Abstract
Annotation of the human serum N-linked glycome is a formidable challenge but is necessary for disease marker discovery. A new theoretical glycan library was constructed and proposed to provide all possible glycan compositions in serum. It was developed based on established glycobiology and retrosynthetic state-transition networks. We find that at least 331 compositions are possible in the serum N-linked glycome. By pairing the theoretical glycan mass library with a high mass accuracy and high-resolution mass spectrometry, human serum glycans were effectively profiled. Correct isotopic envelope deconvolution to monoisotopic masses and the high mass accuracy instruments drastically reduced the amount of false composition assignments. The high throughput capacity enabled by this library permitted the rapid glycan profiling of large control populations. With the use of the library, a human serum glycan mass profile was developed from 46 healthy individuals. This paper presents a theoretical N-linked glycan mass library that was used for accurate high throughput human serum glycan profiling. Rapid methods for evaluating a patient’s glycome are instrumental for studying glycan based markers.
Keywords: Glycomics, Interpretation of Mass Spectra, Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, Serum, Carbohydrates
1 INTRODUCTION
Glycomics involves the extensive and comprehensive study of structure and characterization of glycans. Glycans have complicated structures that are difficult to analyze because they can have varying monosaccharide compositions, degrees of branching, and linkages. Unlike proteins compositions, which are directly related to a linear DNA template from a genome, glycan compositions are determined by a series of often competing enzymatic glycosyltransferase reactions.
N-glycosylation is the addition of an oligosaccharide chain to the asparagine of the peptide backbone. It is further governed by a set of biological rules that determine its position and structural diversity. Changes in N-glycosylation have been clearly established in some diseases such as cancer where the normal glycosylation machinery is affected [1–8]. For this reason, there have been several attempts to use N-linked glycans as markers for diseases [7, 9–23].
N-linked glycans have great diversity but have many characteristics that make them distinct from other glycoconjugates such as O-linked glycans, glycosaminoglycans, and glycolipids. N-linked glycans are found only on asparagine that is adjoined by a variable amino acid followed by serine, threonine, or less commonly cysteine amino acid (Asn-Xaa-Ser/Thr/Cys) [24–26]. Except for proline, the variable amino acid is not specific. Enzymes such as PNGase F can be used to cleave off N-linked glycans under physiological conditions. The N-linked core is comprised of three mannose sugars attached to a chitobiose core (Man3GlcNAc2). N-linked glycans are often differentiated into three main types depending on which monosaccharides are added to the N-linked core. High mannose glycans contain mannose monosaccharides in addition to the N-linked core. Structures containing six additional mannose units to the core are routinely observed. It is possible that some or all of the three terminal glucose units from intact dolichol precursors may still be present and detectable if the typical glycan degradation pathway was circumvented or disrupted. Complex-type glycans are created after the dolichol precursor has been degraded down to the N-linked core and subsequent N-acetylglucosamine, galactose, deoxyhexose (fucose), and sialic acid monosaccharides are enzymatically added. Hybrid type glycans contain elements of both high mannose and complex-type structures.
Although N-glycosylation is the most common form of protein glycosylation in serum, the total number of compositions and structures in the N-linked glycome is unknown. However, knowledge of the scope of the N-linked glycan library will significantly aid in the annotation and the discovery of N-linked glycan markers. This paper presents a new theoretical library of glycan masses designed for human serum glycans for disease marker discovery. The library aids in the interpretation of the mass spectra of glycans and minimize falsely assigned sugar compositions. In addition, it minimizes the need for additional biological filters to constrain the compositions because such considerations are built into the library. Large molecular weight sugars (up to 23 monosaccharides ~4500 Da) were included in the library since they may be present in samples but hard to detect due to metastable ions in the gas phase or low abundance in samples. The theoretical library can be used to rapidly identify sugars present or absent in a sample with a low false positive rate.
There are several existing methods for annotating glycan mass spectra. GlycoMod [27] is a useful algorithm for finding all possible compositions for a glycan given a set of monosaccharides and expected ranges for a given monosaccharide composition. Typically, billions of theoretical oligosaccharide masses are compared and a subset of matches is selected that is within a mass tolerance range specified by the user. Although all possibilities will be found, the prevalence of false positives is common and becomes more abundant with increasing glycan mass, error tolerance, and monosaccharide diversity. The high false positive rate is one of the major drawbacks of numerical combinatorial approaches.
An alternative approach for identifying glycan compositions is GlycoSuiteDB [28]. GlycoSuiteDB is a proprietary, privately curated library derived and sourced from published literature. This approach has the advantage of direct ties to the biological and chemical studies but is limited to retrospective studies. Cartoonist [29] is an algorithm designed to annotate N-linked glycan spectra with the help of an archetype based glycan library. The Cartoonist library is based on 300 empirical archetype glycan structures found in spectra of mammalian samples. By expanding the degree of fucosylation and sialylation, varying the presence or absence of bisecting GlcNAc residues, or substituting NeuGc for NeuAc, the archetype structures expand to a set of 2800 different plausible structures. GlycoWorkbench [30] is a software tool designed for analyzing glycans. Aside from a useful tool for drawing glycans, one feature of GlycoWorkbench is that it has the ability to annotate lists of masses by searching full empirical glycan databases. Without proper restrictions for the searches, however, the data bases have potentially a prohibitively high false positive rate. In contrast, the theoretical N-linked glycan library presented here is focused on human serum and is not limited to retrospective empirical studies. The theoretical library provides a road map for finding well-established and undiscovered glycans.
The utility of the N-linked glycan library in this study was examined by analyzing a series of spectra of N-linked glycans released from human serum samples. Simple modifications to the neutral mass in the glycan library with various charge carrying species allow for direct and expeditious comparison of the library to experimental monoisotopic spectra. High resolution mass spectrometers are able to resolve monoisotopic ions (ions composed of only atoms in their principle isotopic state (e.g. 12C, 14N, 16O etc.) from their isotopologue ions, which can contain a variety of naturally occurring isotopes often occurring with a +1.00866 Da m/z shift [31]. Removing non-monoisotopic peaks from mass spectra is essential for library comparison so that isotopologue peaks are not falsely assigned as monoisotopic glycans.
This report describes the construction of a theoretical glycan library based on well-established biological rules. The library is used for automatically annotating mass spectra of glycans mixtures. The efficacy of the library for annotating mass spectra was evaluated on human serum samples. These samples present a formidable challenge for analysis due to the complexity and biological diversity of the mixture, but their potential for disease marker discovery is readily apparent. Furthermore, enzymatically released N-linked glycans from serum yield abundant and heterogeneous glycan mixtures that are amenable to profiling by mass spectrometry.
2 MATERIALS AND METHODS
Human Serum Samples
Serum samples from control individuals (n = 46) were acquired from the UC Davis Medical Center Clinical Laboratories (Internal Review Board (IRB) approved protocol). Serum samples arrived frozen and were transferred to a −75°C freezer prior to processing.
Enzyme Release of N-Linked Glycans
Peptide N-glycosidase F (PNGase F, 500,000 units/mL, purified from flavobacterium meningosepticum) was obtained from New England BioLabs (Ipswich, MA). In preparation for the enzymatic glycan release, 100 µL serum was dissolved in digestion buffer (pH 7.5, 100 mM ammonium bicarbonate, 5 mM dithiolthreitol) and heated in boiling water for 2 minutes to denature the proteins. After cooling at room temperature, 1.5µL PNGase F (750 units) were added and the mixture was incubated at 37°C for 12 hours. An additional 1.0µL of PNGase F was added and the incubation was continued at 37°C for an additional 12 hours. An 800µL-aliquot of ethanol was then added to help precipitate the peptides and proteins. The solution was frozen in a −75°C freezer for 60 minutes and then centrifuged at 13,300 rotations per minute for 20 minutes (Eppendorf, 5415 D). The supernatant was removed from the precipitate and dried in a centrifugal evaporator (Savant, AES 2010).
Oligosaccharide Purification
PNGase F released oligosaccharides were purified by graphitized carbon cartridge solid phase extraction (GCC-SPE) with procedures adopted from a previous publication [32]. GCC-SPE cartridges (150 mg bed weight, 4 mL cartridge volume) were acquired from Alltech (Deerfield, IL). Prior to use, the cartridge was washed with nanopure water and conditioned with 0.1% trifluoroacetic acid (TFA) in 80% acetonitrile (AcN)/H2O (v/v). Glycan solutions were applied to the GCC cartridge and subsequently washed with several cartridge volumes of nanopure (E-Pure, Barnstead) water at a flow rate of 1 mL/minute for desalting. Glycans were eluted with 10% AcN/H2O (v/v), 20% AcN/H2O (v/v), and 40% AcN/H2O (v/v) with 0.05% TFA. Each fraction was collected and dried in a centrifugal evaporator apparatus. Fractions were reconstituted in nanopure water prior to MS analysis.
Mass Spectrometric Analysis
Mass spectra were recorded on an external source MALDI-FTICR instrument (HiResMALDI, IonSpec Corporation, Irvine, CA) equipped with a 7.0 T superconducting magnet and a pulsed Nd:YAG laser 355 nm. A solution of 2, 5-dihydroxybenzoic acid (DHB) was used as the matrix for all oligosaccharide analyses (0.05 mg/µL in 50% AcN). For negative mode analyses, 0.6 µL of oligosaccharide solution was applied to the MALDI probe, followed by 1 µL of the appropriate matrix solution. The sample was vacuum dried and subjected to mass spectrometric analysis. For positive mode analyses, the same sample preparation was applied, except that 0.3 µL 0.01 M NaCl was added to the matrix-analyte mixture to produce primarily sodiated species.
The three fractions eluted during GCC-SPE (abbreviated as 10%AcN, 20%AcN, 40%AcN) were analyzed with MALDI-FTICR MS and the spectra were recorded. The oligosaccharides were eluted by solvent polarity resulting in the neutral oligosaccharides being detected in the 10%AcN and 20%AcN fractions while the acidic oligosaccharides were detected in the 20%AcN and 40%AcN fractions. The charge polarity of the mass spectrometer was switched between positive mode and negative mode to improve the ion signal and minimize ion suppression of the respective neutral or acidic oligosaccharides. Four spectra were collected for each sample: 10%AcN and 20%AcN fractions in the positive mode and 20%AcN and 40%AcN fractions in the negative mode. The Fourier transformed signal from the mass spectrometer contains ~1 million data points collected with an Analog-to-Digital Converter (ADC) rate of 1MHz. With our sample set of 46 samples, 184 spectra were collected.
Spectrum Data Pre-Processing
A threshold of 6 S/N was used to separate the signal from the noise. The spectra were internally calibrated with Omega 8 software (IonSpec, Irvine, CA) using six known glycan masses present in all spectra. Each spectrum was normalized by the total ion abundance.
3 RESULTS AND DISCUSSION
3.1 Creation of Theoretical Retrosynthetic State Transition Network Glycan Libraries
Retrosynthetic networks were constructed by starting with the largest plausible high mannose, complex, and hybrid type oligosaccharides and sequentially degrading them one monosaccharide at a time until only the N-linked core sugars remained. Highly branched structures were used along with bisecting N-acetylglucosamine (GlcNAc) connected to the core mannose (Man) and sialic acid (N-acetylneuraminic acid, Neu5Ac) residues. Deoxyhexose (fucose) groups were temporally omitted during this step for simplification and were re-added later.
The retrosynthetic state-transition library is limited to the maximum complexity of the starting point glycans. The branching is limited to tetra-antennary oligosaccharides because they are well known through GlcNAc glycosyltransferase IV and V [33], and the species are experimentally found in human serum [12, 22]. Although penta-antennary carbohydrates have been reported in fish eggs [34], chicken ovalbumin [35], and human hepatocellular carcinoma cell line Hep G2 [36], five antennary glycans were not included because they were seldom detected in human serum. Similarly, N-acetyllactosamine extensions on the antennae have been reported for the human Tamm-Horsfall glycoprotein in urine [37] but were not included for the same reason. The current version of the library is bounded by the parent structures, depicted in Figure 1, which restrict the antennae to four. Furthermore, N-acetyllactosamine groups are restricted to one per antenna. Future versions will include penta-antennary branching and poly-N-acetyllactosamine extensions; however, these additions are expected to potentially increase the false positive rate.
Figure 1.
Starting N-linked glycan structures prior to simulated retrosynthetic degradation. High mannose (left), hybrid (center), complex (right). The hexasaccharide N-linked core is boxed in the high mannose diagram.
The sites for possible fucosylation were recorded throughout the creation of the library to facilitate this process. The parent structure was then decomposed by removing one monosaccharide residue at a time from the terminal end. This process was repeated for all of the terminal ends of the parent structure. Since many parent oligosaccharides yield common structures upon degradation, many of the structures in the networks are internally self-consistent. The resulting common structure is established when two or more parent structures degrade to the same degradation product. Upon completion, sequential pathways can be traced to any of the oligosaccharides from the N-linked core. Although pathways of the enzymatic glycosyltransferases may differ because of the orders of addition, the ending sugar composition would have identical molecular masses. Upon completion, individual state transition networks are generated from each of the high mannose, complex, and hybrid glycan parent structures. The state transition networks are included in Figures 2(a)–(c).
Figure 2.
(a). Complex retrosynthetic state-transition network prior to fucose addition. The chitobiose core was omitted in the figure for clarity. Representative bi-antennary, tri-antennary and tetra-antennary glycan degradation families are highlighted.
Figure 2(b). High mannose retrosynthetic state-transition network prior to fucose addition. The chitobiose core was omitted in the figure for clarity.
Figure 2(c). Hybrid retrosynthetic state-transition network prior to fucose addition. The chitobiose core was omitted in the figure for clarity.
Fucose groups were omitted from the state transition networks to simplify the network’s generation process. After the non-fucosylated retrosynthetic state-transition networks were drawn, each structure was converted to a composition, and fucosylation sites were recorded. Because the number of fucosylation sites was documented throughout the library creation process, the degree of fucosylation can be controlled computationally. Similar methods for covering fucosylated and non-fucosylated species have been implemented in the Cartoonist algorithm [29]. The sites used for potential fucosylation were the first GlcNAc on the N-linked core (core fucosylation) and the first GlcNAc on each of the complex or hybrid-type antenna (branch fucosylation) [38]. The addition resulted in a range of fucosylation from zero to five (tetra-antennary branched + N-linked core) based on the degree of branching and the degree of degradation. Although there is some debate as to whether the core fucosylated high mannose type glycans are created biosynthetically or are degradation products from larger glycans, fucosylated high mannose glycans have been observed in recombinant proteins expressed from human embryonic kidney cells that were either β1,2-N-acetylglucosaminyltransferase I deficient or in the presence of swainsonine (a Golgi α-mannosidase II inhibitor) [39]. Similar results were also reported for β1,2-N-acetylglucosaminyltransferase I deficient Chinese hamster ovary cells [40]. Fucosylated high mannose compositions were included because they could be present in serum. Fucose monosaccharides are easily added computationally as long as the potential sites are recorded while the networks are being drawn. After fucosylation, the neutral masses are calculated based on their composition.
When the fucosylated high mannose, complex, and hybrid state-transition networks were complete, they were added together to generate a combined N-linked library. Although the complete library is based on putative structures drawn in the networks, many isomers may exist for a given composition. The final library contains 331 distinct neutral compositions ranging in mass from ~1000–4500 Da. Listed in Supplement Table 1 are the compositions and accurate neutral masses of the components of the library. The monoisotopic masses were calculated using the mass constants from the NIST Physics Laboratory (2006 web update, http://physics.nist.gov), which contain constant values recommended by the Committee on Data for Science and Technology (CODATA) [31].
In order to make the libraries employable for mass spectrometry, the neutral masses need to be turned into ions by adding or subtracting charge carrying species. Protonated and deprotonated versions of the library are the simplest because the neutral masses are adjusted by adding or subtracting a proton. For sodium-doped samples, sodiated cation adducts are often found [M + Na]+ as well as the sodium replacement of acidic protons on the acidic groups of sialic acids [M − H + 2Na]+. The sodium replacement can occur on each of the sialic acid groups yielding a maximum of four substitutions for the largest complex type structure in this study.
3.2 Application of the Theoretical Glycan Library to Human Serum
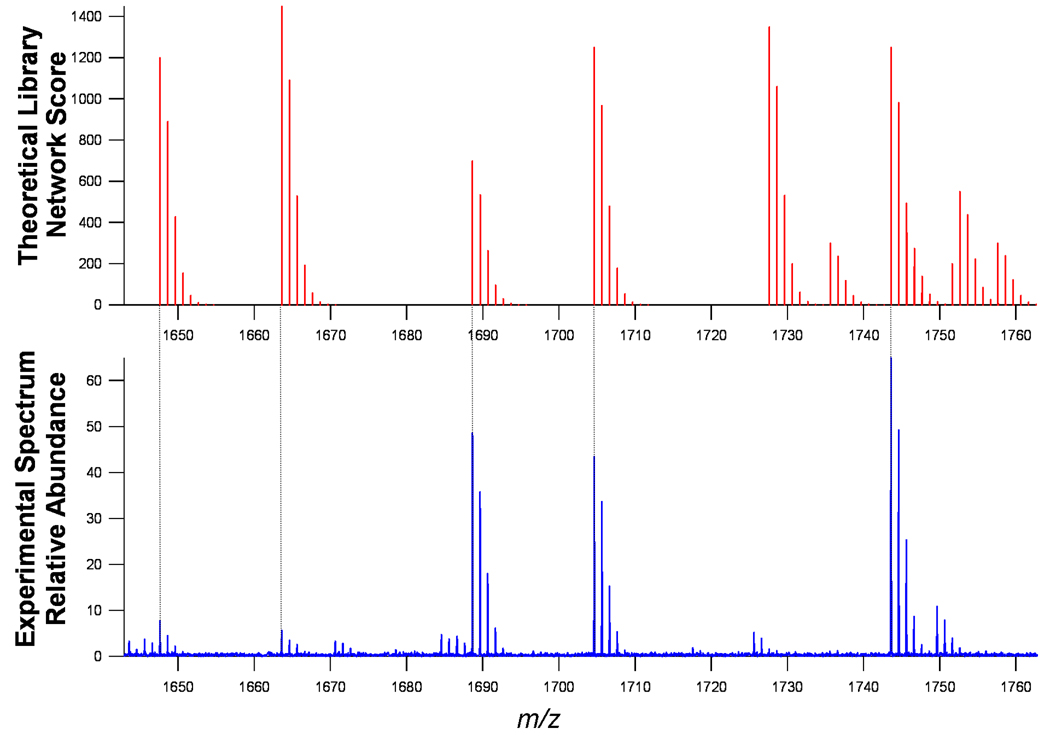
Identifying glycans in the mass spectra simply involves, in principle, superimposing the experimental spectra on the theoretical spectra. An expanded window corresponding to the range m/z 1640–1760 (Figure 3) shows the comparison of the theoretical library with the actual mass spectrum. The tie lines show the correspondence between the monoisotopic peaks indicating the identification of specific compositions. The N-linked glycan masses were then extracted from deconvoluted monoisotopic mass lists from each spectrum using a 15 ppm mass error window. Even though a 15 ppm mass error window was chosen to span inter-spectrum differences in calibration, the mass accuracy of the glycan assignments was 5.5±3 ppm mass error over a mass range of 500–3250 Da.
Figure 3.
Zoom of theoretical spectrum (top) real serum spectrum (bottom). The library was converted to Na+ adducts to compare with the MALDI spectrum of serum glycans.
The isotopic pattern on the theoretical spectra can be turned off as shown in Figure 4. This figure shows a complication where the isotopologue of the experimental spectra can match a monoisotopic ion from the theoretical library. Therefore, before an ion is identified (above the desired statistical signal-to-noise threshold), care must be taken to ensure that the signal corresponds to the monoisotopic peak. Commercial deisotoping software (PeakHunter, IonSpec) was used to generate theoretical isotopic distributions for comparisons to the data.
Figure 4.
A serum mass spectrum superimposed on top of the monoisotopic theoretical library (vertical bars). This figure demonstrates the importance of correct monoisotopic peak assignments prior to glycan assignments. There are three options when an experimental ion is in accord with the theoretical library: (1) The ion is a monoisotopic ion (Dots) and correctly annotated, (2) The ion is an isotope peak (Triangles) and is not annotated or (3) The ion is not detectable above the noise threshold (horizontal dotted line) and is not annotated.
The 331 glycan compositions are sufficiently distinct with only two compositions differing by less than 0.37 Da. Without deisotoping, approximately 64% of the masses overlap an isotopologue of other compositions. The frequency of a composition overlapping with an isotopologue (typically one 13C) is depicted in Supplement Figure A. The smallest difference is calculated to be 0.0134 Da corresponding to the difference between two deoxyhexose and the +1 isotopologue of a Neu5Ac. The overlap can be resolved with a resolution of at least 12500 (m/Δm at half height). The glycan library has the greatest density of masses corresponding to 2500–3500 Da (Supplement Figure A), which also correspond to the region of higher overlap.
A combined list of glycan compositions detected in the human sera is made by extracting theoretical library glycans from each spectrum. The accurate neutral masses, compositions, and other important information are listed in Supplement Table 2. Overall, 98 glycan masses were observed when the positive and negative modes were combined. When the masses are reduced to zero charge species, 78 discrete compositions were detected. The glycans detected in serum represent 24% coverage of the theoretical retrosynthetic library.
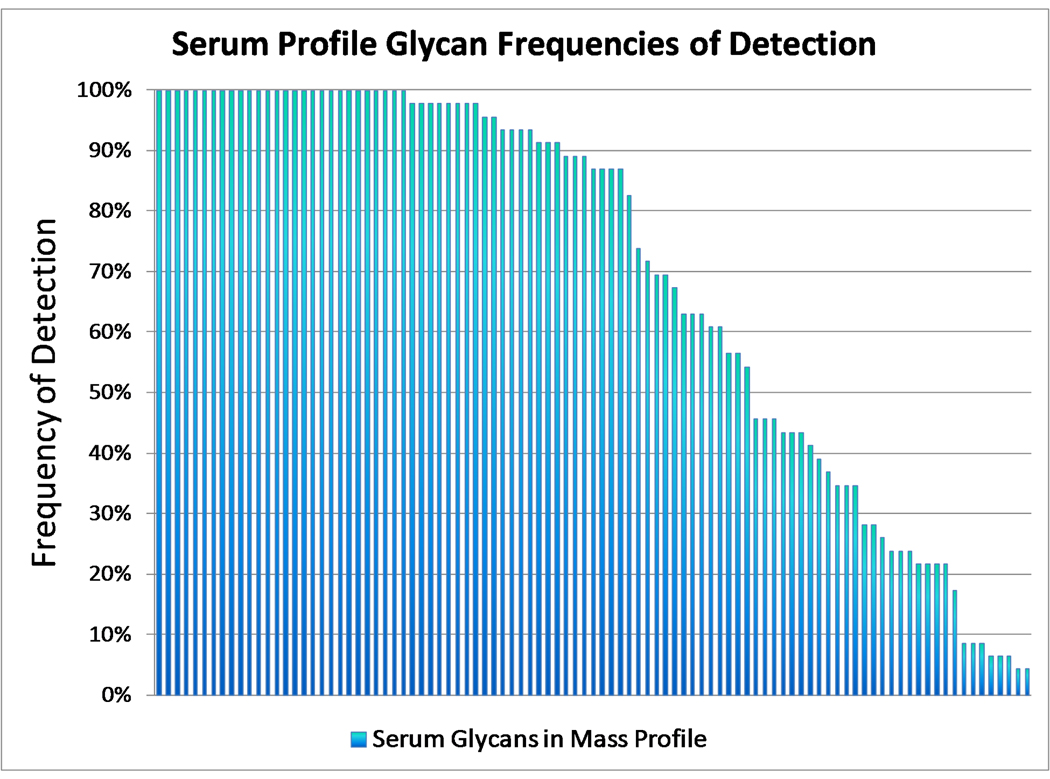
A plot was constructed based on the frequency of detection in the serum samples of each specific composition (Figure 5). There were 28 compositions detected in 100% of the samples. However, not all are the most abundant peaks in the spectra. The relative intensities of the 28 peaks vary from as little as 1% up to the most abundant ion (100%) in the spectra. The average relative abundance was 21%. The most common peaks are generally distributed between the three glycan classes.
Figure 5.
Frequency of detection vs. N-linked glycan mass profile. The y-axis is the frequency of detection of an N-linked glycan mass out of 46 spectra while the x-axis is the glycan mass. The x-axis is sorted from the most frequently detected to the least.
While the high mannose types are compositionally distinct, assigning complex or hybrid glycans is complicated by the overlap in the two classes. The overlap is due to structures with common compositions with multiple putative structures that fall into either of the two classes. In addition, some of the overlap is attributed to degradation structures from the hybrid starting point glycan that is common to those in the complex library. These structures are typically classified as complex-type oligosaccharides and are classified as such here.
A systematic set of guidelines has been developed to separate further the complex only glycans from the truly ambiguous complex/hybrid set. These rules are derived based on the observations of the three classes. An outline of the classification rules is included in Supplement Figure B(1) and B(2). The rules have been used to classify the compositions generated by the library and were found to predict the correct class in all cases. The consistency between the classification rules and the retrosynthetic networks ensures the completeness of library as well as the efficacy of the classification rules.
A pie chart reporting percentages of glycan compositions from each class in the theoretical library is presented in Supplement Figure C(1). Partitioning the theoretical library into the 4 sets (high mannose, complex, hybrid, and complex/hybrid) allows for automated classification of experimental glycan compositions. In the theoretical library, 75% of the structures are complex, 13% of the structures are hybrid, 3% are high mannose, and 9% are indistinguishable. The classification was applied to serum and the resulting assignments are presented in the pie chart in Supplement Figure C(2). Interestingly, the number of structures that are complex and complex/hybrid are in nearly equal proportions (36% and 36%, respectively) while the hybrid was smaller at 17% and the high mannose at 11%. In terms of the most abundant peaks (top 57 relative intensities greater than 1% absolute intensity), about half were complex/hybrid (52%), a quarter were complex (25%), and the rest were roughly split between hybrid and high mannose (14% and 9%, respectively).
The serum glycome can also be partitioned by into sets of glycans that contain fucose or sialic acid. The quantity of glycans in each set, presented as a percentage of all the glycans in the serum library, is presented in Supplement Figure D. The majority of the sugars detected, 88% of all the compositions, across all fractions were either sialylated, fucosylated, or both sialylated and fucosylated. Those that were fucosylated only represented 30% of the structures, sialylated only 15%, and both fucosylated and sialylated 31%. Glycans with neither monosaccharide was only 24%.
4 CONCLUDING REMARKS
This library can be expanded in many ways to improve its utility. It can be increased to include other organisms, or it can be customized for specific individual organisms. It can also be increased when larger or new saccharide modifications are discovered.
The library has several obvious applications. The retrosynthetic state-transition network library is useful for profiling human serum. It can be used to extract glycan masses directly from spectra containing N-linked glycans as long as precautions are used to obtain monoisotopic masses. A list of compositions determined in this way can be used for more effective bioinformatics analysis (e.g. hypothesis testing, bootstrapping, machine learning, etc.). Proper isotope filtering further decreases the number of ions of interest which improves the results from multiple testing algorithms such as Bonferroni testing corrections.
The prescribed putative structures from the retrosynthetic state-transition networks can be fragmented in silico via other software packages to produce a library of fragmentation spectra. The resulting fragmentation spectra could be juxtaposed to experimental spectra for rapid structural analysis. Applying these algorithms to this library can generate a fragmentation library that may be used analogously to peptide fragmentation libraries for elucidating structures with greater certainty.
Finally, the library can be used to estimate the size of the serum N-linked glycome. There are 331 compositions based on the theoretical library. There are a limited number of glycosyltransferases so that a limited number of putative structures are expected. We have recently published a study of the nano liquid chromatography separation of N-linked glycans from serum (Submitted). This study indicated that the number of isomers is generally small, typically less than five. If the average number of isomers per composition is five, then the total number of glycans would be around 1655. This value is roughly consistent with the hundreds of different structures suggested by Morelle et al. [41]. However, that number comes from elution of a single fraction and contains many of the abundant glycans in serum. The relatively small number suggests that elucidating the entire glycome is not an intractable problem and can possibly be solved by persistent structure determination.
This approach addresses the need for gross partitioning the glycome to identify as much components while decreasing ion suppression. Previous studies of glycan profiles have shown that the major components are sialylated oligosaccharides [14]. Separating the glycome into three fractions allows for partitioning into chemically similar glycans. For example, the neutral oligosaccharides elute at a different fraction than the anionic oligosaccharides. This simple fractionation allows one to observe abundant neutral oligosaccharides and anionic oligosaccharides nearly discretely. This capability is important in profiling whole glycomes for high throughput disease marker discovery.
ACKNOWLEDGEMENTS
We gratefully acknowledge financial support provided by the National Institute of Health RO1 GM049077.
Footnotes
CONFLICT OF INTEREST
There are no conflicts of financial/commercial conflicts of interest for this article.
REFERENCES
- 1.Barrabes S, Pages-Pons L, Radcliffe CM, Tabares G, et al. Glycosylation of serum ribonuclease 1 indicates a major endothelial origin and reveals an increase in core fucosylation in pancreatic cancer. Glycobiology. 2007;17:388–400. doi: 10.1093/glycob/cwm002. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekaran EV, Xue J, Neelamegham S, Matta KL. The pattern of glycosyl- and sulfotransferase activities in cancer cell lines: a predictor of individual cancer-associated distinct carbohydrate structures for the structural identification of signature glycans. Carbohydrate Research. 2006;341:983–994. doi: 10.1016/j.carres.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Gornik O, Royle L, Harvey DJ, Radcliffe CM, et al. Changes of serum glycans during sepsis and acute pancreatitis. Glycobiology. 2007;17:1321–1332. doi: 10.1093/glycob/cwm106. [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconjugate Journal. 1997;14:569–576. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]
- 5.Nan BC, Shao DM, Chen HL, Huang Y, et al. Alteration of N-acetylglucosaminyltransferases in pancreatic carcinoma. Glycoconjugate Journal. 1998;15:1033–1037. doi: 10.1023/a:1006950311937. [DOI] [PubMed] [Google Scholar]
- 6.Peracaula R, Royle L, Tabares G, Mallorqui-Fernandez G, et al. Glycosylation of human pancreatic ribonuclease: differences between normal and tumor states. Glycobiology. 2003;13:227–244. doi: 10.1093/glycob/cwg019. [DOI] [PubMed] [Google Scholar]
- 7.Ressom HW, Varghese RS, Goldman L, An Y, et al. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. Journal of Proteome Research. 2008;7:603–610. doi: 10.1021/pr0705237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saldova R, Royle L, Radcliffe CM, Hamid UMA, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 9.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, et al. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. Journal of Proteome Research. 2006;5:1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 10.Comunale MA, Lowman M, Long RE, Krakover J, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. Journal of Proteome Research. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto S, Asao T, Takahashi J, Yagihashi Y, et al. alpha(1)-acid glycoprotein fucosylation as a marker of carcinoma progression and prognosis. Cancer. 2004;101:2825–2836. doi: 10.1002/cncr.20713. [DOI] [PubMed] [Google Scholar]
- 12.Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, et al. Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. Journal of Proteome Research. 2008;7:1109–1117. doi: 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirmiz C, Li BS, An HJ, Clowers BH, et al. A serum glycomics approach to breast cancer biomarkers. Molecular & Cellular Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, et al. Alterations in the serum glycome due to metastatic prostate cancer. Journal of Proteome Research. 2007;6:1822–1832. doi: 10.1021/pr060664t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, et al. Glycomics analysis of serum: a potential new biomarker for ovarian cancer? International Journal of Gynecological Cancer. 2007;18:470–475. doi: 10.1111/j.1525-1438.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu XE, Desmyter L, Gao CF, Laroy W, et al. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology. 2007;46:1426–1435. doi: 10.1002/hep.21855. [DOI] [PubMed] [Google Scholar]
- 17.Peracaula R, Tabares G, Royle L, Harvey DJ, et al. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13:457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 18.Qiu Y, Patwa TH, Xu L, Shedden K, et al. Plasma Glycoprotein Profiling for Colorectal Cancer Biomarker Identification by Lectin Glycoarray and Lectin Blot. Journal of Proteome Research. 2008;7:1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabares G, Radcliffe CM, Barrabes S, Ramirez M, et al. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16:132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 20.Ye JJ, Liu H, Kirmiz C, Lebrilla CB, Rocke DM. On the analysis of glycomics mass spectrometry data via the regularized area under the ROC curve. BMC Bioinformatics. 2007;8:477–489. doi: 10.1186/1471-2105-8-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Patwa TH, Qiu WL, Shedden K, et al. Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. Journal of Proteome Research. 2007;6:1864–1874. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Qiu WL, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. Journal of Proteome Research. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. Journal of Proteome Research. 2006;5:1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 24.Vance BA, Wu WY, Ribaudo RK, Segal DM, Kearse KP. Multiple dimeric forms of human CD69 result from differential addition of N-glycans to typical (Asn-X-Ser/Thr) and atypical (Asn-X-Cys) glycosylation motifs. Journal of Biological Chemistry. 1997;272:23117–23122. doi: 10.1074/jbc.272.37.23117. [DOI] [PubMed] [Google Scholar]
- 25.Miletich JP, Broze GJ. Beta-Protein-C Is Not Glycosylated at Asparagine-329 - the Rate of Translation May Influence the Frequency of Usage at Asparagine-X-Cysteine Sites. Journal of Biological Chemistry. 1990;265:11397–11404. [PubMed] [Google Scholar]
- 26.Titani K, Kumar S, Takio K, Ericsson LH, et al. Amino-Acid-Sequences of Human Vonwillebrand-Factor. Biochemistry. 1986;25:3171–3184. doi: 10.1021/bi00359a015. [DOI] [PubMed] [Google Scholar]
- 27.Cooper CA, Gasteiger E, Packer NH. GlycoMod - A software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 28.Cooper CA, Harrison MJ, Wilkins MR, Packer NH. GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Research. 2001;29:332–335. doi: 10.1093/nar/29.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg D, Sutton-Smith M, Paulson J, Dell A. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics. 2005;5:865–875. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- 30.Ceroni A, Maass K, Geyer H, Geyer R, et al. GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res. 2008 doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 31.Mohr PJ, Taylor BN. CODATA recommended values of the fundamental physical constants: 2002. Reviews of Modern Physics. 2005;77:1–107. doi: 10.1103/RevModPhys.93.025010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancaster KS, An HJ, Li BS, Lebrilla CB. Interrogation of N-linked oligosaccharides using infrared multiphoton dissociation in FT-ICR mass spectrometry. Analytical Chemistry. 2006;78:4990–4997. doi: 10.1021/ac0600656. [DOI] [PubMed] [Google Scholar]
- 33.Taylor ME, Drickammer K. Introduction to Glycobiology. New York: Oxford University Press; 2006. [Google Scholar]
- 34.Taguchi T, Kitajima K, Niimi T, Muto Y, et al. Complete Assignments of C-13 Nmr Resonances to All the Carbon-Atoms of the Trimannosido-Di-N-Acetylchitobiosyl Structure in a Pentaantennary Decasaccharide Glycopeptide. Carbohydrate Research. 1995;275:185–191. doi: 10.1016/0008-6215(95)00144-i. [DOI] [PubMed] [Google Scholar]
- 35.Harvey DJ. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. Journal of the American Society for Mass Spectrometry. 2005;16:647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Campion B, Leger D, Wieruszeski JM, Montreuil J, Spik G. Presence of Fucosylated Triantennary, Tetraantennary and Pentaantennary Glycans in Transferrin Synthesized by the Human Hepato-Carcinoma Cell-Line Hep G2. European Journal of Biochemistry. 1989;184:405–413. doi: 10.1111/j.1432-1033.1989.tb15032.x. [DOI] [PubMed] [Google Scholar]
- 37.van Rooijen JJM, Kamerling JP, Vliegenthart JFG. The abundance of additional N-acetyllactosamine units in N-linked tetraantennary oligosaccharides of human Tamm-Horsfall glycoprotein is a donor-specific feature. Glycobiology. 1998;8:1065–1075. doi: 10.1093/glycob/8.11.1065. [DOI] [PubMed] [Google Scholar]
- 38.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158r–184r. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 39.Crispin M, Harvey DJ, Chang VT, Yu C, et al. Inhibition of hybrid-and complex-type glycosylation reveals the presence of the GlcNAc transferase I-independent fucosylation pathway. Glycobiology. 2006;16:748–756. doi: 10.1093/glycob/cwj119. [DOI] [PubMed] [Google Scholar]
- 40.Lin AI, Philipsberg GA, Haltiwanger RS. Core Fucosylation of High-Mannose-Type Oligosaccharides in Glcnac Transferase I-Deficient (Lec1) CHO Cells. Glycobiology. 1994;4:895–901. doi: 10.1093/glycob/4.6.895. [DOI] [PubMed] [Google Scholar]
- 41.Morelle W, Flahaut C, Michalski JC, Louvet A, et al. Mass spectrometric approach for screening modifications of total serum N-glycome in human diseases: application to cirrhosis. Glycobiology. 2006;16:281–293. doi: 10.1093/glycob/cwj067. [DOI] [PubMed] [Google Scholar]