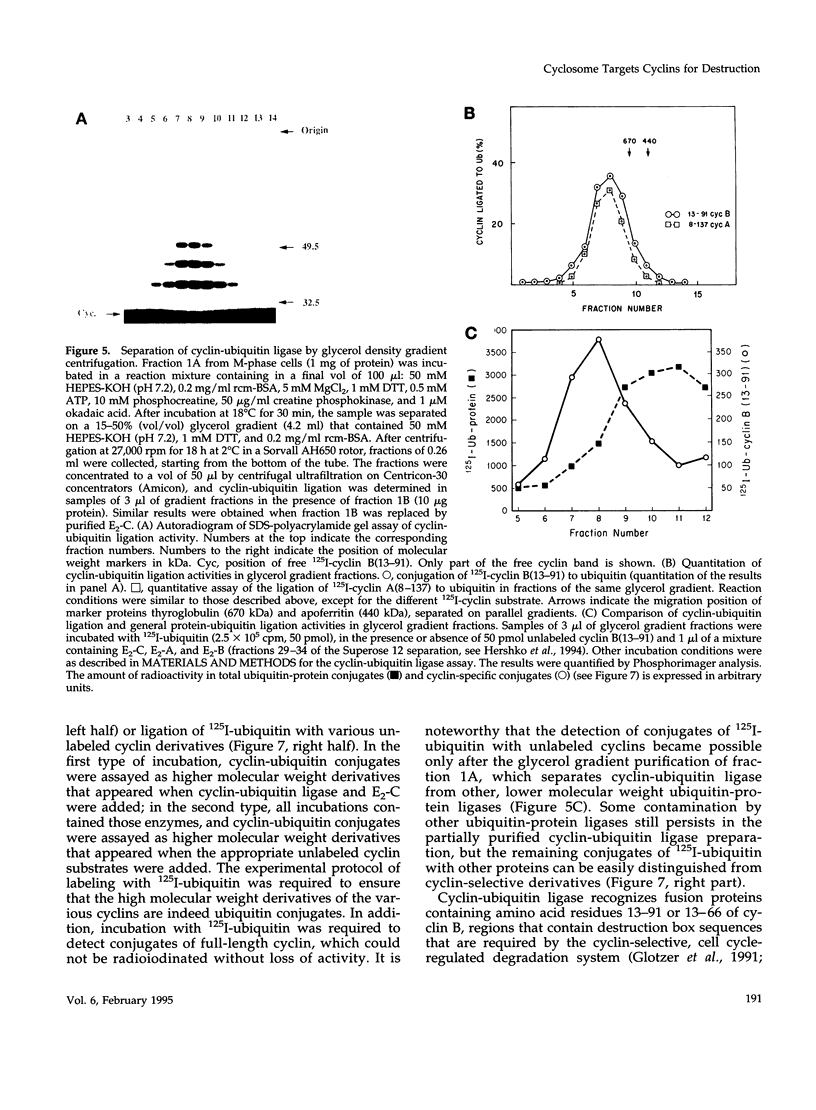
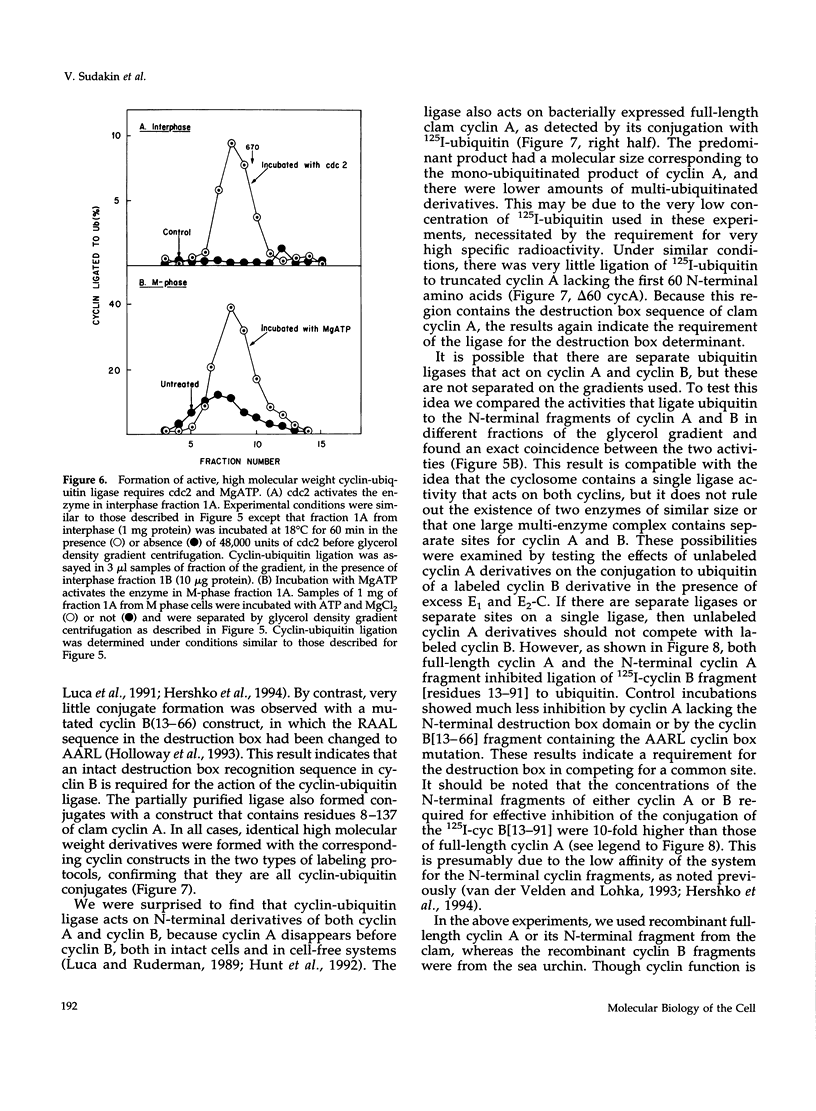
- Hershko A., Ganoth D., Sudakin V., Dahan A., Cohen L. H., Luca F. C., Ruderman J. V., Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994 Feb 18;269(7):4940–4946. [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983 Jul 10;258(13):8206–8214. [PubMed] [Google Scholar]
- Hershko A., Heller H., Eytan E., Reiss Y. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986 Sep 15;261(26):11992–11999. [PubMed] [Google Scholar]
- Hershko A., Rose I. A. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway S. L., Glotzer M., King R. W., Murray A. W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993 Jul 2;73(7):1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Hunt T., Luca F. C., Ruderman J. V. The requirements for protein synthesis and degradation, and the control of destruction of cyclins A and B in the meiotic and mitotic cell cycles of the clam embryo. J Cell Biol. 1992 Feb;116(3):707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Lorca T., Fesquet D., Zindy F., Le Bouffant F., Cerruti M., Brechot C., Devauchelle G., Dorée M. An okadaic acid-sensitive phosphatase negatively controls the cyclin degradation pathway in amphibian eggs. Mol Cell Biol. 1991 Feb;11(2):1171–1175. doi: 10.1128/mcb.11.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Labbé J. C., Devault A., Fesquet D., Strausfeld U., Nilsson J., Nygren P. A., Uhlen M., Cavadore J. C., Dorée M. Cyclin A-cdc2 kinase does not trigger but delays cyclin degradation in interphase extracts of amphibian eggs. J Cell Sci. 1992 May;102(Pt 1):55–62. doi: 10.1242/jcs.102.1.55. [DOI] [PubMed] [Google Scholar]
- Luca F. C., Ruderman J. V. Control of programmed cyclin destruction in a cell-free system. J Cell Biol. 1989 Nov;109(5):1895–1909. doi: 10.1083/jcb.109.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F. C., Shibuya E. K., Dohrmann C. E., Ruderman J. V. Both cyclin A delta 60 and B delta 97 are stable and arrest cells in M-phase, but only cyclin B delta 97 turns on cyclin destruction. EMBO J. 1991 Dec;10(13):4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey D., Yoo Y., Rechsteiner M. Ubiquitin metabolism in cycling Xenopus egg extracts. J Biol Chem. 1993 Oct 5;268(28):21205–21211. [PubMed] [Google Scholar]
- Mayer A. N., Wilkinson K. D. Detection, resolution, and nomenclature of multiple ubiquitin carboxyl-terminal esterases from bovine calf thymus. Biochemistry. 1989 Jan 10;28(1):166–172. doi: 10.1021/bi00427a024. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Kirschner M. W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989 May 25;339(6222):275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989 May 25;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Furuno N., Okazaki K., Tanaka H., Ogawa Y., Sagata N. Degradation of Mos by the N-terminal proline (Pro2)-dependent ubiquitin pathway on fertilization of Xenopus eggs: possible significance of natural selection for Pro2 in Mos. EMBO J. 1993 Oct;12(10):4021–4027. doi: 10.1002/j.1460-2075.1993.tb06080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994 Sep 9;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Rose I. A. Mechanism of ubiquitin carboxyl-terminal hydrolase. Borohydride and hydroxylamine inactivate in the presence of ubiquitin. J Biol Chem. 1986 Aug 5;261(22):10210–10217. [PubMed] [Google Scholar]
- Reiss Y., Heller H., Hershko A. Binding sites of ubiquitin-protein ligase. Binding of ubiquitin-protein conjugates and of ubiquitin-carrier protein. J Biol Chem. 1989 Jun 25;264(18):10378–10383. [PubMed] [Google Scholar]
- Reiss Y., Hershko A. Affinity purification of ubiquitin-protein ligase on immobilized protein substrates. Evidence for the existence of separate NH2-terminal binding sites on a single enzyme. J Biol Chem. 1990 Mar 5;265(7):3685–3690. [PubMed] [Google Scholar]
- Reiss Y., Kaim D., Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988 Feb 25;263(6):2693–2698. [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993 Nov 5;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Schwob E., Böhm T., Mendenhall M. D., Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994 Oct 21;79(2):233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson K. I., Farrell K. M., Ruderman J. V. The clam embryo protein cyclin A induces entry into M phase and the resumption of meiosis in Xenopus oocytes. Cell. 1986 Dec 26;47(6):861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994 Sep 9;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Westendorf J. M., Swenson K. I., Ruderman J. V. The role of cyclin B in meiosis I. J Cell Biol. 1989 Apr;108(4):1431–1444. doi: 10.1083/jcb.108.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden H. M., Lohka M. J. Mitotic arrest caused by the amino terminus of Xenopus cyclin B2. Mol Cell Biol. 1993 Mar;13(3):1480–1488. doi: 10.1128/mcb.13.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]