Abstract
Objective
We have previously shown that TCEAL7 (transcription elongation factor A (SII)-like 7) is epigenetically down-regulated in the majority of epithelial ovarian cancers. We now examine the hypothesis that inherited alterations in TCEAL7 play a role in the etiology of ovarian cancer.
Methods
A two-site case-control study of 930 cases of ovarian cancer and 1037 controls, frequency-matched on residence, age and race, was conducted. Six informative SNPs (tagSNPs and putative-functional SNPs) were genotyped. Logistic regression was used to adjust for potential confounders and determine if inherited variation at this locus was associated with risk of ovarian cancer in general and among cases with invasive disease and serous histology. Gene-level principal component and haplotype analyses were also conducted.
Results
None of the SNPs or haplotypes studied were significantly associated with ovarian cancer risk overall. However, among the 440 invasive serous cases, the minor alleles for three correlated SNPs were significantly associated with reduced risk (p-values < 0.05), summarized gene-level variation was weakly associated with reduced risk (p-value=0.05), and the predominant haplotype was less common among cases than controls (0.36 v 0.40, p-value=0.05), consistent with single-SNP results.
Conclusion
TCEAL7 polymorphisms may play a role in the development of invasive serous ovarian cancers. Follow-up molecular and replication studies are warranted.
Keywords: Polymorphism
Introduction
There are over 205,000 new cases and 125,000 deaths annually from ovarian cancer worldwide [1]; overall five-year survival is around 35%, primarily because 70% of cases are diagnosed in late stages. Elucidating the risks for ovarian cancer may help in early diagnosis and consequently reduce mortality from this disease. Family history is associated with an approximate two-fold increased risk for ovarian cancer, even after accounting for known genetic syndromes due to BRCA1, BRCA2, MLH1, MSH2, and other genes [2], suggesting the existence of additional risk alleles. Although genome-wide association studies (GWAS) are underway to identify newly-associated single-nucleotide polymorphisms (SNPs), the candidate gene approach is a useful complementary mechanism that has been successfully applied in other cancers [3].
Several candidate genes have emerged from accumulating evidence of differential expression or epigenetic silencing in tumors. One down-regulated gene is transcription elongation factor A (SII)-like 7 (TCEAL7) on the X chromosome which encodes a cell death regulatory protein inactivated by methylation [4,5]. Down-regulation of TCEAL7 has been associated with increased NF-κB activity, higher levels of pro-proliferative genes cyclin D1 and c-Myc, and also pro-angiogenic genes IL-6, IL-8, and VEGF [6]. TCEAL7 shares amino acid sequence homology with several other pro-apoptotic proteins [7] and is lost in over 90% of primary ovarian tumors and 100% of cell lines tested compared to adjacent genes on the X chromosome [4]. Furthermore, in immortalized human ovarian epithelial cells, TCEAL7 down-regulation promotes anchorage-independent cell growth, and results suggest that TCEAL7 may limit Myc activity leading to restriction of ovarian epithelial cell transformation [6].
As TCEAL7 is thought to play a role in these critical cancer-related processes, we sought to assess whether inherited variation in this gene was associated with risk of epithelial ovarian cancer. We used a traditional case-control design at two study centers to examine a set of informative TCEAL7 SNPs and here report results of association-testing with the hope that novel risk alleles may help inform on disease biology and risk prediction.
Materials and methods
Study participants
Participants were recruited into two ongoing case-control studies at Mayo Clinic in Rochester, MN and at Duke University in Durham, NC. At Mayo Clinic, cases were women over age 20 years with histologically-confirmed epithelial ovarian cancer living in the Upper Midwest and enrolled within 1 year of diagnosis. Controls without ovarian cancer and who had at least one intact ovary were recruited from among those seen for general medical examinations and frequency-matched to cases on age and region of residence. At Duke University, cases were women aged 20 to 74 years with histologically-confirmed primary epithelial ovarian cancer identified using the North Carolina Central Cancer Registry's rapid case ascertainment system in a 48-county region. Controls without ovarian cancer and who had at least one intact ovary were identified from the same region as the cases using list-assisted random digit dialing and frequency-matched to cases on race and age. Additional participant details are provided elsewhere [8].
Data and biospecimen collection
Information on demographic data and known and suspected ovarian cancer risk factors were collected through in-person interviews at both sites using similar questionnaires. A common data dictionary was developed for covariates to allow combined analysis of data from both sites. Mayo Clinic participants had an extra vial of blood drawn during their scheduled medical visit to be used as a source of genomic DNA, and Duke University participants had a venipuncture at the conclusion of their interview. DNA samples from both sites were extracted from 10 to 15 mL fresh peripheral blood using the Gentra AutoPure LS Purgene salting out methodology (Gentra, Minneapolis, MN) and stored at 4 °C. Due to low quantities of available DNA for the Duke University samples, DNA was transferred to Mayo Clinic for whole-genome amplification (WGA) with the REPLI-G protocol (Qiagen Inc., Valencia CA) which we have previously shown yielded robust and reliable results in these samples [9]. Genomic and WGA DNA concentrations were adjusted to 50 ng/μl before genotyping and verified using a PicoGreen dsDNA quantitation kit (Molecular Probes, Inc., Eugene OR). Samples were bar-coded to ensure accurate processing.
SNP selection
In November 2005, we identified a set of informative SNPs (tagSNPs) in order to maximize “coverage” of underlying genetic variation by capitalizing upon the correlation of SNPs due to linkage disequilibrium (LD) within this gene. We used data from 60 unrelated Utah residents with Northern and Western European Ancestry (CEU) genotyped as part of the international HapMap Consortium (HapMap release 20, NCBI build 35) [10]. For predominantly-Caucasian study populations (89% of current participants are Caucasian), the CEU sample population has proven useful in the identification of several novel genetic associations [11,12]. We found HapMap was more informative for this gene than Perlegen Sciences (which only genotyped one SNP) [13] and Seattle SNPs1 and NIEHS SNPs2 which had not resequenced TCEAL7. Four SNPs within 5 kb of TCEAL7 with minor allele frequency (MAF)≥0.05 were binned using the algorithm of ldSelect [14] based-on pairwise-correlated SNPs at r2≥0.80. rs5945680 (3′ downstream) and rs5945767 (3′ downstream) were binned together, and we selected rs5945680 because of a greater predicted likelihood of genotype success (Illumina-provided SNP_Score, San Diego, CA); rs1045761 (5′ UTR) and rs5945971 (5′ upstream) were selected because they were independent at r2≥0.80. In addition, we identified all putative-functional SNPs (within 1 kb upstream, 5′ UTR, 3′ UTR, or non-synonymous) in Ensembl version 34 with MAF≥0.05 and SNP_Score >0.6. These consisted of rs5987515, rs5987724, and rs17340307 which were all 5′ upstream; these had not been interrogated by HapMap and no LD information was available (Supplemental Table 1).
Genotyping
Genotyping of 1086 genomic and 1282 WGA DNA samples (total=2368 including duplicates and laboratory controls) on 2051 unique study participants was performed at Mayo Clinic for using the Illumina GoldenGate™ BeadArray assay and BeadStudio software for automated genotype clustering and calling according to standard protocols [15], as part of a larger genotyping effort of 1536 SNPs [16,17]. Of 2051 participants genotyped, 74 samples of unacceptable quality (call rate <0.95 or error rate in duplicates >0.01) were excluded, and 10 participants were found to be ineligible (additional pathology review showed non-epithelial or non-ovarian primary) and excluded. Of 1536 assays, 44 SNPs failed, and six SNPs with call rates below 95% and seven monomorphic SNPs were excluded; an additional 51 SNPs failed in WGA samples only. Observed LD between SNPs was estimated using Haploview v. 4.1 [19].
Statistical analysis
Data were summarized using frequencies and percentages for categorical variables, and means and standard deviations for continuous variables. We compared demographic and clinical attributes with case status using chi-square tests and t-tests as appropriate. Association between SNPs and ovarian cancer risk was assessed using logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs). Two sets of analyses were carried out: one comparing the controls to all cases, and one subset to invasive serous cases. Primary tests for association assumed an ordinal (log-additive) genotypic relationship with simple tests for trend, and we estimated risks separately for women with one copy and two copies of the minor allele to women with no copies (referent). In addition, we used a gene-level principal component analysis to create orthogonal (e.g., uncorrelated) linear combinations of the SNP minor allele counts. The component linear combinations that accounted for at least 90% of the variability in TCEAL7 (here, three components) were included in a multivariable logistic regression model and tested using a likelihood ratio test. Also at the gene level, haplotype frequencies were estimated, and a global haplotype score test of no association between haplotypes and risk was evaluated [20]. In addition, individual haplotypes were evaluated against all other haplotypes combined; all models were fit under the assumption of a log-additive haplotype effect. rs17340307 was excluded from gene-level testing due to low observed MAF. All single-SNP, gene-specific, and haplotype-specific analyses described above were adjusted for the design variables of age, race, study site, and geographic region. The following potential confounding variables found to be significantly associated with ovarian cancer risk were also included as covariates: body mass index, postmenopausal hormone use, oral contraceptive use, parity, and age at first birth. No adjustment for multiple testing was conducted; rather, uncorrected p-values are shown and the reader is advised to interpret results conservatively. Statistical analyses used SAS (SAS Institute, Cary, NC, Version 8, 1999), S-Plus (Insightful Corp., Seattle, WA, Version 7.05, 2005), and Haplo.stats3 software programs.
Results
Participants showed the expected distributions of demographic, risk factor, and clinical characteristics. Table 1 includes the distributions of each study population by demographic and lifestyle factors, and Table 2 summarizes histology, stage, and tumor behavior among both case groups. Generally, compared to controls (N=1037), cases (N=930) were more likely to be overweight and have a family history of ovarian cancer, and they were less likely to have used oral contraceptives or given birth. Approximately 60% of cases were of serous histology, and the majority of cases were diagnosed with advanced stage disease. All TCEAL7 SNPs were successfully genotyped with call rates >99.5% and genotypes in Hardy–Weinberg equilibrium among Caucasian controls (p-values >0.05, see Supplemental Table 1) [18]. Concordance was 99.99% between duplicates of genomic DNA, 99.97% between duplicates of WGA DNA, and 99.16% between genomic and WGA DNA, indicating adequate genotyping of WGA DNA [9].
Table 1.
Characteristics of study participants.
| Mayo Clinic |
Duke University |
||||||
|---|---|---|---|---|---|---|---|
| All cases (N=396) | Controls (N=469) | p-value | All cases (N=534) | Controls (N=568) | p-value | ||
| Age, years | Mean (S.D.) | 59.8 (13.3) | 60.1 (13.0) | 0.82 | 54 (11.5) | 54.7 (12.2) | 0.35 |
| Race | Caucasian | 375 (97.2) | 462 (98.5) | 0.73 | 444 (83.3) | 479 (84.3) | 0.77 |
| African American | 3 (0.8) | 2 (0.5) | 70 (13.1) | 74 (13.0) | |||
| Hispanic | 3 (0.8) | 3 (0.8) | 5 (0.9) | 5 (0.09) | |||
| Native American | 0 (0.0) | 0 (0.0) | 5 (0.9) | 6 (1.1) | |||
| Asian | 2 (0.5) | 1 (0.2) | 6 (1.1) | 2 (0.4) | |||
| Other | 3 (0.8) | 1 (0.2) | 3 (0.6) | 2 (0.4) | |||
| Body mass index, kg/m2 | <23 | 79 (20.7) | 110 (25.1) | 0.02 | 132 (25.4) | 139 (25.2) | 0.29 |
| 23–26 | 88 (23.1) | 121 (27.6) | 117 (22.5) | 124 (22.5) | |||
| 26–29 | 98 (25.7) | 112 (25.6) | 106 (20.4) | 136 (24.7) | |||
| ≥29 | 116 (30.4) | 95 (21.7) | 165 (31.7) | 152 (27.6) | |||
| Oral contraceptive use | Never | 176 (47.6) | 166 (38.4) | <0.001 | 182 (34.7) | 181 (32.2) | 0.36 |
| 1–48 months | 98 (26.5) | 92 (21.3) | 158 (30.2) | 160 (28.5) | |||
| ≥48 months | 96 (25.9) | 174 (40.3) | 184(35.1) | 221 (39.3) | |||
| Parity, n/age at first birth, years | Nulliparous | 70 (18.3) | 66 (15.0) | 0.07 | 113 (21.2) | 73 (12.9) | 0.003 |
| 1–2/≤20 years | 29 (7.6) | 25 (5.7) | 73 (13.7) | 69 (12.1) | |||
| 1–2/>20 years | 103 (26.9) | 131 (29.8) | 193 (36.2) | 233 (41) | |||
| ≥3/≤20 years | 73 (19.1) | 64 (14.5) | 81 (15.2) | 93 (16.4) | |||
| ≥3/>20 years | 108 (28.2) | 154 (35.0) | 73 (13.7) | 100 (17.6) | |||
| Ovarian cancer family history | Yes | 51 (13.3) | 33 (7.4) | 0.01 | 42 (7.9) | 25 (4.4) | 0.02 |
| No | 333 (86.7) | 411 (92.6) | 492 (92.1) | 543 (95.6) | |||
| Education achieved | No diploma | 25 (6.9) | 19 (4.3) | <0.001 | 53 (9.9) | 69 (12.1) | 0.40 |
| High school | 136 (37.4) | 117 (26.4) | 153 (28.7) | 149 (26.2) | |||
| Post high school | 203 (55.8) | 307 (69.3) | 327 (61.4) | 350 (61.6) | |||
Data are counts (percentage) unless otherwise indicated. Counts may not total to 1967 subjects due to missing data for some variables. P-values are from t-test for continuous variables and Chi square test for categorical variables; family history indicates first or second degree relative; bold indicates p≤ 0.05.
Table 2.
Clinical characteristics of cases.
| Mayo Clinic |
Duke University |
||
|---|---|---|---|
| All cases (N=396) | All cases (N=534) | ||
| Histology | Serous | 237 (60.0) | 325 (61.2) |
| Mucinous | 28 (7.1) | 64 (12.1) | |
| Endometrioid | 65 (16.5) | 66 (12.4) | |
| Clear cell | 23 (5.8) | 33 (6.2) | |
| Mixed/other | 42 (11.6) | 43 (8.1) | |
| Stage | I | 102 (26.2) | 190 (36.1) |
| II | 29 (7.5) | 40 (7.6) | |
| III | 201 (51.7) | 276 (52.5) | |
| IV | 56 (14.4) | 20 (3.8) | |
| Behavior | Invasive | 334 (84.3) | 405 (76.0) |
| Borderline | 62 (15.7) | 128 (24.0) |
Data are summarized as count (percentage). Counts may not sum to total number of cases due to missing values for some attributes.
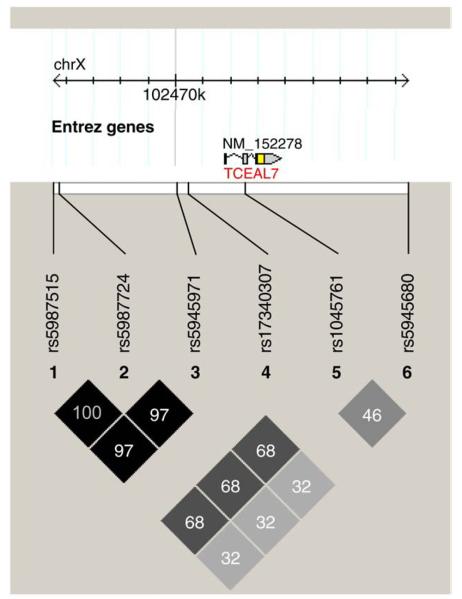
Individual TCEAL7 polymorphisms were not associated with risk of ovarian cancer overall, before or after adjustment for covariates, and gene-level principal component testing indicated that variation in TCEAL7 was not associated with overall risk (p-value=0.06). Because there is evidence that the pathogenesis of ovarian cancer differs by histological subtype, we performed additional analyses restricted to the subset of 440 invasive serous tumors. For each SNP tested, Table 3 shows genotype distributions, ovarian cancer risk associated with heterozygous and minor allele homozygous genotypes, risk associated with each copy of the minor alleles, and p-values reflecting this perallele risk. Minor alleles at 5′ flanking SNPs rs5987515, rs5987724, and rs5945971 were associated with reduced risk (ORs 0.8; 95% CIs 0.7–1.0; p-values <0.05), and, at rs5945680, minor alleles were nonsignificantly associated with increased risk (OR 1.1; 95% CI 0.9–1.3; p-value=0.29). These results appear to represent a common signal due to very strong LD among the three 5′ SNPs (r2 >0.97) and substantial LD among all common SNPs (r2 >0.32) (Fig. 1). Thus, these likely do not reflect independent associations with ovarian cancer risk, but rather, because genotypes across TCEAL7 are correlated, the true causal variant may be any one of the associated SNPs or another ungenotyped, correlated polymorphism. Results of gene-level principal component testing also support an association between these SNPs and risk of serous invasive ovarian cancer (p-value=0.05).
Table 3.
TCEAL7 polymorphisms and risk of ovarian cancer.
| SNP | Genotype | Controls (N=1037) |
All cases (N=930) |
Serous invasive cases (N=440) |
||||
|---|---|---|---|---|---|---|---|---|
| N | N | OR (95% CI) | p-value | N | OR (95% CI) | p-value | ||
| rs5987515 | CC | 377 | 364 | 1.0 (ref) | 177 | 1.0 (ref) | ||
| CA | 476 | 427 | 0.94 (0.77–1.16) | 207 | 0.95 (0.73–1.24) | |||
| AA | 179 | 136 | 0.78 (0.59–1.04) | 60 | 0.64 (0.44–0.94) | |||
| Per A allele | 0.90 (0.78–1.03) | 0.11 | 0.83 (0.70–1.00) | 0.04 | ||||
| rs5987724 | GG | 376 | 365 | 1.0 (ref) | 177 | 1.0 (ref) | ||
| GT | 474 | 428 | 0.95 (0.77–1.17) | 207 | 0.95 (0.73–1.25) | |||
| TT | 179 | 136 | 0.78 (0.59–1.03) | 60 | 0.64 (0.44–0.94) | |||
| Per T allele | 0.90 (0.78–1.02) | 0.11 | 0.83 (0.70–1.00) | 0.05 | ||||
| rs5945971 | GG | 376 | 365 | 1.0 (ref) | 176 | 1.0 (ref) | ||
| GA | 478 | 421 | 0.92 (0.75–1.13) | 205 | 0.94 (0.72–1.23) | |||
| AA | 177 | 139 | 0.80 (0.60–1.05) | 61 | 0.65 (0.44–0.95) | |||
| Per A allele | 0.90 (0.78–1.03) | 0.12 | 0.84 (0.70–1.00) | 0.05 | ||||
| rs17340307 | AA | 1033 | 918 | 1.0 (ref) | 442 | 1.0 (ref) | ||
| AC | 3 | 8 | 2.20 (0.55–8.85) | 1 | 0.66 (0.06–6.95) | |||
| CC | 0 | 0 | N.E. | 0 | N.E. | |||
| Per C allele | 2.20 (0.55–8.85) | 0.27 | 0.66 (0.06–6.95) | 0.73 | ||||
| rs1045761 | CC | 286 | 275 | 1.0 (ref) | 127 | 1.0 (ref) | ||
| CG | 488 | 448 | 0.97 (0.77–1.21) | 226 | 1.10 (0.82–1.48) | |||
| GG | 260 | 204 | 0.82 (0.63–1.07) | 90 | 0.74 (0.52–1.06) | |||
| Per G allele | 0.91 (0.79–1.03) | 0.14 | 0.87 (0.73–1.03) | 0.11 | ||||
| rs5945680 | CC | 520 | 474 | 1.0 (ref) | 219 | 1.0 (ref) | ||
| CT | 432 | 371 | 0.96 (0.79–1.17) | 185 | 1.12 (0.87–1.44) | |||
| TT | 83 | 82 | 1.18 (0.83–1.66) | 39 | 1.21 (0.78–1.89) | |||
| Per T allele | 1.03 (0.89–1.19) | 0.69 | 1.11 (0.92–1.34) | 0.29 | ||||
OR and 95% CI represent odds ratios and 95% confidence intervals from logistic regression analysis, adjusted for age, study site, race, body mass index, hormone therapy use, oral contraceptive use, parity, age at first birth, and area of residence; bold indicates p-value <0.05; N.E. indicates not estimatable.
Fig. 1.
Haploview 4.1 [19] based on Caucasian controls (N=941); r2=0=white and r2=1=black; numbers represent r2* 100, where value is missing r2=1.0; genome build 36.3; rs17340307 excluded from LD calculations due to low MAF.
Consistently, haplotype estimation suggested the existence of a common haplotype (control frequency=0.40) comprised of the minor alleles at rs5987515, rs5987724, and rs5945971 and the major allele at rs5945680. This haplotype is represented by 11110 in Table 4 which lists each haplotype, estimated frequencies in cases and controls, a score statistic (positive suggests increased risk and negative suggests decreased risk) and a corresponding p-value, as well as a global p-value of overall haplotype differences from controls for each case group. Estimated haplotype frequencies were not statistically significantly different between cases and controls in overall global testing (all cases p-value=0.48; invasive serous cases p-value=0.50). However, this particular predominant haplotype was estimated to be less frequent in serous invasive cases (0.36) than in controls (0.40, p-value=0.05). In other words, approximately 40% of all chromosomes studied were estimated to have this configuration of alleles (minor alleles at rs5987515, rs5987724, and rs5945971 and major allele at rs5945680). Compared to the remaining 60% of chromosomes with other configurations, women are estimated to have a borderline decreased risk for serous invasive disease (with carriers of chromosomes with other than the most common haplotype at increased risk). These results are consistent with the single-SNP results and the strong observed LD, and they provide modest evidence for association between variation in the TCEAL7 region and serous invasive ovarian cancer risk.
Table 4.
Estimated TCEAL7 haplotype frequencies among cases and controls.
| Haplotype | Controls | All cases (p-value=0.48) | Serous invasive cases (p-value=0.50) |
||||
|---|---|---|---|---|---|---|---|
| Frequency | Frequency | Score | p-value | Frequency | Score | p-value | |
| 11110 | 0.398 | 0.371 | −1.687 | 0.09 | 0.361 | −1.958 | 0.05 |
| 00001 | 0.289 | 0.286 | 0.282 | 0.77 | 0.293 | 0.612 | 0.54 |
| 00000 | 0.221 | 0.251 | 1.500 | 0.13 | 0.246 | 1.180 | 0.24 |
| 00010 | 0.083 | 0.082 | −0.025 | 0.98 | 0.088 | 0.340 | 0.73 |
| 11010 | 0.003 | 0.003 | −0.092 | 0.93 | 0.003 | 0.305 | 0.76 |
| 00110 | 0.002 | 0.004 | 1.364 | 0.17 | 0.005 | 1.142 | 0.25 |
0 indicates major allele, and 1 indicates minor allele for rs5987515, rs5987724, rs5945971, rs1045761, and rs5945680; adjusted for age, study site, race, parity, region of residence, age at first live birth, oral contraceptive use, hormone therapy use, and body mass index.
Discussion
In a two-study analysis of ovarian cancer cases and controls, we found evidence that correlated SNPs within TCEAL7 (including rs5987515, rs5987724, rs5945971, and rs5945680) differed in frequency among serous invasive ovarian cancer cases and controls. Because proto-oncogenes and tumor suppressor genes are involved in normal cellular proliferation, our hypothesis was that inherited alterations in these genes could initiate tumorigenesis as well as tumor progression. Several oncogenes have been identified in ovarian cancer, and several of these are reasonable candidates for inherited risk [21]. TCEAL7 represses cellular transformation by negatively modulating Myc and NF-κB, and is down-regulated in several cancer cell lines and primary ovarian tumors [6]. It is also down-regulated in breast, brain, and prostate cancer suggesting a significant role in carcinogenesis possibly through uncontrolled expression of cyclin D1 and c-Myc [6]. The suppression of cyclin D1 proteins is most likely mediated through the transcriptional modulation of CCND1 because TCEAL7 associates with its promoter region providing an alternative mechanism of cyclin D1 deregulation apart from gene amplification. TCEAL7 is also believed to suppress the c-Myc target ornithine decarboxylase [6] leading to increased c-Myc and potentially to multi-step carcinogenesis. Furthermore, the loss of TCEAL7 has been shown to promote transcriptional activity of C/EBP, SmadSBE, and Brn-3 that have a role in proliferation, anchorage-independent growth, resistance to growth inhibitors, migratory potential, and tumorigenicity [6]. Finally, a majority of ovarian cancers have shown loss of TCEAL7 [4]. Therefore, we hypothesized that SNPs tagging an underlying variation within this gene may be associated with ovarian cancer risk.
This study used a case-control design in two locales to examine TCEAL7 SNPs and, to our knowledge, is the first examination of the role of these variants in ovarian cancer risk. These results add to a growing body of literature showing associations with SNPs in candidate genes such as cytochrome P450, family 3, subfamily A, polypeptide 4 (CYP3A4), retinoblastoma 1 (RB1), and aurora kinase A (AURKA) [22,23] and SNPs identified in other cancer GWAS [24] and will serve as a complement to the novel loci yet to be reported in ongoing ovarian cancer GWAS. The strengths of this study include a large sample size, control of confounding, and the use of robust genotyping and analytical methods. In addition, the inclusion of putative-functional non-tagging 5′ SNPs as well as tagSNPs facilitated identification of a potential haplotype association among serous invasive cases. The finding of an association in this subset of cases may be due to chance, particularly considering the number of tests performed; however our a priori hypothesis asserted that serous invasive ovarian cancers behaved differently from borderline tumors [25]. Other examples of histology-specific risk alleles have been reported recently in ovarian cancer [22,26].
This analysis leads to future work in the following directions (a) replication among additional invasive serous ovarian cancer study populations, (b) examination of TCEAL7 expression levels by genotype among the current population, (c) denser genotyping to refine the potentially protective haplotype, and (d) analysis of genotypic interactions with closely-related genes. It is known that there are numerous additional genes related to TCEAL7 controlling the interplay of signaling molecules such as c-Myc, cyclin D1, NF-κB, TNF-α, and other inflammatory cytokines. Further examination of this critical gene and those which are biologically related may lead to a better informed biological understanding of serous ovarian cancers, one of the lethal subtypes. In addition, findings such as these will lead to the development of genetic risk prediction panels for eventual classification of women who may most benefit from targeted surveillance or prevention strategies.
Supplementary Material
Acknowledgments
We thank Ms. Elaine Elliott for study management, Ms. Karin Goodman and Ms. Ashley Pitzer for subject recruitment, Dr. Mark Liebow for coordination of control recruitment, and Ms. Kristin White for assistance with table preparation. Financial support was provided by R01 CA88868, R01 CA122443, Fraternal Order of Eagles, and Minnesota Ovarian Cancer Alliance.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ygyno.2009.03.038.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ramus SJ, Harrington PA, Pye C, DiCioccio RA, Cox MJ, Garlinghouse-Jones K, et al. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat. 2007;28(12):1207–15. doi: 10.1002/humu.20599. [DOI] [PubMed] [Google Scholar]
- 3.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 4.Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24(32):5089–100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 5.Brown AL, Kay GF. Bex1, a gene with increased expression in parthenogenetic embryos, is a member of a novel gene family on the mouse X chromosome. Hum Mol Genet. 1999;8(4):611–9. doi: 10.1093/hmg/8.4.611. [DOI] [PubMed] [Google Scholar]
- 6.Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008 doi: 10.1038/onc.2008.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukai J, Hachiya T, Shoji-Hoshino S, Kimura MT, Nadano D, Suvanto P, et al. NADE, a p75NTR-associated cell death executor, is involved in signal transduction mediated by the common neurotrophin receptor p75NTR. J Biol Chem. 2000;275(23):17566–70. doi: 10.1074/jbc.C000140200. [DOI] [PubMed] [Google Scholar]
- 8.Sellers TA, Schildkraut JM, Pankratz VS, Vierkant RA, Fredericksen ZS, Olson JE, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2536–43. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham JM, Sellers TA, Schildkraut JM, Fredericksen ZS, Vierkant RA, Kelemen LE, et al. Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1781–9. doi: 10.1158/1055-9965.EPI-07-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009 doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40(3):316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 13.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307(5712):1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 14.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. BioTechniques. 2002;(Suppl):56–8. 60-1. [PubMed] [Google Scholar]
- 16.Sellers TA, Huang Y, Cunningham J, Goode EL, Sutphen R, Vierkant RA, et al. Association of single nucleotide polymorphisms in glycosylation genes with risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(2):397–404. doi: 10.1158/1055-9965.EPI-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelemen LE, Sellers TA, Schildkraut JM, Cunningham JM, Vierkant RA, Pankratz VS, et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008;68(7):2498–506. doi: 10.1158/0008-5472.CAN-07-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weir BS. Genetic data analysis II: methods for discrete population genetic data. Sinauer Associates, Inc.; Sunderland MA: 1996. [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Andrilli G, Giordano A, Bovicelli A. Epithelial ovarian cancer: the role of cell cycle genes in the different histotypes. Open Clin Cancer J. 2008;2:7–12. doi: 10.2174/1874189400802010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearce CL, Near AM, Van Den Berg DJ, Ramus SJ, Gentry-Maharaj A, Menon U, et al. Validating genetic risk associations for ovarian cancer through the international Ovarian Cancer Association Consortium. Br J Cancer. 2009;100(2):412–20. doi: 10.1038/sj.bjc.6604820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramus SJ, Vierkant RA, Johnatty SE, Pike MC, Van Den Berg DJ, Wu AH, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Intl J Cancer. 2008;123(2):380–8. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H, Koessler T, Ahmed S, Ramus SJ, Kjaer SK, Dicioccio RA, et al. Association study of prostate cancer susceptibility variants with risks of invasive ovarian, breast, and colorectal cancer. Cancer Res. 2008;68(21):8837–42. doi: 10.1158/0008-5472.CAN-08-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr D, Kanitz V, Amann G, Engel J, Burges A, Lohrs U, et al. HER-2/neu gene amplification in ovarian tumours: a comprehensive immunohistochemical and FISH analysis on tissue microarrays. Histopathology. 2006;48(2):149–56. doi: 10.1111/j.1365-2559.2005.02306.x. [DOI] [PubMed] [Google Scholar]
- 26.Pearce CL, Wu AH, Gayther SA, Bale AE, Beck PA, Beesley J, et al. Progesterone receptor variation and risk of ovarian cancer is limited to the invasive endometrioid subtype: results from the ovarian cancer association consortium pooled analysis. Br J Cancer. 2008;98(2):282–8. doi: 10.1038/sj.bjc.6604170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.