Abstract
A number of cycling mammalian cells, such as NIH 3T3, contain abundant subsets of cold-stable microtubules. The origin of such microtubule stabilization in nonneuronal cells is unknown. We have previously described a neuronal protein, stable tubule-only polypeptide (STOP), that binds to microtubules and induces cold stability. We find that NIH 3T3 fibroblasts contain a major 42-kDa isoform of STOP (fibroblastic STOP, F-STOP). F-STOP contains the central repeats characteristic of brain STOP but shows extensive deletions of N- and C-terminal protein domains that are present in brain STOP. These deletions arise from differences in STOP RNA splicing. Despite such deletions, F-STOP has full microtubule stabilizing activity. F-STOP accumulates on cold-stable microtubules of interphase arrays and is present on stable microtubules within the mitotic spindle of NIH 3T3 cells. STOP inhibition by microinjection of affinity-purified STOP central repeat antibodies into NIH 3T3 cells abolishes both interphase and spindle microtubule cold stability. Similar results were obtained with Rat2 cells. These results show that STOP proteins have nonneuronal isoforms that are responsible for the microtubule cold stability observed in mammalian fibroblasts.
Microtubule arrays in mammalian cells are subject to substantial modulation of both assembly state and dynamics (1). In cycling cells, microtubule arrays undergo major changes in morphology and stability during mitosis, necessitating changes in the dynamic behavior of individual microtubules (2–4).
Microtubules assembled from pure tubulin in vitro can exhibit both spontaneous length fluctuations (5) and treadmilling behavior (6). Similar dynamic behaviors are observed in vivo but are clearly regulated by cellular metabolism. For example, many studies have shown that microtubule dynamics in cells or in cell extracts are modulated by protein kinase and protein phosphatase activities (7–11). In the general case, these enzymes do not act directly on tubulin. Protein kinase regulation of microtubule dynamics is largely mediated through action on other proteins that associate with tubulin dimers or with microtubules (1, 12, 13). Such proteins can induce microtubule depolymerization or can promote tubulin assembly by stabilizing assembled microtubules. As examples, microtubule depolymerization is favored by the phosphoprotein stathmin, which binds and sequesters tubulin dimers (13–15), whereas microtubule stabilization is typically mediated by microtubule-associated proteins (MAPs) that associate with polymers (16).
The existence of MAPs with potent microtubule-stabilizing activity in dividing cells has been made evident by cell permeabilization experiments (17). After cell permeabilization, protein kinases are either inhibited or eliminated, and microtubules are resistant to depolymerizing conditions such as dilution of the free tubulin pool and exposure to the microtubule assembly inhibitor nocodazole. Further, in some cell types, including fibroblasts, abundant subsets of microtubules resist exposure to cold temperature.
The microtubule cold stability found in fibroblasts represents an extreme state of polymer stabilization unlikely to be induced by the MAPs identified in cycling cells. These MAPs include E-MAP-115 and MAP4 (18, 19). MAP4 belongs to the MAP2/tau family of microtubule-stabilizing proteins that cannot induce microtubule cold stabilization in vitro or in vivo (20, 21). Overexpression of E-MAP-115 does not induce microtubule cold stability (18). Further, both MAP4 and E-MAP-115 are present in HeLa epithelial cells, which do not exhibit microtubule cold stability. Certain types of cycling cells thus appear to contain hitherto uncharacterized MAPs with potent microtubule stabilizing properties.
We have isolated from brain tissue a microtubule-binding protein, stable tubule-only polypeptide (STOP), that induces complete microtubule stabilization to various destabilizing agents, including exposure to cold temperature (22–25). Despite the apparent neuronal specificity of STOP, its attributes have raised the possibility that MAPs related to STOP may be present in cycling cells exhibiting cold stability (17).
Herein, we have used NIH 3T3 cells to test this possibility. NIH 3T3 cells are of fibroblastic origin and contain cold-stable microtubules. We demonstrate that these cells contain nonneuronal STOP isoforms and that these isoforms are responsible for the observed microtubule cold stability.
MATERIALS AND METHODS
STOP Antibodies.
Various rat brain STOP peptides, homologous between mouse and rat, were used to raise polyclonal antibodies. Polyclonal antibodies 23N and 23C were raised against nonoverlapping peptides corresponding to the N-terminal (23N) or the C-terminal (23C) parts of the brain STOP central repeat motif (25). Immunogenic peptides for the 23N and 23C antibodies were, respectively, PAAGKASGADQRDTRRKAG and TRTEGHEEKPLPPAQSQTQEGG (amino acids 222–240 and 246–267 of the rat brain STOP). Antibodies 136 and 139 were directed against peptides DIKPVKPIKAKPQYKPPDDK and ATKPDDKEQSKEMNNKLAEAK, respectively (amino acids 485–504 and 593–613 of the rat brain STOP). Immunized rabbit serum was affinity-purified against the corresponding peptides. mAb 175 (26) is specific of the brain STOP C-terminal repeat motif (C.B., unpublished data).
Cell Culture and Analysis of Protein Extracts.
HeLa cells were grown in RPMI 1640 medium containing 10% fetal calf serum (FCS). NIH 3T3 and Rat2 cells were grown in DMEM containing 10% FCS. For total cell extracts, NIH 3T3 cells grown to confluence on 100-mm Petri dishes were scraped in 500 μl of boiling 1% SDS and sonicated. For preparation of Triton-soluble cell fractions, NIH 3T3 cells were processed directly or incubated on ice for 30 min before fractionation. Cells were then washed with PBS and extracted for 3 min at the appropriate temperature in 500 μl of lysis buffer (17). For preparation of Triton-insoluble fractions, cells treated as above were subsequently washed with 10 ml of lysis buffer. Cells were then scraped in 500 μl of boiling 1% SDS and sonicated. Brain extracts were prepared as described (26). Protein extracts were then further processed for SDS/PAGE and Western blotting as described (17). STOP antibodies were diluted 1:5,000; goat anti-rabbit (Tago) and anti-mouse (Cappel) horseradish peroxidase-coupled secondary antibodies were diluted to 1:5,000.
Microinjection and Immunofluorescence.
NIH 3T3 or Rat2 cells, grown on coverslips, were injected with affinity-purified 23N or 23C STOP antibodies at 2 mg/ml or with nonimmune purified rabbit IgGs at 2 mg/ml, by using a 5171 micromanipulator and a 5246 transjector (Eppendorf). After a recovery period of 2–16 h, injected cells were directly processed for immunofluorescence or exposed to cold temperature (0°C) for 30 min before processing. Injected cells were fixed in −20°C methanol and then incubated with anti-β-tubulin mAb TUB 2.1 (1:100 dilution, Sigma) followed by an incubation with fluorescein isothiocyanate-coupled goat anti-mouse antibody (1:100 dilution, Jackson ImmunoResearch) and Cy3-coupled goat anti-rabbit antibody (1:1,000, Jackson ImmunoResearch).
For double tubulin/STOP staining, NIH 3T3 or transfected HeLa cells were directly processed for immunofluorescence or cold-treated as above before processing. Cells were permeabilized in lysis buffer for 1 min and further incubated for 1 min in the same buffer containing 10 μM paclitaxel (Taxol, Molecular Probes). Cells were fixed in PBS/4% paraformaldehyde, incubated with mAb TUB 2.1 and affinity-purified STOP 23C antibody (1:50 dilution) and then with secondary antibodies as above. Images were digitalized by using a Princeton RTE-CCD-1317-K/1 camera (Princeton Instruments, Trenton, NJ) and iplab spectrum software (Signal Analytics, Vienna, VA).
Analysis of STOP cDNA and STOP mRNA in NIH 3T3 Cells.
cDNA clones of fibroblastic STOPs were obtained by successive screens of an oligo(dT) and random-primed mouse 3T3 λgt11 cDNA library (CLONTECH) and of an oligo(dT)-primed mouse 3T3 λZAP II cDNA library (Stratagene), with 32P-labeled probes derived from rat brain STOP cDNA (25). Screenings were performed by standard procedures (27). We obtained a total of 19 positive clones, which were sequenced by Genome Express (Grenoble, France). One clone (16C cDNA) contained the entire sequence coding for another STOP isoform (fibroblastic STOP, F-STOP, see text).
For in vitro translation experiments, the ApaI fragment (nucleotides 1,139–2,470) of 16C cDNA, containing the whole coding sequence, was cloned into pBluescript (Stratagene) to obtain plasmid p16C-Apa. This construct was translated in the TnT-coupled reticulocyte lysate kit (Promega). For cDNA transfection experiments, the SpeI–KpnI fragment of p16C-Apa, containing the ApaI fragment, was subcloned into the expression plasmid putSV1 (Eurogentec, Seraing, Belgium). This construction was used to transfect exponentially growing HeLa cells as described (28).
RNA analysis by Northern blotting was performed by standard procedures (27) using 32P-labeled probes derived from 16C cDNA or from STOP genomic clones (see Fig. 2).
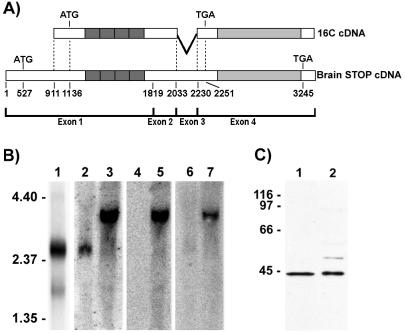
Figure 2.
Characterization of the F-STOP cDNA. (A) Schematic representation showing alignment of 16C cDNA with mouse brain STOP cDNA. Nucleotides are numbered according to the mouse brain STOP cDNA sequence, as deduced from the mouse STOP gene. 16C cDNA lacks nucleotides 1–910 of the brain STOP cDNA and also lacks the entire sequence of exon 3. (B) Northern blot analysis of RNAs from NIH 3T3 cells and from mouse brain. Poly(A)+ RNAs (3 μg) from NIH 3T3 cells were hybridized with 32P-labeled 16C cDNA, and a major 2.4-kb RNA species (lane 1) was detected. Total NIH 3T3 cell RNAs (30 μg; lanes 2, 4, and 6) or total mouse brain RNAs (30 μg; lanes 3, 5, and 7) were hybridized with 32P-labeled probes corresponding to the 3′ region of exon 1 (positions 787–1,818; lanes 2 and 3), to the 5′ region of exon 1 (positions 77–787; lanes 4 and 5), and to exon 3 (positions 2,044–2,181; lanes 6 and 7). As with 16C cDNA, the major STOP mRNA in NIH 3T3 cells lacked nucleotide sequences corresponding to the 5′ end of the brain STOP cDNA and the entire sequence of exon 3. Size markers are in kilobases. (C) Comparison of the translation product of p16C-Apa with F-STOP. Reticulocyte lysate reactions (5 μl; lane 1) and NIH 3T3 cell extracts (30 μg; lane 2) were separated by SDS/PAGE on 10% gels and immunoblotted with 23C STOP antibody. The translation product of p16C-Apa comigrated with F-STOP. Size markers are in kDa.
RESULTS
Immunoblot Analysis and Characterization of STOP Antigens in NIH 3T3 Cells.
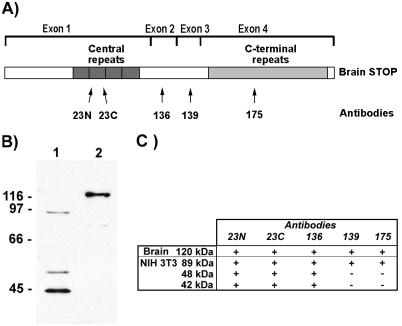
The nucleotide sequence of mouse brain STOP cDNA (GenBank accession no. Y16008) and the amino acid sequence of mouse brain STOP were deduced from analysis of the four exons of the mouse gene (Mtap6, ref. 29). Mouse brain STOP shows extensive homology with rat brain STOP (25). The protein contains two repeat regions, a central repeat region encoded by exon 1 and a C-terminal repeat region encoded by exon 4 (Fig. 1A).
Figure 1.
STOP antigens in NIH 3T3 cell extracts. (A) Schematic representation of mouse brain STOP showing domain structure of the protein. Protein domains encoded by the four exons are indicated. Mouse brain STOP contains a central repeat domain (encoded by exon 1) composed of four repeats and a C-terminal repeat domain (encoded by exon 4) composed of 28 repeats. The localization of the epitopes recognized by STOP antibodies 23N, 23C, 136, 139, and 175 is indicated. (B) Immunoblot analysis of NIH 3T3 cell extract (lane 1) and mouse brain extract (lane 2) using 23C STOP antibody. NIH 3T3 cell extract (40 μg) and brain extract (4 μg) were separated by SDS/PAGE on 8% gels. Size markers are in kDa. (C) Reactivity of brain and NIH 3T3 cell STOP proteins with the various STOP antibodies. For each STOP species, presence (+) or absence (−) of reactivity with the different STOP antibodies is indicated.
We used five STOP antibodies directed against distinct epitopes of the brain protein to probe NIH 3T3 cell extracts for STOP antigens on Western blots. The probes included two antibodies (23C and 23N) directed against two nonoverlapping peptides within the amino acid sequence of the STOP central repeat, two antibodies (136 and 139) that were specific to protein sequences encoded by exons 2 and 3, respectively, and a mAb (mAb 175) recognizing an epitope located in the STOP C-terminal repeat domain (Fig. 1A). NIH 3T3 cell extracts contained a major STOP antigen with an apparent mass of 42 kDa (Fig. 1B, lane 1) that reacted with the two central repeat antibodies (23N and 23C) and with the exon 2 antibody (136) but did not react with the exon 3 antibody (139) or with mAb 175 (Fig. 1C). This major STOP species differed from brain STOP that has a greater mass (Fig. 1B, lane 2) and that reacts with all five antibodies. We chose to call the major NIH 3T3 STOP antigen F-STOP, for fibroblastic STOP.
As a result of screening of two NIH 3T3 cDNA libraries and analysis of 19 overlapping clones, we have determined the cDNA sequence of F-STOP. The longest clone (16C cDNA) had 2,292 bases and terminated with poly(A) tail. A schematic representation of the alignment of 16C cDNA with mouse brain STOP cDNA is shown in Fig. 2A. Compared with the brain STOP cDNA, the 16C cDNA lacked a nucleotide sequence (positions 1–910) corresponding to the 5′ region of exon 1. The first ATG in the 16C cDNA was at position 1,136. The 16C cDNA also lacked the entire nucleotide stretch corresponding to exon 3. The deletion of exon 3 introduced a frameshift that created a stop codon in exon 4 at nucleotide 2,251 of the cDNA, upstream of the sequences coding for the STOP C-terminal repeats.
Northern blot analysis of NIH 3T3 cell poly(A)+ RNAs using 32P-labeled 16C cDNA as a probe showed a major band of 2.4 kb (Fig. 2B, lane 1), similar in size to 16C cDNA. This major NIH 3T3 mRNA hybridized with a probe corresponding to the 3′ region of exon 1 (Fig. 2B, lane 2). In contrast, it did not hybridize with probes corresponding either to the 5′ region of exon 1 or to exon 3 (Fig. 2B, lanes 4 and 6). Brain STOP mRNA was 3.4 kb long and reacted with all probes (Fig. 2B, lanes 3, 5, and 7). These results showed that NIH 3T3 cells contained a major STOP mRNA that was different from brain STOP mRNA but had a domain structure similar to 16C cDNA. This domain structure corroborated the immunoreactivity of F-STOP with the different STOP antibodies (Fig. 1C). Our data provided strong evidence that we had isolated a cDNA corresponding to the major mRNA, containing the complete coding sequence for F-STOP. In agreement with this conclusion, in vitro translation of the 16C cDNA yielded a protein that comigrated with F-STOP in SDS/PAGE (Fig. 2C).
In addition to F-STOP, NIH 3T3 cell extracts contained two minor STOP variants with apparent masses of 48 and 89 kDa, respectively (Fig. 1 B and C). Both variants reacted with the two central repeat antibodies (23N and 23C) and with the exon 2 antibody (136). Moreover, the 89-kDa STOP protein reacted with the exon 3 antibody (139) and with mAb 175. We have not further elucidated the structure of the mRNAs coding for these two minor fibroblastic STOP proteins.
Functional Properties and Subcellular Localization of NIH 3T3 Cell STOP Proteins.
We have tested whether, despite extensive domain deletions, F-STOP had microtubule-stabilizing activity similar to brain STOP. We found that, as brain STOP (25), F-STOP associates with microtubules and induces microtubule cold stabilization when expressed in cells normally devoid of stable polymers (Fig. 3). This result shows that F-STOP has full microtubule-stabilizing activity in vivo.
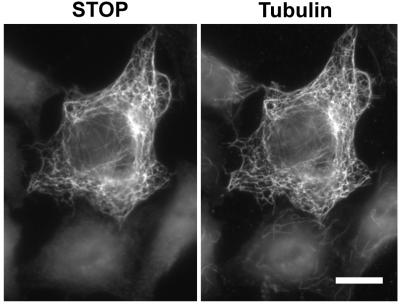
Figure 3.
F-STOP induces microtubule cold stability in HeLa cells. HeLa cells were transfected with cDNA coding for F-STOP. After 24 h, cells were exposed to cold temperature, lysed, and then double-stained with STOP antibody 23C (Left) and with tubulin antibody TUB 2.1 (Right). F-STOP associates with microtubules in transfected cells and F-STOP-associated polymers are resistant to depolymerization induced by cold. (Bar = 10 μm.)
With an antibody against the central repeat region (23C), we analyzed the STOP distribution in NIH 3T3 cells. At physiological temperature, STOP staining was barely detectable on the microtubule network (Fig. 4A, row warm). Remarkably, microtubules in cells exposed to cold temperature before fixation exhibited a bright STOP staining (Fig. 4A, row cold). These results raised the possibility that cell exposure to cold temperature induced a rapid shift of STOP from the unbound pool onto interphase microtubules. The translocation of STOP to microtubules on cold exposure was further confirmed by immunoblot analysis of Triton-soluble and Triton-insoluble protein fractions from cells exposed or not exposed to the cold prior to assay. The bulk of F-STOP protein was present in the Triton-soluble fraction of untreated cells, but a large proportion of F-STOP had shifted to the Triton-insoluble cell fraction after cold exposure (Fig. 4B). These results suggest that F-STOP interaction with interphase microtubules is normally inhibited to a large extent by regulatory controls. F-STOP accumulation on the interphase microtubule array in cold-treated cells may reflect an inhibition of these regulatory controls at cold temperature.
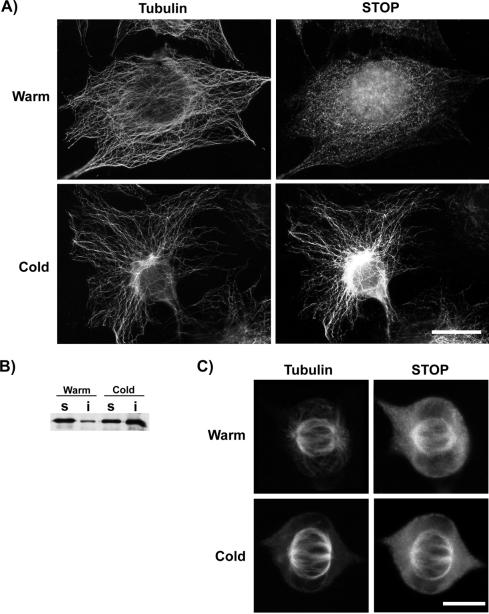
Figure 4.
STOP localization in NIH 3T3 cells. (A) NIH 3T3 cells either were immediately fixed (row warm) or were first exposed to cold temperature for 30 min (row cold). Cells were then double-stained using mAb TUB 2.1 to label tubulin (Left) and 23C antibody to label STOP proteins (Right). STOP staining of microtubules is strongly enhanced in cold-treated cells as compared with untreated cells. (Bar = 10 μm.) (B) Immunoblot analysis of F-STOP presence in Triton-soluble cell fraction (lanes s) and Triton-insoluble cell fraction (lanes i). NIH 3T3 cells were immediately extracted with a Triton-based buffer (lanes warm) or first incubated at 0°C for 30 min (lanes cold). Cell extracts were then separated by SDS/PAGE on 8% gels and assayed for F-STOP content on immunoblots using 23C STOP antibody. In repeated experiments, patterns of F-STOP distribution among soluble and insoluble fractions from the same cells were highly reproducible. The apparent difference in total STOP content between cold-treated and untreated cells reflects random variations between preparations. (C) Mitotic cells analyzed in the same manner as described in A. (Bar = 5 μm.)
STOP regulation is apparently different during mitosis. Examination of STOP distribution in mitotic cells at physiological temperature showed a bright staining of spindle microtubules (Fig. 4C, row warm). This staining was apparently concentrated on kinetochore-to-pole microtubules, whereas astral microtubules were not detectably stained. Exposure of mitotic cells to the cold only moderately enhanced STOP staining on the spindle (Fig. 4C, row cold). As was shown for cells expressing neuronal STOP (30), only the STOP-associated kinetochore-to-pole microtubules were resistant to cold temperature.
These results show that STOP proteins are mainly localized in a soluble pool during interphase and are recruited to the interphase microtubule array on exposure to cold temperature. In contrast, STOP is abundant on spindle microtubules at physiological temperature, indicating that it is subject to mitosis-specific regulatory controls.
STOP Proteins Are Responsible for Microtubule Cold Stability in Mammalian Fibroblasts.
STOP proteins associate with microtubules in NIH 3T3 cells during cold exposure and present a microtubule stabilizing activity (see above). These results suggest a central role of STOP proteins in the induction of polymer resistance to the cold. However, cold stability could also be conferred on microtubules by other factors. To test whether STOP association with microtubules was indeed responsible for microtubule cold stabilization, we examined the effect of STOP inactivation on microtubule stability in NIH 3T3 cells.
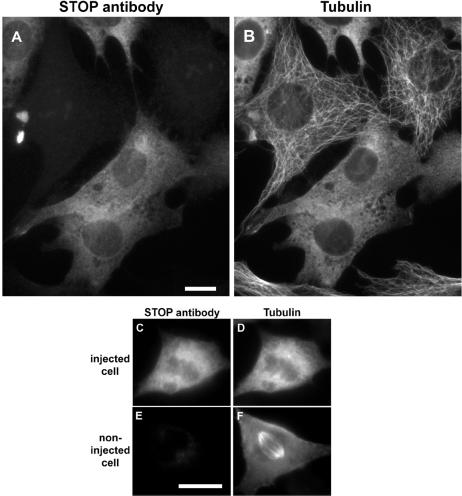
For STOP inactivation, NIH 3T3 cells were injected with affinity-purified 23C STOP antibody. After a recovery period of 2–16 h, cells were either directly fixed or exposed to cold temperature for 30 min before fixation. Cells were then double-stained to detect microinjected STOP antibody and tubulin. Microtubule networks in interphase cells maintained at physiological temperature remained apparently normal after microinjection of the 23C STOP antibody (data not shown). However, when cells were exposed to cold temperature, interphase microtubules in injected cells depolymerized (Fig. 5B), demonstrating a dramatic loss of microtubule cold stability in the presence of microinjected STOP antibody.
Figure 5.
Interphase and spindle microtubule disassembly in cold-treated cells injected with STOP antibody. NIH 3T3 cells were injected with affinity-purified 23C STOP antibody. After a 2-h recovery period, cells were exposed to cold temperature for 30 min. Cells were then fixed and double-stained with Cy3-conjugated anti-rabbit antibody to detect injected 23C antibody (A, C, and E) and mAb TUB 2.1-fluorescein isothiocyanate anti-mouse antibody to detect tubulin (B, D, and F). (A and B) Injected and noninjected interphase cells. (C and D) Injected metaphase cell. (E and F) Noninjected metaphase cell. STOP inhibition abolishes microtubule cold stability. (Bars = 10 μm.)
In the present study, STOP antibody microinjection was performed in interphase cells. Some microinjected cells progressed into mitosis during the recovery period after microinjection. Such mitotic cells exhibited apparently normal spindle microtubules when kept at physiological temperature (data not shown). However, spindle microtubules in microinjected mitotic cells showed complete loss of cold stability (Fig. 5D) as compared with noninjected cells (Fig. 5F). We also observed microinjected cells during later stages of mitosis. At late telophase, midbody microtubules showed a loss of cold stability similar to that of spindle microtubules in metaphase cells (data not shown).
Microinjection experiments performed with 23N STOP antibody instead of 23C antibody gave identical results both in interphase and mitotic cells. We performed control experiments in which NIH 3T3 cells were microinjected with nonimmune purified rabbit IgGs. Such IgGs had no effect on microtubule stability (data not shown). Finally, when injected in other fibroblastic cells showing extensive microtubule cold stability (Rat2 cells), 23N and 23C STOP antibodies could also abolish microtubule cold stability (data not shown).
Thus, these results show that STOP proteins, although not uniquely required for basic microtubule function in cycling cells at warm temperature, are clearly the principal proteins responsible for microtubule cold stability in these cells. Further, their capacity to associate with microtubules appears to be under cell cycle regulation.
DISCUSSION
Cold stability is a remarkable property of microtubules observed in living cells, in sharp contrast with the lability of microtubules assembled from pure tubulin in vitro. Cold-stable microtubules were initially isolated from mammalian brain extracts (31) and this stability property has been shown to arise from the association of microtubules with a specific MAP, designated STOP (23–25). However, microtubule cold stability is not restricted to neuronal cells. It has also been observed in other cell types such as mammalian fibroblasts (17, 32, 33). A widespread view in the microtubule field has been that microtubule cold stability in dividing cells may result from a complex set of interactions between microtubules and other cell components. For instance, cytoplasmic microtubules are connected to intermediate filaments and this affects their stability (34). However, the demonstration that STOP alone can induce microtubule cold stability raised the alternative possibility that microtubule cold stability in nonneuronal cells arises from the action of a single class of MAPs related to neuronal STOP. Herein we show that isoforms of STOP are indeed present in NIH 3T3 fibroblasts. These STOP proteins associate with cold-stable microtubules in interphase cells and in the mitotic spindle. Further, STOP inhibition by microinjected STOP antibodies abolishes microtubule cold stability in two fibroblastic cell types. This analysis demonstrates that microtubule cold stability is induced by the association of STOP proteins with microtubules in fibroblasts. We believe that an important implication of these results is that microtubule stability in cells is apparently controlled by a limited number of specific effectors and not by ill-defined cumulative factors. Given the central role of microtubule dynamics in vital cell functions, it seems logical that only a few cell components that are subject to physiological regulation can interfere massively with microtubule stability.
There are major structural differences between STOP proteins in NIH 3T3 cells and the mouse brain STOP. Brain STOP is encoded by four exons. The major NIH 3T3 cell isoform F-STOP contains part of the STOP domain encoded by exon 1 and the STOP domain encoded by exon 2 but elsewhere shows extensive deletions of protein domains compared with brain STOP. Despite such deletions, F-STOP has full microtubule-stabilizing activity in vivo. The absence of exon 1 sequences in the F-STOP mRNA apparently derives from RNA splicing within a nucleotide sequence that is translated in brain. Such RNA splicing within an exon sequence is curious but not unprecedented (35). We do not know the functional significance of the observed variations of STOP structure between neuronal and fibroblastic cells. An attractive possibility is that the various STOP domains interact with different proteins in a tissue-specific manner. We are currently investigating this possibility.
As STOP is a potent microtubule-stabilizing protein, the generation of active microtubule dynamics in interphase cells must involve rapid regulation of STOP activity. Such regulation is evident from the observation that the bulk of STOP proteins does not bind to microtubules in interphase cells. We have provided (17) evidence that microtubule stability in interphase NIH 3T3 cells was modulated by protein kinases. The apparent inhibition of STOP interaction with microtubules in interphase cells at physiological temperature probably results from STOP phosphorylation by protein kinases, whose activity is inhibited in the cold. Interestingly, STOP proteins accumulate on spindle microtubules both at physiological temperature and in the cold (this study and ref. 30). This suggests a role of STOP in the maintenance or function of the mitotic spindle. It is of interest to note that kinetochore-to-pole microtubules are dynamic, fluxing rapidly poleward (36), despite their association with STOP proteins.
In microinjection experiments, STOP-antibody-injected cells exhibited apparently normal microtubule arrays in interphase and were capable of progression through mitosis despite extensive inhibition of STOP activity. Thus, STOP function seems largely expendable in cells grown in vitro. This could be anticipated, because certain cycling cells such as HeLa substantially lack STOP proteins (25) and nevertheless show apparently normal microtubular function. One possibility is that STOP function in cultured cells is redundant with that of other MAPs. Precedence for functional redundancy between MAPs was already suggested from inactivation experiments of MAP4 and tau (37, 38). However, STOP proteins are unique among the MAPs with respect to their microtubule-stabilizing activity and the STOP system may be required for microtubule functions that we have not assayed. One way to look for STOP function at the cellular level will be to search for conditions other than exposure to the cold that induce STOP recruitment on interphase microtubules. Such experiments may reveal hitherto unsuspected function for cytoskeletal stabilization. Other clues to the understanding of STOP function should arise from the elucidation of the molecular identity of STOP regulators and from the study of STOP function in the context of the tissues and organs in which it is expressed. A STOP gene knock-out in mice should thus be of great interest to assess the consequences of STOP inactivation in those tissues that normally contain STOP proteins.
Acknowledgments
We thank Dr. L. Lafanechère for help in raising STOP antibodies and D. Proietto for technical assistance. This work was supported in part by grants from the French Ministère de l’Enseignement Supérieur et de la Recherche (MesR ACC-SV N° 5), the Commission of the European Communities (Grant CHRX CT94.0642), the Ligue Nationale Contre le Cancer (La Ligue), the Association pour la Recherche sur le Cancer, and the Glaxo–Wellcome Laboratories (France).
ABBREVIATIONS
- STOP
stable tubule-only polypeptide
- F-STOP
fibroblastic STOP
- MAP
microtubule-associated protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y16032).
References
- 1.McNally F J. Curr Opin Cell Biol. 1996;8:23–29. doi: 10.1016/s0955-0674(96)80044-5. [DOI] [PubMed] [Google Scholar]
- 2.Salmon E D, Leslie R J, Saxton W M, Karow M L, McIntosh J R. J Cell Biol. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxton W M, Stemple D L, Leslie R J, Salmon E D, Zavortink M, McIntosh J R. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmont L D, Hyman A A, Sawin K E, Mitchison T J. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison T J, Kirschner M. Nature (London) 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 6.Margolis R L, Wilson L. Cell. 1978;13:1–8. doi: 10.1016/0092-8674(78)90132-0. [DOI] [PubMed] [Google Scholar]
- 7.Bershadsky A D, Gelfand V I. Proc Natl Acad Sci USA. 1981;78:3610–3613. doi: 10.1073/pnas.78.6.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Brabander M, Geuens G, Nuydens R, Willebrords R, De Mey J. Cold Spring Harbor Symp Quant Biol. 1981;46:227–240. doi: 10.1101/sqb.1982.046.01.026. [DOI] [PubMed] [Google Scholar]
- 9.Verde F, Labbé J-C, Dorée M, Karsenti E. Nature (London) 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh Y, Nishida E, Matsuda S, Shiina N, Kosako H, Shiokawa K, Akiyama T, Ohta K, Sakai H. Nature (London) 1991;349:251–254. doi: 10.1038/349251a0. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson J E, Brautigan D L, Vallee R, Olmsted J, Fujiki H, Goldman R D. Proc Natl Acad Sci USA. 1992;89:11093–11097. doi: 10.1073/pnas.89.22.11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drewes G, Ebneth A, Preuss U, Mandelkow E M, Mandelkow E. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 13.Belmont L D, Mitchison T J. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 14.Marklund U, Larsson N, Melander Gradin H, Brattsand G, Gullberg M. EMBO J. 1996;15:5290–5298. [PMC free article] [PubMed] [Google Scholar]
- 15.Jourdain L, Curmi P, Sobel A, Pantaloni D, Carlier M F. Biochemistry. 1997;36:10817–10821. doi: 10.1021/bi971491b. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa N. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 17.Lieuvin A, Labbé J-C, Dorée M, Job D. J Cell Biol. 1994;124:985–996. doi: 10.1083/jcb.124.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masson D, Kreis T E. J Cell Biol. 1993;123:357–371. doi: 10.1083/jcb.123.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapin S J, Bulinski J C. J Cell Sci. 1991;98:27–36. doi: 10.1242/jcs.98.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Pirollet F, Job D, Fischer E H, Margolis R L. Proc Natl Acad Sci USA. 1983;80:1560–1564. doi: 10.1073/pnas.80.6.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baas P W, Pienkowski T P, Cimbalnik K A, Toyama K, Bakalis S, Ahmad F J, Kosik K S. J Cell Sci. 1994;107:135–143. doi: 10.1242/jcs.107.1.135. [DOI] [PubMed] [Google Scholar]
- 22.Job D, Fischer E H, Margolis R L. Proc Natl Acad Sci USA. 1981;78:4679–4682. doi: 10.1073/pnas.78.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Job D, Rauch C T, Fischer E H, Margolis R L. Biochemistry. 1982;21:509–515. doi: 10.1021/bi00532a015. [DOI] [PubMed] [Google Scholar]
- 24.Margolis R L, Rauch C T, Job D. Proc Natl Acad Sci USA. 1986;83:639–643. doi: 10.1073/pnas.83.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosc C, Cronk J D, Pirollet F, Watterson D M, Haiech J, Job D, Margolis R L. Proc Natl Acad Sci USA. 1996;93:2125–2130. doi: 10.1073/pnas.93.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirollet F, Rauch C T, Job D, Margolis R L. Biochemistry. 1989;28:835–842. doi: 10.1021/bi00428a064. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denarier E, Aguezzoul M, Jolly C, Vourc’h C, Roure A, Andrieux A, Bosc C, Job D. Biochem Biophys Res Commun. 1998;243:791–796. doi: 10.1006/bbrc.1998.8179. [DOI] [PubMed] [Google Scholar]
- 30.Margolis R L, Rauch C T, Pirollet F, Job D. EMBO J. 1990;9:4095–4102. doi: 10.1002/j.1460-2075.1990.tb07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb B C, Wilson L. Biochemistry. 1980;19:1993–2001. doi: 10.1021/bi00550a041. [DOI] [PubMed] [Google Scholar]
- 32.Bershadsky A D, Gelfand V I, Svitkina T P, Tint I S. Cell Biol Int Rep. 1979;3:45–50. doi: 10.1016/0309-1651(79)90067-5. [DOI] [PubMed] [Google Scholar]
- 33.Moskalewski S, Thyberg J, Friberg V. Cell Tissue Res. 1980;210:403–415. doi: 10.1007/BF00220198. [DOI] [PubMed] [Google Scholar]
- 34.Goldman R D, Khuon S, Chou Y H, Opal P, Steinert P M. J Cell Biol. 1996;134:971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayer K U, Löhler J, Harbers K. Mol Cell Biol. 1996;16:29–36. doi: 10.1128/mcb.16.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchison T J. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda T, Hirokawa N. Nature (London) 1994;369:488–491. doi: 10.1038/369488a0. [DOI] [PubMed] [Google Scholar]
- 38.Wang X M, Peloquin J G, Zhai Y, Bulinski J C, Borisy G G. J Cell Biol. 1996;132:345–357. doi: 10.1083/jcb.132.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]