Abstract
Homologous recombination in Saccharomyces cerevisiae depends critically on RAD52 function. In vitro, Rad52 protein preferentially binds single-stranded DNA (ssDNA), mediates annealing of complementary ssDNA, and stimulates Rad51 protein-mediated DNA strand exchange. Replication protein A (RPA) is a ssDNA-binding protein that is also crucial to the recombination process. Herein we report that Rad52 protein effects the annealing of RPA–ssDNA complexes, complexes that are otherwise unable to anneal. The ability of Rad52 protein to promote annealing depends on both the type of ssDNA substrate and ssDNA binding protein. RPA allows, but slows, Rad52 protein-mediated annealing of oligonucleotides. In contrast, RPA is almost essential for annealing of longer plasmid-sized DNA but has little effect on the annealing of poly(dT) and poly(dA), which are relatively long DNA molecules free of secondary structure. These results suggest that one role of RPA in Rad52 protein-mediated annealing is the elimination of DNA secondary structure. However, neither Escherichia coli ssDNA binding protein nor human RPA can substitute in this reaction, indicating that RPA has a second role in this process, a role that requires specific RPA–Rad52 protein interactions. This idea is confirmed by the finding that RPA, which is complexed with nonhomologous ssDNA, inhibits annealing but the human RPA–ssDNA complex does not. Finally, we present a model for the early steps of the repair of double-strand DNA breaks in yeast.
In the yeast Saccharomyces cerevisiae, homologous recombination typically initiates at double-strand DNA breaks (DSBs). At an early step of meiosis, DSBs are introduced at specific loci in the chromosome; these breaks serves as sites for initiation of recombination, and loci with a high frequency of DSBs are recombination hot spots (1–4). In addition to its intimate association with the meiotic process, homologous recombination is responsible for the repair of DSBs that are introduced by DNA alkylating reagents, ionizing-radiation, and specific endonucleases (5, 6). A group of genes, referred to as the RAD52 epistasis group, are involved in both homologous recombination and the repair of DSBs (7, 8). Analyses of mutants within this group of genes showed that the RAD52 gene is critically important for both recombination and resistance to x-rays (9–11). Although the RAD52 gene does not show any obvious homology to the known recombination proteins of Escherichia coli, it is conserved in yeast, human, and mice (12, 13), suggesting that the Rad52 protein is unique to eukaryotic organisms. Purified Rad52 protein binds both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) and can anneal complementary ssDNA oligonucleotides (14, 15). More recent reports show that Rad52 protein stimulates DNA strand exchange mediated by Rad51 protein, which is a homologue of the E. coli RecA protein (16–19). In the yeast system, stimulation is due to Rad52 protein interaction with, and alleviation of an inhibitory consequence of ssDNA binding by, replication protein A (RPA), which is the homologue of ssDNA binding protein (SSB) protein (16–18).
RPA is a heterotrimeric complex consisting of polypeptides with molecular masses of 70.4, 29.9, and 13.8 kDa (20, 21). The primary structure of RPA is well-conserved in yeast, human, mice, and Drosophila (22). Studies using primarily human proteins showed that RPA is required for cell-free DNA replication (23) and DNA excision repair (24, 25). The human RPA (hRPA) interacts with some of the proteins that are components of these systems, including replication proteins simian virus 40 large tumor antigen and DNA polymerase α-primase (26), tumor suppresser protein p53 (27), and DNA excision repair proteins XPA (28, 29) and XPG (28). Presumably, these interactions define important cellular functions for RPA.
Less is known about yeast RPA (yRPA). Because each subunit is essential for yeast cell viability, yRPA is believed to be essential for DNA replication (20, 21). An in vitro DNA excision repair system from yeast also requires RPA (30, 31). In addition, genetic analyses particularly of two mutant alleles of the gene encoding the large subunit of RPA (rfa1–44 and rfa1-D228Y), showed that RPA is involved in homologous recombination (32–34). The fact that the phenotype of rfa1–44 mutation is suppressed by overexpression of Rad52 protein (33) suggests that these proteins are involved in the same genetic pathway in homologous recombination and that they may work in concert at the molecular level. In agreement with these yeast data, recent in vitro work in the human system showed that RPA directly interacts with Rad52 protein (35). Furthermore, yRPA stimulates the DNA strand exchange and ATPase activities of Rad51 protein, provided that it is introduced to preassembled Rad51 nucleoprotein complexes (36, 37), but this latter restriction is alleviated by Rad52 protein (16–18). Therefore, RPA and Rad52 protein may have multiple functions in homologous genetic recombination. In this report, we examined the biochemical characteristics of DNA annealing by Rad52 protein and the effect of RPA and other single-stranded DNA-binding proteins on this reaction. We find that the RPA–ssDNA complex is the preferred substrate for the Rad52 protein-mediated annealing of native ssDNA, a result that agrees with the experiments of Shinohara et al. (38). These finding have direct bearing on the mechanism of recombination between directly repeated DNA sequences, which occurs by a ssDNA annealing mechanism, and have implications for the role of Rad52 protein in other recombination processes.
METHODS
Proteins.
The three subunits of RPA, cloned on three separate plasmids (provided by R. Kolodner, University of California, San Diego), were coinduced in yeast and the RPA heterotrimer was prepared as described (39). The Rad52 protein, which was translated from the third start codon in the originally reported ORF (40), was purified as described (17). E. coli SSB protein was expressed and purified as described (41, 42). The concentrations of RPA, Rad52 protein, and SSB protein were determined by using extinction coefficients of 8.8 × 104, 2.4 × 104, and 3.0 × 104 M−1⋅cm−1 at 280 nm, respectively. Recombinant hRPA (43), which was expressed and purified from E. coli, was provided by M. Wold (University of Iowa, Iowa City, IA).
DNA.
Two complementary 48-mer oligonucleotides, oligo-25 (5′-GCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA) and oligo-26 (5′-TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGC), were purchased from Operon and purified by electrophoresis using 10% polyacrylamide gels containing 8 M urea. Plasmid pBluescriptSK+ (Stratagene) was linearized by digestion with ScaI and denatured by incubation at 95°C for 4 min and then quickly chilled on ice. Poly(dT) and poly(dA) were purchased from Pharmacia. The nucleotide concentrations of oligo-25, oligo-26, poly(dT), and poly(dA) were measured by using extinction coefficients of 1.0 × 104, 9.6 × 103, 7.3 × 103, and 8.6 × 103 M−1⋅cm−1 at 260 nm, respectively. Plasmid DNA concentration was measured before denaturation by using an extinction coefficient of 6.5 × 103 M−1⋅cm−1 at 260 nm. Plasmid DNA was prepared and manipulated as described (44). DNA was stored in 10 mM Tris⋅HCl, pH 7.5/1 mM EDTA (TE buffer). All DNA concentrations are expressed as mol of nucleotide. 32P-labeled oligo-26 was produced with T4 polynucleotide kinase (New England Biolabs).
DNA Annealing.
Annealing of DNA was continuously monitored as follows: the indicated amounts of DNA and protein were added in the order given to a cuvette containing 400 μl of 0.2 μM 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes), 30 mM Tris acetate (pH 7.5), 5 mM magnesium acetate, and 1 mM DTT, kept at 30°C. The stock concentration of DAPI was measured by using an extinction coefficient of 3.3 × 104 M−1⋅cm−1 at 345 nm. The binding of DAPI to dsDNA increased the fluorescence of DAPI, which was monitored with an SLM8000 spectrofluorimeter set to excitation and emission wavelengths of 345 and 467 nm, respectively. The slit widths for excitation and emission light were 1 and 4 mm, respectively. At this concentration of DAPI, the fluorescence was proportional to dsDNA concentration up to 10 μM.
Annealing of 32P-labeled oligo-26 and unlabeled oligo-25 was performed under the same conditions as for fluorimetric analysis. At the times indicated, 2 μl of reaction mixture were treated by addition of 2.5 μl of stop buffer containing 6.4 μM unlabeled oligo-26, 0.67% SDS, proteinase K (0.96 mg/ml), and 33 mM EDTA for 15 min at 30°C. For the zero-time sample, proteins and DNA were mixed in the stop buffer. Radioactive products were separated by PAGE on 10% gels in 40 mM Tris acetate, pH 8.1/2 mM EDTA and were both visualized and quantified with a Molecular Dynamics Storm 840 phospho-imager with image-quant software.
RESULTS
Annealing of Complementary Oligonucleotides Can Be Monitored Fluorimetrically.
Mortensen et al. (14) reported that Rad52 protein promotes annealing of oligonucleotides in vitro. To facilitate real-time and quantitative analysis of DNA annealing by Rad52 protein, we used a fluorescent dsDNA-binding dye, DAPI, to monitor dsDNA formation (Fig. 1A). The fluorescence of DAPI is enhanced by its specific binding to the minor groove of dsDNA (45, 46); therefore, formation of dsDNA from complementary ssDNA produces a corresponding increase in DAPI fluorescence (Fig. 1 A and B). This dye has been used to follow, in real-time, the enzymatic unwinding of dsDNA by E. coli RecBCD enzyme (47) and dsDNA-binding by E. coli RecA protein (48).
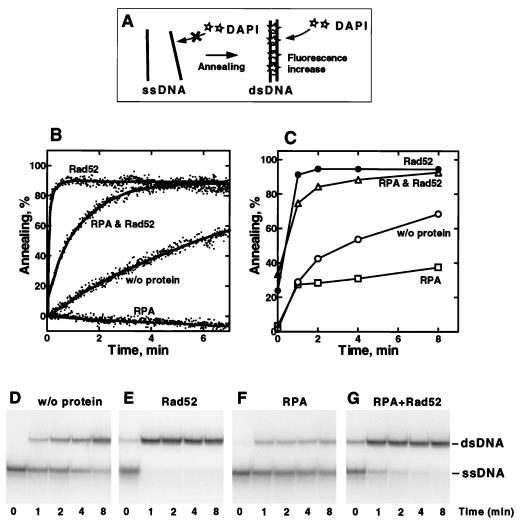
Figure 1.
Rad52 protein-mediated annealing of oligonucleotides. (A) Schematic illustration of the fluorescent annealing assay. DAPI does not bind to ssDNA but binds to dsDNA produced by annealing, resulting in an enhancement of DAPI fluorescence. (B) The 48-mer oligonucleotide (oligo-25; 200 nM) and a complementary oligonucleotide (oligo-26; 200 nM) were incubated in 400 μl of reaction buffer containing 0.2 μM DAPI without protein or in the presence of Rad52 protein, RPA, or both RPA and Rad52 protein. Reactions were started by addition of oligo-26, except in the experiment containing both proteins, which was started by addition of Rad52 protein to preformed RPA–oligonucleotides complexes. The final concentrations of Rad52 protein and RPA were 20 nM and 30 nM, respectively. The fluorescence of DAPI was continuously monitored, and the extent of annealing is expressed as a percentage of dsDNA formed, based on the DAPI fluorescence. (C–G) The same reactions as in B were performed with 32P-labeled oligo-26 and unlabeled oligo-25, and the products were analyzed by PAGE (D–G). (C) Percentages of DNA annealing that are calculated based on radioactivity of bands in D (○), E (•), F (□), and G (▵).
When a 48-mer oligonucleotide (oligo-25; 200 nM) and its complement (oligo-26; 200 nM) were mixed without protein, the fluorescence increased slowly (Fig. 1B, trace w/o protein), showing that these oligonucleotides spontaneously annealed under these conditions. When oligo-26 was added to a preincubated mixture of oligo-25 and 20 nM of Rad52 protein (Fig. 1B, trace Rad52), the DAPI fluorescence increased significantly faster than in the absence of Rad52 protein. In this case, the plateau of fluorescence indicates that approximately 85% of the DNA was annealed in 1 min. No significant fluorescence change was observed when the two oligonucleotides were not complementary; in addition, oligo-25, oligo-26, or Rad52 protein alone did not cause a significant increase in DAPI fluorescence (data not shown). Therefore, we conclude that the increase in fluorescence shown in Fig. 1B measures DNA annealing between oligo-25 and oligo-26 and that Rad52 protein accelerates this process. These results are consistent with a previous report (14), showing that Rad52 protein can promote annealing between complementary oligonucleotides; furthermore, kinetic analyses confirmed that the reaction is second order in ssDNA concentration (data not shown).
Rad52 Protein Can Mediate the Annealing of RPA–Oligonucleotide Complexes.
In the presence of RPA (30 nM), the sequential addition of oligo-25 and oligo-26 to reaction buffer did not show a significant change in fluorescence (Fig. 1B, trace RPA), indicating that RPA inhibited spontaneous annealing of the oligonucleotides. This inhibition is likely the result of a stable ssDNA–RPA complex (39), and not from RPA-mediated displacement of DAPI from dsDNA, because addition of RPA to dsDNA resulted in no fluorescence change (data not shown). In contrast, when Rad52 protein was added to the complementary oligonucleotide–RPA complexes (Fig. 1B, trace RPA and Rad52), DAPI fluorescence rapidly increased, showing that Rad52 protein can promote DNA annealing even when the DNA was previously coated with RPA. Moreover, Fig. 1B also shows that the annealing rate of the RPA-coated DNA by Rad52 protein was significantly higher than the rate of spontaneous annealing but lower than that of free DNA with Rad52 protein; the relative stimulatory effect of Rad52 protein in experiments with RPA versus its absence may be comparable, but because we cannot measure a reliable rate for annealing in the presence of RPA, quantitative comparison is not possible.
To confirm the fluorescence results, we performed the same reactions shown in Fig. 1B, using 32P-labeled oligo-26 and unlabeled oligo-25 and analyzing the labeled annealing products by PAGE (Fig. 1 D–G). Quantitative analyses of these gels (Fig. 1C) show that the results from the fluorescence assay generally parallel the gel electrophoresis assay except for the slightly higher yield observed with RPA.
Because annealing of oligonucleotides can occur spontaneously and both Rad52 protein and RPA can bind ssDNA, the rate of annealing in the presence of these proteins might be elevated simply due to an increased local concentration of oligonucleotide resulting from aggregation of the protein–DNA complexes. To eliminate this possibility, we centrifuged the reactions shown in Fig. 1 D–G at 13,000 × g for 15 min to remove any protein–DNA aggregate, but little radioactivity was precipitated, and it was independent of the extent of annealing (data not shown). This suggests that aggregation is not responsible for the stimulation of annealing mediated by Rad52 protein.
RPA Stimulates Annealing by Eliminating Secondary Structure within ssDNA.
We next examined Rad52 protein-mediated annealing of longer DNA. Fig. 2 shows the results of annealing experiments using linear heat-denatured plasmid dsDNA (length = 2,961 bp). Unexpectedly, Rad52 protein alone failed to promote the annealing of the denatured plasmid DNA (Fig. 2B, trace Rad52). This result is in striking contrast to the annealing of oligonucleotides (Fig. 1), which Rad52 protein promoted quite efficiently.
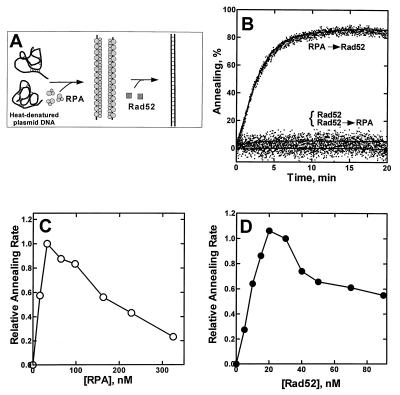
Figure 2.
RPA is needed for Rad52 protein-mediated annealing of heat-denatured plasmid DNA. (A) A schematic illustration for the annealing of linear heat-denatured plasmid DNA. (B) RPA (65 nM) and Rad52 protein (30 nM) were added to linear heat-denatured pBluescriptSK+ dsDNA (800 nM) in the indicated order, and annealing was monitored fluorimetrically. Two curves, Rad52 and Rad52 → RPA, are overlaid. The contribution of DNA secondary structure was subtracted. No significant spontaneous annealing occurred in the absence of protein (data not shown). The effect of RPA concentration (C) was examined under the same conditions used in B, and the effect of Rad52 protein concentration (D) was examined under the same conditions, except that RPA concentration was 30 nM. Relative annealing initial rates were determined from the slopes of the fluorescence–time curves.
When Rad52 protein was added to a preincubated complex of RPA and denatured DNA (Fig. 2B, trace RPA → Rad52), 85% of the DNA annealed quite efficiently. This result again shows that the ssDNA–RPA complex is a good substrate for Rad52 protein-mediated annealing. Furthermore, in contrast to the annealing of oligonucleotides, RPA greatly stimulates the Rad52 protein-dependent annealing of heat-denatured plasmid DNA. However, when RPA was added about 10 min after the addition of Rad52 protein to the DNA (Fig. 2B, trace Rad52 → RPA), annealing was not stimulated. Thus, RPA supports the Rad52 protein-mediated annealing of this longer DNA only when it is first complexed with ssDNA, demonstrating that the RPA–ssDNA complex, itself, is the target for Rad52 protein action.
An RPA titration of the reaction (Fig. 2C) shows that the optimal amount of RPA needed to stimulate the reaction is approximately 30 nM, which corresponds to a ratio of 26–27 nucleotides of ssDNA per RPA heterotrimer. Because this ratio is, within experimental error, the same as the ssDNA binding stoichiometry of our RPA preparation (T.S. and S.C.K., unpublished observations), the optimal substrate for Rad52 protein-mediated annealing appears to be ssDNA that is completely covered with RPA. Irrespective of the rate at which annealing occurred, each reaction in Fig. 2C produced a similar amount of dsDNA product (80–85%; data not shown). Fig. 2C also shows that excess RPA inhibits annealing. This inhibition might result from the binding and sequestration of Rad52 protein by the excess free RPA, a possibility that is examined below. A titration of the reaction with Rad52 protein at the optimal RPA concentration shows that the optimal Rad52 protein concentration is between 20 and 30 nM (Fig. 2D). Because this optimum is close to that of RPA (30 nM), protein–protein interactions between Rad52 protein and RPA may mediate annealing. The reason for the inhibition by excess Rad52 protein is not clear.
An obvious difference between the short oligonucleotide and the plasmid-sized ssDNA substrates is that the longer ssDNA has relatively stable secondary structure that could interfere with efficient annealing. RPA might stimulate Rad52 protein-mediated DNA annealing by eliminating this DNA secondary structure. To test this possibility, we examined annealing of poly(dT) and poly(dA) (Fig. 3), substrates that are 325 nt long on average but are devoid of secondary structure. Though these substrates anneal spontaneously at a significant rate, Rad52 protein permits a more efficient reaction; also, as before, RPA alone completely inhibits annealing. As predicted, RPA did not stimulate Rad52 protein-mediated annealing of these DNA substrates. This finding indicates that one role of RPA in Rad52 protein-mediated annealing is to eliminate secondary structure within native ssDNA.
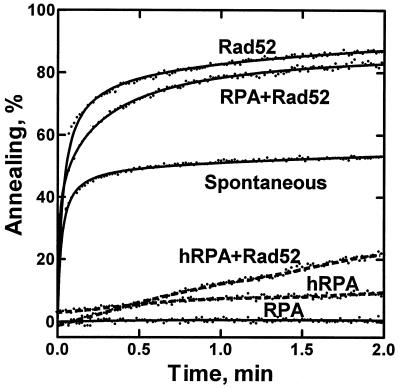
Figure 3.
Rad52 protein-mediated annealing of poly(dT) and poly(dA) is essentially unaffected by RPA. Annealing was performed with poly(dT) (400 nM) and poly(dA) (400 nM) and Rad52 protein (25 nM) in either the presence or absence of yRPA (65 nM) or hRPA (40 nM). The Rad52 protein-mediated reaction with each RPA (traces RPA+Rad52 and hRPA+Rad52) was started by addition of Rad52 protein to preincubated RPA–DNA complexes. Reactions with only RPA (traces RPA and hRPA), with only Rad52 protein (trace Rad52) and without protein (trace Spontaneous) were started by addition of poly(dT) to a preincubated (30 sec) mixture of the other components.
Rad52 Protein-Mediated Annealing of RPA–ssDNA Complexes Is Species-Specific.
Because previous studies suggested a direct protein–protein interaction between Rad52 protein and RPA in both the yeast and human systems (33, 35), we wished to determine whether species specificity is manifest in the DNA annealing reaction. For this reason, E. coli SSB protein and hRPA were tested in the Rad52 protein-dependent annealing reaction (Fig. 4A). Interestingly, Rad52 protein did not anneal DNA that was complexed with either E. coli SSB protein or hRPA. Only yRPA could support efficient DNA annealing by yeast Rad52 protein. The same species specificity was also observed in the annealing of oligonucleotides (data not shown) and of poly(dA) and poly(dT) (Fig. 3, traces hRPA and hRPA+Rad52). In Fig. 4A, each single-stranded DNA binding protein was present at a concentration approximately 1.5-fold higher than that required to cover all DNA molecules based on site sizes of E. coli SSB protein [16 nt per monomer (49, 50)], hRPA [30 nt per heterotrimer (51)], and yRPA (20 nt per heterotrimer (37); T.S. and S.C.K., unpublished results). Because our RPA preparation shows a smaller site size than that of hRPA, we used more yRPA than hRPA in Fig. 4A. However, the lack of stimulation was independent of hRPA concentration (data not shown).
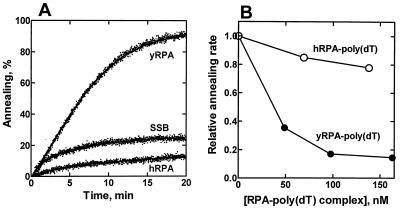
Figure 4.
Neither E. coli SSB protein nor hRPA can substitute for yRPA. (A) Annealing experiments were performed with linear heat-denatured pBluescriptSK+ plasmid DNA (1.6 μM) preincubated with 80 nM hRPA, 150 nM E. coli SSB protein, or 120 nM yRPA. Each ssDNA binding protein was preincubated with the ssDNA for 2.5 min, and the reaction was started by addition of Rad52 protein (20 nM). (B) The yRPA–ssDNA complex can sequester Rad52 protein. Heat-denatured pBluescriptSK+ plasmid DNA (0.8 μM) was preincubated with yRPA (65 nM). Annealing reactions were started by the simultaneous addition of Rad52 protein (20 nM) and various amounts of competitor. The competitor was either yRPA or hRPA that had been preincubated with a stoichiometric amount of poly(dT) for 3 min at 30°C. The rates of annealing, which were calculated from the slopes of fluorescence–time curves at approximately 100 sec after Rad52 protein addition, are expressed as relative values. The concentrations of the competitors are expressed in RPA concentration.
To eliminate the possibility that the inhibition of annealing by hRPA was nonspecific and also to further define the significance of the RPA–Rad52 protein interaction, we examined the effect of complexes of RPA and nonhomologous ssDNA on the annealing reaction (Fig. 4B). Various amounts of RPA or hRPA were preincubated with a stoichiometric amount of poly(dT) and then added to an annealing reaction at the same time as Rad52 protein. The yRPA–poly(dT) complex inhibited the rate of annealing in a concentration-dependent manner, suggesting that the yRPA–poly(dT) complex sequesters the Rad52 protein. On the other hand, the hRPA–poly(dT) complex showed only slight inhibition. These results support the idea that a direct RPA–Rad52 protein interaction is involved in the annealing of RPA-coated ssDNA by Rad52 protein.
DISCUSSION
Rad52 protein was previously shown to catalyze the annealing of two complementary oligonucleotides (14). Herein we report the effect of RPA and other ssDNA binding proteins on the Rad52 protein-mediated annealing of complementary oligonucleotides (Fig. 1), heat-denatured plasmid DNA (Fig. 2), and poly(dT) and poly(dA) (Fig. 3). Direct monitoring of DNA annealing by the dsDNA-specific fluorescent dye DAPI provides some substantial advantages in the study of this process. This method measures annealing in real time and eliminates the possible artifacts of assays, such as gel electrophoresis, in which samples are deproteinized, thereby potentially permitting spontaneous annealing in the absence of protein. For annealing of oligonucleotides, the fluorimetric assay and the gel assay showed parallel results (Fig. 1 B and C). However, in each case, the gel assay shows a slightly higher annealing rate than that of the fluorimetric assay. In addition, there is an apparent difference in results of the two assays performed in the presence of RPA alone: the gel assay shows a substantial amount of dsDNA product (approximately 30%), whereas the fluorimetric assay shows no significant annealing (compare the RPA curves in Fig. 1 B and C). We believe that this difference is caused by spontaneous annealing in the stop buffer used for the gel assay and not by limitations of the fluorimetric assay. For the RPA-only experiments, we presume that within the RPA–oligonucleotide complex, a region of several nucleotides is free to anneal. After deproteinization, the partially paired DNA can fully anneal to make complete dsDNA product; ssDNA is not deproteinized in the fluorescence method and, consequently, this assay is not prone to such an artifact.
Our results indicate that RPA, which has a strong ssDNA binding activity, is inhibitory to spontaneous DNA annealing. In the presence of Rad52 protein, however, annealing of all RPA–ssDNA complexes occurs. Interestingly, the effect of RPA on the efficiency of annealing is dramatically different depending on the DNA substrate. RPA reduces the rate of annealing of short oligonucleotides but is required for the Rad52 protein-mediated annealing of plasmid-length DNA; in contrast, annealing of poly(dT) and poly(dA) is hardly affected by RPA (Fig. 3). These results indicate that one role of RPA is to eliminate the secondary structure of ssDNA. Additionally, because Rad52 protein can promote the annealing of fortuitous regions of intramolecular complementarity that interfere with extended dsDNA formation, RPA permits extensive dsDNA formation by disrupting such weak intramolecular annealing. Our result also shows that hRPA, which is approximately 45% similar to yRPA in amino acid sequence, cannot support DNA annealing mediated by the yeast Rad52 protein (Fig. 4A). The species specificity of this reaction suggests a second role for RPA requiring an RPA–Rad52 protein interaction. This idea is confirmed by the observation that the reaction is inhibited by excess RPA (Fig. 2C) and by the RPA–poly(dT) complex (Fig. 4B). Furthermore, the Rad52 protein-mediated stimulation of DNA strand exchange by Rad51 protein also requires a species-specific interaction with RPA (17, 18) These collective observations suggest that the Rad52 protein-mediated annealing of ssDNA is biologically important.
RPA also affects the functions of another recombination protein, Rad51 protein. RPA stimulates the DNA strand exchange and ssDNA-dependent ATPase activities of Rad51 protein (36, 37). Therefore, RPA can stimulate the activities of both Rad51 and Rad52 proteins. In both cases, a primary role of RPA is to eliminate secondary structure within ssDNA. With Rad51 protein, RPA helps to produce a contiguous Rad51 protein–ssDNA presynaptic filament; specific heterologous protein–protein interactions are not required for this activity. Indeed, E. coli SSB protein can substitute for RPA in this reaction (37). Furthermore, stimulation of enzymatic activity is more effective when RPA is added after an initial Rad51 protein–ssDNA complex has been formed (37). In contrast, with Rad52 protein, a specific protein–protein interaction with RPA is required for stimulation of ssDNA annealing (Figs. 3 and 4); other ssDNA binding proteins cannot substitute for RPA. In addition, stimulation occurs only when Rad52 protein is introduced to RPA–ssDNA complex. Therefore, our results argue that the effects of RPA on Rad51 protein are mediated primarily through interaction of RPA with ssDNA whereas for Rad52 protein, RPA acts through both ssDNA- and protein–protein interactions. These differences suggest alternative mechanisms for the RPA-mediated stimulation of Rad51 protein function versus Rad52 protein function, and they highlight the multiple roles of RPA in genetic recombination, as well as in other cellular processes.
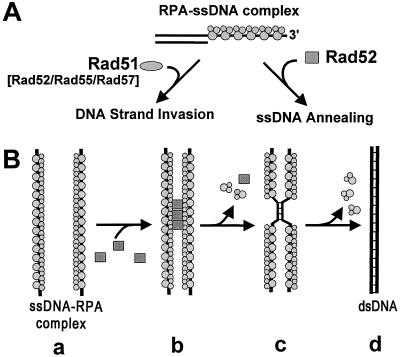
At meiosis, 5′-end-specific resection of DSBs produces 3′ ssDNA tails (52). Several lines of evidence indicate that this ssDNA is acted upon by RPA, Rad51, Rad52, Rad55, and Rad57 proteins as an early step of homologous recombination (16–18, 36, 37, 53–55). On the basis of the results in this and recent papers, we present a model for the repair of the resected DSBs (Fig. 5A). First, RPA, a protein that is abundant in the yeast cell, binds the ssDNA produced by resection. Although resection is likely to be concurrent with assembly of the recombination apparatus, RPA plays an essential early step because assembly of the recombination proteins is typically kinetically slower than the binding of a ssDNA binding protein; also, because physical analysis demonstrates that these ssDNA tails are approximately 1 kb long, RPA will be needed to prevent formation of intramolecular ssDNA secondary structure, which is inhibitory to both Rad51 and Rad52 protein function. This RPA–ssDNA complex undergoes one of two alternative fates. One fate is replacement of RPA by Rad51 protein, aided by Rad52 and Rad55–Rad57 proteins (16–18, 53), to produce a contiguous Rad51 protein–ssDNA filament (Fig. 5A, left pathway). This Rad51 protein–ssDNA complex would then invade homologous dsDNA to produce homologously paired joint molecules with the associated exchange of DNA strands (36, 37, 54, 55). An alternative fate is the annealing of the RPA–ssDNA complex to complementary ssDNA by Rad52 protein (Fig. 5A, right pathway). In this process, as we showed in this article, Rad52 protein interacts with RPA via protein–protein interaction (Fig. 5B). Interestingly, Rad52 protein, as well as RPA, is required for both these processes. This coincides with the fact that RAD52 function is required for most homologous recombination in S. cerevisiae.
Figure 5.
Model for the repair of DSBs by yeast recombination proteins. (A) There are two biochemical pathways of recombination: in both pathways, RPA binds the 3′ ssDNA tail produced by resection of the DSB. In the first (leftward arrow), in the presence of Rad52 protein and/or the Rad55–Rad57 protein complex, Rad51 protein displaces the RPA to form the presynaptic filament needed for homologous pairing and DNA strand exchange. In the alternative pathway (rightward arrow), Rad52 protein anneals the RPA–ssDNA complex with complementary ssDNA provided (presumably) by the other end of DSB. (B) The mechanism of Rad52 protein-mediated annealing of ssDNA. RPA eliminates the secondary structure within ssDNA (a), and Rad52 protein interacts with the RPA bound to the ssDNA (b) and mediates annealing (c and d).
Genetic analyses in S. cerevisiae show that one of the mechanisms for homologous recombination between direct-repeat sequences involves ssDNA annealing (56–59). A particular rfa1 mutant strain, rfa1-D228Y, shows elevated frequencies of direct-repeat recombination that appear to result from a reduced cellular concentration of RPA (34). This genetic finding can be explained by our in vitro observations that excess RPA inhibits Rad52 protein-mediated ssDNA annealing and that, at all concentrations, RPA inhibits spontaneous ssDNA annealing in the absence of Rad52 protein. Thus, a lower RPA concentration could increase recombination between direct-repeat sequences, a view that was espoused earlier (J. Smith and R. Rothstein, personal communication).
Acknowledgments
We thank Dr. Richard Kolodner for the RPA overproducing strain; Dr. Mark Wold for hRPA; Dr. Elena Zaitseva for the Rad52 overexpressing plasmid; Dr. Akira Shinohara for communication of results prior to publication; and the members of our laboratory, Dan Anderson, Deana Arnold, Piero Bianco, Frederic Chedin, Julie Kleiman, Frank Harmon, Noriko Kantake, Alex Mazin, and Erica Seitz for comments on the manuscript. This work was supported by Grants AI-18987 from the National Institute of Health and RG63 from the Human Frontier Science Program to S.C.K.
ABBREVIATIONS
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- SSB
single-stranded DNA binding protein
- RPA
replication protein A
- DAPI
4′,6-diamidino-2-phenylindole
- DSB
double-strand break
- hRPA
human RPA
- yRPA
yeast RPA
References
- 1.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 2.Wu T C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 3.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 4.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resnick M A. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 6.Thaler D S, Stahl F W. Annu Rev Genet. 1988;22:169–97. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- 7.Game J C. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 8.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Broach J R, Jones E, Pringle J, editors. I. Press, Plainview, NY: Cold Spring Harbor Lab; 1991. pp. 407–521. [Google Scholar]
- 9.Rattray A J, Symington L S. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rattray A J, Symington L S. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. Nature (London) 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 12.Muris D F, Bezzubova O, Buerstedde J M, Vreeken K, Balajee A S, Osgood C J, Troelstra C, Hoeijmakers J H, Ostermann K, Schmidt H, et al. Mutat Res. 1994;315:295–305. doi: 10.1016/0921-8777(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 13.Park M S. J Biol Chem. 1995;270:15467–15470. doi: 10.1074/jbc.270.26.15467. [DOI] [PubMed] [Google Scholar]
- 14.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman E H, Ogawa H. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- 16.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 17.New J H, Sugiyama T, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 19.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 20.Brill S J, Stillman B. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 21.Heyer W-D, Rao M R S, Erdile L F, Kelly T J, Kolodner R D. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 23.Fairman M, Prelich G, Tsurimoto T, Stillman B. Biochim Biophys Acta. 1988;951:382–387. doi: 10.1016/0167-4781(88)90110-8. [DOI] [PubMed] [Google Scholar]
- 24.Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 25.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 26.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta A, Ruppert J M, Aster J C, Winchester E. Nature (London) 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 28.He Z, Henricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Masutani C, Sugasawa K, Hanaoka F, et al. J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- 30.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 31.He Z, Wong J M S, Maniar H S, Brill S J, Ingles C J. J Biol Chem. 1996;271:28243–28249. doi: 10.1074/jbc.271.45.28243. [DOI] [PubMed] [Google Scholar]
- 32.Longhese M P, Plevani P, Lucchini G. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firmenich A A, Elias-Arnanz M, Berg P. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith J, Rothstein R. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park M S, Ludwig D L, Stigger E, Lee S H. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 36.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 37.Sugiyama T, Zaitseva E M, Kowalczykowski S C. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 38.Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S. & Ogawa, T. (1998) Genes Cells, in press. [DOI] [PubMed]
- 39.Alani E, Thresher R, Griffith J D, Kolodner R D. J Mol Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 40.Adzuma K, Ogawa T, Ogawa H. Mol Cell Biol. 1984;4:2735–2744. doi: 10.1128/mcb.4.12.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeBowitz J. Ph.D. thesis. Baltimore, MD: Johns Hopkins University; 1985. [Google Scholar]
- 42.Kowalczykowski S C, Bear D G, von Hippel P H. In: The Enzymes. Boyer P D, editor. New York: Academic; 1981. pp. 373–442. [Google Scholar]
- 43.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 45.Kubista M, Akerman B, Norden B. Biochemistry. 1987;26:4545–4553. doi: 10.1021/bi00388a057. [DOI] [PubMed] [Google Scholar]
- 46.Kapuscinski J, Szer W. Nucleic Acids Res. 1979;6:3519–3534. doi: 10.1093/nar/6.11.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eggleston A K, Rahim N A, Kowalczykowski S C. Nucleic Acids Res. 1996;24:1179–1186. doi: 10.1093/nar/24.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaitsev E N, Kowalczykowski S C. Nucleic Acids Res. 1998;26:650–654. doi: 10.1093/nar/26.2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohman T M, Overman L B, Datta S. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 50.Bujalowski W, Lohman T M. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 51.Kim C, Wold M S. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 52.Sun H, Treco D, Szostak J W. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 53.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 54.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 55.Tracy R B, Baumohl J K, Kowalczykowski S C. Genes Dev. 1997;11:3423–3431. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein H L. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiestl R H, Prakash S. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas B J, Rothstein R. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozenberger B A, Roeder G S. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]