Abstract
We have presented a new protein–protein docking approach to model heterodimeric structures based on the conformations of the monomeric units. The conventional modeling method relies on superimposing two monomeric structures onto the crystal structure of a homologous protein dimer. The resulting structure may exhibit severe backbone clashes at the dimeric interface depending on the backbone dissimilarity between the target and template proteins. Our method overcomes the backbone clashing problem and requires no a priori knowledge of the dimeric structure of a homologous protein. Here we used human Cystic Fibrosis Transmembrane conductance Regulator (CFTR), a chloride channel whose dysfunction causes cystic fibrosis, for illustration. The two intracellular nucleotide-binding domains (NBDs) of CFTR control the opening and closing of the channel. Yet, the structure of the CFTR’s NBD1–NBD2 complex has not been experimentally determined. Thus, correct modeling of this heterodimeric structure is valuable for understanding CFTR functions and would have potential applications for drug design for cystic fibrosis treatment. Based on the crystal structure of human CFTR’s NBD1, we constructed a model of the NBD1–NBD2 complex. The constructed model is consistent with the dimeric mode observed in the crystal structures of other ABC transporters. To verify our structural model, an ATP substrate was docked into the nucleotide-binding site. The predicted binding mode shows consistency with related crystallographic findings and CFTR functional studies. Finally, genistein, an agent that enhances CFTR activity, though the mechanism for such enhancement is unclear, was docked to the model. Our predictions agreed with genistein’s bell-shaped dose-response relationship. Potential mutagenesis experiments were proposed for understanding the potentiation mechanism of genistein and for providing insightful information for drug design targeting at CFTR. The method used in this study can be applied to modeling studies of other dimeric protein structures.
Keywords: Molecular modeling, Molecular docking, CFTR
1. Introduction
Many proteins form dimers as functional units. One such example is the Cystic Fibrosis Transmembrane conductance Regulator (CFTR). CFTR is a chloride channel in the apical membrane of epithelial cells that is regulated by cAMP-dependent protein kinase (PKA) and ATP [1]. The 1480-amino acid protein belongs to the superfamily of ATP-binding cassette (ABC) transporters. CFTR consists of two transmembrane domains (TMDs), two intracellular nucleotide-binding domains (NBD1 and NBD2) and one unique intracellular regulatory (R) domain. It has been proposed that the ATP-driven dimerization of NBD1 and NBD2 leads to the opening of the channel, and the subsequent hydrolysis of ATP at NDB2 results in dimer dissociation which is responsible for the closing of the channel (for review, see Refs. [2–4]).
Mutations in the CFTR gene cause cystic fibrosis (CF), the most common fatal, inherited disease in Caucasian populations [5]. CFTR is therefore an important therapeutic target for CF treatment. Despite significant advances in synthesizing novel compounds that can modulate the activity of CFTR, there is still no cure for CF (see [6–8] for review). The three-dimensional structure of CFTR is highly desirable for functional studies and therapeutic design regarding CFTR.
A recent breakthrough in the CF field is the determination of the crystal structures of mouse and human CFTR’s NBD1 [9–11]. These structures allow computational modeling of the unknown structure of the NBD1–NBD2 heterodimer of human CFTR.
A few efforts have recently been devoted to constructing the human CFTR’s NBD1–NBD2 structures [12–14] by utilizing the crystal structures of the NBD homodimers of other members in the ABC transporter superfamily such as MJ0796 [15]. Despite providing new insights into CFTR’s structural features, these modeled structures contain limitations, such as the low levels of sequence identity in regions between human CFTR and other ABC transporters that will directly affect the accuracy of those structures modeled by homology [12]. Another limitation is that the superimposed NBD dimer can yield severe backbone clashes at the NBD1–NBD2 interface which need to be removed by deleting some protein segments [13]. These maneuvers could potentially introduce some artifacts into the modeled dimeric structure. Lastly, this type of overlying methods requires the dimeric structure of a homologous protein, restricting their application to more general cases.
In the present work, we proposed to use a new protein–protein docking approach to construct the human CFTR’s NBD1–NBD2 heterodimeric complex based on the crystal structure of human CFTR’s NBD1 and a modeled structure of human CFTR’s NBD2 built by using NBD1 as a mold. The method requires no a priori knowledge of any dimeric structure of homologous proteins. It also overcomes the potential severe backbone clashing problem in the conventional overlying method. The constructed NBD1–NBD2 model was consistent with the experimentally observed NBD dimers of ABC transporters (see [16] for review). To further validate our model, we docked ATP molecules to the CFTR’s NBD1–NBD2 dimer, and reproduced the observed ATP binding mode in the NBD1 monomer. Lastly, we used the modeled NBD heterodimer to investigate the interaction between genistein and human CFTR’s NBD dimer by molecular docking. Genistein is an isoflavonoid found in soybeans and soy products that has a prominent effect in potentiating CFTR channel activity [17–24]. It is regarded as the gold standard for drug screening programs spearheaded by the Cystic Fibrosis Foundation. However, the molecular mechanism of genistein’s activation remains unclear. Based on our computational results, the putative binding sites of genistein on CFTR were identified, showing consistency with experimental findings.
2. Methods
2.1. Model human NBD2 structure by homology modeling
The structure of the human CFTR’s NBD2 was modeled using the crystal structure of the human CFTR’s NBD1 as a template (PDB code: 1XMI) [11]. The sequence alignment, giving a sequence identity of 24.8% and a similarity of 43.6% between NBD1 and NBD2, is shown in Fig. 1. Before modeling, the bound ATP was removed from the template. Based on the crystal structure of NBD1, the MODELLER program [25] was used to generate the three-dimensional structure of NBD2. Finally, AMBER force field minimization [26] was performed to optimize the constructed NBD2 model.
Fig. 1.
Sequence alignment of the NBD1 and NBD2 of human CFTR, obtained by slightly adjusting the sequence alignment presented by Lewis and coworkers [9] based on the crystal structure of human CFTR’s NBD1. The alignment shows a sequence identity of 24.8% and a similarity of 43.6% between NBD1 and NBD2. The “*” symbols represent identical amino acid residues, and the “+” symbols show residue pairs with positive BLOSUM62 matrix scores, representing similar sequence alignment. The prominent consensus sequences among the ABC transporter superfamily are highlighted. Among them, Walker A and Walker B are highly conserved nucleotide binding motifs in many proteins. The Signature sequence is a unique sequence that characterizes the ABC transporters. See Ref. [4] for detailed explanations.
2.2. Construct human NBD1–NBD2 heterodimer by protein–protein docking
Instead of using the conventional superimposition method described in Section 1, we proposed to use a protein–protein docking approach for model construction. We performed two types of docking calculations with ATP-free and ATP-bound NBD monomeric structures. In both cases, we obtained the biologically correct NBD1–NBD2 heterodimeric structures. Considering the critical role of ATP molecules in NBD1–NBD2 dimerization [2–4], we focused on the docking study with ATP-bound NBD monomeric structures. Specifically, NBD1 and NBD2 monomers were treated as two different proteins. The ZDOCK (version 2.1) protein–protein docking program [27] was used to search globally all possible binding configurations between NBD1 and NBD2. During protein–protein docking with human NBDs, the default parameters of ZDOCK were used. Namely, NBD1 was fixed and NBD2 was uniformly sampled with an Euler angle interval of 15° in the entire rotational space, yielding a total of 3600 rotations. For each rotation of NBD2, the complete search over the translational space was performed by the Fast Fourier Translational (FFT) algorithm with a grid spacing of 1.2 Å, and only the top translation of NBD2 with the best shape complementarity according to the scoring function in ZDOCK was kept. This yielded a total of 3600 putative NBD1–NBD2 binding modes in a global search. By default, the top 2000 putative NBD1–NBD2 binding configurations were retained in the ZDOCK final results. These 2000 NBD1–NBD2 binding configurations from ZDOCK were further refined and rescored using a more accurate scoring function, ITScore-PP, recently developed in our group [28]. The top NBD1–NBD2 binding mode that has the lowest ITScore-PP score was chosen. This predicted structure was then optimized via AMBER force field minimization. The optimized NBD1–NBD2 structure was used as the protein target for docking ATP and genistein.
It should be noted that the C-terminal extension of NBD1 into the R domain (i.e., the last 31 residues at the C-terminus) was not included in our modeling study for two reasons. First, this segment was found to exhibit prominent flexibility in crystallographic studies [9,11]. Second, our functional studies with ΔR-CFTR suggest that a deletion of the R domain has little effect on CFTR gating [29–32]. It should also be noted that residues 412–428 were missing in the crystal structure (1XMI) and therefore were not included in the present modeling study.
2.3. Molecular docking against NBD1–NBD2 heterodimer
The MDock docking program was used to identify feasible binding sites of ATP and genistein on the modeled CFTR’s NBD1–NBD2 dimer. MDock integrates the efficient ITScore scoring functions and fast ensemble docking algorithm we recently developed [33–36]. The accuracy of MDock has been extensively tested in terms of ligand binding mode identification, apparent affinity prediction, as well as virtual database screening with diverse test sets [33,34]. The source codes and executables of MDock are available free of charge for academic users (http://zoulab.dalton.missouri.edu/). During the docking calculations, the NBD1–NBD2 heterodimer was treated as a rigid body while the ligand was allowed to be flexible. Ligand flexibility was represented by docking multiple conformers of a ligand generated by OMEGA (OpenEye Scientific Software, Santa Fe, NM). The bound ATP molecules were removed from the constructed ATP-bound NBD1–NBD2 complex. The protein and ligand were saved in SYBYL-style mol2 format (Tripos Inc., St. Louis, MO) using UCSF Chimera [37]. The DOCK sphere points, representing all possible binding sites on the protein, were generated using the SPHGEN program [38] in the DOCK software package [39]. About 100 sphere points were then selected around the NBD1–NBD2 interface, representing the putative ligand binding sites, to be used by MDock for orientational searches for ATP and genistein molecules. The default docking parameters in MDock were used except for “maximum orientations” which was set to 2000 in the present study. Namely, the ligand orientations were generated by automatically matching the ligand atoms to the sphere points in the binding site. Atom clash check was performed for each generated ligand orientation. Only those orientations with less than four atom clashes between protein and ligand were valid. The orientation search continued until 2000 valid orientations were generated or the matching process was exhausted. The obtained valid ligand orientations were then scored by the ITScore scoring function and ranked from low to high according to their binding scores. The top 100 ligand orientations were retained for each conformer of the ligand. Finally, all the ligand binding orientations for different conformers of the ligand were reranked according their scores. The top 500 binding orientations were kept for further analysis.
3. Results
3.1. The modeled structure of human CFTR’s NBD1–NBD2 dimer
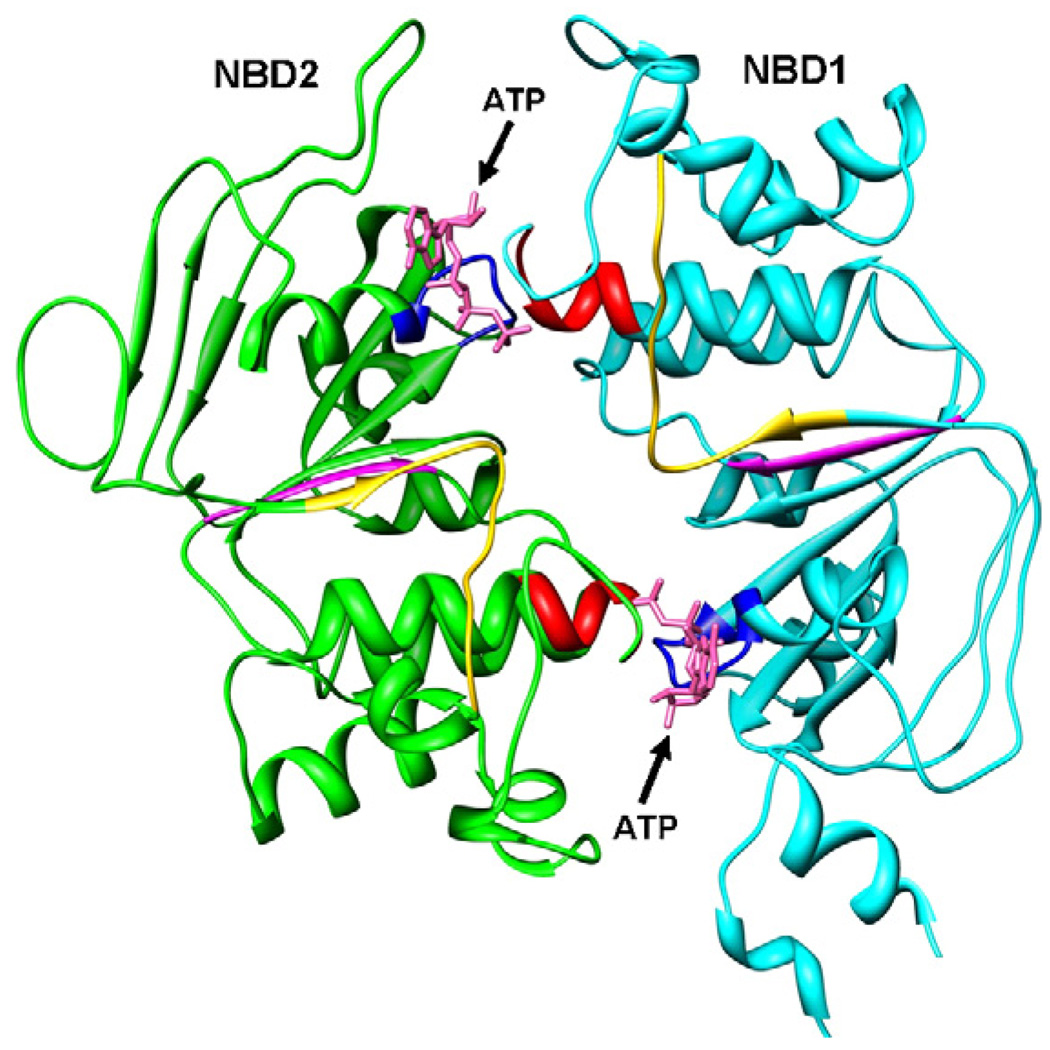
The top human NBD1–NBD2 dimer structure predicted by our protein–protein approach is shown in Fig. 2. A notable feature of the predicted structure is the head-to-tail conformation in which the Walker A and B motifs of one NBD and the signature motif of the other NBD form the ATP-binding sites (see Fig. 1 for the definition of the motifs). The same unique dimeric mode is observed in the recently solved crystal structures of the other ABC transporters (e.g., [40–44,15,45]). It should be emphasized that the protein–protein docking process automatically predicted the heterodimer as opposed to manually overlying other ABC transporters [13,12,14]. In other words, the head-to-tail conformation of NBDs is the most stable binding orientation in terms of general protein–protein interactions.
Fig. 2.
The modeled human CFTR’s NBD1–NBD2 heterodimer by protein–protein docking (top view). The NBD1 and NBD2 are represented by ribbons and colored in cyan (NBD1) and green (NBD2), including signature motif (red), Walker A motif (blue), Walker B motif (magenta), and Q-loop (yellow). The ATP molecules are shown in stick mode for reference (pink). The figure was prepared by UCSF Chimera [37].
In addition to the top binding mode of head-to-tail conformation, we also investigated the top 50 NBD1–NBD2 complexes constructed from our protein–protein docking approach by clustering the docking solutions with a RMSD cutoff of 5.0 Å. It was found that the top head-to-tail binding mode belongs to the largest cluster that includes 10 similar NBD1–NBD2 dimeric structures. This indicates that the head-to-tail conformation of NBDs is also thermodynamically stable for protein interactions.
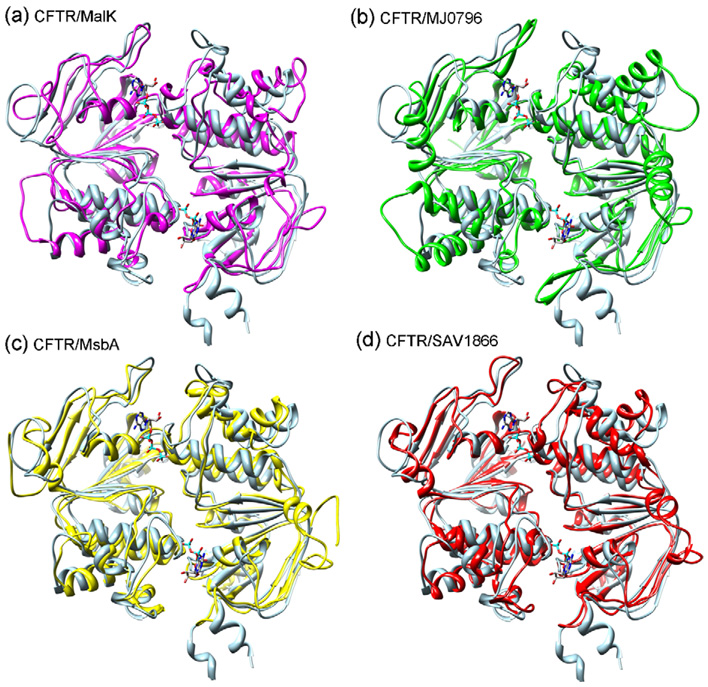
Fig. 3 showed a comparison of the modeled human CFTR’s NBD1–NBD2 complex with four homologous dimers in the ABC transporter family, MalK (PDB code: 1Q12) [44], MJ0796 (PDB code: 1L2T) [15], MsbA (PDB code: 3B60) [45] and SAV1866 (PDB code: 2HYD) [42], respectively. It can be seen from the figure that two major structural similarities are shared among the five complexes. First, they all adopt the head-to-tail conformations. Second, the dimer interface including the ATP binding site is rather conserved and has much less conformational changes than the rest of the protein. Despite the similarities among five NBD1–NBD2 complexes, there exist significant differences in their conformations (Fig. 3 and Table 1). The backbone RMSDs between the human NBD1–NBD2 complex and the four homologous dimers vary from 3.48 to 6.67 Å for the whole protein, from 1.72 to 3.90 Å for the NBD1–NBD2 interface, and from 1.78 to 3.50 Å for the ATP binding sites, respectively (shown in Table 1). The comparison in Table 1 also suggested that MsbA and SAV1866 may be slightly better templates than MalK and MJ0796 if using the conventional overlying method to construct human CFTR’s NBD1–NBD2 structure, because MsbA and SAV1866 have smaller backbone RMSD (i.e., conformational difference) at their interfaces. Indeed, as reported in the work by Moran et al. [13], the MJ0796-based human CFTR’s NBD1–NBD2 dimeric structure using the overlying method still experienced severe backbone clashes at its interface after extensive energy minimization. A total of 38 residues from NBD1 and 37 residues from NBD2 were deleted to remove the clashes [13].
Fig. 3.
Comparison of the modeled human CFTR’s NBD1–NBD2 heterodimer and four homologous NBD dimers of MalK [44](a), MJ0796 [15](b), MsbA [45](c), and SAV1866 [42](d) in the ABC transporter superfamily. The NBD1 and NBD2 are represented by ribbons. The ATP molecules are colored by atom type and shown in stick mode for reference. The homologous dimers are superimposed onto the human CFTR’s NBD1–NBD2 heterodimer by using UCSF Chimera [37]. The CFTR NBD1–NBD2 is colored in light blue, and the four homologous dimers are colored in magenta (a, Malk), green (b, MJ0796), yellow (c, Msba) and red (d, SAV1866), respectively. In each panel, the monomers on the right stand for NBD1 and the monomers on the left for NBD2. The figure was prepared by UCSF Chimera [37].
Table 1.
Root mean square deviation (RMSD) of the NBD dimers between human CFTR and four homologous ABC transporters.
| Dimer (Å) | Dimer interface a(Å) | ATP binding sites b(Å) | |
|---|---|---|---|
| MalK [44] | 6.67 | 3.90 | 3.50 |
| MJ0796 [15] | 4.95 | 2.23 | 2.41 |
| MsbA [45] | 4.70 | 1.72 | 1.78 |
| SAV1866 [42] | 3.43 | 1.74 | 1.81 |
The RMSD was calculated based on the backbone atoms after superimposing the homologous dimers onto human CFTR’s NBD1-NBD2 complex using UCSF Chimera [37].
If any atom of a residue in the NBD1 (NBD2) is less than 5.0 Å from any atom of the NBD2 (NBD1) or the ATP molecules, the residue is defined to belong to the dimer interface.
The ATP binding sites stand for those residues of the NBDs that are within 5.0 Å from the ATP molecules.
3.2. The binding mode of ATP in the human CFTR’s NBD1–NBD2 dimer
To further verify our constructed dimeric structure, we used the molecular docking approach described in Section 2 to dock the two substrate molecules (i.e., ATP) to the modeled CFTR’s NBD1–NBD2 dimer. We obtained almost the same binding mode of ATP as that observed in the crystal structure of ATP-bound NBD1 monomer of human CFTR.
For comparison, we also constructed an NBD1–NBD2 heterodimer structure by superimposing the human CFTR’s NBD1 and NBD2 monomers onto the MJ0796 dimer, as used in the previous models [12,13]. It was found that the ATP binding sites are blocked in the constructed NBD1–NBD2 dimer structure because of the severe atomic clashes at the dimer interface (data not shown). These results suggest that the constructed human CFTR’s NBD1–NBD2 heterodimer by protein–protein docking is perhaps structurally more reasonable than those generated by simply fitting NBDs onto known NBD dimers.
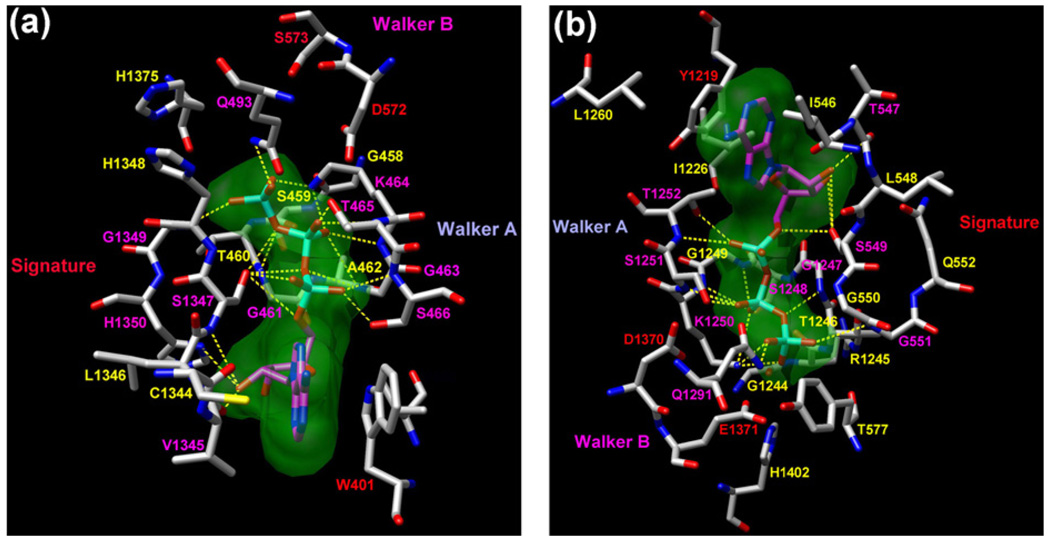
The predicted ATP binding mode in the CFTR’s NBD1–NBD2 dimer is illustrated in Fig. 4 and the corresponding scores are shown in Table 2. Fig. 4(a) shows the ATP binding site consisting of Walker A and B motifs of NBD1 and the signature motif of NBD2, referred to as the NBD1 site. Fig. 4(b) shows the NBD2 site, which is defined similarly. Residues that have any atom within 5 Å from the ATP molecules are displayed, except S573 in Fig. 4(a). S573 is displayed despite being 5.7 Å away from the ATP, because it is a distinct residue in the Walker B motif of CFTR’s NBD1. The corresponding residue on NBD2 of CFTR and NBDs of many other ABC transporters is a glutamate (e.g., E1371 for CFTR’s NBD2) which plays a crucial role in ATP hydrolysis. The replacement of glutamate by serine at this location results in the incapability of CFTR’s NBD1 in hydrolyzing ATP [46–48]. In Fig. 4(a), residues at the NBD1 site that form hydrogen bonds with the ATP molecule include G461, G463, K464, T465, S466 (Walker A of NBD1), Q493 (Q loop of NBD1),V1345,S1347,G1349 and H1350 (signature motif of NBD2). Many of these residues are functionally important in CFTR channel regulation (see the reviews in Refs. [2] and [4]). Mutations at most of these residues are CF-associated mutations (see http://www.genet.sickkids.on.ca/cftr). D572 of Walker B of NBD1 is supposed to coordinate MgATP. W401 of NBD1 forms ring–ring stacking with the adenine ring of ATP, which is consistent with our experimental finding that this residue coordinates the bound ATP at the NBD1 site [32]. In Fig. 4(b), residues at the NBD2 site that exhibit hydrogen bonding with the second ATP molecule include G1247, S1248, K1250, S1251, T1252 (Walker A of NBD2), Q1291 (Q loop of NBD2), T547, S549, and G551 (signature motif of NBD1). D1370 and E1371 (Walker B of NBD2) are expected to coordinate MgATP. Y1219 of NBD2 coordinates the adenine ring of ATP through ring–ring stacking, again consistent with our experimental results [32]. These predicted interactions corroborate what have been observed in the crystal structures solved for the NBD dimers of other ABC transporters [16].
Fig. 4.
Interactions between the docked ATP molecules and the modeled CFTR’s NBD1–NBD2 dimer at the NBD1 site (a) and NBD2 site (b). The two panels show all the NBD residues that have atoms within 5 Å from the ATP molecules, except S573 in panel (a). Both the NBD residues and the ATP molecules are plotted in stick mode and colored by atom type. For clarity, each ATP is enclosed by a molecular surface. The potential hydrogen bonds between the ATP and NBDs are shown in yellow dashed lines. The residues forming hydrogen bonds with ATP are labeled in magenta. The other important residues reported in literature are labeled in yellow. The figure was prepared by UCSF Chimera [37].
Table 2.
Predicted binding energy scores of genistein and ATP molecules bound to human CFTR NBD1–NBD2 using MDock (see Fig. 6 for the corresponding orientations).
| Genistein | ATP | |
|---|---|---|
| Site 1a | −51.7 | −103.3 |
| Site 1b | −36.9 | −73.7 |
| Site 2a | −49.2 | - |
| Site 2b | −38.4 | - |
| Site 3 | −45.0 | - |
The binding sites are defined in Figure 5.
3.3. Probing the binding sites of genistein on the NBD heterodimer of human CFTR
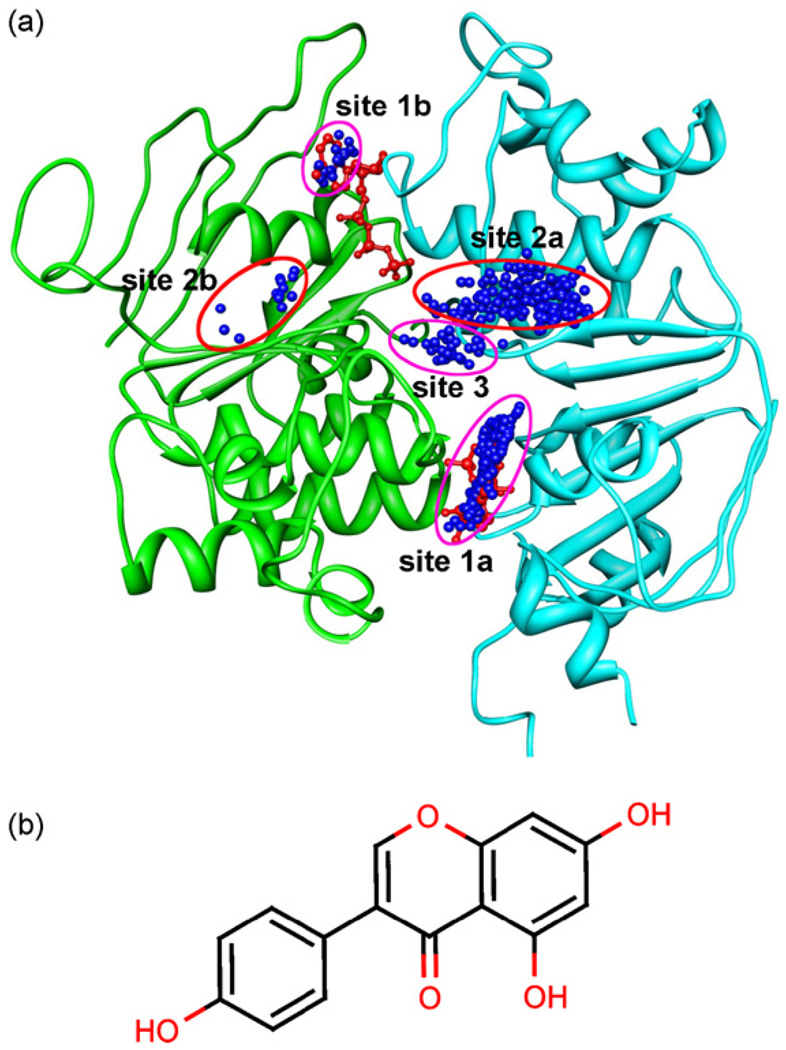
Investigating genistein’s binding sites may help explain the action mechanism of genistein and other potentiators and thus contributes to drug design for CF treatment. With this in mind, we performed a docking calculation of genistein by running the MDock program on the modeled CFTR’s NBD1–NBD2 heterodimer, as described in the Methods section. The top 500 docked orientations of genistein were shown in Fig. 5. It can be seen from the figure that the 500 ligand orientations of genistein roughly form five clusters, corresponding to five putative binding sites on the NBD1–NBD2 dimer. According to their binding scores, the top orientations of genistein in each of five cluster were selected to represent five putative binding sites on the NBD1–NBD2 dimer, respectively, which are displayed in Fig. 6. The corresponding binding scores are listed in Table 2. These five binding sites can be grouped into three types. The first type of binding site (sites 1a and 1b) is located at the ATP binding site, where bindings of genistein and ATP are competitive. As with ATP, one aromatic ring of genistein stacks well with the residue W401 on NBD1 (site 1a) or Y1219 on NBD2 (site lb). Noticeably, the calculated affinity scores for genistein at sites la and lb (−51.7 and −36.9, respectively) are significantly worse than the affinity scores for ATP (−103.3 and −73.7, respectively). This finding may explain early experimental results that elevated concentrations of genistein inhibit CFTR [20,21].
Fig. 5.
(a) The top 500 putative binding modes of genistein on the human CFTR’s NBD1–NBD2 heterodimer predicted by molecular docking (top view), in which each docked orientation is represented by a blue dot for better illustration. The NBD1 and NBD2 are represented by ribbons, and colored in cyan (NBD1) and green (NBD2), respectively. ATP molecules are shown in ball-and-stick mode for reference (red). The 500 ligand orientations roughly form five clusters as indicated by circles, representing five putative binding sites. The figure was prepared by UCSF Chimera [37]. (b) Two-dimensional structure of genistein. The figure was produced by MarvinView (ChemAxon Ltd., http://www.chemaxon.com/marvin).
Fig. 6.
A stereo view of the interactions between genistein and human CFTR’s NBD1–NBD2 heterodimer at the five putative binding sites (top view). NBD1 and NBD2 are represented by ribbons, and colored in cyan (NBD1) and green (NBD2), respectively. The genistein molecules are shown in ball-and-stick mode (blue), and the ATP molecules are shown in stick mode for reference (yellow). Several residues that are important for interacting with genistein are displayed in stick mode (magenta). The figure was prepared by UCSF Chimera [37].
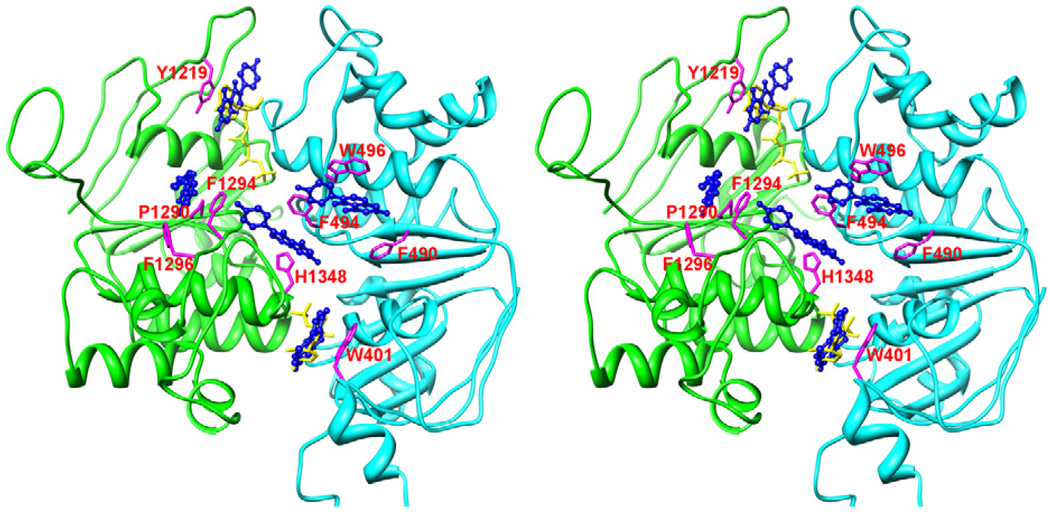
The second type of binding site is located almost symmetrically on NBD1 (site 2a) and NBD2 (site 2b), respectively. These two ligand orientations interact with residues F490, F494 and W496 on NBD1 and residues P1290, F1294 and F1296 on NBD2, respectively. Although this binding site is not located at the dimer interface, genistein may affect the stabilization of the NBD1–NBD2 dimer by interacting with interface residues such as F494 and F1294 or by inducing NBDs’ conformational changes. The third binding site (site 3) is located at the central NBD1–NBD2 interface coordinated by the residues F494 of NBD1 and F1294 and H1348 of NBD2. Binding of genistein at this site can directly stabilize the NBD1–NBD2 dimer.
Several reasons make sites 2a, 2b and 3 interesting. First, with multiple aromatic amino acids involved in coordinating genistein, these sites may assume a higher binding affinity for genistein. Second, the three sites are physically close to the highly conserved glutamine residue in the Q-loop of respective NBD (Q493 for NBD1 and Q1291 for NBD2). The interactions between the Q-loop and signature motif may explain why G551D and G1349D, the CF-associated mutations in the signature motif, influence genistein’s effects [49,13]. Third, binding genistein at any of the three sites helps stabilize the NBD1–NBD2 dimer because genistein at these sites can directly interact with the residues from one or both sides at the NBD1–NBD2 interface, as discussed before (Fig. 6). Noticeably, our previous studies suggest that stabilizing the NBD heterodimer will keep the channel open and enhance CFTR current [30–32]. Therefore, genistein binding to one of the three sites may explain the potentiation effect of genistein. These predictions on genistein’s binding sites need to be further verified by future mutagenesis experiments.
4. Conclusion
In the present study, we have used NBD1–NBD2 heterodimer of human CFTR as an example to present a protein–protein docking approach for modeling dimeric structures based on the monomeric structures without a priori structural knowledge about homologous protein dimers. It also avoids the problem of severe backbone clashes at the interface, another potential disadvantage of the conventional overlying method.
The dimeric mode of our constructed human CFTR’s NBD1–NBD2 structure is consistent with the unique head-to-tail dimeric modes found in the crystal structures of other members in the ABC transporter superfamily. The predicted binding mode of the substrate (i.e., ATP) also shows consistency with the binding mode observed in the crystal structures of other ABC transporters and in functional studies of CFTR. In contrast, direct super-imposition of the monomeric structures onto the crystal structure of the NBD homodimer of another member in the ABC transporter family may cause severe backbone clashes at the dimeric interface and leave insufficient space for ATP to bind.
Based on our modeled structure, we performed molecular docking to predict putative binding sites of genistein on the dimer. Our prediction of multiple binding sites for gensitein corroborates the measured bell-shaped dose–response relationship of genistein that at low concentrations, genistein enhances the CFTR current [20,21,13](presumably due to genistein’s binding to sites 2 and/or 3) whereas at high concentrations, CFTR current decreases [20,21](presumably due to genistein’s competition with ATP at site 1). The modeling results suggest that an efficient, ideal potentiator should bind to site 3 at the NBD interface but not to site 1 at ATP binding sites, a theory that may be used in the practice of drug design targeting at CFTR.
To clarify the potentiation mechanism of genistein, we propose point mutations at the following residues to identify the binding site(s) of genistein: F490, F494 and W496 on NBD1, and P1290, F1294, F1296, and H1348 on NBD2 (see Fig. 6). Converting one of these residues to glycine or alanine is expected to remove the corresponding ring–ring stacking interaction between genistein and CFTR at sites 2 or 3 and therefore reduce binding tightness. Our preliminary study with F494G-CFTR using whole-cell electrophysiological recording showed that indeed the apparent affinity of genistein was lowered to 25.3 µM, compared to 4.4 µM for wild-type CFTR (M. Li and T.-C. Hwang, unpublished data). Future whole-cell dose–response measurements and further single-channel studies on these mutated CFTR channels together with control experiments would clarify whether genistein binds to either or both sites 2 and 3. The findings would provide valuable information for rational drug design targeting at CFTR for the treatment of cystic fibrosis.
Acknowledgements
Support to XZ from OpenEye Scientific Software Inc. (Santa Fe, NM) and Tripos, Inc. (St. Louis, MO) is gratefully acknowledged. Zou is supported by the Cystic Fibrosis Foundation grant ZOU07I0, NIH grant DK61529, and the Research Board Award of the University of Missouri RB-07-32. Hwang is supported by NIH R01DK55835 and NIH R01HL53445. The support from Cystic Fibrosis Foundation Therapeutics, Inc. CLARKE06XXO grant is gratefully acknowledged. The work is also supported by Federal Earmark NASA Funds for Bioinformatics Consortium Equipment and additional financial support from Dell, SGI, Sun Microsystems, TimeLogic, and Intel. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
References
- 1.Riordan J, Rommens J, Kerem B, Alon N, Rozhamel R, Grzeleczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Chen T-Y, Hwang T-C. CLC-0 and CFTR: Chloride channels evolved from transporters. Physiol. Rev. 2008;88:351–387. doi: 10.1152/physrev.00058.2006. [DOI] [PubMed] [Google Scholar]
- 3.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou X, Hwang T-C. ATP hydrolysis-coupled gating of CFTR chloride channels: Structure and function. Biochemistry. 2001;40:5579–5586. doi: 10.1021/bi010133c. [DOI] [PubMed] [Google Scholar]
- 5.Welsh MJ, Smith AE. Molecular mechanisms of the CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 6.Amaral MD, Kunzelmann K. Molecular targeting of CFTR as a therapeutic approach to cystic fibrosis. Trends Pharmacol. Sci. 2007;28:334–341. doi: 10.1016/j.tips.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Becq F. On the discovery and development of CFTR chloride channel activators. Curr. Pharm. Des. 2006;12:471–484. doi: 10.2174/138161206775474459. [DOI] [PubMed] [Google Scholar]
- 8.Hwang T-C, Sheppard DN. Molecular pharmacology of the CFTR Cl− channel. Trends Pharmacol. Sci. 1999;20:448–453. doi: 10.1016/s0165-6147(99)01386-3. [DOI] [PubMed] [Google Scholar]
- 9.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat. Struct. Mol. Biol. 2004;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, Maloney PC, Guggino WB, Hunt JF, Emtage S. Impact of the delta-F508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 12.Callebaut I, Eudes R, Mornon JP, Lehn P. Nucleotide binding domains of human cystic fibrosis transmembrane conductance regulator: Detailed sequence analysis and three dimensional modeling of the heterodimer. Cell. Mol. Life Sci. 2004;61:230–242. doi: 10.1007/s00018-003-3386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran O, Galietta LJ, Zegarra-Moran O. Binding site of activators of the cystic fibrosis transmembrane conductance regulator in the nucleotide binding domains. Cell. Mol. Life Sci. 2005;62:446–460. doi: 10.1007/s00018-004-4422-3. [DOI] [PubMed] [Google Scholar]
- 14.Mense M, Vergani P, White DM, Altberg G, Nairn AC, Gadsby DC. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 2006;25:4728–4739. doi: 10.1038/sj.emboj.7601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith PC, Karpowich N, Milien L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenstein K, Dawson RJP, Locher KP. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Illek B, Fischer H, Santos GF, Widdicombe JH, Machen TE, Reenstra WW. cAMP-independent activation of CFTR Cl channels by the tyrosine kinase inhibitor genistein. Am. J. Physiol. 1995;268:C886–C893. doi: 10.1152/ajpcell.1995.268.4.C886. [DOI] [PubMed] [Google Scholar]
- 18.Illek B, Fischer H, Machen TE. Alternate stimulation of apical CFTR by genistein in epithelia. Am. J. Physiol. 1996;270:C265–C275. doi: 10.1152/ajpcell.1996.270.1.C265. [DOI] [PubMed] [Google Scholar]
- 19.Hwang T-C, Wang F, Yang IC-H, Reenstra WW. Genistein potentiates wild-type and delta F508-CFTR channel activity. Am. J. Physiol. 1997;273:988–998. doi: 10.1152/ajpcell.1997.273.3.C988. [DOI] [PubMed] [Google Scholar]
- 20.Weinreich F, Wood PG, Riordan JR, Nagel G. Direct action of genistein on CFTR. Pflugers Archiv. 1997;434:484–491. doi: 10.1007/s004240050424. [DOI] [PubMed] [Google Scholar]
- 21.Wang F, Zeltwanger S, Yang I, Nairn A, Hwang T-C. Action of genistein on cystic fibrosis transmembrane conductance regulator gating: evidence of two binding sites with opposite effects. J. Gen. Physiol. 1998;111:477–490. doi: 10.1085/jgp.111.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illek B, Zhang L, Lewis NC, Moss RB, Dong JY, Fischer H. Defective function of the cystic fibrosis-causing missense mutation G551D is recovered by genistein. Am. J. Physiol. 1999;277:C833–C839. doi: 10.1152/ajpcell.1999.277.4.C833. [DOI] [PubMed] [Google Scholar]
- 23.Randak C, Auerswald EA, Assfalg-Machleidt I, Reenstra WW, Machleidt W. Inhibition of ATPase, GTPase and adenylate kinase activities of the second nucleotide-binding fold of the cystic fibrosis transmembrane conductance regulator by genistein. Biochem. J. 1999;340:227–235. [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Nakkash L, Hu SH, Li M, Hwang T-C. A common mechanism for cystic fibrosis transmembrane conductance regulator protein activation by genistein and benzimidazolone analogs. J. Pharmacol. Exp. Ther. 2001;296:464–472. [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Stuart A, Fiser A, Sá nchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 26.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Dauan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA., III . AMBER 7. San Francisco, CA: University of California; 2002. [Google Scholar]
- 27.Chen R, Weng ZP. A novel shape complementarity scoring function for protein–protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 28.Huang S-Y, Zou X. An iterative knowledge-based scoring function for protein–protein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 29.Bompadre SG, Ai T, Cho JH, Wang X, Sohma Y, Li M, Hwang T-C. CFTR gating I: Characterization of the ATP-dependent gating of a phosphorylation-independent CFTR channel (ΔR-CFTR) J. Gen. Physiol. 2005;125:361–375. doi: 10.1085/jgp.200409227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bompadre SG, Cho JH, Wang X, Zou X, Sohma Y, Li M, Hwang T-C. CFTR gating II: Effects of nucleotide binding on the stability of open states. J. Gen. Physiol. 2005;125:377–394. doi: 10.1085/jgp.200409228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Wang X, Li M, Sohma Y, Zou X, Hwang T-C. High affinity ATP/ADP analogues as new tools for studying CFTR gating. J. Physiol. 2005;569:447–457. doi: 10.1113/jphysiol.2005.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Wang X, Liu H-Y, Zou X, Li M, Hwang T-C. The two ATP binding sites of Cystic Fibrosis Transmembrane conductance Regulator (CFTR) play distinct roles in gating kinetics and energetics. J. Gen. Physiol. 2006;128:413–422. doi: 10.1085/jgp.200609622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein–ligand interactions. I. Derivation of interaction potentials. J. Comput. Chem. 2006;27:1866–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 34.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein–ligand interactions. II. Validation of the scoring function. J. Comput. Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- 35.Huang S-Y, Zou X. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 36.Huang S-Y, Zou X. Efficient molecular docking of NMR structures: Application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 39.Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput. -Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 40.Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 41.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 42.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 43.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabo K, Szakacs G, Hegedus T, Sarkadi B. Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator: Different patterns in the two nucleotide binding domains. J. Biol. Chem. 1999;274:12209–12212. doi: 10.1074/jbc.274.18.12209. [DOI] [PubMed] [Google Scholar]
- 47.Aleksandrov L, Aleksandrov AA, Chang X-B, Riordan JR. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J. Biol. Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 48.Basso C, Vergani P, Nairn AC, Gadsby DC. Prolonged nonhydrolytic interaction of nucleotide when CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J. Gen. Physiol. 2003;122:333–348. doi: 10.1085/jgp.200308798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derand R, Bulteau-Pignoux L, Becq F. The cystic fibrosis mutation G551D alters the non-Michaelis-Menten behavior of the cystic fibrosis transmembrane conductance regulator (CFTR) channel and abolishes the inhibitory genistein binding site. J. Biol. Chem. 2002;277:35999–36004. doi: 10.1074/jbc.M206121200. [DOI] [PubMed] [Google Scholar]