Abstract
We and others have conducted targeted genetic association analyses of ABCA1 in relation to Alzheimer disease risk with a resultant mixture of both support and refutation, but all previous studies have been based upon only a few markers. Here, a detailed survey of genetic variation in the ABCA1 region has been performed in a total of 1567 Swedish dementia cases (including 1275 with Alzheimer disease) and 2203 controls, providing evidence of association with maximum significance at marker rs2230805 (OR = 1.39; 95% CI 1.23–1.57, P = 7.7 × 10−8). Haplotype-based tests confirmed association of this genomic region after excluding rs2230805, and imputation did not reveal additional markers with greater support. Significantly associating markers reside in two distinct linkage disequilibrium blocks with maxima near the promoter and in the terminal exon of a truncated ABCA1 splice-form. The putative risk allele of rs2230805 was also found to be associated with reduced cerebrospinal fluid levels of β-amyloid. The strongest evidence of association was obtained when all forms of dementia were considered together, but effect sizes were similar when only confirmed Alzheimer disease cases were assessed. Results further implicate ABCA1 in dementia, reinforcing the putative involvement of lipid transport in neurodegenerative disease.
Keywords: ABCA1, Alzheimer, dementia, SNP, amyloid
Introduction
Cholesterol metabolism is an intriguing biological target for study in relation to dementia risk. The original discovery of genetic association of APOE (MIM# 107741) and Alzheimer disease (AD) risk served as the first strong indication of a link between lipids and neurodegeneration [Strittmatter et al., 1993]. Sequence variants of APOE contribute to variability in cholesterol and phospholipid levels [Viiri et al., 2005; Yue et al., 2005] which is likely to be a primary pathogenic mechanism leading to AD [Corder et al., 1993; Poirier et al., 1993; Strittmatter et al., 1994]. Apart from directly influencing plasma lipid levels [Beekman et al., 2004] APOE also appears to strongly affect β-amyloid (Aβ) deposition in the brain [Beffert et al., 1999], suggesting a connection between Aβ, the main component of plaques, and apolipoprotein metabolism. In further support of this, another prominent phenotypic consequence of APOE polymorphism is to increase variance in cerebrospinal fluid (CSF) levels of the 42 amino acid fragment of β-amyloid (Aβ1–42) [Prince et al., 2004]. Importantly, other genes that are proven to lead to rare familiar forms of AD (APP, PSEN1, and PSEN2) [Goate et al., 1991; Rogaev et al., 1995; Sherrington et al., 1995] have also been shown to be involved in β-amyloid metabolism [Scheuner et al., 1996]. On this basis, the identification of genes that contribute to variance in both Aβ and lipid processing may be of aid in understanding the biological mechanisms underlying AD.
Genetic association studies of cholesterol related genes in relation to AD are abundant but have not yet produced any highly replicable novel candidates apart from APOE. One gene however that has received more attention than others is ABCA1, encoding the ATP binding cassette A1 transporter (MIM# 600046). ABCA1 has been studied extensively in relation to cholesterol metabolism following the discovery that mutations in ABCA1 are the primary cause of Tangier disease, for which a key biological feature is a reduction in plasma HDL levels [Bodzioch et al., 1999; Brooks-Wilson et al., 1999; Rust et al., 1999; Clee et al., 2001; Tregouet et al., 2004]. ABCA1 facilitates the unidirectional efflux of free cholesterol and phospholipids from cells and the lipidation of most of the exchangeable apolipoproteins (including APOA1 and APOE), enabling the formation of nascent HDL molecules [Yancey et al., 2003]. Studies have shown that deficiency of ABCA1 in the brain leads to a specific decrease in APOE levels [Hirsch-Reinshagen et al., 2004; Wahrle et al., 2004]. Interestingly ABCA1, like APOE, appears to be involved in Aβ metabolism [Fukumoto et al., 2002], a result that fits with evidence suggesting a relationship between cholesterol metabolism and Aβ metabolism [Simons et al., 1998; Kojro et al., 2001]. Emerging data thus suggest that ABCA1 plays an important role in lipid metabolism in both peripheral tissues and the central nervous system (CNS), and like APOE, may influence Aβ. Genetic association studies of ABCA1 and AD risk or AD-related phenotypes have produced a mixture of positive findings [Wollmer et al., 2003; Katzov et al., 2004; Sundar et al., 2007] and negative reports [Li et al., 2004; Shibata et al., 2006]. All previous studies however have focused on a few genetic markers. In the present study we have attempted to rectify this by conducting a more thorough genetic analysis of the ABCA1 region and broadening our search to include both dementia and AD risk.
Materials and Methods
Human Samples
Samples used in the present study were derived primarily from the population-based Swedish Twin Registry [Lichtenstein et al., 2002; Pedersen et al., 2002]. An independent non-twin case-control Swedish AD sample was also included. In total, DNA was available for 1567 dementia cases and 2203 controls (1275 had a possible or probable AD diagnosis). Among dementia cases, 1233 were unrelated. There were 990 men and 1213 women in the control group, and 598 men and 969 women in the dementia group. Average age-at-sampling for controls was 77.7 ± 8.7 (SD) years and age-at-onset for dementia/AD cases was 75.3 ± 8.3 (SD) years. Samples are described in more detail below.
SATSA
The Swedish Adoption/Twin Study of Aging (SATSA) sample comprises all pairs of twins who indicated that they had been separated before the age of 11 and reared apart and a sample of twins reared together matched on gender and date and county of birth [Pedersen et al., 1984]. Participants were aged 50 and older at first cognitive testing (Median=65 years). SATSA twins aged 50 and older were invited to participate in-person testing sessions (IPT) in which cognitive functioning and performance measures were administered (Median age =65 years). Altogether 858 twins have been followed longitudinally up to six IPT sessions across 19 years. Blood samples for DNA extraction and genotyping for the current analyses were collected at the third IPT. Individuals in SATSA were screened for cognitive dysfunction by telephone using the TELE interview [Gatz et al., 1995] or by in-person MMSE performance [Gatz et al., 1997; Pedersen et al., 2004]. Those who screened positive were assessed clinically, including physical and neurological examinations, neuropsychological assessment, and neuro-imaging. Diagnoses were made by consensus conference with a multidisciplinary team, following DSMIII-R criteria and NINCDS-ADRDA criteria for AD. In addition, other dementias were assessed, including vascular dementia (VaD), mixed Alzheimer's and vascular dementia, dementia due to a particular cause (e.g., hydrocephalus or Parkinson's disease) or dementia unspecified. There were 65 pairs (aged 55+ yrs) in which one or both were demented as assessed by AG 08724/01-07. Surviving SATSA twins were again evaluated for dementia by AG 08724/08-12 (HARMONY), described below. The current study included 625 individuals with DNA.
OCTO-twin
The OCTO-Twin sample began with 351 twin pairs (149 MZ and 202 like-sex DZ pairs) aged 80 years or older (i.e., birth years 1913 or earlier). Average age was 83.57 and 67% of the sample were female. Up to five waves of longitudinal follow-ups at two year intervals were conducted on all living twins who agreed to participate, irrespective of co-twin status. Blood samples for subsequent extraction of DNA were collected between the first and second in person assessment. One hundred seventy five (175) dementia cases were identified by a comparable process to SATSA but were followed in the framework of the OCTO-Twin schedule. Surviving OCTO-Twin participants were again evaluated for dementia by HARMONY. The current study included 443 individuals with DNA.
GENDER
All living pairs of unlike-sex twins born between 1906 and 1925 were identified through the Swedish Twin Registry. A total of 605 twin pairs were sent surveys assessing health and other factors [Gold et al., 2002]. A subset of this population-based sample completed in-person assessments consisting of a short cognitive battery and the same health assessments as in SATSA and OCTO-Twin. A total of 488 individuals agreed to give blood samples in the in-person assessment. Members of this sample who were demented have been formally diagnosed through HARMONY and after the third wave of in-person assessments by GENDER, similar to the process in SATSA. The current study included 411 individuals with DNA.
HARMONY
All twins from the Swedish Twin Registry aged 65 and older were screened by telephone for cognitive dysfunction [Gatz et al., 2005]. This included any surviving twins from the SATSA, OCTO-Twin and GENDER studies described above. The response rate to the screening was 71% out of a total population of 20,269 [Gatz et al., 2005]. A total of 14,435 twins were screened by telephone, seen in person and/or screened by proxy (56% female). The protocol resulted in 11.5% screening positive for cognitive dysfunction. Clinical diagnoses are available for 1488 twins not identified and worked-up by the earlier SATSA and OCTO-Twin studies. Diagnoses were made by consensus conference, following DSM IV criteria and NINCDS/ADRDA criteria for AD. In addition, other dementias were assessed (e.g., vascular dementia -VaD; mixed Alzheimer's and vascular dementia) using established criteria (see Gatz et al., 2005). The current study included 1147 individuals with DNA.
The Swedish non-twin case-control sample consists of 896 late-onset Alzheimer disease (LOAD) patients and 248 controls. These individuals were recruited from three prospective longitudinal studies of patients with dementia from Mölndal, Piteå, and Malmö, Sweden. In the AD cohort, 803 had a clinical and 93 a neuropathological diagnosis. All clinically diagnosed AD patients underwent a thorough investigation, which included a medical history, physical, neurological and psychiatric examination, screening laboratory tests, ECG, X-ray of the chest, EEG, and computerized tomography (CT) of the brain. MMSE was administered by a trained nurse and recorded in journals. When the study was completed and AD cases identified, journals were reviewed for the latest MMSE score. Clinical AD diagnoses were made according to the NINCDS-ADRDA criteria. All neuropathologically diagnosed AD patients also fulfilled the clinical NINCDS-criteria for probable AD and met the neuropathological CERAD criteria for definitive AD. Among controls, 140 were healthy volunteers without history, symptoms or signs of psychiatric or neurological disease, malignant disease, or systemic disorders. Cognitive status was examined using MMSE, and individuals with scores below 28 were not included as controls. There were 108 autopsy controls consisting of patients who had died from cardiac disease or malignant disease. Their medical records revealed no history of dementia or other psychiatric or neurological diseases. Post-mortem examination revealed no macroscopic infarcts. All autopsy individuals (AD and control) were matched by age at death and all clinically diagnosed AD cases and healthy volunteers were matched by age-at-onset/age-at exam respectively.
An additional reference Swedish population was included consisting of 3013 men from the CAPS (Cancer Prostate in Sweden) study. Subjects with prostate cancer were identified and recruited from four of the six regional cancer registries in Sweden diagnosed between 2001 and 2003. Among 3648 identified subjects with prostate cancer, 3161 (87%) agreed to participate. Control subjects, who were recruited concurrently with case subjects, were randomly selected from the Swedish Population Registry and matched according to the expected age distribution of cases (groups of 5-year intervals) and geographic region. A total of 2149 of 3153 control subjects (68%) who were invited subsequently agreed to participate in the study. DNA samples from blood were available for 2893 case subjects (92%) and 1781 control subjects (83%). For the present study a total of 3013 men (1025 control subjects and 1988 case subjects) from the CAPS population with genome-wide SNP data available were selected as a reference population.
Marker Selection
Genetic markers around ABCA1 (including 20kbp upstream of the transcription start site and 10kbp downstream of the transcription end site) were selected with previous findings, functional candidature, LD, and Illumina SNP design score as criteria (the full final list is shown in Supp. Table S1). Illumina scores were calculated by an algorithm developed by the company that predicts success of the assay for the marker. Markers rs4149268 and rs3890182 were included since these were identified to associate with lipid level in recent genome-wide studies [Kathiresan et al., 2008; Willer et al., 2008]. Based on the genotype data for CEU samples of HapMap Release 22, all polymorphic markers in the dataset were taken into consideration. At first, LD blocks were searched with Haploview 4.0 [Barrett et al., 2005]. Prioritization included markers in exons, within 80bp of exon boundaries, and within 1kb upstream from the 1st exon or downstream from the last exon of any predicted gene according to the UCSC genome browser, and SNPs that can tag the LD blocks. Among the markers outside of LD blocks, those which could be prioritized by the same scheme were included. After selection, Illumina scores for all markers were calculated and those that didn’t satisfy the criteria for Illumina probe chemistry were replaced with an SNP in perfect LD if available (r2 = 1) or other tagging SNP.
Genotyping
Genotyping was performed using the Illumina GoldenGate assay system on Illumina BeadStation 500GX equipment, currently housed and implemented at the Uppsala University SNP Technology Platform. A detailed description of the technology, the staff and the genotyping facility can be found online at (http://www.medsci.uu.se/molmed/snpgenotyping/methods.htm). Prior to use on the Illumina system, all samples were subjected to Whole Genome Amplification (WGA) using standard kits involving Phi29 DNA polymerase (Amersham). For three markers (rs2230806, rs2230808 and rs4149313) genotyping of all samples prior to whole genome amplification was also conducted using Dynamic Allele Specific Hybridization [Prince et al., 2001]. We note that markers rs2230806, rs2230808, rs4149313, rs2066718, and rs4149312 were previously reported on in a subset of 537 cases and 511 Swedish controls that overlap with the present study [Katzov et al., 2004].
CSF Biomarkers
CSF samples were obtained in the AD case-control study by lumbar puncture in the L3/L4 or L4/L5 inter-space. Further details of CSF collection can be found elsewhere [Andreasen et al., 1999]. CSF Aβ42 was determined using a sandwich enzyme-linked immunosorbent assay (ELISA) (Innotest b-amyloid (1–42), Innogenetics, Ghent, Belgium) constructed to specifically measure Aβ42 [Vanmechelen and Vanderstichele, 1998]. The microtubule-associated protein tau, a CSF marker of neuronal degeneration, was determined using a sandwich ELISA (Innotest hTAU-Ag, Innogenetics, Ghent, Belgium) constructed to measure total tau, i.e., all isoforms of tau irrespective of phosphorylation state [Blennow et al., 1995].
Statistics
Hardy-Weinberg equilibrium for individual loci was assessed using the Pearson χ2 statistic. Initial tests of association between individual markers and dementia risk were performed without considering family structure using Haploview v4.1 [Barrett et al., 2005]. Odds ratios presented using Haploview were based upon allele counts. To account for relatedness, secondary analyses were conducted where only one twin was included among MZ pairs, both members of DZ pairs and all non-twin case-control participants. Haplotype background testing around rs2230805 was performed using the proxy association function implemented in PLINK [Purcell et al., 2007]. Haplotypes were further estimated after LD block definition in individual blocks using Haploview v4.1 [Barrett et al., 2005]. Tests of genotypes versus quantitative traits (age-at-onset, CSF tau, and CSF Aβ42) were conducted using ANOVA.
Results
Through marker prioritization (see Methods) a list of 52 SNPs was decided upon for which genotyping assays were designed and all DNA samples subjected to (1567 dementia cases and 2203 controls). Genotypes were obtained for 46 markers with success rates > 90% for all individuals. Genotyping success rates for all markers were equivalent in men and women and in cases and controls. Across these markers, genotyping was attempted in duplicate in 100 individuals, for which there were no discordant calls. From the outset, we were particularly interested in rs2230806 (R219K), rs2230808 (R1587K), and rs4149313 (I883M), since these are the only known common non-synonymous variants (nsSNPs) in the region. Prior to genotyping on the Illumina platform and prior to WGA, DASH [Prince et al., 2001] had also been used previously for these markers in all samples (previously unpublished – see methods). Genotyping concordance between the two platforms was in excess of 98% for all three markers (not shown). Of note, rs2230806 was not in Hardy-Weinberg equilibrium (HWE), but the deviation was equivalent using both of the above genotyping platforms. We explored this more closely by stratifying both by disease, gender, and sample origin (twins or non-twin case-control study – see methods) and noted that distortion of HWE was only evident in women. Three additional markers in LD with rs2230806; rs2230805 (r2 = 0.80), rs2472386 (r2 = 0.38), and rs2065412 (r2 = 0.15) also deviated significantly from HWE, but this was again restricted to women only. The strongest deviation was noted for rs2230805 (p = 4.5 × 10−9, N = 2144). For the other markers, in each case the significance of the deviation in women decreased with decreasing LD from rs2230805 (p = 7.4 × 10−5 for rs2230806; p = 0.0025 for rs2472386; p = 0.044 for rs2065412). We considered whether this could be due to non-specific amplification of homologous X-chromosome regions, but BLAST searches failed to identify sequence similarities. There were no confirmed polymorphisms (including structural variants) adjacent to either rs2230805 or rs2230806 that could be interfering with PCR or genotyping probe chemistry. The significance of the deviation from HWE for rs2230805 was p = 0.29 and p = 5.7 × 10−6 in men and women from the twin sample, respectively, and p = 0.06 and p = 1.3 × 10−4 in men and women from the non-twin case-control sample, respectively. In both cases the deviation was on the order of 2–3% decreased heterozygosity, with an elevation of both rare and common-allele homozygotes. To evaluate if the deviation from HWE in women was age dependent, we stratified the sample at the median age into younger (age < 78) and older (age ≥ 78) groups. The strongest deviation was found in the elderly group of women and this is shown in Table 1.
Table 1.
Deviation from Hardy-Weinberg equilibrium in young and elderly women for rs2230805
| age group | genotypes | HWE χ2 | p value | |||
|---|---|---|---|---|---|---|
| AA | AG | GG | ||||
| < 78 | observed | 63 | 300 | 663 | 12.7 | 3.65E-04 |
| expected | 44 | 338 | 644 | |||
| ≥ 78 | observed | 74 | 301 | 680 | 23.2 | 1.46E-06 |
| expected | 48 | 353 | 653 | |||
Age groups were defined by the median of the population including both cases and controls.
An initial screen of all 46 markers in the total sample in relation to dementia risk was performed using Haploview v4.1. This indicated strong association of both rs2230805 (OR = 1.39; 95% CI 1.23–1.57, P = 7.7 × 10−8) and rs2230806 (OR 1.25, CI 1.12–1.40, P = 5.9 × 10−5), with several additional markers exceeding an uncorrected p-value threshold of 0.05 (see Supp. Table S1). For both rs2230805 and rs2230806 (both are G/A polymorphisms), the common 'G' allele was associated with increased risk. Testing rs2230805 across gender indicated similar effect sizes in both men (OR = 1.40, 95% CI 1.15–1.71, p = 4.0 × 10−4) and women (OR = 1.40, CI 1.19–1.60, p = 2.3 × 10−5), suggesting that the previously observed HWE distortion was not a contributing factor. The common allele of marker rs2230805 was significantly enriched when only possible and probable AD cases were considered (OR = 1.41, CI 1.23–1.61, p = 3.1 × 10−7). The original study design for this project was to have case-control studies from twin and non-twin samples as two independent replication materials, but in the interest of power we considered all samples together for primary analyses. Nonetheless, we did note that for rs2230805, the common allele was significantly enriched in twin (OR = 1.30, CI 1.10–1.53, p = 1.4 × 10−3) and non-twin (OR = 1.57, CI 1.21–2.03, p = 4.0 × 10−4) samples when considered separately. Although we didn’t specifically genotype and report on rs2230805 previously [Katzov et al., 2004], we had tested other markers in ABCA1 in a subset of the present study. Removal of the 537 AD cases and 511 controls previously reported provided an OR estimate for rs2230805 of 1.33, CI 1.16–1.52, p = 15 2.4 × 10−5. The effect size estimate in those 537 cases and 511 controls was OR 1.40, CI 1.10–1.77, p = 0.006.
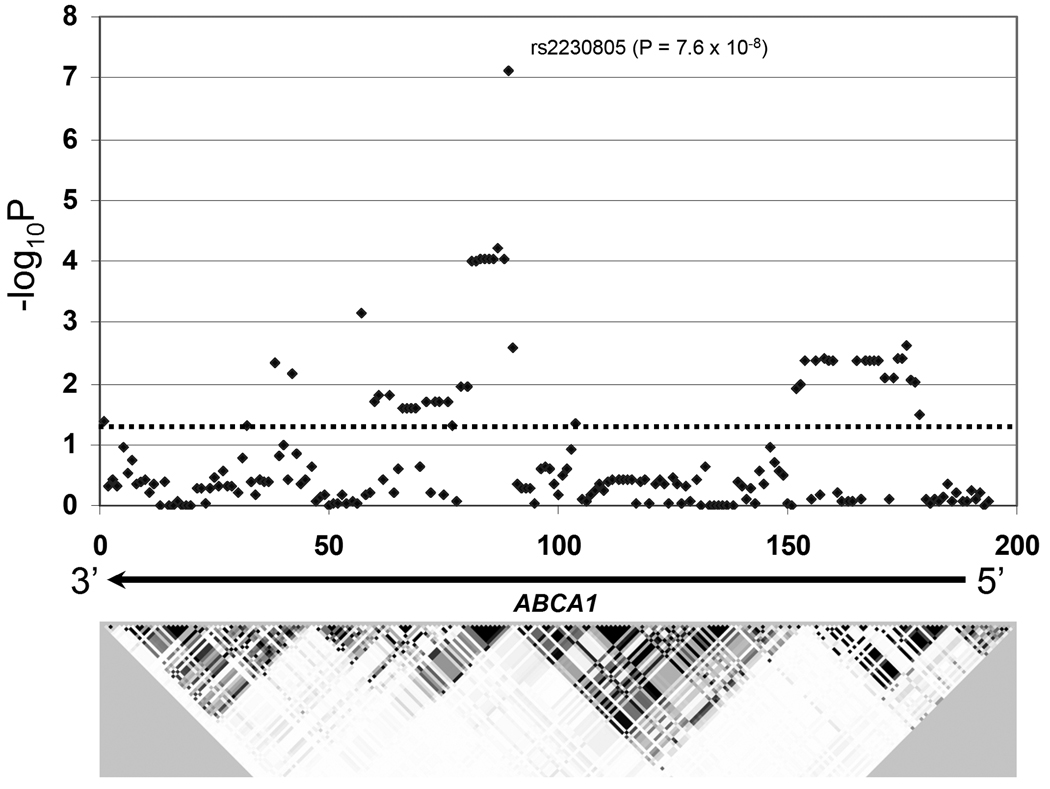
We used IMPUTE [Marchini et al., 2007] to probabilistically infer additional genotypes from the ABCA1 region. With 10kb windows on either side of the directly typed region, this resulted in a total of 148 additional markers with >90% genotype calls across individuals at a posterior probability in excess of 90%. The results of all 194 markers for dementia risk in all samples are shown in Figure 1. No additional markers exceeded the statistical significance of either rs2230805 or rs2230806. Figure 1 in particular highlights two possible independent LD blocks with significant signals. Genotypes of rs2230805 (the marker exhibiting the maximum significance of the first LD block) and rs12350560 (the maximum for a directly genotyped marker in the second LD block) were entered as terms in a logistic regression model and this also supported their independence (not shown). A second question is whether epistasis occurs between these two sites and thus a genotype by genotype interaction term was also tested with logistic regression, but this was not significant. Finally, we evaluated the joint effects of these two markers, again using logistic regression and establishing risk scores based upon number of risk alleles from each of the two loci (rs2230805 and rs12350560). This is shown in Table 2, where the result was highly significant (OR 1.35; 95% CI 1.23 – 1.50, p = 1.6 × 10−9), but was not consistent with additivity.
Figure 1.
Schematic overview of the ABCA1 region from marker rs363717 to marker rs3887137 (106.68 Mb to 106.73 Mb on chromosome 9q) depicting LD structure represented by r2 values (darker shading = higher LD). Unadjusted allelic association of −log10P (Y-axis) values for 194 genetic markers (46 directly genotyped and 148 imputed) in the X-axis in relation to the full dementia versus control sample are shown with a maximum for rs2230805. ABCA1 is transcribed from right to left in the above scheme and extends from approximately marker 185 to 1. The dashed horizontal line reflects an unadjusted p = 0.05 threshold.
Table 2.
Joint effects of markers rs2230805 and rs12350560
| Genotype at rs2230805 | Genotype at rs12350560 | Score | Nr Cases (%) | Nr Controls (%) |
|---|---|---|---|---|
| AA | AA | 0 | 0 | 1 |
| AA | AG | 1 | 8 (0.5) | 25 (1.2) |
| AG | AA | 1 | " | " |
| AG | AG | 2 | 118 (8.4) | 247 (12) |
| AA | GG | 2 | " | " |
| GG | AA | 2 | " | " |
| AG | GG | 3 | 459 (32.7) | 788 (38.5) |
| GG | AG | 3 | " | " |
| GG | GG | 4 | 819 (58.3) | 989 (48.3) |
Logistic regression was used to evaluate significance based upon a cumulative allele risk score. Four groups (1–4) were generated by combining groups with equivalent risk scores. The "zero" score group was removed since only one observation was available. The full additive model result was OR 1.35; 95% CI 1.23 – 1.50, p = 1.6 × 10−9.
We used the proxy association function that is provided with PLINK [Purcell et al., 2007] to assess the haplotype background around rs2230805. This provided evidence of association with haplotypes constructed using adjacent markers in the absence of rs2230805 (omnibus p = 0.016 considering all haplotypes and with p = 3.8 × 10−5 for the best single haplotype). Further tests of haplotypes using only the original set of 46 directly typed variants were conducted with Haploview 4.1, with significant haplotype differences between cases and controls being noted for multiple blocks (full results are shown in Supp. Table S2).
We evaluated the allele frequencies of directly genotyped markers against a relatively large independent and distinct Swedish reference population that had been genotyped as part of a genome-wide association study (see materials). This was done in order to assess if the dementia cases or the controls deviated from the population frequencies. In comparing non-demented controls against this reference population, there was only one instance of a significant difference from among thirteen markers that overlapped between the samples (rs4149271, p = 0.031). Of particular note, the minor-allele frequency (MAF) for rs2230805 in the reference population was 0.222, versus 0.228 in the non-demented control population (p = 0.36), supporting that it is the allele frequencies of dementia group that differ from the Swedish population average.
Markers rs2230805, rs2230806, and rs10512338 were tested for association with additional AD/dementia-related quantitative traits, specifically age-at-onset, and CSF measures of Aβ42 and tau. Only AD cases were included in this analysis. Across these traits and markers there was evidence that rs2230805 influenced variance in CSF Aβ42 levels (F2,624 = 5.04, p = 0.0066). Post-hoc pair-wise tests suggested this was mainly driven by a difference between the heterozygote group and common allele homozygotes (F1,601 = 8.3, p = 0.0042) whereby common homozygotes had lower average Aβ42 levels (A/G = 556 ± 234 (SD) vs. G/G = 503 ± 182 (SD) in pg/ml CSF). This is consistent with this allele increasing risk, considering that AD cases have lower Aβ42 levels than controls and that APOE ε4 is associated with low CSF Aβ42 levels [Prince et al., 2004].
Marker rs2230805 is located in the last exon of the only putative alternative splice-form of ABCA1 (BC034824). This truncated form comprises 5 exons and its function is presently unknown. The program F-SNP [Lee and Shatkay, 2008] was used to assess the alternative alleles of this marker for potential functional effects, but no important differences were noted. In order to explore for effects of genetic variants on ABCA1 expression, we extracted genetic and expression data from previous genome-wide studies that had both sources of information [Dixon et al., 2007; Myers et al., 2007]. Of the genetic markers in the latter study, 55 SNPs around ABCA1 were tested for association with a single ABCA1 target transcript expression in 193 brain samples using PLINK. There were no variants showing significant association with P-values < 0.05 after Bonferroni correction for 55 tests. Likewise, we investigated association between ABCA1 expression in lymphocytes of 400 individuals and genetic variants using SNP Browser 1.0.1 [Dixon et al., 2007], but no significant effects were noted.
Twin-pair adjusted tests were conducted for rs2230805 to account for dependencies that may impact standard errors and thus significance levels. Alternating logistic regression (ALR) was used to account for pair dependency, allowing both members of the pair to enter in the analyses while accounting for MZ and DZ pair correlation structures [Carey et al., 1993; Jansson et al., 2003]. The total N was 3,682 with 2,679 clustered pairs, where those in the case-control sample were treated as members of incomplete DZ pairs. The ALR procedure resulted in an odds ratio (OR) and a 95% confidence interval for estimating the prediction of rs2230805 on dementia.
Covariates included sex. Statistical analyses were performed in SAS 9.1 using the GENMOD procedure (SAS Institute, Inc., Raleigh, NC). Correcting for sex, the effect of rs2230805 was significant at p = 7.8 ×10−7 (OR=1.34, 95% CI=1.19–1.50). The equivalent logistic model not accounting for twin-ness (recoding genotypes as 0, 1, and 2) was significant at p = 2.3 × 10−7 (OR 1.34, 95% CI = 1.19–1.49). One final model was also fitted for rs2230805 adjusting for the known association with APOE, by including e4 carrier status as a covariate. This significant at p = 3.2 × 10−7 (OR 1.36; 95% CI = 1.21–1.55).
Discussion
A survey of genetic variation spanning the ABCA1 locus was performed in relation to dementia and Alzheimer disease risk. Using a combination of direct genotyping and imputation, strong evidence of association was obtained for multiple SNPs in the region, with a focal point around markers rs2230805 and rs2230806. The latter marker has been extensively studied in relation to both AD risk and numerous other phenotypes, in particular those related to cardiovascular disease and plasma lipid traits (e.g. Clee et al., 2001). With special regard to rs2230806, there have been some important contingencies, with one report emphasizing strong genetic association in a gender specific manner [Sundar et al., 2007]. Estimates of effect sizes however in the present study suggest equivalence in both women and men. Additionally, there has been a recent report of potential modification of ABCA1 association with AD by APOE, but no such evidence of interaction was observed in the present data set [Rodríguez-Rodríguez et al., 2007].
Support that ABCA1 may be involved in dementia risk is also provided by the observation of a modest, but significant effect of the key associated variant (rs2230805) on CSF levels of Aβ42. The putative risk allele was associated with lower CSF levels, this being consistent with the effect of the ε4 allele of APOE, which is also associated with both increased risk and reduced CSF Aβ42. The effect size for rs2230805 was however much lower than for APOE, with the former contributing to approximately 1.6% of the variance in CSF Aβ42, in contrast to 8.1% for APOE [Prince et al., 2004]. If an effect on Aβ by ABCA1 is etiological, then these effect sizes may be seen to mirror well the putative disease risk increase by the respective genes (OR 1.35 for ABCA1, and OR 6~8 for APOE). In terms of additional AD/dementia related phenotypes, no effects of disease-associated markers were noted with CSF levels of tau, or with AD or dementia age-at-onset as previously reported [Wollmer et al., 2003].
The key disease-associated marker, rs2230805, is a synonymous coding variant, but it is unclear if its alternative alleles differentially affect the binding of common splicing factors. This might however be supported in that mRNA sequencing efforts have identified a possible ABCA1 splice-form that is truncated just after the exon in which rs2230805 occurs. We also used a relatively new resource that combines information from multiple prediction tools for the functional consequences of SNPs [Lee and Shatkay, 2008]. This did highlight a potential change for rs2230805 for exonic-splicing regulatory (ESR) sequences, but this was only evident in 1 of 4 databases and so in our view remains to be proven experimentally. An intriguing finding in the present study was that significant signals may be occurring in two separate LD blocks, with an additional associated locus (apart from that containing rs2230805) residing nearer to the gene promoter. Thus if allelic heterogeneity is present for this locus, there may be two alternative splice forms, each susceptible to variable expression levels due to promoter variation. This may be one explanation to why our in silico search for variants affecting expression levels was negative. Each of these possible mechanisms may be targets for cell-based assays and/or estimates of splice-form levels in human samples.
The deviation from Hardy-Weinberg equilibrium for key associated variants is an important issue in this study. The arguments that this may be a real observation are that this is exhibited by multiple markers in LD and for one marker (rs2230806) genotyping was confirmed with a second distinct methodology. The most intriguing aspect is that the deviation is apparent only in women, and seen to be replicating in two independent samples. Thus, for genotyping error to be the cause would require the selective miscalling of genotypes in women, as well as miscalling of 4 adjacent markers that happen to be in LD. For the latter, the markers are sufficiently distant from one another that genotyping chemistry is not likely to be a factor. We consider two possible explanations for these observations. The first is that it is not genotyping error for a single marker, but rather distortion of haplotype frequencies in the region. The second, which is not exclusive from the first, is that haplotype frequencies are changing with age in women and at advanced age (women represented in this sample) it can be detected as a deviation from Hardy-Weinberg. This is supported in that the deviation from HWE is more severe in this sample in elderly individuals. Genotyping in additional large samples of both young and elderly women will be necessary to validate this.
In summary, we present further evidence of genetic association of ABCA1 with both AD and dementia in general for a Swedish population. For these particular samples, the interval possibly containing functional variation is narrowed with the marker showing the best evidence being located in exon 5. There was however an indication that additional functional variation might be occurring in the promoter and thus allelic heterogeneity may be an important consideration in further replication efforts. The issue of Hardy-Weinberg disequilibrium in women is intriguing and strongly deserving of further study.
Supplementary Material
Acknowledgments
We are grateful for generous funding from the US National Institutes of Health (grants AG028555, AG08724, and AG08861) and the Swedish Medical Research Council (grant 2007–2722).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Andreasen N, Minthon L, Clarberg A, Davidsson P, Gottfries J, Vanmechelen E, Vanderstichele H, Winblad B, Blennow K. Sensitivity, specificity, and stability of CSF-tau in AD in a community-based patient sample. Neurology. 1999;53:1488–1494. doi: 10.1212/wnl.53.7.1488. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beekman M, Posthuma D, Heijmans BT, Lakenberg N, Suchiman HE, Snieder H, de Knijff P, Frants RR, van Ommen GJ, Kluft C, Vogler GP, Slagboom PE, Boomsma DI. Combined association and linkage analysis applied to the APOE locus. Genet Epidemiol. 2004;26:328–337. doi: 10.1002/gepi.10318. [DOI] [PubMed] [Google Scholar]
- Beffert U, Aumont N, Dea D, Lussier-Cacan S, Davignon J, Poirier J. Apolipoprotein E isoform-specific reduction of extracellular amyloid in neuronal cultures. Brain Res Mol Brain Res. 1999;68:181–185. doi: 10.1016/s0169-328x(99)00073-x. [DOI] [PubMed] [Google Scholar]
- Blennow K, Wallin A, Agren H, Spenger C, Siegfried J, Vanmechelen E. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol Chem Neuropathol. 1995;26:231–245. doi: 10.1007/BF02815140. [DOI] [PubMed] [Google Scholar]
- Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Genest J, Jr, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- Carey V, Zeger SL, Diggle P. Modelling multivariate binary data with alternating logistic regressions. Biometrika. 1993;80:517–526. [Google Scholar]
- Clee SM, Zwinderman AH, Engert JC, Zwarts KY, Molhuizen HO, Roomp K, Jukema JW, van Wijland M, van Dam M, Hudson TJ, Brooks-Wilson A, Genest J, Jr, Kastelein JJ, Hayden MR. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation. 2001;103:1198–1205. doi: 10.1161/01.cir.103.9.1198. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Deng A, Irizarry MC, Fitzgerald ML, Rebeck GW. Induction of the cholesterol transporter ABCA1 in central nervous system cells by liver X receptor agonists increases secreted Abeta levels. J Biol Chem. 2002;277:48508–48513. doi: 10.1074/jbc.M209085200. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds C, Nikolic J, Lowe B, Karel M, Pedersen N. An empirical test of telephone screening to identify potential dementia cases. Int Psychogeriatr. 1995;7:429–438. doi: 10.1017/s1041610295002171. [DOI] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- Gatz M, Fratiglioni L, Johansson B, Berg S, Mortimer JA, Reynolds CA, Fiske A, Pedersen NL. Complete ascertainment of dementia in the Swedish Twin Registry: the HARMONY study. Neurobiol Aging. 2005;26:439–447. doi: 10.1016/j.neurobiolaging.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Gold CH, Malmberg B, McClearn GE, Pedersen NL, Berg S. Gender and health: a study of older unlike-sex twins. J Gerontol B Psychol Sci Soc Sci. 2002;57:S168–S176. doi: 10.1093/geronb/57.3.s168. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J Biol Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- Jansson M, Gatz M, Berg S, Johansson B, Malmberg B, McClearn GE, Schalling M, Pedersen NL. Association between depressed mood in the elderly and a 5-HTR2A gene variant. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:79–84. doi: 10.1002/ajmg.b.20016. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, Brookes AJ, Blennow K, Kehoe PG, Prince JA. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci U S A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Lau K, Catanese J, Sninsky J, Nowotny P, Holmans P, Hardy J, Powell J, Lovestone S, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of ABCA1 with late-onset Alzheimer's disease is not observed in a case-control study. Neurosci Lett. 2004;366:268–271. doi: 10.1016/j.neulet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, De Faire U, Floderus B, Svartengren M, Svedberg P, Pedersen NL. The Swedish Twin Registry: a unique resource for clinical, epidemiological and genetic studies. J Intern Med. 2002;252:184–205. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Friberg L, Floderus-Myrhed B, McClearn GE, Plomin R. Swedish early separated twins: identification and characterization. Acta Genet Med Gemellol (Roma) 1984;33:243–250. doi: 10.1017/s0001566000007285. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Lichtenstein P, Svedberg P. The Swedish Twin Registry in the third millennium. Twin Res. 2002;5:427–432. doi: 10.1375/136905202320906219. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Gatz M, Berg S, Johansson B. How heritable is Alzheimer's disease late in life? Findings from Swedish twins. Ann Neurol. 2004;55:180–185. doi: 10.1002/ana.10999. [DOI] [PubMed] [Google Scholar]
- Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer's disease. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- Prince JA, Feuk L, Howell WM, Jobs M, Emahazion T, Blennow K, Brookes AJ. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res. 2001;11:152–162. doi: 10.1101/gr.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince JA, Zetterberg H, Andreasen N, Marcusson J, Blennow K. APOE epsilon4 allele is associated with reduced cerebrospinal fluid levels of Abeta42. Neurology. 2004;62:2116–2118. doi: 10.1212/01.wnl.0000128088.08695.05. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez E, Mateo I, Llorca J, Sánchez-Quintana C, Infante J, García-Gorostiaga I, Sánchez-Juan P, Berciano J, Combarros O. Association of genetic variants of ABCA1 with Alzheimer's disease risk. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:964–968. doi: 10.1002/ajmg.b.30552. [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, Mar L, Sorbi S, Nacmias B, Piacentini S, Amaducci L, Chumakov I, Cohen D, Lannfelt L, Fraser PE, Rommens JM, George-Hyslop PHS. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shibata N, Kawarai T, Lee JH, Lee HS, Shibata E, Sato C, Liang Y, Duara R, Mayeux RP, St George-Hyslop PH, Rogaeva E. Association studies of cholesterol metabolism genes (CH25H, ABCA1 and CH24H) in Alzheimer's disease. Neurosci Lett. 2006;391:142–146. doi: 10.1016/j.neulet.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K. Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci U S A. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Goedert M, Weisgraber KH, Dong LM, Jakes R, Huang DY, Pericak-Vance M, Schmechel D, Roses AD. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91:11183–11186. doi: 10.1073/pnas.91.23.11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer's disease. Neurobiol Aging. 2007;28:856–862. doi: 10.1016/j.neurobiolaging.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Tregouet DA, Ricard S, Nicaud V, Arnould I, Soubigou S, Rosier M, Duverger N, Poirier O, Mace S, Kee F, Morrison C, Denefle P, Tiret L, Evans A, Deleuze JF, Cambien F. In-depth haplotype analysis of ABCA1 gene polymorphisms in relation to plasma ApoA1 levels and myocardial infarction. Arterioscler Thromb Vasc Biol. 2004;24:775–781. doi: 10.1161/01.ATV.0000121573.29550.1a. [DOI] [PubMed] [Google Scholar]
- Vanmechelen E, Vanderstichele H. Towards an earlier diagnosis of Alzheimer's disease. J Biotechnol. 1998;66:229–231. doi: 10.1016/s0168-1656(98)00169-2. [DOI] [PubMed] [Google Scholar]
- Viiri LE, Loimaala A, Nenonen A, Islam S, Vuori I, Karhunen PJ, Lehtimaki T. The association of the apolipoprotein E gene promoter polymorphisms and haplotypes with serum lipid and lipoprotein concentrations. Atherosclerosis. 2005;179:161–167. doi: 10.1016/j.atherosclerosis.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmer MA, Streffer JR, Lutjohann D, Tsolaki M, Iakovidou V, Hegi T, Pasch T, Jung HH, Bergmann K, Nitsch RM, Hock C, Papassotiropoulos A. ABCA1 modulates CSF cholesterol levels and influences the age at onset of Alzheimer's disease. Neurobiol Aging. 2003;24:421–426. doi: 10.1016/s0197-4580(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Yancey PG, Bortnick AE, Kellner-Weibel G, de la Llera-Moya M, Phillips MC, Rothblat GH. Importance of different pathways of cellular cholesterol efflux. Arterioscler Thromb Vasc Biol. 2003;23:712–719. doi: 10.1161/01.ATV.0000057572.97137.DD. [DOI] [PubMed] [Google Scholar]
- Yue P, Isley WL, Harris WS, Rosipal S, Akin CD, Schonfeld G. Genetic variants of ApoE account for variability of plasma low-density lipoprotein and apolipoprotein B levels in FHBL. Atherosclerosis. 2005;178:107–113. doi: 10.1016/j.atherosclerosis.2004.06.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.