Abstract
The antioxidant defense system involves complex functional coordination of multiple components in different organelles within the plant cell. Here, we have studied the Arabidopsis thaliana early response to the generation of superoxide anion in chloroplasts during active photosynthesis. We exposed plants to methyl viologen (MV), a superoxide anion propagator in the light, and performed biochemical and expression profiling experiments using Affymetrix ATH1 GeneChip® microarrays under conditions in which photosynthesis and antioxidant enzymes were active. Data analysis identified superoxide-responsive genes that were compared with available microarray results. Examples include genes encoding proteins with unknown function, transcription factors and signal transduction components. A common GAAAAGTCAAAC motif containing the W-box consensus sequence of WRKY transcription factors, was found in the promoters of genes highly up-regulated by superoxide. Band shift assays showed that oxidative treatments enhanced the specific binding of leaf protein extracts to this motif. In addition, GUS reporter gene fused to WRKY30 promoter, which contains this binding motif, was induced by MV and H2O2. Overall, our study suggests that genes involved in signalling pathways and with unknown functions are rapidly activated by superoxide anion generated in photosynthetically active chloroplasts, as part of the early antioxidant response of Arabidopsis leaves.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-007-9274-4) contains supplementary material, which is available to authorized users.
Keywords: Antioxidant response, Chloroplast, Hsp, Oxidative stress, WRKY
Introduction
Reactive oxygen species (ROS) are continuously produced during photosynthesis in the chloroplasts by partial reduction of oxygen molecules or energy transfer to them. The major site of superoxide anion (O•−2) production is the thylakoid membrane-bound primary electron acceptor of photosystem I (PSI). This O•−2 is further converted to hydrogen peroxide (H2O2) within the chloroplast mostly by CuZn–superoxide dismutase (SOD) (Asada 1994). At the same time, excited triplet chlorophyll molecules in photosystem II (PSII) interact with O2 to generate singlet oxygen (Macpherson et al. 1993). Plants had to evolve efficient strategies to cope with the accumulation of these potentially toxic compounds that are integral components of oxygenic photosynthesis (Mittler 2002).
In recent years, it is increasingly clear that ROS also function as signalling molecules to control various processes in plants outside of oxidative stress (Apel and Hirt 2004), and a transcriptomic analysis of the specific signalling capacities of different ROS in plants was reported (Gadjev et al. 2006). In addition, signals from damaged chloroplasts that coordinate nuclear gene expression have been described (Koussevitzky et al. 2007). However, our knowledge on the signalling pathways arising from ROS generated in chloroplasts is still incomplete.
Efforts to identify ROS-responsive genes on a global scale in Arabidopsis thaliana have provided significant progress in surveying gene expression in response to singlet oxygen (op den Camp et al. 2003) and H2O2 (Desikan et al. 2000; Vandenabeele et al. 2004). Arabidopsis mutants deficient in the ROS scavenging system or external application of H2O2 to Arabidopsis plants and cell cultures, where H2O2 is expected to diffuse into the cell, provided comprehensive analyses of the H2O2 transcriptional gene networks (Desikan et al. 2000; Mittler et al. 2004; Vandenabeele et al. 2004; Davletova et al. 2005). To attribute a signalling role to a specific ROS it is important to confine its generation to a definable intracellular compartment, in short time and in a way that the intensity of the stress is severe but not strong enough to cause collateral effects. In chloroplasts, it is possible to generate O•−2 steadily in the light by treating plants with methyl viologen (MV) or paraquat (Mehler 1951). Bipyridyl compounds such as MV are redox-active molecules that are taken up by the cell, undergo univalent reduction and subsequently transfer their electrons to oxygen, forming the superoxide anion and regenerating oxidized MV, that may engage in successive rounds of redox cycling (reviewed in Asada and Takahashi 1987; Halliwell and Gutteridge 1989). Although MV can be reduced by a number of enzymes and electron transfer systems of the plant cell, photoreduction in chloroplasts represents the most efficient pathway. During illumination, electrons are donated to MV from PSI, ensuring that superoxide is formed primarily in the chloroplasts. MV can also mediate toxic effects in the dark, albeit to a lesser extent; in this case, the major electron donor may be a microsomal NADPH-cytochrome P-450 reductase, as it is in animal cells (Halliwell and Gutteridge 1989). The effects of MV in the dark are much less dramatic (Tsang et al. 1991). Therefore, plants grown photoautotrophically harboring well developed thylakoid membranes are needed to generate O•− 2at significant rate in the chloroplast. An analysis of expression data obtained from Arabidopsis seedlings, grown heterotrophically on MS medium containing sucrose and exposed to exogenously applied 10 μM MV (available at http://www.arabidopsis.org/info/expression/ATGenExpress.jsp), revealed that the plant’s response to MV was delayed when compared with the response to other ROS (Gadjev et al. 2006). The authors attributed this difference to the longer time needed for MV to penetrate the leaves and perform its action.
Early-response genes are expected to encode signalling or regulatory proteins such as protein kinases/phosphatases and transcription factors. Such genes respond quickly with a change in transcript level after stress treatment. Transcriptional profiling of Arabidopsis leaves after wounding revealed clear examples of early response genes whose protein products could function to regulate late response gene expression (Cheong et al. 2002).
Here we evaluate the expression of more than 22,000 Arabidopsis thaliana genes during early generation of O•−2 in the chloroplasts of soil-grown plants using high-density oligonucleotide probe microarrays. Our goal was to study the influence of early chloroplastic O•−2 generation at the transcriptional level and chloroplastic components, to shed some light on the role that O•−2 might play in the signalling pathways connecting chloroplasts and nuclei. Considering that all antioxidant genes known to date are nuclear-encoded (Grene 2002), it is essential for the photoautotrophic life-style of plants to coordinate nuclear gene expression with chloroplast function. Arabidopsis plants grown for two weeks on soil were subjected to a 2 h MV treatment in the light. Genes showing expression changes after the oxidative challenge were assigned to functional categories and biochemical tests of antioxidant or stress-related proteins were performed in combination with the expression profiling. Resulting data were compared with the global changes of gene expression in Arabidopsis seedlings in response to different types of ROS and environmental stresses.
Materials and methods
Plant growth conditions and treatments
Arabidopsis thaliana (L.) Heynh. Col-0 plants were grown on soil in 10-cm pots (50 per flat) under 120 μmol m−2 s−1 with a 16-h period at 23°C, and relative humidity of 65–70% for 2 weeks. Plants were carefully removed from soil and floated with adaxial surfaces facing the light source in 50 μM MV, 0.01 % (v/v) Tween 20 solution for 2 h at 120 μmol m−2 s−1 and 23°C. Control plants were incubated in parallel, and handled as treated plants, except that MV was omitted. After treatment, aerial parts were separated from the roots, and immediately processed for RNA isolation. For testing ROS induction of WRKY30 promoter, transgenic Arabidopsis plants described below were grown for 3–4 weeks, removed from soil, vacuum infiltrated and then floated in 5 × 10−3 % (v/v) Silwet L-77 (Lehle Seeds, Round Rock, TX, USA) solution supplemented with either 50 μM MV or 20 mM H2O2.
Determination of pigment content
Pigments were extracted from leaves with 96 % (v/v) ethanol and the supernatant was used to measure contents of chlorophyll a and b, and carotenoids (Lichtenthaler 1987).
Electrolyte leakage
Aerial parts from two-week-old Arabidopsis plants were thoroughly rinsed with de-ionized water and incubated in a solution containing 5 × 10−3% (v/v) Silwet L-77 (control) or supplemented with 50 μM MV for different periods of time. The conductivity of the suspending solution was measured with a conductance meter (Twin Compact Meter-Horiba, Norhampton, UK) after incubation, and again after autoclaving at 120°C for 30 min to release all electrolytes. Electrolyte leakage was expressed as a percentage of total electrolytes. Measurements were carried out in triplicate.
Chlorophyll fluorescence
Two-week-old Arabidopsis plants were dark-adapted for 30 min prior to measurements. Chlorophyll fluorescence was measured at 23°C employing a Qubit Systems Inc. pulse-modulated fluorometer (Kingston, Canada). Photosynthetic parameters (maximum quantum efficiency, Fv/Fm; quantum yield,ΦPSII; non-photochemical quenching, NPQ; and 1—photochemical quenching parameter, 1−qP) were calculated as described (Baker and Rosenqvist 2004).
Oxidative treatment and extraction of soluble proteins
Two-week-old Arabidopsis seedlings were subjected to 50 μM MV treatment in a time-dependent manner, harvested, and the tissue was homogenized with 50 mM potassium phosphate pH 7.5, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM ascorbate, 15 % (v/v) glycerol and 2 % (w/v) polyvinyl polypyrrolidone for ascorbate peroxidase (APX), glutathione reductase (GR) and catalase (CAT) activity assays. Proteins for glutamine synthetase (GS) activity determination were extracted with 100 mM imidazol-HCl pH 7.5, 0.5% (v/v) β-mercaptoethanol, 1 mM MnS04, 20% (v/v) glycerol and 1 mM PMSF. Extraction of SOD was carried out according to published procedure (Gupta et al. 1993). In all cases the supernatant, recovered at 4°C by centrifugation (20,000 × g, 20 min), was used for native PAGE, activity assay or SDS-PAGE.
Native PAGE and activity staining
Equal amounts of soluble protein (25 μg) from leaf extracts were subjected to native PAGE. GS activity was assayed in the gel according to Scarpeci et al. (2007). In-gel activities of antioxidant enzymes were determined as previously described for SOD (Beauchamp and Fridovich 1971), and for CAT and peroxidase (POD) (Kang et al. 1999).
Enzyme activity assays
APX was determined by the decrease in A290 of an assay mixture containing 0.5 mM ascorbate (extinction coefficient: 2.8 mM−1 cm−1) (Gupta et al. 1993). GR activity was measured by following the decrease in A340 (extinction coefficient: 6.22 mM−1 cm−1) due to NADPH oxidation (Gupta et al. 1993). CAT was determined by the decrease in A240 of an assay mixture containing 10 mM H2O2 (extinction coefficient: 0.0394 mM−1cm−1), 50 mM potassium phosphate pH 7.8, 1 mM EDTA at 25°C as previously described (Kang et al. 1999).
Protein blot analysis
Leaf proteins were resolved on 12 % SDS-PAGE, transferred to nitrocellulose membranes and immunoreacted with polyclonal antibodies raised against GS (Scarpeci et al. 2007), cytosolic and chloroplastic heat shock proteins (Hsp) 70 (Rial et al. 2006), 2-Cys peroxiredoxin (2-Cys Prx) and Rubisco large subunit.
Detection of O•−2 and H2O2
Leaves were floated in 50 mM sodium phosphate pH 7.5, 0.2% (w/v) nitroblue tetrazolium (NBT) according to Jabs et al. (1996). When the pale yellow NBT reacts with O•−2, a dark blue insoluble formazan compound is produced (Maly et al. 1989). Detection of H2O2 was performed by infiltrating MV-treated leaves with a solution of 1 mg/ml 3,3′-diaminobenzidine (DAB) essentially as described by Thordal-Christensen et al. (1997). H2O2 is visualized as a reddish-brown colouration. Prior to imaging, chlorophyll was removed from leaves with 70 % (v/v) ethanol.
RNA isolation
Total RNA was isolated using TRIzol (Invitrogen Life Technologies, Karlsruhe, Germany) following the manufacturer’s procedure. Quality and quantity of RNA were monitored spectrophotometrically at 260 and 280 nm. RNA with an A260/A280 ratio higher than 1.2 was used for further analysis.
Northern blot hybridization
Twenty μg of total RNA from control and MV-treated aerial tissue were fractionated on 1.5% (w/v) agarose, 15 % (v/v) formaldehyde gels in a buffer containing 20 mM MES, 5 mM sodium acetate, 1 mM EDTA, pH 7. After electrophoresis, equal loading of the gel was confirmed by staining with ethidium bromide and checking the UV fluorescence of rRNA using a transilluminator. Then, the gel was washed twice with sterile distilled water and once with 10× SSC (Sambrook and Russell 2001), and blotted by capillary transfer to Hybond N+ nylon membrane (Amersham Biosciences, Buenos Aires, Argentina) for 12 h at 20°C in the same buffer. Membranes were pre-hybridized and hybridized in 6× SSC, 0.1% (w/v) BSA, 0.1% (w/v) Ficoll, 0.1% (w/v) polyvinyl pyrrolidone 400, 0.5% (w/v) SDS at 60°C for 2 and 15 h, respectively. Probes (100–150 ng DNA) were purified from agarose gels using Gel Extraction Kit (Qiagen, Hilden, Germany), and 32P-labelled by random priming using the Klenow fragment of DNA polymerase and [α32P]-dATP (Prime-a-gene Labeling System, Promega, Madison, USA). After labeling, the unincorporated [α32P]-dATP was removed by filtration of probes through Sephadex G-50 (Nick column, Pharmacia, Munich, Germany).
Expression profiling
Samples of total RNA from two independent biological replicates from the aerial parts of 2 h MV-treated and non-treated control seedlings (25 specimens in each sample) were sent to the German Resource Center for Genomic Research (RZPD, Berlin) for probe preparation and Arabidopsis GeneChip hybridization (Affymetrix Full Genome Array ATH1, Affymetrix Inc., Santa Clara, CA). RNA yield and purity were determined spectrophotometrically, and RNA integrity was checked using an Agilent 2100 Bioanalyser (Agilent Technologies, Böblingen, Germany). Twenty μg of total RNA were used as template for double-stranded cDNA synthesis (SuperScript Choice system, Gibco/BRL, Carlsbad, California). Biotin-labelled cRNA was synthesized using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Life Sciences, Inc, Farmingdale, NY). Affymetrix GeneChip hybridization experiments, including washing and scanning, were performed as described in the Affymetrix technical manual.
Data analysis
Data were normalized using robust multiarray analysis (RMA, R-package; BioConductor, http://www.bioconductor.org/) for quantile normalization and background adjustment. Expression analysis was performed using the Microarray Suite software (MAS version 5.0) with standard parameters (Affymetrix GeneChip Expression, Analysis Technical Manual; Affymetrix). Comparisons were performed for each of the two replicate experiments independently. Genes with “absent” calls in replicate experiments were removed from further analysis leaving 16,063 probe sets called “present” by MAS 5.0. Background adjustment was performed using RMA. Log ratios were calculated from normalized, log-transformed RMA outputs (=MV-treated – control). For statistical analysis, the data were subjected to the parametric tests using all available error estimates as implemented in the Genespring software and had to pass the false discover Benjamini-Hochberg correction. A P cut-off value of 0.05 was chosen. Genes were considered to be differentially expressed when exhibiting at least an average 3-fold difference in normalized signals across all replicate experiments. Expression values were obtained by averaging the normalized values of the two independent repeats. Data were transferred into Microsoft Access and Excel for further analysis.
Functional analysis of data was performed using the MapMan software (Thimm et al. 2004).
Data obtained from GENEVESTIGATOR
Within the different tools of GENEVESTIGATOR V3 (https://i.genevestigator.ethz.ch/), the Stimulus Viewer and hierarchical clustering were used to reveal the response profiles of genes to abiotic stress treatment (Zimmermann et al. 2004). Data from experiments of cold, heat, oxidative, osmotic, salt, drought, genotoxic, and wounding treatments of high quality ATH1: 22k arrays of AtGenExpress repository (AT-120: Kudla/Puchta/Bartels/Harter/Nover) were retrieved for comparison with our data or selected genes.
Promoter analysis
Promoter sequences spanning 1 kb upstream of the translation initiation codon were downloaded from the TAIR database (ftp://ftp.arabidopsis.org/Sequences/blast_onlydatasets/). The upstream regions were analyzed employing the MEME protocol (http://meme.sdsc.edu) and the PLACE database of regulatory elements (Higo et al. 1999). For MEME, a fixed minimum motif length of 5 and a maximum of 10 was set, and 50 motifs were requested using the zero or one occurrence per sequence model. Sequence logos for the conserved motifs in promoters of O•−2 induced genes were created using a web-based application (http://weblogo.berkeley.edu/logo.cgi).
Electrophoretic mobility shift assays
For total leaf protein extracts, tissues were homogenized using liquid nitrogen and a buffer containing 20 mM HEPES, 40 mM KCl, 1 mM DTT, 0.2 mM PMSF and 20% (v/v) glycerol. The homogenate was centrifuged (20,000 × g, 20 min, 4°C) and the supernatant was kept at −80°C.
Double-stranded DNA probes were formed by mixing equal amount of oligonucleotides W-box (5′-CGTTCTTCAGTCAAAAAGTCAAACTATCTC-3′) or mW-box (5′-CGTTCTTCAGGTCAAAAGGTCAACTATCTC-3′) and their corresponding complementary oligonucleotides. The mix was kept at 80°C for 5 min and then left to reach room temperature. Double-stranded DNA fragments were radio-labelled at their 5′-ends using T4 polynucleotide kinase to achieve specific activities of approximately 105 cpm ng−1. The 32P-labelled probes were purified from unincorporated nucleotides with Sephadex-G50 spin columns.
Binding buffer contained 3 μg poly(dIdC) (Sigma, St Louis, MO, USA), 1 U heparin, 20 mM HEPES, 40 mM KCl, 10% (v/v) glycerol, 4 mM MgCl2, 1 mM DTT and protein extract in a final volume of 20 μl. The binding reaction was carried out using 0.1 pmol of double-stranded oligo DNA (105 cpm) and 40 μg of total Arabidopsis leaf protein. DNA–protein complexes were allowed to form at 20°C for 10 min and then at 4°C for 30 min, and were resolved on 5% polyacrylamide gels in 0.5× TBE, 4 mM MgCl2 and 5% (v/v) glycerol. Gels were pre-run for 90 min at 100 V using 0.5× TBE and 4 mM MgCl2. After sample loading, electrophoresis was allowed to proceed until the bromophenol dye had migrated three-fourth through the gel. Following electrophoresis, gels were fixed with 10 % (v/v) acetic acid for 5 min, dried onto 3 MM Whatman paper at 80°C for 30 min and radioactivity was detected using a Storm 840 apparatus (Amersham Biosciences, Buenos Aires, Argentina).
Construction of promoter-GUS fusion lines
A 1.96-kb 5′ genomic fragment upstream of the ATG initiation codon of WRKY30 gene was amplified by PCR using the reverse primer 5′-CGGGATCCTACGTTCAAAGAGTGG-3′ (added BamHI restriction site underlined) and the forward primer 5′-CGTCGACGACCGGTATCGCAACAC-3′ (added SalI restriction sites underlined). The promoter fragment was cloned into the polylinker of plasmid pBI101.1 (Clontech, Heidelberg, Germany) at the BamHI and SalI restriction sites and thereby fused to the Escherichia coliuidA reporter gene. Correctness of the construct was verified by sequencing. The construct was introduced into Agrobacteriumtumefaciens strain GV3101 pMP90 by electroporation, and Arabidopsis (Col-0) plants were transformed by floral dip infiltration (Clough and Bent 1998).
GUS assay
After the treatments, tissues were collected and rinsed in 50 mM sodium phosphate pH 7.2, 10 mM EDTA, 0.33 mg ml−1 potassium ferricyanide, and then transferred to the same solution containing in addition 0.5 mg ml−1 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc; Gold Biotechnology, St Louis, MO, USA). Tissues were vacuum-infiltrated for 3 min and then incubated at 37°C for 24 h. Tissues were destained by soaking in 70% (v/v) ethanol.
Results
Generation of O•−2, photosynthetic activity and antioxidative response of Arabidopsis plants
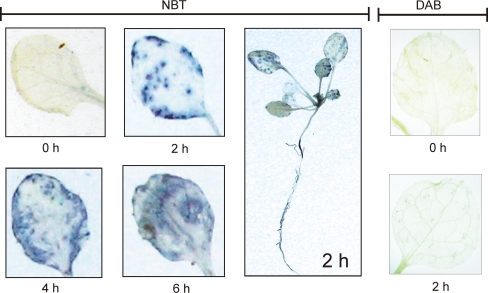
Two-week-old Arabidopsis seedlings grown on soil were treated with MV in the light followed by NBT infiltration to visualize O•−2 formation. Superoxide anion is the major oxidant species responsible for reducing NBT to insoluble formazan (Maly et al. 1989), which is observed as a purple deposit. Numerous localized spots were observed in the leaf after a 2 h MV treatment, and they spread all over the leaf by longer MV incubations (Fig. 1). A representative Arabidopsis seedling at 2 h MV treatment after NBT staining is also shown (Fig. 1). By contrast, plants subjected to the same MV treatment for 2 h did not accumulate H2O2 at a detectable level as revealed by DAB staining (Fig. 1).
Fig. 1.
Superoxide generation by MV treatment in illuminated Arabidopsis seedlings. Two-week-old Arabidopsis seedlings grown on soil were subjected to 50 μM MV treatment in the light (120 μmol photons m−2 s−1) for the duration indicated below each panel. Leaf samples were stained for O•−2 accumulation by the NBT staining after MV treatment, and a complete Arabidopsis seedling is also shown. Generation of H2O2 after identical MV treatment were followed by DAB staining and the results are shown in the right panels
To test the functional performance of the photosynthetic apparatus after MV treatment, we measured the chlorophyll fluorescence emitted by Arabidopsis leaves after saturating pulses of halogen actinic light. These pulses fully reduce photosynthetic electron carriers and several parameters can be measured, such as Fv/Fm, indicating the reduction state of the primary electron acceptor of PSII (QA); qP, reflecting the PSII operating efficiency; and NPQ, which represents the energy dissipated as heat related to energization of the thylakoid membrane due to lumen acidification during photosynthesis. Table 1 shows that the photosynthetic performance of leaves of soil-grown Arabidopsis plants was better than that of plants cultured on solid MS medium plus sucrose. When plants grown on soil were subjected to MV treatment Fv/Fm decreased steadily with time after the MV treatment, indicating a change of the electron transfer capacity. Concomitantly with this, the ΦPSII of electron transfer at PSII, which indicates the photosynthetic efficiency of PSII, was markedly reduced at 3 h MV treatment. The PSII redox state (1−qP) was higher, to various degrees, in all MV-treated plants compared to non-treated plants, and markedly increased at 3 h MV treatment (Table 1), suggesting that the reduced state of QA diminished under prolonged MV treatment. Within the first hour of MV exposure, the NPQ of these leaves was 22% higher than in control leaves, tending to decrease after several hours of MV treatment, possibly due to decreased electron transfer capacity.
Table 1.
Photosynthetic performance of Arabidopsis plants grown on solid MS medium or on soil and subjected to MV treatment
| Fv/Fm | ΦPSII | NPQ | 1−qP | |
|---|---|---|---|---|
| Growth condition | ||||
| MS, control | 0.56 ± 0.02 | 0.44 ± 0.02 | 0.36 ± 0.02 | 0.11 ± 0.01 |
| Soil, control | 0.64 ± 0.03 | 0.79 ± 0.03 | 0.44 ± 0.03 | 0.08 ± 0.01 |
| Time after MV treatment of soil-grown plants | ||||
| 1 h | 0.59 ± 0.03 | 0.33 ± 0.02 | 0.51 ± 0.03 | 0.15 ± 0.01 |
| 2 h | 0.49 ± 0.03 | 0.30 ± 0.02 | 0.33 ± 0.02 | 0.18 ± 0.01 |
| 3 h | 0.39 ± 0.02 | 0.17 ± 0.02 | 0.23 ± 0.01 | 0.31 ± 0.02 |
Two-week-old Arabidopsis plants were grown autotrophically on soil or in MS medium plus 3 % (w/v) sucrose. Chlorophyll fluorescence was measured in attached Arabidopsis leaves and the following parameters were calculated: Fv/Fm, ΦPSII, NPQ and 1−qP. Each given value ± SD represents the average of four identical experiments. The leaf temperature was 23°C
Electrolyte leakage, indicative of membrane integrity, and photosynthetic pigments were measured at different time points after MV exposure and the results are shown in Supplementary Fig. 1. From the beginning of the MV treatment, ion leakage was detectable to different extents in comparison to control Arabidopsis leaves, increasing markedly at the end of the treatment (70 % at 7 h) (Supplementary Fig. 1A). Total chlorophyll and carotenoid contents were not affected within 4 h of MV challenge (Supplementary Fig. 1B and C).
The antioxidant response of Arabidopsis plants under MV treatment was followed by determining SOD, CAT and POD activities in native protein gels. Two-week-old Arabidopsis plants were exposed to 50 μM MV for up to 6 h, and leaf proteins extracted at different time points. SOD activity was relatively constant during the oxidative challenge (Supplementary Fig. 2). After 6 h MV treatment CAT activity was invariable, while POD activity decreased in leaf extracts (Supplementary Fig. 2). APX, GR and CAT activities were assayed in solution (Supplementary Fig. 3) and no variations were observed between control and 2 h MV treated plants. After longer MV exposure times there was a significant drop in APX activity, remaining a total activity of 74 % at 4 and 6 h comparing with that of control plants. On the other hand, GR and CAT were unaffected in response to longer MV exposure. These results suggest that the antioxidant machinery of Arabidopsis leaves was largely unaltered by 2 h MV treatment, although at longer exposure times, peroxidases were the most affected enzymes.
Consequences of MV treatment of illuminated Arabidopsis seedlings on protein levels
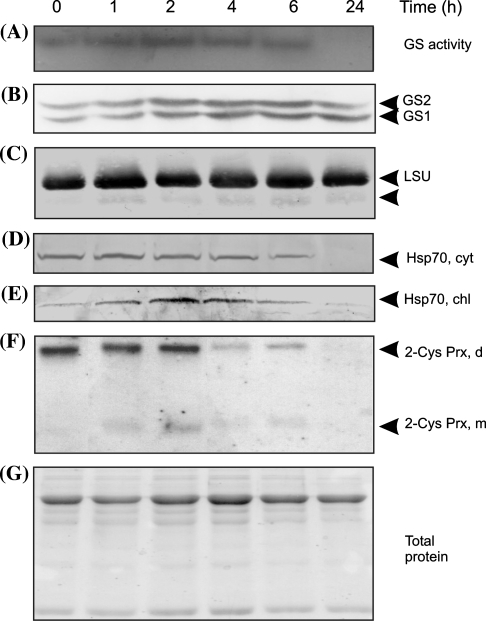
Chloroplast early ROS targets such as GS and Rubisco (Palatnik et al. 1999) were followed in MV-treated plants by GS in-gel activity measurements and Western blot analysis (Fig. 2). GS protein and activity remained constant during 6 h of MV treatment, but after 24 h of oxidative challenge no GS activity was detected with no significant modification of GS protein level (Fig. 2a, b). Rubisco large subunit and presumably a degradation product visible since 1 h MV exposure remained constant over 24 h of oxidative treatment (Fig. 2c).
Fig. 2.
GS activity and Western blot analysis of GS, Rubisco, Hsp70 and 2-Cys Prx after oxidative treatment of Arabidopsis leaves. Time courses of (a) GS activity (25 μg), (b) GS protein (80 μg), (c) Rubisco large subunit (LSU) (10 μg), (d) cytoplasmic (cyt) and (e) chloroplastic (chl) Hsp70 (80 μg), (f) 2-Cys Prx (15 μg) in total leaf extracts following 50 μM MV plant treatment with light. Leaf protein extracts (7 μg) were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue (g). Arrows indicate: cytosolic (GS1) and chloroplastic (GS2) GS isoforms (panel b), Rubisco large subunit degradation product (panel c) and 2-Cys Prx dimer (d) and monomer (m) (panel f)
Two nuclear-encoded chloroplast proteins such as Hsp70 and 2-Cys Prx and a cytosolic Hsp70 were also analyzed by immunoblotting based on the observations that some members of the Hsp70 family are stress-inducible (Boston et al. 1996), and that 2-Cys Prx prevents oxidative damage of chloroplast proteins (Konig et al. 2003). Chloroplastic and cytosolic Hsp70 were specifically detected using different antibodies that discriminate between them (Rial et al. 2003). Cytosolic Hsp70 remained unaltered for up to 6 h of MV treatment in the light (Fig. 2d), while chloroplastic Hsp70 was clearly induced at 2–4 h of MV treatment (Fig. 2e). After 24 h of MV treatment, Hsp70 declined in both compartments (Fig. 2d, e). On the other hand, the levels of 2-Cys Prx were constant for the first 2 h of oxidative treatment, after which the enzyme decreased markedly becoming undetectable at 24 h (Fig. 2f). The 2-Cys Prx monomer was visible at 1–2 h of oxidative stress, probably arising from overoxidation of 2-Cys-Prx monomer as it was reported in barley plants subjected to drought or low temperatures (Konig et al. 2003).
These results show that a 2 h MV treatment of Arabidopsis leaves in the light generates only mild changes in the levels of proteins under analysis, suggesting that main cell functions are largely preserved under this experimental condition.
Transcriptional response of Arabidopsis seedlings to MV treatment in the light
As shown, several hours of steady generation of O•−2 in illuminated chloroplasts of Arabidopsis seedlings caused severe oxidative damage. We wanted to analyze transcriptional changes occurring at earlier time points in response to chloroplastic O•−2 generation, i.e., when inhibition of photosynthesis is incipient (Table 1) and the level and activity of chloroplastic (Fig. 2) and antioxidant proteins (Supplementary Figs. 2 and 3) are not affected. To this end, two-week-old Arabidopsis seedlings were removed from soil and floated for 2 h in the light in a solution containing 50 μM MV (in the presence of 0.01% (v/v) Tween 20 to facilitate cellular uptake of MV). Control plants were incubated in the same way with the exception that MV was omitted. RNAs were isolated from the aerial parts of MV-treated and control seedlings, and subjected to transcript profiling employing Affymetrix GeneChip array ATH1 (for more details of array experimental design, data normalization and analysis see “Materials and methods”). The microarray data have been deposited in the public repository ArrayExpress (www.ebi.ac.uk/arrayexpress) under the accession number E-ATMX-28. From all genes analyzed, the vast majority exhibited increased abundance taking a minimal 3-fold difference between MV-treated and control samples as a threshold (Supplementary Table 1).
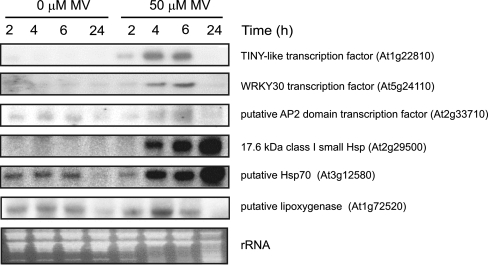
Microarray data were verified by Northern blot analysis (Fig. 3) of a selection of six genes strongly induced in the profile such as transcription factors and Hsp genes. In all cases, a clear increase in transcript abundance was observed at 2 to 4 h of MV treatment, which is in agreement with the expression data obtained through the gene chip hybridization experiments. Except for Hsps, all transcripts assayed were down-regulated after 24 h of MV treatment.
Fig. 3.
Analysis of MV up-regulated genes by Northern blot hybridization. Twenty micrograms of total RNA from control and MV-treated seedlings harvested at the indicated times after the onset of MV treatment were fractionated on formaldehyde–agarose gels and transferred to nylon membranes. Membranes were hybridized with 32P-labelled cDNA fragments of the indicated genes and their MIPS identifiers are indicated
Functional analysis of genes early induced by MV treatments
The potential functions of early induced genes by MV in different biological processes were assessed by the MapMan software. A large fraction of genes exhibiting an at least 3-fold change in expression level were found to be up-regulated after the MV treatment (Table 2). Sixty-eight of the MV-induced genes have no ontology assigned or unknown function (Table 2). Several of the MV-affected genes belong to categories that function in abiotic or biotic stress responses, that encode enzymes that have a role in transcriptional regulation or are involved in signalling events (Table 2, Supplementary Fig. 4 and 5). Among the genes belonging to abiotic stress category, those encoding Hsp were the most abundant (Table 3 and Supplementary Fig. 4). From the 18 highly induced Hsp genes, ten belong to the small Hsp family, five members co-localize in the cytosol, and nine genes still have undetermined subcellular location (Table 3). Protein kinases, protein phosphatases, and receptor kinases (mainly leucine rich repeat) were highly induced among the genes involved in signalling processes (Supplementary Fig. 5). Importantly, the expression level of various transcription factors, in particular those of the WRKY family, increased after a short-term (2 h) MV treatment (Table 4).
Table 2.
Potential functions of genes affected by O•−2 generated in chloroplasts.
| Category | Ratio (log2 scale) | |
|---|---|---|
| > 1.66 | < −1.66 | |
| Abiotic stress | 20 | 0 |
| Biotic stress | 16 | 0 |
| Cell cycle | 2 | 0 |
| Cell organization | 3 | 0 |
| Development | 10 | 2 |
| Enzyme families | 30 | 2 |
| Hormone metabolism | 16 | 4 |
| Metal handling | 2 | 0 |
| Not assigned | ||
| Ontology | 25 | 0 |
| Unknown function | 43 | 5 |
| Protein degradation | 7 | 1 |
| Protein posttranslational modification | 7 | 0 |
| Protein synthesis | 2 | 0 |
| Redox regulation | 4 | 0 |
| Regulation of transcription | 30 | 3 |
| Signalling | 23 | 0 |
| Transport | 10 | 1 |
Functional categories of genes whose expression ratios between the MV-treated and the non-treated control samples varied by at least 3-fold were identified using the MapMan software, as described in “Materials and methods”. Ratios of expression are indicated on a log2 scale. Genes with a P value smaller than 0.05 were categorized
Table 3.
Gene members of heat shock proteins induced by 2 h MV treatment in the light.
| Gene category | Locus identifier | Ratio (log2) | Subcellular location |
|---|---|---|---|
| Heat shock proteins | |||
| Hsp17.6A | At5g12030 | 5.21 | cytosol |
| Hsp 17.6A-CI | At1g59860 | 4.63 | ND |
| Hsp17.6B-CI | At2g29500 | 4.48 | ND |
| Hsp17.4-CI | At3g46230 | 4.28 | ND |
| Hsp17.4-CIII | At1g54050 | 4.08 | ND |
| Hsp17.6-CII | At5g12020 | 4.04 | ND |
| Hsp101 | At1g74310 | 3.55 | cytosol |
| DNAJ heat shock family protein | At2g20560 | 3.45 | ND |
| Hsp70, putative | At1g16030 | 3.28 | cytosol |
| Hsp70, putative | At3g12580 | 3.27 | cytosol |
| Hsp17.6-CI | At1g53540 | 3.07 | ND |
| HsfA2 | At2g26150 | 2.85 | nuclear |
| Hsp81-1 | At5g52640 | 2.38 | cytosol |
| DNAJ heat shock N-terminal domain-containing protein | At1g71000 | 2.16 | ND |
| HsfA4A | At4g18880 | 2.13 | nuclear |
| Hsp23.5-M | At5g51440 | 2.02 | mitochondria |
| HSP17.5-CI | At5g37670 | 1.850 | mitochondria |
| Hsp70 | At2g32120 | 1.69 | ND |
TAIR (www.arabidopsis.org) information was the source for subcellular location of genes. ND means not determined
Table 4.
Transcription factor families induced or repressed by MV
| Transcription factor family | Locus identifier | Ratio (log2) |
|---|---|---|
| AP2-ERF | ||
| TINY | At1g22810 | 3.98 |
| DREB2A | At5g05410 | 3.44 |
| Subfamily B-4 | At2g33710 | 2.51 |
| Subfamily B-3 | At2g31230 | 1.83 |
| bHLH | ||
| bHLH Pfam domain protein | At4g36540 | −2.05 |
| C2H2 | ||
| AN1-like | At3g28210 | 3.74 |
| ZAT11 | At2g37430 | 2.84 |
| Zn finger Pfam domain | At5g04340 | 1.81 |
| ZAT12 | At5g59820 | 1.77 |
| C3H | ||
| Zn finger (C3HC4-type RING finger) | At1g14200 | 3.29 |
| HB | ||
| BEL1 | At5g41410 | −1.74 |
| Hsf | ||
| HsfA2 | At2g26150 | 2.85 |
| HsfA4A | At4g18880 | 2.13 |
| NAC | ||
| NAM | At1g01720 | 2.22 |
| NAM | At1g77450 | 2.21 |
| NAM | At5g63790 | 1.98 |
| NAM | At3g49530 | 1.70 |
| PHOR1 | ||
| U-box domain | At3g11840 | 2.15 |
| U-box domain | At5g64660 | 1.66 |
| WRKY | ||
| WRKY6 | At1g62300 | 3.12 |
| WRKY30 | At5g24110 | 3.00 |
| WRKY46 | At2g46400 | 2.73 |
| WRKY11 | At4g31550 | 2.09 |
| WRKY33 | At2g38470 | 2.02 |
| WRKY40 | At1g80840 | 1.94 |
| WRKY48 | At5g49520 | 1.82 |
| WRKY28 | At4g18170 | 1.79 |
| WRKY22 | At4g01250 | 1.61 |
Only a small number of genes encoding proteins of redox-related metabolism, including glutaredoxin family proteins, and mono and dehydroascorbate reductases, were found to be affected by MV under our experimental conditions (Supplementary Fig. 4). These data are consistent with a preserved antioxidant response in Arabidopsis leaves at 2 h MV treatment, as suggested by the enzyme activity measurements (Supplementary Fig. 2 and 3).
Of the few genes exhibiting a minimum of 3-fold reduced expression upon MV treatment (Supplementary Table 1), five encode proteins of unknown function, four encode auxin-induced proteins (Supplementary Fig. 5A), two encode transcription factors (Table 4 and Supplementary Table 3), and two encode PSI and PSII proteins. These data are in accordance with the observed decrease in the photosynthetic electron transport activity of Arabidopsis leaves (Table 1). On the other hand, transcript of 9-kD protein of PSI showed an opposite pattern in a 24 h MV transcriptomic footprint (Gadjev et al. 2006), probably reflecting an adaptation of plants to long term MV treatment. Other genes involved in photosynthesis-related processes such as the Calvin cycle or photorespiration were not affected by the 2 h MV treatment (data not shown).
Down-regulation of genes related to the photosynthetic electron transport system were also observed when Arabidopsis plants were exposed to high light intensity (Kimura et al. 2003). This condition leads to the production of ROS in the chloroplast (Niyogi 1999). To investigate the transcriptional response of Arabidopsis under situations where ROS are produced in the chloroplast, we performed comparative analysis of the transcriptome of Arabidopsis seedlings subjected to 2 h MV treatment and high light, using publicly available microarray experiments (Rossel et al. 2002; Kimura et al. 2003; Vanderauwera et al. 2005). At a threshold of 2.8-fold change, Hsps were the most abundant up-regulated genes in common (Supplementary Table 2). Other common genes up-regulated codify for the cytosolic APX1 (At1g07890) and the transcription factors DREB2A (At5g05410) and the NAC domain protein (At1g01720). DREB2A is a key regulator of drought response, therefore 61 genes responsive to high light and drought (Kimura et al. 2003) were additionally compared with MV-regulated genes. Only three genes were in common among high light, drought and MV treatment of Arabidopsis seedlings encoding: an arabinogalactan-protein AGP2 (At2g22470), an Hsp70 (At3g12580) and a putative protein phosphatase 2C (At3g62260).
We further analyzed the expression of all 2 h MV induced Hsp genes (Table 3) under other abiotic stress conditions such as salt, drought, wounding or cold using GENESVESTIGATOR tools (Supplementary Fig. 6). Two Hsp70 (At1g16030 and At2g32120) and two small Hsps (At1g54050 and At5g51440) were not significantly induced by any stress conditions other than late oxidative or heat stress. On the other hand, the HsfA4A (At4g18880) was only up-regulated more than 3-fold by osmotic stress (Supplementary Fig. 6).
Transcription factors induced or repressed by 2 h MV treatment in the light
Several members of common transcription factor families such as AP2-ERF, C2H2, Hsf, NAC and WRKY were highly induced by MV (Table 4). MV-dependent up-regulation of members of the AP2-ERF (At1g22810 and At2g33710) and WRKY (At5g24110) families was confirmed by Northern blot analysis (Fig. 3). The highest number of MV-affected transcription factor genes was found in the WRKY family, with nine members being up-regulated at least 3-fold. Among the WRKY family members, WRKY6, WRKY30 and WRKY46 were highly stimulated (Table 4). Comparing the profiles of these transcription factors (Table 4) with transcription factor genes up-regulated by H2O2 exposure (Davletova et al. 2005) or generated in CAT-deficient Arabidopsis mutants exposed to high light conditions (Vanderauwera et al. 2005), rendered an overlap of eight genes in all three conditions (Supplementary Table 3). Several members belonging to the WRKY family were expressed in two out of three experiments and only WRKY6 was induced by the three oxidative treatments. WRKY46, WRKY28 and WRKY22 were up-regulated exclusively by MV treatment.
As mentioned above, only a relatively small number of genes was found to be down-regulated by MV treatment (Table 2). They include two transcription factors belonging to basic helix-loop-helix (bHLH) (At4g36540) and HB families, an homeotic protein BEL1 homolog (At5g41410) (Table 4).
Identification of conserved motifs in promoters of MV induced genes
Promoters of genes strongly up-regulated after MV treatment were selected for a MEME search to identify potential transcription factor binding sites. This computational tool discovers motifs (highly conserved regions) in groups of DNA or protein sequences, i.e., in promoters of genes selected according to a common expression pattern (Bailey and Elkan 1994).
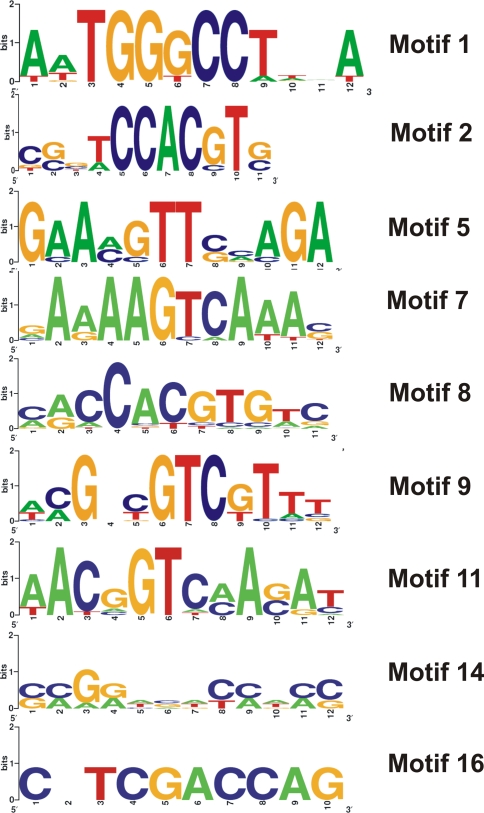
For this analysis we selected all genes that exhibited at least 4-fold induction after MV treatment (168 genes in total). One-kb long promoter regions located 5´ upstream of the predicted ATG initiation codons of these genes were analyzed using the MEME software tool. Sixteen motifs were found and represented with different incidences in the dataset and in the genome (Supplementary Table 4). We could not identify with confidence known transcription factor binding motifs in the promoters of MV-inducible genes (Supplementary Table 4), except for the core ABRE element (ACGT) (Leung and Giraudat 1998), an abscisic acid-dependent cis-acting regulatory element, found at high frequency in motif 2 and less frequently also in motif 9 (Fig. 4). Instead, different motifs were found to be highly over-represented in genes encoding transcription factors, Hsps or Hsfs, and proteins involved in signalling cascades (Supplementary Table 4).
Fig. 4.
Sequence logos of relevant motifs conserved in promoters of MV induced genes. The relative height of each letter indicates the relative abundance of the corresponding nucleotide at that position in each motif
Motif 2 (CGGTCCACGTG) was found in eight genes encoding a putative Hsp70, a small Hsp17.4-CIII, WRKY6, a cytochrome P450 enzyme, a methyl transferase, an ethylene-responsive binding protein, and a Zn finger protein. Two of these genes contain additional motifs, i.e., the Zn finger protein gene (At5g27420) has motifs 1 and 7, and the methyl transferase gene (At1g21110) has motifs 7 and 16. Interestingly enough, this methyl transferase promoter shares motifs 2, 7 and 16 with the promoters of WRKY6, WRKY30 and WRKY46, respectively. Motif 9 (ACGACGTCGTTT) was found in the promoters of 23 genes encoding Hsp81, HsfA4A and several signal transduction components such as protein phosphatase 2C (also containing motifs 8 and 11), MAPK, and a leucine-rich repeat transmembrane protein kinase (Supplementary Table 4).
Motif 1 (AATGGGCCTTAA) or motif 5 (GAAAGTTCCAGA) was highly represented in promoters of Hsp genes (Supplementary Table 4), and both motifs were detected in small Hsps genes (At3g46230 and At1g54050). Motif 5 contained the consensus sequence ‘nGAAn’, a highly conserved heat shock element found in the promoters of many genes involved in cell protection such as protein folding, detoxification, and maintenance of cell wall integrity in Saccharomyces cerevisiae (Yamamoto et al. 2005).
Motifs 7, 8 and 11 were found in several transcription factors belonging to different families (WRKY, AP2 and NAM) (Supplementary Table 4). Remarkably, the four WRKY transcription factors highly induced by MV did not share common motifs in this analysis. The four different motifs that could be distinguished in the promoters of WRKY6 (motif 2, CGGTCCACGTC), WRKY11 (motif 14, CCGGTCACCTCC), WRKY30 (motif 7, GAAAAGTCAAAC) and WRKY46 (motif 16, CGTCGACCAG) showed p-values lower than 10−6 (Supplementary Table 4), indicating a low probability that a single random subsequence matches along the length of the motif. WRKY factors bind to W boxes (C/T)TGAC(T/C), which are present in the promoters of many plant genes that are associated with defense (Eulgem et al. 2000). Motif 7 (GAAAAGTCAAAC), which at high frequency contains the GTCA core element (Fig. 4) that resembled the inverted consensus W box sequence, was found to be present in the WRKY30 promoter (Supplementary Table 4). Motif 11 (AACGGTCAAGAT), shared by the promoters of three different transcription factors, also contains the GTCA element at high frequency (Fig. 4).
Leaf proteins bind to the GAAAAGTCAAAC motif
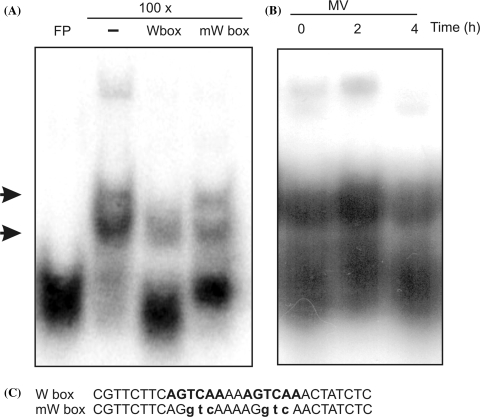
The presence of WRKY proteins in MV-treated Arabidopsis leaf extracts was assayed by gel retardation experiments. To this end, a 30-mer double-stranded oligonucleotide (W box) containing the consensus motif 7 was used as a probe (Fig. 5). Several retarded bands were observed when leaf protein extracts were tested for W box binding (Fig. 5a). Competition experiments using unlabelled W box and mutated W box (mW box) probes revealed that two of the retarded bands were due to specific binding of WRKY proteins present in leaf extracts (Fig. 5a). Specific binding activity to motif 7 increased after a 2 h MV treatment (Fig. 5b), indicating that WRKY proteins were rapidly induced by the oxidative treatment.
Fig. 5.
DNA binding activity of protein extracts of MV-treated Arabidopsis leaves. (a) Competition experiments were carried out with Arabidopsis leaf protein extracts and a 32P-labelled W box probe. As competitors, 100-fold unlabelled W box or mW box probes were added. Arrows indicate specific retarded bands. (b) DNA binding activity of Arabidopsis leaf protein extracts from control or MV-treated leaves. (c) Nucleotide sequences of probes used for the DNA binding assays. W box contains a double W box (AGTCAA), which was mutated into AGGTCA in the mW box. FP: free probe
Rapid response of WRKY30 promoter containing the GAAAAGTCAAAC motif to oxidative treatments
To assess in vivo functionality of motif 7, we produced transgenic Arabidopsis plants expressing the GUS reporter gene under the control of 1.96-kb WRKY30 promoter (PrAtWRKY30::GUS lines), which contains this motif. WRKY30 expression was clearly induced by MV in the light (Fig. 3), as several other WRKY family members (Table 4). Therefore, we analyzed the response of WRKY30 to different oxidative treatments. PrAtWRKY30::GUS lines were subjected in the light to MV and to externally added hydrogen peroxide and the GUS staining pattern was recorded (Fig. 6). GUS gene under the control of WRKY30 promoter was highly and rapidly induced by MV, while 24 h of hydrogen peroxide treatment was necessary to detect GUS activity (Fig. 6).
Fig. 6.
Response of WRKY30 expression to oxidative stress in Arabidopsis leaves. PrAtWRKY30::GUS lines were exposed to 50 μM MV or 20 mM H2O2 for the times indicated. A control leaf is shown in the right panel
Identification of MV-specific responsive genes in Arabidopsis leaves
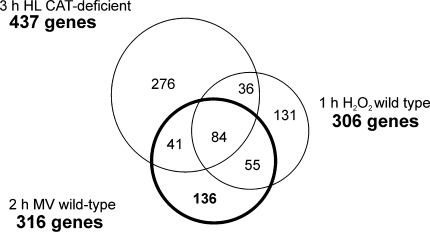
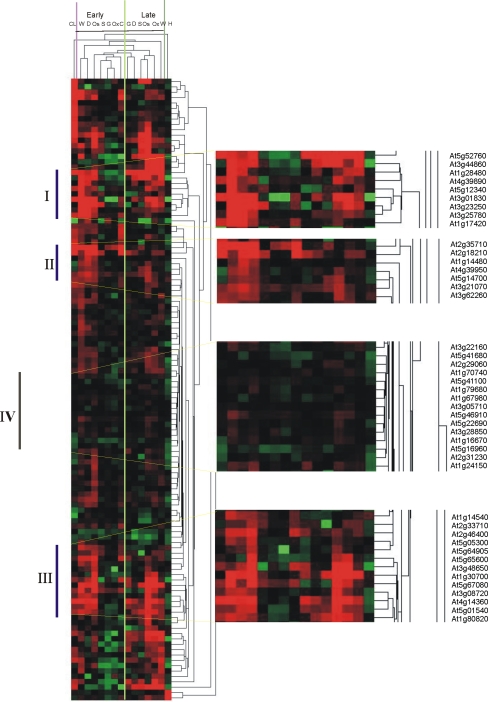
In illuminated leaves, MV propagates superoxide anion in the chloroplast (Mehler 1951; Asada and Takahashi 1987), which subsequently dismutates to H2O2 either spontaneously or enzymatically via SODs (Tsang et al. 1991). To present more insight into the MV-specifically regulated genes in Arabidopsis leaves, we compared the 2 h MV expression profile with the one obtained after 1 h H2O2 treatment (20 mM), which was exogenously added to 5-day-old wild-type Arabidopsis (Davletova et al. 2005) or H2O2 internally generated in CAT-deficient plants subjected for 3 h to high light (Vanderauwera et al. 2005), following the threshold of 3-fold change. Figure 7 presents the overlap of the rapidly MV-regulated genes with H2O2-regulated genes. Eighty four genes were responsive to MV and H2O2 internally or externally generated. Within these 84 commonly regulated genes, several transcription factors could be identified (Supplementary Table 3) such as WRKY6, ZAT12, and DREB2A, being the last one also in common with high light stress (Supplementary Table 2). After identification of all genes in common among these stress conditions, we extracted the genes that were exclusively regulated by MV. We identified 136 genes, within them 117 genes were up-regulated, and nineteen genes encoded unknown proteins (Supplementary Table 5). We further compared the responsiveness of these up- and down-MV-regulated genes to various environmental stimuli by hierarchical average linkage clustering separately (Fig. 8 and Supplementary Fig. 7). Several groups of co-expressed genes were observed at diverse early and late stress conditions. Three clusters of early transcriptional changes (Fig. 8 I, II and III) could be distinguished for up-regulated genes in wounding, drought and osmotic stress, partially overlapping with late gene clusters responsive to wounding, oxidative and osmotic stresses. These clusters are dominated by genes encoding proteins of unknown functions. The expression profile of 2 h MV responsive genes was not affected by early salt, oxidative and genotoxic stresses. A remarkable group of genes is the one strongly regulated by 2 h MV but not responsive to any other environmental stress (Fig. 8, IV and Supplementary Fig. 7). This group is represented by fifteen genes up-regulated and seven genes down-regulated by MV. From the up-regulated genes not responsive to any other stress, there are six genes encoding protein of unknown functions, a quinone oxidoreductase-like protein, a putative S-adenosyl-l-methionine:trans-caffeoyl-Coenzyme A 3-O-methyltransferase, a wall-associated kinase 2, a disease resistance protein-like, three genes related with signal transduction pathways, an ethylene response factor-like AP2 domain transcription factor, and a putative SCARECROW gene regulator (Fig. 8). From the genes down-regulated by MV but not responsive to any other stress there are three which encode proteins of unknown functions, the PSI 9kDa protein and an homeotic transcription factor BEL1 (Supplementary Fig. 7).
Fig. 7.
ROS-regulated genes within oxidative and high light conditions in Arabidopsis. Wild-type plants treated with 20 mM H2O2 for 1 h (Davletova et al. 2005), 50 μM MV for 2 h (present work) and CAT- deficient Arabidopsis mutant subjected for 3 h to high light (HL) (Vanderauwera et al. 2005) are indicated together with the total number of gene input of each stress condition. For comparison, genes whose expression ratios varied at least 3-fold between treated and control samples were considered. In the Venn diagram, numbers of genes are given that are unique to that gene set or in common sections between sets
Fig. 8.
Hierarchical clustering of combined abiotic stress data sets with the identified 2 h MV up-regulated genes. Clusters of early and late responsive genes as well as a group of unresponsive genes are indicated. Locus identification numbers are shown for representative clusters containing functionally related genes involved in cold (C), drought (D), genotoxic (G), heat (H), osmotic (Os), oxidative (Ox), salt (S), and wounding (W). I, II and III indicate clusters of early transcriptional changes and IV a group of genes responsive to 2 h MV but not to any other environmental stress. CL means cold late. Red and green indicate up- and down-regulation, respectively. The full-color range represents log base 2 ratios of −2.0 to 2.0
Discussion
MV is an effective agent that produces O•−2 in the light, amply used as herbicide in different plant systems. Steady O•−2 generation in the chloroplast is dependent on photosynthetic electron transport activity. Under the experimental conditions described in “Materials and methods” the Fv/Fm of two-week-old Arabidopsis plants grown on soil usually range between 0.6 and 0.7. Arabidopsis plants grown heterotrophically on solid MS media showed lower photosynthetic activity (Table 1), therefore plants grown on soil were selected for further analysis. Superoxide production in Arabidopsis leaves after a 2 h MV treatment in the light had only marginal effect on photosynthesis (Table 1), membrane integrity and photosynthetic pigments (Supplementary Fig. 1), various antioxidant systems (Supplementary Figs. 2 and 3) and integrity of chloroplastic proteins (Fig. 2). Functional analysis of genes that changed under ROS generated in the chloroplast revealed that several members of plant transcription factor families were up-regulated (Table 4), with the WRKY superfamily being the most represented. Eighteen members of the Hsp family were highly induced in this expression profile (Table 3, Supplementary Fig. 4) with nine of them classified as small Hsps (Boston et al. 1996). Small Hsp genes are not expressed in vegetative tissue under non-stress conditions (Volkov et al. 2006), and they are the most dominant proteins produced in response to heat stress (Sun et al. 2002). A common feature of small Hsps is their ability to form large oligomers, and changes in the oligomeric state are associated with the chaperone activity of these proteins. In addition to heat stress, plant small Hsps are also induced under other stress conditions (Sun et al. 2002), suggesting that they may play an important role in stress tolerance. In Arabidopsis cell culture exposure to H2O2 was reported to induce expression of Hsp17.6, Hsp18.2 (Volkov et al. 2006), Hsp17.6B-CI and Hsp17.7-CII (Nishizawa et al. 2006), which are highly expressed in our profile (Table 3). In addition, Hsp17.6B-CI was dramatically induced in Arabidopsis plants subjected to 1 or 3 h high light irradiation regime (Supplementary Table 2). Except HsfA4A (At4g18880), all other Hsp genes listed in Table 3 were up-regulated in CAT-deficient Arabidopsis plants within 8 h of high light irradiation (Vanderauwera et al. 2005). It seems likely that the increase in transcript level of these small Hsp genes is mediated by the increase in cellular H2O2 accumulation under several environmental stresses in Arabidopsis plants. On the other hand, the small Hsp17.4-CIII (At1g54050), highly induced in 2 h MV treated Arabidopsis (Table 3), was not expressed in leaves under a variety of environmental stresses (Supplementary Fig. 6), suggesting a role of Hsp17.4-CIII in oxidative stress generated in the chloroplast. We also observed a clear and rapid increase of chloroplast Hsp70s by MV, whilst cytosolic Hsp70 isoforms were unaffected (Fig. 2), suggesting a role of chloroplast Hsp70 proteins under O•−2 generation. According to current knowledge, Hsp70 proteins are recruited to prevent unfolding, stabilizing partially unfolded intermediates and refolding stress-denatured proteins (Sung et al. 2001). There is increasing evidence for cross-protection against different environmental stresses. Heat shock and drought were shown to induce several Hsp genes in Arabidopsis (Rizhsky et al. 2004b), being twelve of them in common with our experiment. The expression of Hsps is controlled by the activity of heat shock transcription factors (Nover et al. 2001). Our expression profile showed an induction of HsfA2 (Table 3), that was reported to be a regulator of a subset of genes in the heat stress response in Arabidopsis (Schramm et al. 2006). Expression of six putative HsfA2 target genes (Nishizawa et al. 2006) were observed in this profile, including Hsp101, Hsp70 and small Hsps. In addition, the yeast heat shock factor (Hsf) 1 binding sequence nTTCn (or nGAAn) (Yamamoto et al. 2005) was highly represented in motif 5 and to a lesser extent in motif 1 (Fig. 4), both sequences found in many Hsp promoters (Supplementary Table 4), suggesting the hypothesis that this heat shock element could be functional in oxidative stress-specific transcription in Arabidopsis.
Overlapping transcription footprints of genes induced during eight different ROS-generating conditions with genes responsive to short MV challenge (2 h) revealed few common candidates. They include ferritin and glutaredoxin, both proteins related to oxidative stress in plants (Mittler et al. 2004). Constitutive expression of ferritin was shown to enhance the tolerance of transgenic plants to abiotic and biotic stresses (Rizhsky et al. 2004a). Several genes encoding chloroplast protein were down-regulated in our experiments, such as those encoding PSI (AtCg01060) or PSII (AtCg00680) components. These results suggest that the photosynthetic machinery is progressively shut down in order to arrest the electron transport capacity of the chloroplast to avoid photoinhibition. On the other hand, chloroplast signalling to the nucleus is known to change nuclear gene expression according to environmental cues (Fey et al. 2005). In view of the fact that the cytoplasms of all living cells are highly reducing environments, it appears likely that redox signals arising from chloroplasts are translated into other types of signals either through redox-sensitive kinases (Aro and Ohad 2003) or by activation of other signalling mechanisms (Koussevitzky et al. 2007). Interestingly, the bHLH transcription factor At4g36540, which is strongly down-regulated by MV, was previously reported to be rapidly (within 30 min) induced by treatment with brassinosteroids (BR). Hence it was named BEE2 for BR Enhanced Expression 2 (Friedrichsen et al. 2002). BEE2, together with its closely related bHLH proteins BEE1 and BEE3, is required for a full BR response. Our observations indicate a possible regulatory link between chloroplast-generated O•−2 and BR signalling in plants.
Among the MV-affected signal transduction-related genes, many encode transcriptional regulators, including several WRKY transcription factors (Table 4), receptor-like kinases, and proteins that regulate protein turnover (Supplementary Fig. 5). Most WRKY genes appear to have a function in pathogen-triggered signalling cascades. In fact, WRKY6, highly induced in this profile, has previously been reported to play a role during senescence and in defense-related processes (Robatzek and Somssich 2001). In this work, WRKY30 and WRKY46 genes were also strongly up-regulated. The WRKY proteins encoded by these genes have similar molecular masses and structures (group III of the WRKY superfamily) (Eulgem et al. 2000), and they are expressed at the same growth-stage as revealed by Gene Chronologer analysis (Zimmermann et al. 2004). One additional WRKY transcription factor, i.e., WRKY48 belonging to group IIc, was newly identified as a MV-induced gene, which together with WRKY30 and WRKY46 are likely to be involved in the plant antioxidant response. WRKY40 has been reported to be functionally related to WRKY18 or WRKY60 (Xu et al. 2006). However, in this early O•− 2-generation expression profile only induction of WRKY40 was observed (Table 4).
A search to identify potential transcription factor binding sites in the promoter of genes that exhibited at least 4-fold induction after MV treatment revealed enrichment of the sequence GAAAAGTCAAAC (motif 7) contained in 12 of 168 genes (7.1%), relative to the entire genome (0.09%) (Supplementary Table 4), suggesting a functional role of this sequence. Motif 7 contains the W box consensus binding site of WRKY transcription factors. The early induction of WRKY transcription factors by MV in the light was further evidenced in vitro and in vivo. The first evidence was the enhanced binding activity to motif 7 of protein extracts from Arabidopsis leaves treated for 2 h with MV in the light (Fig. 5). Alternatively, pre-existing WRKY proteins could be chemically modified by oxidative conditions, resulting in higher affinity of protein binding to motif 7. Further evidences come from analysis of the response of a particular WRKY gene, WRKY30, which showed an induction during MV treatment by Northern blotting (Fig. 3). Furthermore, PrAtWRKY30::GUS line was highly responsive to MV and, to a lesser extent, to externally added hydrogen peroxide (Fig. 6). In accordance with it, analysis of transcription factors that are up-regulated by 2 h MV treatment and common with H2O2 experiments in Arabidopsis revealed that the induction of WRKY30 was less than 3-fold in 5-day-old wild-type Arabidopsis seedlings treated with 20 mM H2O2 for 1 h (Davletova et al. 2005), but it was highly expressed in CAT-deficient Arabidopsis mutants under high light regime, that is, when H2O2 is internally generated (Supplementary Table 3). WRKY48 showed a similar pattern of induction to that of WRKY30, indicating that both genes are responsive to ROS generated intracellularly.
Expression patterns of genes specifically induced by MV were compared with a selection of relevant abiotic stresses (Fig. 8 and Supplementary Fig. 7). In general we observed clusters of genes early or late up-regulated in response to wounding, drought and osmotic stress in common with MV and partially overlapping regions. A cluster of seven early up-regulated genes was represented by four proteins of unknown function, which is a challenge for further research. In addition WRKY46 (At2g46400) was found in another cluster of early up-regulated genes suggesting that ROS generated in chloroplast trigger common signalling pathways with these stresses. A relevant group of genes up-regulated by 2 h MV that showed no variation under other stress condition include those encoding O-methyltransferase and wall-associated kinase, both proteins related to the formation of cell wall or signalling continuum between the cell wall and the cytoplasm, respectively. Caffeoyl Coenzyme A 3-O-methyltransferase is involved in the biosynthesis of monolignol (Raes et al. 2003), a precursor of lignin, which is form in many plants in response to wounding and infection to protect plant tissues against invading pathogens. Several MV-specific genes encode disease resistance proteins (Supplementary Table 5). Recently it was reported that pathogen-responsive protein kinase cascade plays an active role in promoting ROS generation in chloroplasts and inducing a hypersensitive response-like cell death (Liu et al. 2007). It is likely that ROS generated in chloroplasts by MV treatment could share a signalling pathway with the pathogen-induced hypersensitive response.
In summary, the early response of Arabidopsis leaves to chloroplast-generated ROS includes a strong up-regulation of genes belonging to categories functioning in abiotic stress responses and transcription, expressed proteins of unknown function, and down-regulation of few genes such as those encoding PSI and PSII components. Common expressed genes in Arabidopsis response pathways to ROS early generated in the chloroplast by MV and other environmental stresses include many small Hsps and several WRKY transcription factors, from which WRKY46, 28 and 22 had not been detected before by transcription profiling. In addition, several 2 h MV responsive genes identified for the first time in this study encode proteins of unknown function. Discovering functions of these proteins represents great challenges for future research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by a grant from the Bilateral Cooperation Project AL-PA02-B1-019 SECYT-BMBF (Argentina-Germany). Bernd Mueller-Roeber thanks the Fond der Chemischen Industrie for funding (No. 0164389). Telma E. Scarpeci was supported by short-term DAAD and long-term CONICET doctoral scholarships.
For the generous gift of antisera we wish to thank Dr. Ricardo Wolosiuk, Instituto Leloir, Argentina (2-Cys Prx), Dr. Eduardo Ceccarelli (cytosolic and chloroplastic Hsp70) and Dr. Alejandro Viale (Rubisco), both from IBR. We thank Dr. Karin Koehl and her colleagues from the Max-Planck Institute of Molecular Plant Physiology, Golm, for expert plant care.
Abbreviations
- 2-Cys Prx
2-Cys peroxiredoxin
- APX
Ascorbate peroxidase
- bHLH
Basic helix-loop-helix
- CAT
Catalase
- DAB
3,3′-diaminobenzidine
- GS
Glutamine synthetase
- GR
Glutathione reductase
- Hsp
Heat shock protein
- Hsf
Heat shock transcription factor
- H2O2
Hydrogen peroxide
- Fv/Fm
Maximum quantum efficiency
- MV
Methyl viologen
- NBT
Nitroblue tetrazolium
- NPQ
Non-photochemical quenching
- PMSF
Phenylmethylsulfonyl fluoride
- POD
Peroxidase
- PSI
Photosystem I
- PSII
Photosystem II
- qP
Photochemical quenching
- QA
Primary quinone acceptor of the PSII reaction center
- ΦPSII
Quantum yield
- ROS
Reactive oxygen species
- O•−2
Superoxide anion
- SOD
Superoxide dismutase
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-007-9274-4) contains supplementary material, which is available to authorized users.
References
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399 [DOI] [PubMed]
- Aro EM, Ohad I (2003) Redox regulation of thylakoid protein phosphorylation. Antioxid Redox Signal 5:55–67 [DOI] [PubMed]
- Asada K (1994) Production and action of active oxygen species in photosynthetic tissues. In: Foyer C, Mullineaux P (eds) Photooxidative stresses in plants: causes and amelioration. CRC Press Inc., BocaRaton, pp 77–104
- Asada K, Takahashi M (1987) Production and scavenging of active oxygens in photosynthesis. In: Kyle DJ, Osmond CB, Arntzen DJ (eds) Photoinhibition. Elsevier/North Holland Biomedical Press, Amsterdam, pp 227–280
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36 [PubMed]
- Baker NR, Rosenqvist E (2004) Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J Exp Bot 55:1607–1621 [DOI] [PubMed]
- Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287 [DOI] [PubMed]
- Boston RS, Viitanen PV, Vierling E (1996) Molecular chaperones and protein folding in plants. Plant Mol Biol 32:191–222 [DOI] [PubMed]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129:661–677 [DOI] [PMC free article] [PubMed]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856 [DOI] [PMC free article] [PubMed]
- Desikan R, Neill SJ, Hancock JT (2000) Hydrogen peroxide-induced gene expression in Arabidopsis thaliana. Free Radic Biol Med 28:773–778 [DOI] [PubMed]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206 [DOI] [PubMed]
- Fey V, Wagner R, Brautigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmuller R, Pfannschmidt T (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280:5318–5328 [DOI] [PubMed]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162:1445–1456 [DOI] [PMC free article] [PubMed]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141:436–445 [DOI] [PMC free article] [PubMed]
- Grene R (2002) Oxidative stress and acclimation mechanisms in plants. In: Somerville C, Meyerowitz E (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0009, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Gupta AS, Webb RP, Holaday AS, Allen RD (1993) Overexpression of superoxide dismutase protects plants from oxidative stress (Induction of ascorbate peroxidase in superoxide dismutase-overexpressing plants). Plant Physiol 103:1067–1073 [DOI] [PMC free article] [PubMed]
- Halliwell B, Gutteridge JMC (1989) Free radicals in biology and medicine. Oxford University Press, Oxford
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300 [DOI] [PMC free article] [PubMed]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273:1853–1856 [DOI] [PubMed]
- Kang KS, Lim CJ, Han TJ, Kim JC, Jin CD (1999) Changes in the isozyme composition of antioxidant enzymes in response to aminotriazole in leaves of Arabidopsis thaliana. J Plant Biol 42:187–193 [DOI]
- Kimura M, Yamamoto YY, Seki M, Sakurai T, Sato M, Abe T, Yoshida S, Manabe K, Shinozaki K, Matsui M (2003) Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol 77:226–233 [DOI] [PubMed]
- Konig J, Lotte K, Plessow R, Brockhinke A, Baier M, Dietz KJ (2003) Reaction mechanism of plant 2-Cys peroxiredoxin. Role of the C terminus and the quaternary structure. J Biol Chem 278:24409–24420 [DOI] [PubMed]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316:715–719 [DOI] [PubMed]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222 [DOI] [PubMed]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Packer L, Douce R (eds) Methods Enzymol 148:350–382
- Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S (2007) Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J 51:941–954 [DOI] [PubMed]
- Macpherson AN, Telfer A, Barber J, Truscott TG (1993) Direct detection of singlet oxygen from isolated Photosystem II reaction centres. Biochim Biophys Acta 1143:301–309 [DOI]
- Maly FE, Nakamura M, Gauchat JF, Urwyler A, Walker C, Dahinden CA, Cross AR, Jones OT, De Weck AL (1989) Superoxide-dependent nitroblue tetrazolium reduction and expression of cytochrome b-245 components by human tonsillar B lymphocytes and B cell lines. J Immunol 142:1260–1267 [PubMed]
- Mehler AH (1951) Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch Biochem 33:65–77 [DOI] [PubMed]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410 [DOI] [PubMed]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498 [DOI] [PubMed]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48:535–547 [DOI] [PubMed]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50:333–359 [DOI] [PubMed]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6:177–189 [DOI] [PMC free article] [PubMed]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332 [DOI] [PMC free article] [PubMed]
- Palatnik JF, Carrillo N, Valle EM (1999) The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol 121:471–478 [DOI] [PMC free article] [PubMed]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071 [DOI] [PMC free article] [PubMed]
- Rial DV, Ottado J, Ceccarelli EA (2003) Precursors with altered affinity for Hsp70 in their transit peptides are efficiently imported into chloroplasts. J Biol Chem 278:46473–46481 [DOI] [PubMed]
- Rial DV, Arakaki AK, Almara AM, Orellano EG, Ceccarelli EA (2006) Chloroplast Hsp70s are not involved in the import of ferredoxin-NADP+ reductase precursor. Physiol Plant 128:618–632 [DOI]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004a) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279:11736–11743 [DOI] [PubMed]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004b) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134:1683–1696 [DOI] [PMC free article] [PubMed]
- Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 28:123–133 [DOI] [PubMed]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130:1109–1120 [DOI] [PMC free article] [PubMed]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. CSHL Press, Cold Spring Harbor, New York
- Scarpeci TE, Marro ML, Bortolotti S, Boggio SB, Valle EM (2007) Plant nutritional status modulates glutamine synthetase levels in ripe tomatoes (Solanum lycopersicum cv. Micro-Tom). J Plant Physiol 164:137–145 [DOI] [PubMed]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Doring P (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60:759–772 [DOI] [PubMed]
- Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577:1–9 [DOI] [PubMed]
- Sung DY, Vierling E, Guy CL (2001) Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 126:789–800 [DOI] [PMC free article] [PubMed]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939 [DOI] [PubMed]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194 [DOI]
- Tsang EW, Bowler C, Herouart D, Van Camp W, Villarroel R, Genetello C, Van Montagu M, Inze D (1991) Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3:783–792 [DOI] [PMC free article] [PubMed]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inze D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39:45–58 [DOI] [PubMed]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139:806–821 [DOI] [PMC free article] [PubMed]
- Volkov RA, Panchuk II, Mullineaux PM, Schoffl F (2006) Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol 61:733–746 [DOI] [PubMed]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310–1326 [DOI] [PMC free article] [PubMed]
- Yamamoto A, Mizukami Y, Sakurai H (2005) Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem 280:11911–11919 [DOI] [PubMed]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.